Abstract
The purpose of the study was to investigate the role of salicylic acid (SA) signalling in Ny-1-mediated hypersensitive resistance (HR) of potato (Solanum tuberosum L.) to Potato virus Y (PVY). The responses of the Ny-1 allele in the Rywal potato cultivar and transgenic NahG-Rywal potato plants that do not accumulate SA were characterized at the cytological, biochemical, transcriptome, and proteome levels. Analysis of noninoculated and inoculated leaves revealed that HR lesions started to develop from 3 d post inoculation and completely restricted the virus spread. At the cytological level, features of programmed cell death in combination with reactive oxygen species burst were observed. In response to PVY infection, SA was synthesized de novo. The lack of SA accumulation in the NahG plants led to the disease phenotype due to unrestricted viral spreading. Grafting experiments show that SA has a critical role in the inhibition of PVY spreading in parenchymal tissue, but not in vascular veins. The whole transcriptome analysis confirmed the central role of SA in orchestrating Ny-1-mediated responses and showed that the absence of SA leads to significant changes at the transcriptome level, including a delay in activation of expression of genes known to participate in defence responses. Moreover, perturbations in the expression of hormonal signalling genes were detected, shown as a switch from SA to jasmonic acid/ethylene signalling. Viral multiplication in the NahG plants was accompanied by downregulation of photosynthesis genes and activation of multiple energy-producing pathways.
Key words: Plant–pathogen interactions, Potato virus Y, Potyviridae, salicylic acid, Solanum tuberosum, whole transcriptome analysis.
Introduction
Potato (Solanum tuberosum L.) is the fourth-most-important food crop in the world, with a yield of 324 million tonnes in 2010, and a continuous increase in the world production of tubers (http://faostat.fao.org/). Potato virus Y (PVY) is a member of the Potyviridae family, and it is currently considered as the most economically harmful virus for cultivated potatoes. PVY has a wide host range, mainly within the Solanaceae family, and it is distributed worldwide. Isolates of PVY species are highly variable at the biological, serological, and molecular levels (Scholthof et al., 2011). Strain PVYN-Wi, which was originally described in the Polish cv. Wilga (Chrzanowska, 1991), belongs to the N strain group, although it is serologically related to PVY° (Singh et al., 2008). In the current context of a highly competitive international potato market, the weaknesses in both the knowledge of PVY–host interactions and the diagnostic tools have led to a situation in which necrotic PVY isolates are still potentially responsible for huge agronomic and economic losses (Scholthof et al., 2011).
The ability of viruses to cause disease is determined by molecular interactions between the host plant and virus factors. These interactions directly affect virus replication and movement, symptom development, and host defence responses, leading to either compatible (sensitive) or incompatible (resistant) interactions (Whitham et al., 2006). In potato, there are two main types of resistance to PVY: extreme resistance, conferred by the Ry genes, and hypersensitive resistance (HR), conferred by the Ny genes (reviewed in Kogovšek and Ravnikar, 2013). The potato cv. Rywal carries the Ny-1 gene, and it develops HR that is manifested as necrotic lesions on inoculated leaves 3 d post inoculation (dpi) with various PVY strains (PVY°, PVYN, PVYN–Wi, PVYNTN). Development of HR restricts virus multiplication and spreading. However, the response is temperature dependent, as growth at a higher temperature (28 °C) prevents HR and allows the virus to spread systemically (Szajko et al., 2008).
Salicylic acid (SA) is a key component in the orchestration of the events during and following HR (Lewsey et al., 2009). Deficiency in SA signalling leads to an impairment of the defence responses and susceptibility to pathogen attack (Takahashi et al., 2004; Sanchéz et al., 2010; Jovel et al., 2011). In the case of compatible interactions, SA can still improve the immunity by delaying the onset of viral replication and disease development, through mediation of the expression of a cohort of defence-related genes that induce a defence-like response (Huang et al., 2005; Baebler et al., 2011). Previous studies have suggested significant differences in the possible mechanism of SA action in potato defences in comparison to the model pathosystems of Arabidopsis thaliana and tobacco. In potato, basal levels of salicylates are 10–100-fold higher than in Arabidopsis or tobacco (Navarre and Mayo, 2004), and the increase in SA levels after pathogen treatment is relatively moderate (Krečič-Stres et al., 2005; Halim et al., 2007). Therefore, it has been suggested that potato has poor SA sensitivity and that, rather than biosynthesis, enhancement of its effectiveness is responsible for activating the SA-dependent signal transduction pathway after infection (Yu et al., 1997). It has been proposed that the inactive salicylate glycosylates (SAGs) are the storage form of salicylates that, when necessary, can be converted into the free and active SA (Hennig et al., 1993). Plants that are unable to accumulate SA due to the expression of salicylate hydroxylase (NahG), which catalyses the conversion of SA to catechol, have been widely used to study the role of SA in potato–pathogen interaction (Navarre and Mayo, 2004; Halim et al., 2007; Yu et al., 1997; Sanchéz et al., 2010; Baebler et al., 2011).
The understanding of the mechanisms that underlie resistance responses to viruses is still incomplete. The network of factors that can interact to establish resistance has to be of great complexity to provide a plant with effective weaponry to fight the variety of pathogens. Genomic tools have allowed the assessment of gene expression at a genome-wide scale, which has provided unprecedented views of host–virus interactions (Rodrigo et al., 2012). Compared to numerous whole transcriptome studies on compatible plant–virus interactions, which were reviewed and subjected to meta-analysis by Postnikova and Nemchinov (2012) and Rodrigo and coworkers (2012), studies of incompatible interactions are scarce (Ishihara et al., 2004; Marathe et al., 2004; Baebler et al., 2009; Vuorinen et al., 2010; Pacheco et al., 2012). As far as is known, there has been only one whole-transcriptome approach to investigate the involvement of SA in the response of A. thaliana to a set of compatible viruses (Huang et al., 2005), while the role of SA in potato–virus interactions has not been investigated yet at the whole-transcriptome level.
The aim of the present study was to investigate the role of SA signalling in Ny-1-mediated HR. The responses to PVYN–Wi infection were studied in a Ny-1 background in nontransgenic plants, plants impaired in SA accumulation, and plants with compromised resistance due to its temperature-dependent nature. Cytological and biochemical characterization combined with transcriptome and proteome analyses have revealed that SA is a crucial factor for inhibition of the spread of PVY in parenchymal tissue, while a lack of SA results in delayed early transcriptional events, which can lead to inefficient defence responses and disease development.
Materials and methods
Plant material
Potato (S. tuberosum ssp. tuberosum; 2n=4x=48) cv. Rywal plants were obtained from the Institute of Plant Breeding and Acclimatisation—National Research Institute, Młochów (Poland). Plants expressing the NahG transgene (NahG-Rywal) were generated using the binary plasmid pCIB containing salicylate hydroxylase (NahG), which was generously provided by Syngenta Biotechnology. The plasmid was introduced into Agrobacterium tumefaciens LBA4404 and used for potato leaf disc transformation (Chen et al., 1994). Transgenic potato plants were regenerated as described by Mac et al. (2004). Plants of both genotypes were grown for 4 weeks in soil under controlled environmental conditions (16/8 light/dark cycle, 20 °C) as described previously (Szajko et al., 2008). PVY strain N-Wilga (PVYN–Wi; accession no. EF558545) was derived from potato cv. Wilga and was multiplied and maintained in tobacco plants, Nicotiana tabacum cv. Samsun.
PVY inoculation was performed on 4-week-old potato plants. Three bottom leaves were dusted with carborundum powder and rubbed with cheesecloth dipped in a sap prepared from the leaves of the PVY-infected tobacco plants. After 10min, the leaves were washed liberally with tap water. In mock inoculations, water was used instead of the sap.
Viral amplification was monitored by semiquantitative reverse-transcription PCR as described before (Szajko et al., 2008), but by running only 22 cycles. Separate PCR reactions of potato elongation factor 1-α (EF-1) expression were performed as loading control (Varet et al., 2002).
Quantification of free salicylic acid and salicylic acid glycosylates
Free SA was extracted and quantified from noninoculated and inoculated leaves at several times after mock or viral inoculation, as described previously (Malamy et al., 1990) and with modifications introduced by Krzymowska et al. (2007). HPLC analysis was performed and SAGs were quantified as described previously (Malamy et al., 1992). Total salicylates were calculated as the sum of the free SA and SAGs.
Callose and hydrogen peroxide staining
Visualization of callose deposition in leaf discs was performed according to Peterhansel et al. (1997) and Vogel and Somerville (2000). Hydrogen peroxide (H2O2) was visualized using 1mg ml–1 3,3′-diaminobenzidine-HCl, pH 3.8 (DAB), as described previously (Thordal-Christensen et al., 1997; Talarczyk et al., 2002). Microscopic observations and documentation were performed using Microphot-SA fluorescence microscope (Nikon Corporation) with a OCRA–ER digital camera (Hamamatsu Photonics).
Light and transmission electron microscopy
For morphological and ultrastructural observations, fragments of the leaves were dissected from mock-inoculated and PVY-inoculated potato plants, at 0, 1, 3, and 6 dpi. The leaf pieces were fixed in freshly made Karnovsky fixative for 2h under reduced pressure (0.4 atm). They were then post-fixed in 2% (w/v) osmium tetroxide in 0.05M sodium cacodylate buffer for 2h and embedded in EPON epoxy resin (Fluka). Semithin sections were stained with crystal violet (1% w/v) and inspected under an Olympus AX70 microscope, with the images captured with an Olympus DP50 digital camera. For ultrastructure observations, ultra-thin sections were stained with uranyl acetate and lead citrate, and examined with a JEM 1200 EX transmission electron microscope (JEOL).
The presence of viral particles in inoculated leaves of cv. Rywal was examined 6 dpi by the negative-staining method under transmission electron microscopy (Supplementary Fig. S1, available at JXB online).
Microarray analysis
Total RNA from the inoculated leaves was extracted, treated, purified, and quality controlled as described previously (Baebler et al., 2009). The custom 60-mer oligo microarrays (4×44; design ID 015425; Agilent) were designed by the Potato Oligo Chip Initiative consortium (Kloosterman et al., 2008). For microarray hybridization, 1 μg RNA was spiked with 500ng One-Color Spike-Mix (Agilent), amplified and labelled with Cy-5 using MessageAmp II aRNA amplification kit (Ambion), and hybridized using Gene Expression Hybridization kits (Agilent), according to manufacturer protocols. The dried microarrays were scanned with an LS200 Array Scanner (Tecan) and the images were analysed and data extracted with ArrayPro (Media Cybernetics), according to Baebler et al. (2009).
The data were analysed in the R environment (R Core Team, 2012) with the Bioconductor limma package (Smyth, 2005). The background signal on all of the microarrays was uniform and low and was hence ignored in further calculations. The raw data was quantile normalized. Features below background intensity (log2A <5) in more than 95% of samples were excluded from further analysis. The correlation coefficients between biological replicates ranged from 0.97 to 1.00, indicating their high similarities. Differentially expressed genes (Benjamini and Hochberg corrected P-value ≤0.05, fold change ≥2) between virus and mock-inoculated plants at each time point and for each genotype were identified using the empirical Bayes method (Smyth, 2005). Functional analysis of differentially expressed genes was performed using MapMan ontology (Usadel et al., 2009), adapted for potato (Rotter et al., 2007). Gene set enrichment analysis (GSEA; Subramanian et al., 2005) was performed to search for groups of genes involved in the same processes that were significantly (false discovery rate corrected Q-value < 0.05) altered by virus inoculation, using MapMan ontology as the source of the gene sets.
The data discussed in the present study have been deposited in NCBI Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE46180.
To validate the microarray results, the expression of the genes involved in carbohydrate metabolism, hormonal signalling, and defence responses was analysed by quantitative PCR in the same RNA samples as for microarray analysis (Supplementary Table S1).
Two-dimensional differential gel electrophoresis
To complement the data at the transcriptome level, proteome analyses were performed on cv. Rywal, PVY, and mock-inoculated leaves, collected 1 and 3 dpi, and taken from the same plants (four biological replicates), as for the transcriptome analysis. Protein extraction and separation were carried out as described previously (Renaut et al., 2008) using 300mg fresh leaf material. A statistical analysis of spot abundances was performed using the same software: t-test with P<0.05 and abundance variation (infected/mock) of at least 1.3-fold; two-way ANOVA was performed to find proteins differentially expressed in time after inoculation (P<0.05, PVY infection as one factor, time as the second factor). Differentially expressed spots were excised from gels, digested with trypsin, and spotted on MALDI targets, as described previously (Renaut et al., 2008). Proteins were identified by searching against the NCBI nr (Viridiplantae) and potato EST databases using a MASCOT server (Matrix Science, London; www.matrixscience.com). Searches were carried out using a mass window of 150 ppm for the precursor and 0.75Da for the fragments. The search parameters allowed for carboxyamidomethylation of cysteine, oxidation of methionine, tryptophan to kynurenine, and double oxidation of tryptophan (N-formylkynurenine). The search results were evaluated based on the peptide scores. Homology identification was retained with a probability set at 95%. Proteins were identified with at least two unique peptides with a significant score.
Results
Morphological characterization of HR response to PVY
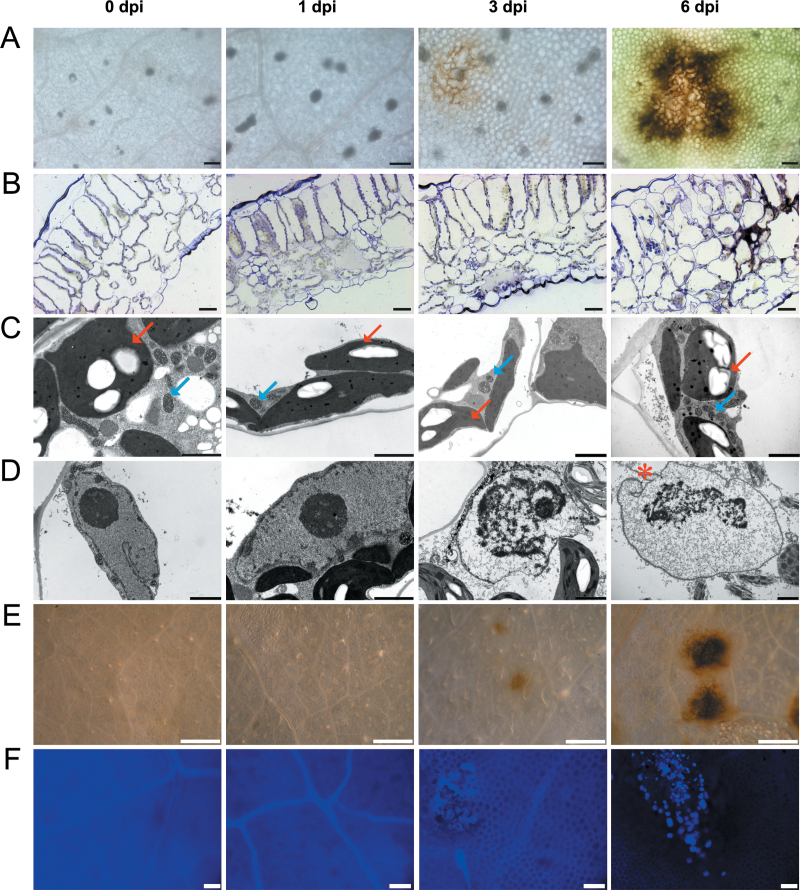
To understand HR response observed in cv. Rywal, lesion development was examined at the cytological level. Lesions started to appear on the abaxial side of the leaf in mesophyll cells at 3 dpi and became fully developed at 6 dpi. Deposition of brown-coloured material, probably containing phenolic compounds (e.g. lignins) was observed at the lesion edge at 6 dpi (Fig. 1A). Disorganization of tissue architecture at the site of the lesion was observed under light microscopy (Fig. 1B). Analysis of the cells in this area by electron microscopy revealed no significant changes in the sizes and shapes of the chloroplasts or mitochondria (Fig. 1C, arrows). In contrast, the nuclei and nucleoli underwent gradual degradation, which was already visible at 3 dpi and was even more pronounced at 6 dpi (Fig. 1D). At 3 dpi, the nucleoli were significantly smaller in comparison to the 0 and 1 dpi samples, and at 6 dpi the nucleoli appeared to be partially degraded. At 3 and 6 dpi, the chromatin was partially condensed. At 6 dpi, nuclear envelope invaginations were visible (Fig. 1D, red asterisk).
Fig. 1.
Analysis of the hypersensitive response to PVY infection at the macroscopic, light and electron microscopy levels in tissue samples from PVY-inoculated cv. Rywal leaves at 0, 1, 3, and 6 dpi. (A) Chlorophyll-removed tissue under the dissecting microscope. (B) Cross-sections of leaf tissue fixed for electron microscopy. (C, D) Transmission electron micrographs. (E) DAB-stained tissue under the dissecting microscope. (F) Callose-stained tissue observed under the fluorescence microscope with a DAPI filter. Arrows in C, chloroplasts (red), mitochondria (blue); asterisk in D, nuclear envelope invagination. Bars: 100 μm (A, F), 20 μm (B), 2 μm (C, D), 1mm (E).
DAB staining revealed high levels of H2O2 in the areas covering and surrounding the lesions (Fig. 1E, 3 and 6 dpi), which suggests the involvement of reactive oxygen species (ROS) in this HR process. Also, callose deposits (detected by aniline blue staining) were present in the lesion areas at 3 dpi and were even more abundant at 6 dpi (Fig. 1F).
Negative-staining electron microscopy detected the presence of PVY viral particles in the extracts of the inoculated leaves at 6 dpi, as isolated from the lesion areas (Supplementary Fig. S1), but not in extracts from the surrounding tissues. This confirms that the HR completely restricted the viral spread.
Dynamics of SA accumulation in HR response to PVY
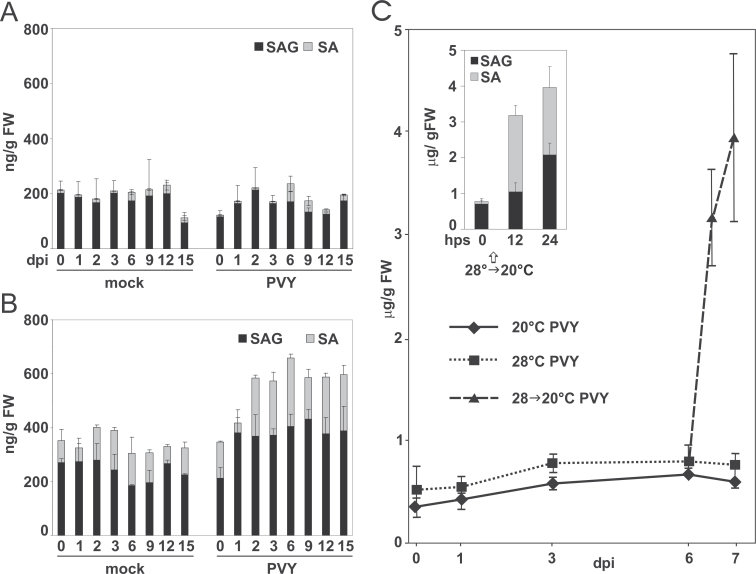
To study the role of SA in the response of the Rywal plants, the contents of free SA and its conjugates, SAGs, were measured in upper noninoculated leaves and in inoculated leaves following the mock and virus treatments. The level of salicylates in the mock-inoculated leaves showed no significant changes throughout the study, with fluctuations between 300 and 400ng (g fresh weight)–1 (Fig. 2B, left). In contrast, in the PVY-inoculated leaves, the salicylate levels rose between 1 and 2 dpi, reaching approximately 600ng (g fresh weight)–1; they then remained in that range until at least 15 dpi (Fig. 2B, right; Supplementary Table S2). Both SA and SAGs increased c.2-fold, which indicated de novo SA synthesis. The amounts of free SA remained constantly at about half those of SAGs. In contrast, there were no significant changes in SA or SAG levels in upper noninoculated leaves (Fig. 2A). To confirm that SA is synthesized de novo during PVY infection, temperature-shift conditions were applies. Similarly to that described previously (Szajko et al., 2008), in PVY-infected Rywal plants maintained at the higher temperature (28 °C), the total SA levels increased c.2-fold between 1 and 3 dpi (Fig. 2C). However, by transferring these plants from 28 to 20 °C at 6 dpi, there was a significant 5-fold increase in the total SA level 24h after the temperature shift (Fig. 2C, Supplementary Table S2), accompanied by rapid lesion development (data not shown). Interestingly, at 12h post temperature shift, there was a 27-fold increase in the free SA form, followed by conversion to SAGs at 24h post temperature shift (Fig. 2C). No changes in SA levels were detected in mock-treated plants after the temperature shift. This clearly indicates that, after viral infection, SA is synthesized de novo and further converted into the SAG storage form.
Fig. 2.
Changes in SA levels after PVY inoculation. (A, B) Changes in SA and SAG levels after mock or PVY inoculation in upper noninoculated (A) and inoculated (B) leaves of potato cv. Rywal from 0 to 15 d post infection (dpi). (C) Total salicylates in PVY-inoculated leaves, at stable temperatures (20 and 28 °C) and with temperature shift from 28 °C to 20 °C at 6 dpi. Inset: free SA and SAG at 0, 12, and 24h post temperature shift (hps). Data are mean ± standard deviation (n=5). SA, salicylic acid; SAG, salicylate glycosylates. Statistical significance of data is presented in Supplementary Table S1.
Role of SA in viral spread and disease development in PVY-infected Rywal plants
To further investigate the role of SA in the Rywal response to PVY infection, plants expressing the NahG transgene that encodes a salicylate hydroxylase were generated. Expression of the NahG transgene was checked in 10 selected transgenic lines using Northern analysis (Supplementary Fig. S2) and four were selected for further analyses. In contrast to potato NahG plants of cv. Pentland Ivory (Sanchéz et al., 2010), and similar to NahG plants of cv. Désirée (Halim et al., 2007), NahG-Rywal plants showed no yellowing or spontaneous lesion formation.
To determine the effects of NahG expression on the accumulation of SA, a control line (transformed with an empty vector) and plants of four selected lines with high levels of NahG mRNA were inoculated with PVY and the SA levels were measured 6 dpi. In the control line, the SA and SAG levels and fold of induction were similar to the nontransformed cv. Rywal (Fig. 2), whereas NahG plants did not accumulate SA or SAGs to detectable levels before or after PVY inoculation (Supplementary Fig. S3), as previously reported for potato NahG plants in response to pathogens (Halim et al., 2007; Sanchéz et al., 2010).
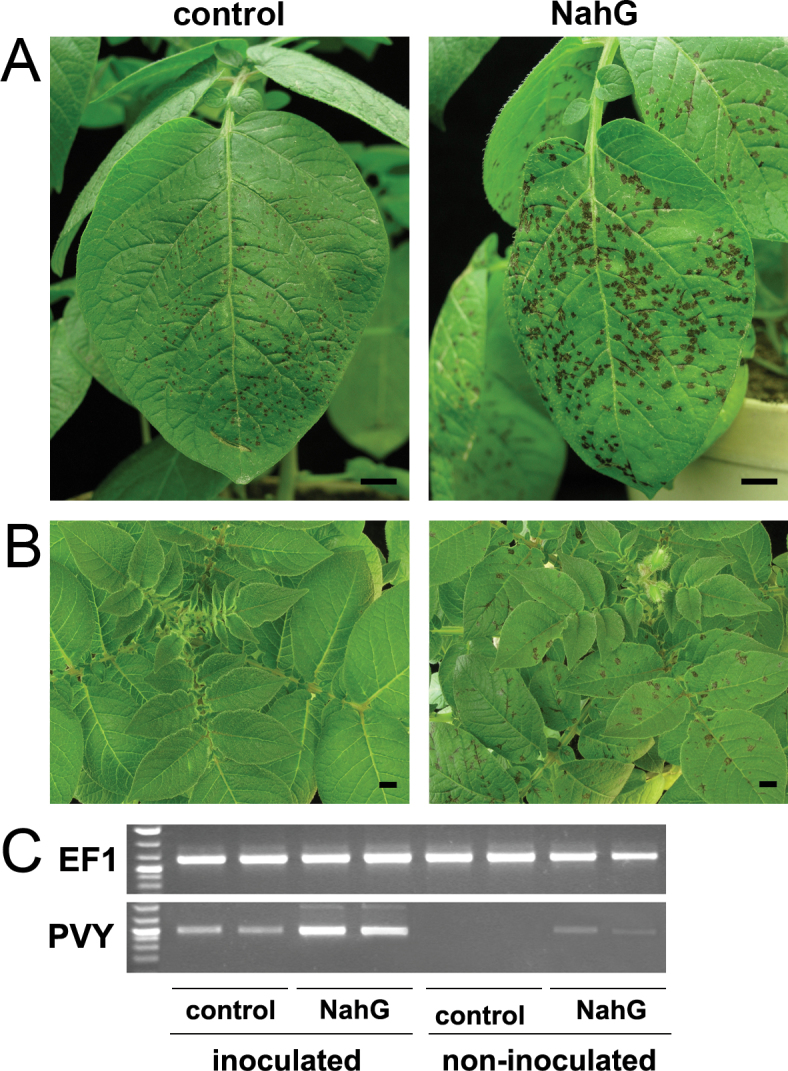
PVY infection of the control and NahG plants resulted in different phenotypes. In both genotypes, lesions on the inoculated leaves started to be visible by 3 dpi and developed similarly to 6 dpi. Afterwards, the lesions on the control plants remained unchanged, while those on the NahG plants continuously expanded, becoming approximately 2-fold larger at 10 dpi, in comparison to the control plants (Fig. 3A). At this time, the first signs of lesions developing on upper noninoculated leaves were observed in the NahG plants, with lesions reaching their full size at 14 dpi (Fig. 3B). At 10 dpi, the viral RNA levels were 3–4-fold higher in the inoculated leaves of the NahG plants, and at 14 dpi, viral RNA was detected in the upper noninoculated leaves of the NahG plants but not of the control plants (Fig. 3C).
Fig. 3.
Phenotypes and viral RNA accumulation in PVY- inoculated Rywal and NahG-Rywal plants. (A, B) Symptom development on inoculated leaves in Rywal (pROK42, control) and NahG-Rywal plants at 10 d post infection (dpi) (A) and on upper noninoculated leaves at 14 dpi (B). (C) Semiquantitative reverse-transcription PCR detection of PVY coat protein RNA and EF1 expression (loading control) in inoculated (6 dpi) and noninoculated (14 dpi) leaves, respectively. Bars: 1cm.
To determine whether the observed changes in the sizes of the lesions and the systemic virus spreading were due to the presence or absence of SA, leaves of PVY-inoculated NahG plants were sprayed with benzothiadiazole, a functional analogue of SA. This treatment restored the normal size of the lesions, as observed for the wild-type plants (Supplementary Fig. S4A), which confirmed that the phenotype observed in the NahG plants was due to lack of SA. Moreover, to confirm that the observed enhanced susceptibility of the NahG plants was not derived from the accumulation of the SA degradation product catechol, as proposed previously (van Wees and Glazebrook, 2003), nontransformed plants were sprayed with 1mM catechol solution prior to PVY inoculation. This treatment neither changed the size of lesions nor caused lesion formation in noninoculated leaves (Supplementary Fig. S4A), which indicates that catechol itself does not induce susceptibility to PVY. The observed phenotype effects were consistent with the presence of viral RNA, which after both chemical treatments was detected only in the inoculated leaves (Supplementary Fig. S4B).
To further investigate the role of SA in inhibition of systemic viral spread, a grafting experiment was performed with three types of grafts (Fig. 4A). When a nontransgenic scion was grafted onto a NahG stock (NT:NahG graft), systemic symptoms in the scion were observed after stock inoculation with the virus. Similarly, when a NahG plant served both as scion and rootstock (NahG:NahG graft), systemic symptoms were observed after inoculation; however, no systemic symptoms were observed when nontransgenic Rywal plants were used as rootstock (NahG:NT graft; Fig. 4B). The presence of the viral RNA in scions of NahG:NahG and NT:NahG grafts was confirmed by semiquantitative reverse-transcription PCR, whereas no virus was detected in scions of NahG:NT grafting (Fig. 4C).
Fig. 4.
Grafting of Rywal and NahG-Rywal plants. The rootstocks of the grafted plants were inoculated with PVY. (A) Schematic representation of the grafts. (B) Scions 21 d post rootstock infection (C) Semiquantitative reverse-transcription PCR detection of PVY coat protein RNA and EF1 expression (loading control) in all grafts. For each graft type, plant material from two independent scions was analysed. NT, nontransgenic Rywal plants; nahG, NahG-Rywal plants. Bar: 1cm.
Transcriptome profiling of HR response to PVY infection
To better understand the role of SA in the Ny-1-mediated HR response described above, high-throughput transcriptome analysis was performed, using leaf material collected from mock- and virus-inoculated plants of cv. Rywal and NahG-Rywal at three time points (1, 3, and 6 dpi).
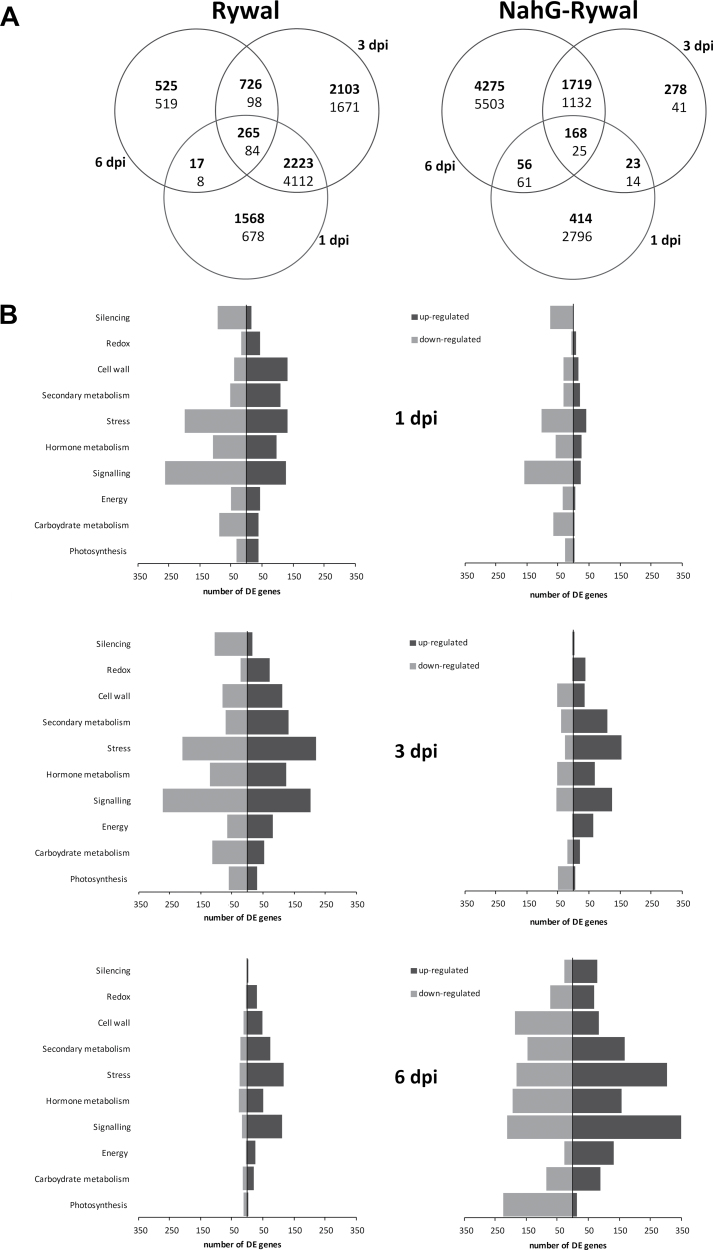
The plants responded to the viral inoculation by transcriptional reprogramming. In the Rywal plants, the most pronounced changes were observed at 1 and 3 dpi (Fig. 5A, left). More than 6000 genes were differentially expressed at both time points, indicating the continuity of response. Among the functional classes of the upregulated genes in Rywal at 1 dpi, there were cell-wall-, stress-, and secondary-metabolism-associated gene groups. At the same time, groups of genes associated with carbohydrate metabolism, signalling, and RNA silencing were mostly downregulated (Fig. 5B). A strong decrease in the total number of differentially expressed genes was observed at 6 dpi, where induction prevailed over repression (Fig. 5).
Fig. 5.
Differentially expressed (DE) genes in virus vs. mock treated plants of cv. Rywal (left) and NahG-Rywal (right) at 1, 3, and 6 d post infection (dpi). (A) Venn diagram of all of the DE genes (bold, upregulated; regular, downregulated). (B) Representation of numbers of DE genes (black, upregulated; grey, downregulated) in the selected functional groups; MapMan ontology bins: photosynthesis (bins 1, 19), carbohydrate metabolism (minor CHO, major CHO, gluconeogenesis; 2, 3, 6), energy (glycolysis, fermentation, OPP, TCA/organic transformation, mitochondrial electron transport/ATP synthesis; 4, 5, 7, 8, 9), signalling (30), hormone metabolism (17), stress (20), secondary metabolism (16), cell wall (10), redox (21), and silencing (27.5).
In contrast, in NahG plants at 1 dpi, the response was very weak, and the differentially expressed genes were mostly downregulated (e.g. stress, carbohydrate metabolism, signalling; Fig. 5). At 3 dpi, there were more upregulated genes (e.g. secondary metabolism, stress, signalling), with the exception of photosynthesis-related genes, the expression of which was strongly downregulated (Fig. 5B). Similar functional groups were differentially expressed at 6 dpi. Differential regulation of more than a quarter of all genes (Fig. 5A) could be related to disease development and multiplication of the virus at 6 dpi (Fig. 3B, C).
The microarray data were validated (correlation coefficient=0.78) by analysing 10 biologically relevant genes, involved in photosynthesis, carbohydrate, hormone metabolism, and defence response by quantitative PCR (Supplementary Table S3).
Early activation of genes involved in defence responses
A number of genes involved in plant stress and defence responses were differentially expressed during the entire experiment in both genotypes. However, significant differences in kinetics and quantity were detected between the analysed genotypes. Induction of genes that code for pathogenesis-related proteins (PR proteins; proteinase inhibitors, thaumatin, chitinases) and genes involved in the production of ROS (mostly peroxidases), was observed at 1 dpi in both genotypes (Fig. 5B, Supplementary Table S4). However, fewer genes were differentially expressed and the differences compared to mock-inoculated plants were smaller in the SA-deficient NahG plants. On the other hand, genes coding for enzymes involved in cell-wall rearrangements and synthesis of secondary metabolites (terpenoids, flavonoids, alkaloids, simple phenolic substances) were upregulated in Rywal at 1 dpi, while in the NahG-Rywal these genes were downregulated or upregulated at later time points (Fig. 5B, Supplementary Table S4). This suggests a strong delay in gene activation in the case of SA deficiency.
Surprisingly, genes involved in signalling (several receptors and MAP kinases) were downregulated at 1 and 3 dpi in both genotypes. Moreover, the silencing components were strongly downregulated in cv. Rywal at 1 and 3 dpi (Fig. 5B, Supplementary Table S4).
Changes in hormone signalling pathways
The shift in the overall hormonal signalling network in response to the virus in two genotypes can be best demonstrated by GSEA data (Table 1, Supplementary Table S4). In Rywal plants, the genes coding for enzymes involved in SA synthesis were upregulated at the first two time points, which correlated with increased SA (Fig. 2B). At 6 dpi, strong upregulation of genes of the jasmonic acid (JA) synthesis pathway was observed (Table 1), accompanied by upregulation of some of the genes involved in ethylene (ET) synthesis and signal transduction pathways. In the NahG plants, after the transient induction of SA synthesis pathway genes at 1 dpi, there was strong activation of the genes of both JA and ET pathways at 3 and 6 dpi. In both genotypes, there was strong inhibition of genes of the auxin signal transduction pathway (auxin responsive factors), while no changes in auxin synthesis genes were detected. Genes involved in ABA synthesis and degradation were downregulated in the NahG plants at 3 and 6 dpi. Moreover, the genes of the gibberellin and brassinosteroid pathways were induced at 1 and 3 dpi in Rywal plants, while the genes of the latter pathway were also downregulated in the NahG plants at 6 dpi.
Table 1.
Changes in hormonal signalling Upregulated (+) and downregulated (–) MapMan functional groups identified by gene set enrichment analysis (false discovery rate corrected Q < 0.05) in potato cv. Rywal and NahG-Rywal at 1, 3, and 6 dpi after PVY inoculation. dpi, Days post infection.
| Functional group | Rywal | NahG-Rywal | ||||
|---|---|---|---|---|---|---|
| 1 dpi | 3 dpi | 6 dpi | 1 dpi | 3 dpi | 6 dpi | |
| Salicylic acid metabolism | + | + | + | |||
| Jasmonate metabolism | + | + | – | |||
| Ethylene metabolism | + | + | ||||
| Auxin signal transduction | – | – | – | – | ||
| ABA synthesis/degradation | – | – | ||||
| Gibberellin metabolism | + | + | ||||
| Brassinosteroid synthesis/degradation | + | + | – | |||
Repression of photosynthesis and activation of energy-producing pathways in the disease response of the NahG plants
In Rywal plants at 1 dpi, genes involved in photosynthesis were upregulated or downregulated. Among the upregulated group, there were genes involved in light reactions (PS I and II polypeptide units) and the Calvin cycle (RuBisCO small subunit). At 3 dpi, more photosynthesis-related genes were downregulated, while at 6 dpi, only a small number of genes were differentially expressed, again mostly downregulated. The highest number of changes in gene expression were observed at 6 dpi in the NahG plants (Fig. 5A), while downregulation of photosynthesis-related genes was observed as early as 1 dpi, and these effects became significant at 3 dpi (Supplementary Table S4), and even more pronounced at 6 dpi, when many genes were downregulated (Fig. 5B). The downregulation of photosynthesis genes coincided with downregulation of tetrapyrole biosynthesis genes.
At 6 dpi in the NahG plants, there were most changes in gene expression (Fig. 5A), while the genes related to photosynthesis, the primary source of carbohydrates, were downregulated (Fig. 5B). There was strong induction of most of the genes coding for enzymes involved in glycolysis, as well as enzymes catalysing the release of glucose and fructose from sucrose (e.g. vacuolar, cytosolic, and apoplastic invertases). Phosphorylated glucose from glycolysis can enter the oxidative pentose phosphate pathway, the genes for which were also upregulated both at 3 and 6 dpi (Supplementary Table S4). The gene encoding pyruvate dehydrogenase, a regulatory point for pyruvate flux, was highly upregulated. Genes of another pathway that contributes to substrate production, lipid β-oxidation, were significantly upregulated at 3 and 6 dpi in the NahG plants (Supplementary Table S4). Strong demand for energy can also mobilize amino acids into energy pathways; not surprisingly then, there was strong upregulation of the gene encoding glutamate dehydrogenase, a connector between amino acids and the citric acid cycle (Bolton, 2009).
Analysis of HR response to PVY at the proteome level
To complement the knowledge of the HR response of potato to PVY inoculation at the proteome level, this work analysed samples of cv. Rywal prior to the appearance of HR lesions (1 dpi, 3 dpi). Using a 2D differential gel electrophoresis approach, c.1200 protein spots were detected. Fifteen spots were differentially expressed after infection with PVY, in the comparison of PVY- versus mock-inoculated samples either at both time points or for individual time points. In three spots (1, 5, 9), the MS analyses indicated the presence of multiple proteins (at least one peptide matched a second nonrelated protein), and so these were excluded from further analysis. The differentially expressed proteins identified (Table 2) are involved in different metabolic pathways (e.g. in photosynthesis and different stress-response-related processes). These groups of proteins have been commonly detected as differentially expressed in previously studied plant–virus interactions (reviewed by Di Carli et al., 2012). Additionally, three differentially expressed spots were detected for which the MS analysis produced only one significant peptide match (spots 8, 12) or no significant match (spot 6), and could thus not be unambiguously identified.
Table 2.
Differentially expressed proteins in the hypersensitive resistance response of potato to infection with PVY Proteins are grouped according to their functional ontology and, for each spot, the hits identified through the NCBI databases (nr taxonomy: Viridiplantae) are given. Values are average ratio (log2) of the protein abundance (PVY vs. mock all together and separately for 1 and 3 dpi). dpi, Days post infection; NS, no significant change compared to control.
| Spot no. | NCBI ID | Description | PVY/mock | 1 dpi | 3 dpi |
|---|---|---|---|---|---|
| Photosynthesis | |||||
| 2 | gi|108773138 | RuBisCO large subunit | NS | NS | –0.8 |
| 3 | gi|108773138 | RuBisCO large subunit | NS | NS | –0.7 |
| 4 | gi|108773138 | RuBisCO large subunit | NS | NS | –0.8 |
| 13 | gi|417593 | RuBisCO small chain 2A | NS | NS | –0.5 |
| 15 | gi|197132134 | photosystem I subunit VII | NS | NS | –0.6 |
| Plant defence (PR proteins) | |||||
| 7 | gi|1705808 | Endochitinase 4 | –2.2 | NS | NS |
| Oxidation–reduction process (oxidative stress response) | |||||
| 10 | gi|228249640 | Fe superoxidase dismutase | –0.7 | –0.5 | –0.8 |
| 14 | gi|38073257 | Superoxidase dismutase | NS | NS | 0.4 |
| Protein folding | |||||
| 11 | gi|7331143 | Chaperonin 21 precursor | 0.6 | NS | NS |
| Unknown function | |||||
| 6 | Unidentified protein | –0.7 | NS | NS | |
| 8 | Unidentified protein | –0.7 | NS | NS | |
| 12 | Unidentified protein | NS | –0.6 | NS | |
This work next compared the protein abundance data to the transcriptome data (Supplementary Table S5). Two superoxide dismutases were identified as differentially abundant at the protein level: one increased, while the other decreased. This result corresponds to the changes detected at the transcriptome level, albeit the response is stronger at the gene expression level (Supplementary Table S5). Similarly, protein abundance of RuBisCO small chain 2A, endochitinase 4 and chaperonin 21 matched the results at the transcript level, but the response was less intensive at the protein level and in some cases more rapid at the transcriptome level. Discrepancies in these comparisons of proteome and transcriptome data might be the consequence of time delays between the transcript and protein synthesis, or of posttranscriptional modifications that give rise to different spots on 2D gels (Yang et al., 2011; Vogel and Marcotte, 2012). RuBisCO large subunit (gi|108773138) was identified in three different spots, and at the protein and transcript levels the results were diametrically opposed (i.e. upregulation at the transcript level, and downregulation at the protein level), which might be a consequence of posttranslational modifications of the protein.
Discussion
To study the role of SA signalling in Ny-1-mediated HR responses to PVY, a comprehensive analysis of cv. Rywal and SA-depleted NahG-Rywal plants at the cytological, biochemical, transcriptome, and proteome levels was performed.
Programmed cell death in the hypersensitive response to PVY
The HR is characterized by rapid and precisely localized cell death at the site of pathogen entry (Mur et al., 2008). In PVY-inoculated cv. Rywal, lesions efficiently restricted the viral movement. At the cytological level, features of programmed cell death were observed. Some of these might be connected with autophagy, which is hypothesized to form a negative feedback loop in the modulation of SA signalling, to negatively regulate senescence and programmed cell death (Yoshimoto et al., 2009). Other changes in ultrastructure, like chromatin condensation, have been observed previously in plant–pathogen interactions (Krzymowska et al., 2007). However, no qualitative or quantitative changes in mitochondria or chloroplasts were observed in the present study (Fig. 1C), which is in contrast to previous studies (Hinrich-Berger, 1999; Pompe-Novak et al., 2001; Krzymowska et al., 2007).
ROS are one of the major groups of programmed cell death effectors (Mur et al., 2008). In plant pathosystems, they are also involved in strengthening of the host cell walls and in cellular signalling and have thus been shown to be key elements in successful pathogen arrest (reviewed by O’Brien et al., 2012). Detection of large amounts of H2O2 from 3 dpi (Fig. 1E) and the strong upregulation of peroxidase and other redox-related genes from 1 dpi, (Fig. 5B, Supplementary Tables S3 and S4) suggest that a burst of ROS has an important role in the defence response in this pathosystem.
Callose deposition has also been shown to have an important role in plant–virus interactions (Iglesias and Meins, 2000; Dobnik et al., 2013; Zavaliev et al., 2013). In the present study, callose deposition appeared to demarcate the lesion cells from the surrounding tissue from 3 dpi (Fig. 1F) which can be linked to the upregulation of several cell-wall-related genes as early as 1 dpi (Fig. 5B, Supplementary Table S4).
SA is synthesized de novo in the Ny-1-dependent HR
In agreement with previous studies (Yu et al., 1997), high basal levels of SA, mostly in the form of SAGs were observed in this study. In contrast to the hypothesis of Yu et al. (1997), that it is an enhancement of SA perception in response to pathogen attack that triggers the defence response in potato, rather than de novo SA synthesis, the increases in both SA forms after viral infection (Fig. 2B) suggest de novo synthesis of SA. This was further confirmed in temperature-shift experiments, where the increased number of virus-infected cells triggered massive de novo synthesis of the free form of SA (Fig. 2C). Increase of SA (Fig. 2C) at elevated temperatures, when the virus spreads systemically, suggests that SA alone is not sufficient for an effective defence, but requires additional R-gene-mediated signalling. SNC1 and N-mediated resistance has been shown to be compromised at increased temperature (Zhu et al., 2010). Increased levels of salicylates have been detected in PVY-infected susceptible potato genotypes (Krečič-Stres et al., 2005) and in susceptible Arabidopsis ecotypes upon infection with different viruses (Chandra-Shekara et al., 2006; Palukaitis and Carr, 2008), which, together with the present data, could indicate that SA synthesis is, to some extent, independent of R-mediated resistance responses.
SA deficiency causes viral spread and disease development
In agreement with previous studies on compatible (Baebler et al., 2011) and incompatible (Sanchéz et al., 2010) interactions between potato and viruses, the inability of Rywal plants to accumulate SA resulted in a more susceptible phenotype (Fig. 3). The most pronounced phenotypic feature of the NahG-Rywal response to viral infection was the enlargement of the lesions until at least 14 dpi. The inability of the plant to restrict the virus led to the systemic spread of the virus (Fig. 3C), which was detected also in the next generation of plants derived from the tubers of the infected plants (data not shown). Larger lesions have been reported for NahG potato plants infected with Potato virus X (Sanchéz et al., 2010) and NahG tobacco plants infected with Tobacco mosaic virus (Mur et al., 1997), although these studies did not shown systemic spreading of these viruses. However, SA depletion has been shown to allow Plum pox virus spread in tobacco (Alamillo et al., 2006). In contrast, Wang and coworkers (2011) showed reductions in symptoms and viral multiplication in NahG Arabidopsis plants, which were possibly due to inhibition of ROS responses by higher levels of ascorbic acid and glutathione.
The data suggest that inhibition of systemic viral movement occurs at the level of cell-to-cell movement and phloem loading, as once a virus is present in the phloem, it can spread systemically even in the presence of elevated levels of SA in the plant (Fig. 4). Together with plasmodesmata-located proteins, SA has been implicated in cell-to-cell communication upon bacterial infection (Wang et al., 2013). Thus, it can be concluded that the formation of lesions is a SA-independent process, although SA is a crucial element for inhibition of virus multiplication and spreading, both locally and systemically.
Lack of SA results in quantitative differences in the transcriptome response to PVY
The whole-transcriptome analysis performed in the present study confirms the role of SA in orchestrating defence reactions. Recognition of an invading pathogen results in reprogramming of gene expression, to promote specific defence reactions in the infected plant.
The abundance and continuity of the Rywal transcriptome response at the first two time points suggests that an effective response exists already at 1 dpi, which it maintained to 3 dpi and diminished by 6 dpi (Fig. 5A). Host responses in incompatible virus interactions are more robust than those in compatible ones, most likely due to the delay in pathogen recognition or the temporal suppression of plant defence mechanisms in the compatible interactions (Jones and Dangl, 2006; Pacheco et al., 2012). In NahG plants, where the same signal was sent from Ny-1, a weak expression response was observed at 1 dpi (Fig. 5). The lack of both the basic SA level and its increase, which can potentiate defence responses, is the most probable reason for the observed quantitative differences in the NahG response to PVY and its subsequent failure to stop viral replication and spread. Plants have different mechanisms that regulate hormone balance (Robert-Seilaniantz et al., 2011). Therefore, if SA does not accumulate, then the SA-dependent mechanisms are not potentiated and the triggered response is ineffective, which leads to a shift to a different type of response in the NahG plants (Fig. 5). Differences in the extent of the gene expression changes between the genotypes (Fig. 5) can also be explained by spatiotemporal separation of the responses. As the greater part of the defence response is triggered in virus-infected tissue (Yang et al., 2007), it can be speculated that the SA-driven response is localized to the lesion area, and is mostly diminished when the lesions are fully developed and the viral spread has stopped.
Plants resist pathogen infection via activation of physical and biochemical defences, such as cell-wall thickening, lignin deposition, generation of ROS, and accumulation of secondary metabolites or antimicrobial proteins, such as the PR proteins (van Loon et al., 2006). Many of these processes have been shown to be SA dependent (Whitham et al., 2006; Wang et al., 2011). Observed differences in the defence-related responses between the genotypes used in the present study were quantitatively lower induction of PR proteins and delayed activation of cell-wall- and secondary-metabolism-related genes in the NahG-Rywal genotype, which confirms the role of SA in potentiating R-mediated responses (Shirasu and Lambas, 1997). Downregulation of cell-wall-modifying enzymes has been reported as a common feature of different plant–virus interactions (Yang et al., 2007; Pacheco et al., 2012). In the present study, the phenomenon has been only observed in the diseased stage (NahG.Rywal, 6 dpi). It appears that the rapid activation of cell-wall-modifying enzymes has a crucial role in preventing viral spread. The delay of the induction of the peroxidase genes (PR-9) might be connected with the disabled response in the NahG plants, as was shown for NahG tobacco plants (Mur et al., 2000). The basal level of PR-1 expression was two orders of magnitude lower in the NahG plants (data not shown), which indicates that SA is necessary for the maintenance of basal levels of the PR proteins, which was shown previously for NahG-Désirée potato plants (Halim et al., 2007; Baebler et al., 2011).
Perturbation of hormonal signalling in the absence of SA
Complex hormonal signalling networks and their crosstalk enable efficient defence responses. It is expected that, by compromising one component, the whole hormonal regulation network might be disturbed, which was shown in this study. While the observed changes (Table 1) mostly follow the classical binary model of JA/ET- and SA-signalling antagonism (Takahashi et al., 2004), the simultaneous upregulation of individual SA- and ET-signalling genes in Rywal at 3 dpi indicates possible synergistic interactions (Halim et al., 2009). This also suggests that both SA- and JA/ET-dependent responses are activated upon PVY infection, but the latter are activated later and are inhibited in Rywal plants.
Auxins are known to be involved in hormone crosstalk during pathogenesis. Downregulation of several genes involved in auxin signal transduction in the present study indicates antagonistic interactions between SA and the auxins in this pathosystem. In Arabidopsis, auxin-responsive factors have been shown to be the major targets of viral silencing suppressors, leading to developmental abnormalities (Jay et al., 2011). In contrary to the suggestion of positive correlation of JA and auxin signalling (Robert-Seilaniantz et al., 2011), genes coding for auxin-responsive factors were inhibited in the NahG plants, possibly due to the synergistic action of SA and JA discussed above. Moreover, SA has been shown to negatively regulate auxin transport through inhibition of clathrin-mediated endocytosis (Du et al., 2013), which might explain the downregulation of some of the auxin transport proteins (e.g. PIN7).
Activation of energy-producing pathways in the disease response of the NahG-Rywal plants
The allocation of resources at the onset of defence reactions results in a demand for energy in the infected tissue, as the production of defence-related compounds becomes ‘top priority’ for the plant, and the photosynthetic rates are reduced until the pathogenic growth has been terminated (Bolton, 2009). This appears to be the case in the NahG plants, where the photosynthesis- and chlorophyll-biosynthesis-related genes were strongly downregulated (Fig. 5B), which is in agreement with the transcriptome data from various biotic stress studies (reviewed by Bilgin et al., 2010). Interestingly, upregulation of genes involved in photosynthesis was observed at 1 and 3 dpi in Rywal plants. Local (Chou et al., 2000; Berger et al., 2004) and transient (Baebler et al., 2009) increases in photosynthetic rates and gene expression, respectively, were observed earlier. It appears that a transient activation of photosynthesis is triggered to provide energy for initiation of the resistance response.
The collapse of photosynthesis activity inevitably leads to metabolic transition from source to sink in infected tissue. The resulting demand for carbohydrates and energy is compensated for through increased activities of hexose transporters, cell-wall invertases, the oxidative pentose phosphate pathway, and respiratory metabolism, the genes for which were all upregulated in the NahG plants at 3 and 6 dpi. This reprogramming might further enhance the expression of defence-related genes and the production of secondary metabolites (Bolton, 2009). The oxidative pentose phosphate pathway is a major source of NADPH in nonphotosynthetic cells, and NADPH oxidases are the main contributors to ROS synthesis during HR (Mur et al., 2008). Interestingly, no dramatic changes in expression of genes in any of the energy-connected pathways were detected in the nontransgenic plants of cv. Rywal (Supplementary Table S4), which suggests that the response of this genotype is localized and precise, it does not lead to reprogramming of the entire plant metabolism, and it ceases when the virus is arrested.
Conclusions
Data presented here show that Ny-1-mediated resistance responses can completely restrict PVY to the site of the lesion. Recognition of PVY by Ny-1 generates a signal that initiates the plant defence response. This requires an increase in SA levels, to induce rapid changes in gene expression and to limit the viral spread. Virus inoculation induces de novo SA synthesis, which has a crucial role in the inhibition of PVY spreading into the parenchymal tissue. However, once the virus enters the plant vascular system, elevated levels of SA are not sufficient to stop it from spreading. SA deficiency of NahG plants results in attenuation of the plant defence responses and the virus can spread systemically.
The transcriptome analysis followed in the present study demonstrates many fascinating possibilities for interactions between both recognized and potential components of plant responses to viral infection. However, the exact signalling pathways and the effector components responsible for the changes observed still need to be elucidated.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Table S1. Quantitative PCR primers and probes used for microarray validation
Supplementary Table S2. Significance of changes in salicylates contents
Supplementary Table S3. Validation of microarray results by quantitative PCR.
Supplementary Table S4. Results of the gene set enrichment analysis (available as separate.xls file)
Supplementary Table S5. Comparison of transcriptome and proteome results.
Supplementary Fig. S1. PVY particles in lesion extracts
Supplementary Fig. S2. The expression of NahG in NahG-Rywal transgenic and control potato lines
Supplementary Fig. S3. Accumulation of salicylic acid and its glucoside in NahG lines
Supplementary Fig. S4. Symptom development and viral RNA accumulation after PVY inoculation and treatment with benzothiadiazole or catechol
Acknowledgements
The authors would like to acknowledge Mirek Sobczak, Neža Turnšek, and Sébastien Planchon for their excellent technical assistance, Magda Krzymowska for critically reading of the manuscript, and Chris Berrie for language revision. This study was partially conducted at the Centre for Functional Genomics and Bio-Chips, Ljubljana, Slovenia and was supported by the Slovenian Research Agency (programme P4-0165 and project J4-4165), the Polish National Science Centre (project no. N N302 114737), the Slovenian-Polish bilateral project (2010–2011), short-term FEBS and CEEPUS (2008-5463-BILATERAL) fellowships to KW, and COST actions (FA0806 and FA0603).
References
- Alamillo JM, Saénz P, García JA. 2006. Salicylic acid-mediated and RNA-silencing defense mechanisms cooperate in the restriction of systemic spread of plum pox virus in tobacco. The Plant Journal 48, 217–227 [DOI] [PubMed] [Google Scholar]
- Baebler Š, Krečič-Stres H, Rotter A, et al. 2009. PVYNTN elicits a diverse gene expression response in different potato genotypes in the first 12h after inoculation. Molecular Plant Pathology 10, 263–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baebler Š, Stare K, Kovač M, et al. 2011. Dynamics of responses in compatible potato–Potato virus Y interaction are modulated by salicylic acid. PloS One 6, e29009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S, Papadopoulos M, Schreiber U, Kaiser W, Roitsch T. 2004. Complex regulation of gene expression, photosynthesis and sugar levels by pathogen infection in tomato. Physiologia Plantarum 122, 419–428 [Google Scholar]
- Bilgin DD, Zavala JA, Zhu J, Clough SJ, Ort DR, DeLucia EH. 2010. Biotic stress globally downregulates photosynthesis genes. Plant, Cell and Environment 33, 1597–1613 [DOI] [PubMed] [Google Scholar]
- Bolton MD. 2009. Primary metabolism and plant defense-fuel for the fire. Molecular Plant–Microbe Interactions 22, 487–497 [DOI] [PubMed] [Google Scholar]
- Chandra-Shekara AC, Gupte M, Navarre D, Raina S, Raina R, Klessig D, Kachroo P. 2006. Light-dependent hypersensitive response and resistance signaling against Turnip Crinkle Virus in Arabidopsis . The Plant Journal 45, 320–334 [DOI] [PubMed] [Google Scholar]
- Chen H, Nelson RS, Sherwood JL. 1994. Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16, 664–8, 670. [PubMed] [Google Scholar]
- Chou HM, Bundock N, Rolfe SA, Scholes JD. 2000. Infection of Arabidopsis thaliana leaves with Albugo candida (white blister rust) causes a reprogramming of host metabolism. Molecular Plant Pathology 1, 99–113 [DOI] [PubMed] [Google Scholar]
- Chrzanowska M. 1991. New isolates of the necrotic strain of Potato virus Y (PVYN) found recently in Poland. Potato Research 34, 179–182 [Google Scholar]
- Di Carli M, Benvenuto E, Donini M. 2012. Recent Insights into plant–virus interactions through proteomic analysis. Journal of Proteome Research 11, 4765–4780 [DOI] [PubMed] [Google Scholar]
- Dobnik D, Baebler Š, Kogovšek P, Pompe-Novak M, Štebih D, Panter G, Janež N, Morisset D, Žel J, Gruden K. 2013. β-1,3-glucanase class III promotes spread of PVYNTN and improves in planta protein production. Plant Biotechnology Reports 7, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Tejos R, Beck M, Himschoot E, Li H, Robatzek S, Vanneste S, Friml J. 2013. Salicylic acid interferes with clathrin-mediated endocytic protein trafficking. Proceedings of the National Academy of Sciences, USA 110, 7946–7951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim VA, Altmann S, Ellinger D, Eschen-Lippold L, Miersch O, Scheel D, Rosahl S. 2009. PAMP-induced defense responses in potato require both salicylic acid and jasmonic acid. The Plant Journal 57, 230–242 [DOI] [PubMed] [Google Scholar]
- Halim VA, Eschen-Lippold L, Altmann S, Birschwilks M, Scheel D, Rosahl S. 2007. Salicylic acid is important for basal defense of Solanum tuberosum Against Phytophthora infestans . Molecular Plant–Microbe Interactions 20, 1346–1352 [DOI] [PubMed] [Google Scholar]
- Hennig J, Malamy J, Grynkiewicz G, Indulski J, Klessig DF. 1993. Interconversion of the salicylic acid signal and its glucoside in tobacco. The Plant Journal 4, 593–600 [DOI] [PubMed] [Google Scholar]
- Hinrich-Berger J. 1999. Cytological responses of susceptible and extremely resistant potato plants to inoculation with Potato virus Y. Physiological and Molecular Plant Pathology 55, 143–150 [Google Scholar]
- Huang Z, Yeakley JM, Garcia EW, Holdridge JD, Fan JB, Whitham SA. 2005. Salicylic acid-dependent expression of host genes in compatible Arabidopsis–virus interactions. Plant Physiology 137, 1147–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias VA, Meins F., Jr 2000. Movement of plant viruses is delayed in a beta-1,3-glucanase-deficient mutant showing a reduced plasmodesmatal size exclusion limit and enhanced callose deposition. The Plant Journal 21, 157–166 [DOI] [PubMed] [Google Scholar]
- Ishihara T, Sakurai N, Sekine KT, Hase S, Ikegami M, Shibata D, Takahashi H. 2004. Comparative analysis of expressed sequence tags in resistant and susceptible ecotypes of Arabidopsis thaliana infected with cucumber mosaic virus. Plant Cell Physiology 45, 470–480 [DOI] [PubMed] [Google Scholar]
- Jay F, Wang Y, Yu A, Taconnat L, Pelletier S, Colot V, Renou JP, Voinnet O. 2011. Misregulation of AUXIN RESPONSE FACTOR 8 underlies the developmental abnormalities caused by three distinct viral silencing suppressors in Arabidopsis . PLoS Pathogens 7, e1002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444, 323–329 [DOI] [PubMed] [Google Scholar]
- Jovel J, Walker M, Sanfaçon H. 2011. Salicylic acid-dependent restriction of tomato ringspot virus spread in tobacco is accompanied by a hypersensitive response, local RNA silencing, and moderate systemic resistance. Molecular Plant–Microbe Interactions 24, 706–718 [DOI] [PubMed] [Google Scholar]
- Kloosterman B, De, Koeyer D, Griffiths R, et al. 2008. Genes driving potato tuber initiation and growth: identification based on transcriptional changes using the POCI array. Functional and Integrative Genomics 8, 329–340 [DOI] [PubMed] [Google Scholar]
- Kogovšek P, Ravnikar M. 2013. Physiology of the potato–Potato virus Y interaction. In: Lüttge U, Beyschlag W, Francis D, Cushman J, editors, Progress in Botany 74. Berlin, Heidelberg: Springer Berlin Heidelberg [Google Scholar]
- Krečič-Stres H, Vučak C, Ravnikar M, Kovač M. 2005. Systemic Potato virus YNTN infection and levels of salicylic and gentisic acids in different potato genotypes. Plant Pathology 54, 441–447 [Google Scholar]
- Krzymowska M, Konopka-Postupolska D, Sobczak M, Macioszek V, Ellis BE, Hennig J. 2007. Infection of tobacco with different Pseudomonas syringae pathovars leads to distinct morphotypes of programmed cell death. The Plant Journal 50, 253–264 [DOI] [PubMed] [Google Scholar]
- Lewsey M, Palukaitis P, Carr JP. 2009. Plant–virus interactions: defence and counter-defence. In: Parker J, editor, Molecular aspects of plant disease resistance. Wiley-Blackwell, Oxford: pp 134–176 [Google Scholar]
- Mac A, Krzymowska M, Barabasz A, Hennig J. 2004. Transcriptional regulation of the gluB promoter during plant response to infection. Cellular and Molecular Biology Letters 9, 843–853 [PubMed] [Google Scholar]
- Malamy J, Carr JP, Klessig DF, Raskin I. 1990. Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250, 1002–1004 [DOI] [PubMed] [Google Scholar]
- Malamy J, Hennig J, Klessig DF. 1992. Temperature-dependent induction of salicylic acid and its conjugates during the resistance response to tobacco mosaic virus infection. The Plant Cell 4, 359–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marathe R, Guan Z, Anandalakshmi R, Zhao H, Dinesh-Kumar SP. 2004. Study of Arabidopsis thaliana resistome in response to cucumber mosaic virus infection using whole-genome microarray. Plant Molecular Biology 55, 501–520 [DOI] [PubMed] [Google Scholar]
- Mur LA, Brown IR, Darby RM, Bestwick CS, Bi YM, Mansfield JW, Draper J. 2000. A loss of resistance to avirulent bacterial pathogens in tobacco is associated with the attenuation of a salicylic acid-potentiated oxidative burst. The Plant Journal 23, 609–621 [DOI] [PubMed] [Google Scholar]
- Mur L, Kenton P, Lloyd AJ, Ougham H, Prats E. 2008. The hypersensitive response; the centenary is upon us but how much do we know? Journal of Experimental Botany 59, 501–520 [DOI] [PubMed] [Google Scholar]
- Mur LAJ, Bi YM, Darby RM, Firek S, Draper J. 1997. Compromising early salicylic acid accumulation delays the hypersensitive response and increases viral dispersal during lesion establishment in TMV-infected tobacco. The Plant Journal 12, 1113–1126 [DOI] [PubMed] [Google Scholar]
- Navarre DA, Mayo D. 2004. Differential characteristics of salicylic acid-mediated signaling in potato. Physiological and Molecular Plant Pathology 64, 179–188 [Google Scholar]
- O’Brien J, Daudi A, Butt VS, Bolwell GP. 2012. Reactive oxygen species and their role in plant defence and cell wall metabolism. Planta 236, 765–779 [DOI] [PubMed] [Google Scholar]
- Pacheco R, García-Marcos A, Manzano A, de Lacoba MG, Camañes G, García-Agustín P, Díaz-Ruíz JR, Tenllado F. 2012. Comparative analysis of transcriptomic and hormonal responses to compatible and incompatible plant–virus interactions that lead to cell death. Molecular Plant–Microbe Interactions 25, 709–723 [DOI] [PubMed] [Google Scholar]
- Palukaitis P, Carr JP. 2008. Plant resistance responses to viruses. Journal of Plant Pathology 90, 153–171 [Google Scholar]
- Peterhansel C, Freialdenhoven A, Kurth J, Kolsch R, Schulze-Lefert P. 1997. Interaction analyses of genes required for resistance responses to powdery mildew in barley reveal distinct pathways leading to leaf cell death. The Plant Cell 9, 1397–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompe-Novak M, Wrischer M, Ravnikar M. 2001. Ultrastructure of chloroplasts in leaves of potato plants infected by Potato virus YNTN . Phyton 41, 215–226 [Google Scholar]
- Postnikova O, Nemchinov LG. 2012. Comparative analysis of microarray data in Arabidopsis transcriptome during compatible interactions with plant viruses. Virology Journal 9, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team 2012. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- Renaut J, Hausman JF, Bassett C, Artlip T, Cauchie HM, Witters E, Wisniewski M. 2008. Quantitative proteomic analysis of short photoperiod and low-temperature responses in bark tissues of peach (Prunus persica L. Batsch). Tree Genetics and Genomes 4, 589–600 [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. 2011. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annual Review of Phytopathology 49, 317–343 [DOI] [PubMed] [Google Scholar]
- Rodrigo G, Carrera J, Ruiz-Ferrer V, del Toro FJ, Llave C, Voinnet O, Elena SF. 2012. A meta-analysis reveals the commonalities and differences in Arabidopsis thaliana response to different viral pathogens. PloS One 7, e40526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotter A, Usadel B, Baebler Š, Stitt M, Gruden K. 2007. Adaptation of the MapMan ontology to biotic stress responses: application in solanaceous species. Plant Methods 3, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchéz G, Gerhardt N, Siciliano F, Vojnov A, Malcuit I, Marano MR. 2010. Salicylic acid is involved in the Nb-mediated defense responses to Potato virus X in Solanum tuberosum . Molecular Plant–Microbe Interactions 23, 394–405 [DOI] [PubMed] [Google Scholar]
- Scholthof KB, Adkins S, Czosnek H, et al. 2011. Top 10 plant viruses in molecular plant pathology. Molecular Plant Pathology 12, 938–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K, Lambas C. 1997. Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. The Plant Cell 9, 261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RP, Valkonen JPT, Gray SM, Boonham N, Jones RAC, Kerlan C, Schubert J. 2008. Discussion paper: The naming of Potato virus Y strains infecting potato. Archives of Virology 153, 1–13 [DOI] [PubMed] [Google Scholar]
- Smyth GK. 2005. Limma: linear models for microarray data. In: Gentelman R, Carey S, Dudoit S, Irizarry R, Huber W, editors, Bioinformatics and computational biology solutions using R and Bioconductor , New York: Springer, pp. 397–420 [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences, USA 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajko K, Chrzanowska M, Witek K, Strzelczyk-Żyta D, Zagórska H, Gebhardt C, Hennig J, Marczewski W. 2008. The novel gene Ny-1 on potato chromosome IX confers hypersensitive resistance to Potato virus Y and is an alternative to Ry genes in potato breeding for PVY resistance. Theoretical and Applied Genetics 116, 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kanayama Y, Zheng MS, Kusano T, Hase S, Ikegami M, Shah J. 2004. Antagonistic interactions between the SA and JA signaling pathways in Arabidopsis modulate expression of defense genes and gene-for-gene resistance to cucumber mosaic virus. Plant Cell Physiology 45, 803–809 [DOI] [PubMed] [Google Scholar]
- Talarczyk A, Krzymowska M, Borucki W, Hennig J. 2002. Effect of yeast CTA1 gene expression on response of tobacco plants to tobacco mosaic virus infection. Plant Physiology 129, 1032–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. 1997. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley–powdery mildew interaction. The Plant Journal 11, 1187–1194 [Google Scholar]
- Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M. 2009. A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species, Maize. Plant, Cell and Environment 32, 1211–1229 [DOI] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CMJ. 2006. Significance of inducible defense-related proteins in infected plants. Annual Review of Phytopathology 44, 135–162 [DOI] [PubMed] [Google Scholar]
- van Wees SC, Glazebrook J. 2003. Loss of non-host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. The Plant Journal 33, 733–742 [DOI] [PubMed] [Google Scholar]
- Varet A, Parker J, Tornero P, Nass N, Nurnberger T, Dangl JL, Scheel D, Lee J. 2002. NHL25 and NHL3, two NDR1/HIN1–1ike genes in Arabidopsis thaliana with potential role(s) in plant defense. Molecular Plant–Microbe Interactions 15, 608–616 [DOI] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM. 2012. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nature Reviews in Genetics 13, 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Somerville S. 2000. Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proceedings of the National Academy of Sciences, USA 97, 1897–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuorinen AL, Gammerlgard E, Auvien P, Somervuo P, Dere S, Valkonen JPT. 2010. Factors underpinning the responsiveness and higher levels of virus resistance realised in potato genotypes carrying virus-specific R genes. Annals of Applied Biology 157, 229–241 [Google Scholar]
- Wang SD, Zhu F, Yuan S, et al. 2011. The roles of ascorbic acid and glutathione in symptom alleviation to SA-deficient plants infected with RNA viruses. Planta 234, 171–181 [DOI] [PubMed] [Google Scholar]
- Wang X, Sager R, Cui W, Zhang C, Lu H, Lee JY. 2013. Salicylic acid regulates plasmodesmata closure during innate immune responses in Arabidopsis . The Plant Cell 25, 2315–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham SA, Yang C, Goodin MM. 2006. Global impact: elucidating plant responses to viral infection. Molecular Plant–Microbe Interactions 19, 1207–1215 [DOI] [PubMed] [Google Scholar]
- Yang C, Guo R, Jie F, Nettleton D, Peng J, Carr T, Yeakley JM, Fan JB, Whitham SA. 2007. Spatial analysis of Arabidopsis thaliana gene expression in response to turnip mosaic virus infection. Molecular Plant–Microbe Interactions 20, 358–370 [DOI] [PubMed] [Google Scholar]
- Yang H, Huang Y, Zhi H, Yu D. 2011. Proteomics-based analysis of novel genes involved in response toward soybean mosaic virus infection. Molecular Biology Reports 38, 511–521 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K. 2009. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis . The Plant Cell 21, 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DQ, Liu YD, Fan BF, Klessig DF, Chen ZX. 1997. Is the high basal level of salicylic acid important for disease resistance in potato? Plant Physiology 115, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavaliev R, Levy A, Gera A, Epel BL. 2013. Subcellular dynamics and role of Arabidopsis beta-1,3-glucanases in cell-to-cell movement of tobamoviruses. Molecular Plant–Microbe Interactions 26, 1016–30 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Qian W, Hua J. 2010. Temperature modulates plant defense responses through NB-LRR proteins. PLoS pathogens 6, e1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.