Summary
A chloroplast-localized tetratricopeptide repeat-containing protein, SG1, was identified through a slow-greening mutant in Arabidopsis. SG1 is required for proplastid to chloroplast transition and its mutation disrupted the transcriptions of chloroplast-related genes. It also genetically interacts with GUN1 or GUN4.
Key words: Albino, Arabidopsis thaliana, chloroplast development, proplastid to chloroplast transition, slow greening, tetratricopeptide repeat-containing protein.
Abstract
A new gene, SG1, was identified in a slow-greening mutant (sg1) isolated from an ethylmethanesulphonate-mutagenized population of Arabidopsis thaliana. The newly formed leaves of sg1 were initially albino, but gradually became pale green. After 3 weeks, the leaves of the mutant were as green as those of the wild-type plants. Transmission electron microscopic observations revealed that the mutant displayed delayed proplastid to chloroplast transition. The results of map-based cloning showed that SG1 encodes a chloroplast-localized tetratricopeptide repeat-containing protein. Quantitative real-time reverse transcription–PCR data demonstrated the presence of SG1 gene expression in all tissues, particularly young green tissues. The sg1 mutation disrupted the expression levels of several genes associated with chloroplast development, photosynthesis, and chlorophyll biosynthesis. The results of genetic analysis indicated that gun1 and gun4 partially restored the expression patterns of the previously detected chloroplast-associated genes, thereby ameliorating the slow-greening phenotype of sg1. Taken together, the results suggest that the newly identified protein, SG1, is required for chloroplast development in Arabidopsis.
Introduction
The chloroplast is a crucial organelle in higher plants. It is essential for fixation of CO2 and also for biosynthesis of carbon skeletons, fatty acids, pigments, and amino acids from inorganic nitrogen (Staehelin, 2000). Plastids are generally believed to have originated from a unicellular photosynthetic bacterium, which was incorporated into a eukaryotic host cell (Dyall, 2004). During evolution, most of the genes encoded by the bacterial ancestor were transferred to the host nuclear genome. For example, the plastid genome of Arabidopsis thaliana encodes ~100 proteins; however, >2000 proteins are encoded by the nuclear genes that function in the chloroplast (Abdallah et al., 2000; Richly and Leister, 2004; Cui, 2006). Consequently, normal plastid development depends on the coordination of nuclear and plastid signals. This coordination is accomplished by nuclear signals that regulate the expression of plastid-encoded and nuclear-encoded plastid proteins, and also by signals sent from the developing plastids to the nucleus.
Plastids send signals to the nucleus via retrograde signalling, which operates through four distinct signal transduction pathways that are dependent on tetrapyrrole biosynthesis, plastid gene expression, the plastid redox state, or reactive oxygen species (ROS) (Surpin et al., 2002; Nott et al., 2006; Kakizaki et al., 2009). The identification and characterization of the genomes uncoupled (gun) mutants (gun1–gun6), in which the developmental status of plastids and the nuclear-encoded chloroplast genes are uncoupled, have considerably enhanced understanding of retrograde signalling (Susek et al., 1993; Mochizuki et al., 2001; Larkin et al., 2003; Woodson et al., 2011). An example of this uncoupling is the abnormally high expression level of light-harvesting chlorophyll a/b-binding protein 1 (LHCB1.1) in gun mutants when chloroplast development is blocked by the herbicide norflurazon {4-chloro-5-(methylamino)-2-[3-(trifluoromethyl) phenyl]-3-(2H)-pyridazinone} (Susek et al., 1993; Mochizuki et al., 2001).
The process of chloroplast biogenesis is a complex plastic process, involving the interaction of environmental, cellular, and temporal factors (Pogson and Albrecht, 2011). The most influential environmental factor is light—in the absence of light, proplastids change into etioplasts (Robertson and Laetsch, 1974). Cellular factors include factors inside and outside the plastids. Most of the identified cellular factors are chloroplast-localized proteins involved in protein import, chloroplast gene transcription, RNA maturation, and protein translation and assembly (Pogson and Albrecht, 2011). Temporal factors such as embryo maturation can also influence chloroplast development (Albrecht et al., 2008; Kim et al., 2009; Pogson and Albrecht, 2011).
In flowering plants, the development and activity of chloroplasts differ between cotyledons and true leaves. In cotyledons, plastids partially develop during embryogenesis; however, their development is arrested during seed maturation and dormancy. Cotyledons serve primarily as storage organs until the seedling becomes autotrophic; the cotyledons may then develop chloroplasts (Mansfield and Briarty, 1996; Charuvi et al., 2012). In contrast, the chloroplasts of true leaves differentiate directly from the proplastid present in the shoot apex, and their primary function is photosynthesis (Charuvi et al., 2012). In developed leaves, chloroplasts are further propagated by fission, similar to that observed in bacteria (Pyke, 1997; Leon et al., 1998; Pogson and Albrecht, 2011). These differences have been examined in mutants having chloroplast defects restricted either to the cotyledons or to the true leaves. For instance, the snowy cotyledon (sco) mutant group has chlorotic or bleached cotyledons but green true leaves (Albrecht et al., 2006, 2008, 2010; Shimada et al., 2007). Conversely, immutans (im) and variegated2 (var2) mutants have green cotyledons but chlorotic true leaves (Aluru et al., 2006; Kato et al., 2007; Liu et al., 2010).
In the past decade, studies of chloroplast development mutants have enhanced our understanding of the transition from proplastids to chloroplasts within true leaves. A useful and well-characterized mutant of Arabidopsis, var2, has green cotyledons and true leaves with green and white areas. The phenotype is the result of deactivation of a metalloprotease, FtsH2. The chloroplasts in the green sectors are normal; however, the white sectors contain undifferentiated plastids, indicating the blockage of proplastid to chloroplast transition (Kato et al., 2007; Sakamoto et al., 2009). The occurrence of such variegated mutants has also been reported in maize (Zea mays) and tomato (Solanum lycopersicum) (Han et al., 1992; Carol and Kuntz, 2001). In Arabidopsis, the delayed greening1 mutant (dg1) has a delayed-greening phenotype, caused by postponed chloroplast development (Chi et al., 2008). Many albino mutants have also been reported, for example csr1-1 in Z. mays and atecb2 in Arabidopsis (Asakura et al., 2004; Yu et al., 2009). Most of these mutations are lethal, because they prevent the development of mature chloroplasts.
In the present study, a tetratricopeptide repeat (TPR)-containing protein, slow green1 (SG1), was identified that affects chloroplast development in Arabidopsis. The sg1 mutant had disturbed expression of chloroplast-related genes and displayed delayed chloroplast differentiation. The findings suggest that SG1 is required for chloroplast development in Arabidopsis.
Materials and methods
Plant materials and growth conditions
Arabidopsis ecotype Columbia (Col) was used as the wild type (WT). T-DNA insertion lines SALK_046229C and SALK_026339 were obtained from the ABRC (Ohio State University). The seeds of gun1-1, gun4-1, and gun5-1 were kindly provided by Professor Enrique Lopez Juez (University of London, UK). All plants were grown at room temperature (22–25 °C) under long-day conditions (16h light/8h dark). To inhibit photoinhibition, sg1 was grown under 80 mmol m–2 s–1 white light. For phenotype identification, all plants were grown under conditions similar to that used for sg1. Otherwise, the plants were grown under 125 mmol m–2 s–1 white light.
Mutant isolation and mapping
The sg1 mutant was isolated from an ethylmethanesulphonate (EMS)-mutagenized M2 population with a Col background. The SG1 locus was mapped by using individuals of an F2 population derived from a cross between sg1 and WT Ler (Landsberg erecta). After PCR amplification, genetic markers were scored as simple sequence length polymorphism (SSLPs) or cleaved amplified polymorphic sequences (CAPSs) (Michaels and Amasino, 1998). The markers and restriction enzymes used to reveal the polymorphisms are detailed in Supplementary Tables S3 and S4 available at JXB online. MIE15 was a CAPS marker cleaved by the MseI-restricted enzyme. Map distances were calculated according to Kosambi (1943).
Transmission electron microscopy
Immediately after harvest, the sixth leaves from plants at different growth stages were cut into small pieces, and fixed in 2.5% glutaraldehyde in phosphate buffer (pH 7.4) for 4h at 4 °C. The samples were rinsed and incubated in 1% OsO4 for 12h at 4 °C. Then the samples were rinsed with phosphate buffer (pH 7.0), infiltrated with a graded series of epoxy resin in epoxy-propane, and then embedded in Epon 812 resin. Thin sections (~50nm) were obtained by using an ultramicrotome (Leica). The sections were stained in 2% uranyl acetate (pH 5.0), followed by 10mM lead citrate (pH 12), and viewed under a transmission electron microscope (JEM-1230). All images in this study were processed using Adobe Photoshop and Image J software.
Complementation test and overexpression of SG1
For the complementation test, the region between ~1500bp upstream of SG1 and the entire genomic fragment of SG1 was PCR-amplified by using the primers SG1-P1F (ACG CGT CGA CGT CTT GGC CTT TTA GTA GTT TAA TG) and SG1-P1R (GG GGT ACC GTT CTC CTC ACT ACC AC), and cloned into the binary vector pCAMBIA1300 by using the SalI and KpnI enzymes (underlined regions indicate the introduced SalI and KpnI sites, respectively). For overexpression of SG1, the full-length genomic DNA of SG1 (introns are absent in SG1) was amplified by using the primers SG1-P2F (GC TCT AGA GCA TGA TTT CGT CTC TCT CAG) and SG1-P1R, and cloned into the binary vector pCAMBIA1300-GFP (green fluorescent protein) by using the XbaI and KpnI enzymes. The resulting plasmids pSG1::SG1::GFP and p35S::SG1::GFP were transferred into Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986), and transformed into sg1 or WT plants by using the floral dip method (Clough and Bent, 1998). Transformed plants were selected on Murashige and Skoog (MS) medium containing 25mg l–1 hygromycin.
Subcellular localization of GFP proteins
The constructed p35S::SG1::GFP plasmid was transformed into Arabidopsis protoplasts to observe the transient expression of the fusion protein. Meanwhile the p35S::GFP vector was transformed as a control. The procedures for protoplast isolation and plasmid transformation were described by Kim and Somers (2010). The transformed protoplasts were observed by using confocal microscopy. A 488nm argon ion laser line was used for excitation of GFP and chlorophyll, while 505–515nm and 650nm emission filters were used for simultaneously capturing GFP and chlorophyll fluorescence, respectively, by using an Olympus FV1000MPE2 confocal microscope.
Chlorophyll detection
Total chlorophyll was determined in triplicate according to the method described by Lichtenthaler and Wellburn (1983). Extracts were obtained from the sixth leaves or seedlings at different growth stages. Approximately 0.2g of fresh tissue was homogenized in 5ml of 80% acetone for 12h in darkness. Spectrophotometric quantification was carried out in a Gene Quant spectrophotometer (GE Healthcare), using the following calculations: Chl a=12.21×A663–2.81×A646, and Chl b=20.13×A646–5.03×A663 (μg ml–1).
Real-time reverse transcription–PCR
Total RNA was extracted from 0.5g of plant tissue by using the E.Z.N.A Plant RNA Kit (Omega) according to the manufacturer’s instructions, with the addition of an RNase-free DNase I treatment (Omega). The cDNAs were synthesized from 1 μg of total RNA using the Prime Script™ RT Reagent Kit (Perfect Real Time; Takara). All of the quantitative real-time reverse transcription–PCR (qRT–PCR) measurements were performed using an MX 3000 Real-time PCR system (Stratagene) with SYBR Premix Ex Taq (Takara, Japan), according to the manufacturer’s instructions. The housekeeping gene β-tubulin was used as a normalization control. The relative expression was calculated by using the formula 2–ΔΔCt. All the experiments were performed for each biological replicate. The primer sequences for qRT–PCR are provided in Supplementary Table S5 available at JXB online.
Protein extraction and SDS–PAGE
Seedlings or different growth stage leaves harvested were immediately frozen in liquid nitrogen and pulverized. Total proteins were extracted in extraction buffer (0.1% Triton X-100, 0.1% SDS, 0.01M EDTA, 0.01M β-mercaptoethanol, 0.05M Na2HPO4, pH 7.0). The homogenates were centrifuged at 12 000rpm for 15min at 4 °C. The protein concentrations of the supernatants were determined by using western blotting of β-tubulin. Total proteins were separated by 12% SDS–PAGE. For each experiment, a minimum of three independent replicates were performed.
Results
Identification of a slow-greening mutant
To identify the genes involved in chloroplast development, a slow-greening mutant, designated sg1, was isolated from an EMS-mutagenized population of Arabidopsis. The initial rosette leaves of sg1 were completely albino, but gradually became green (Fig. 1A). At ~3 weeks post-emergence, the leaves of the mutant were as green as those of the WT (Fig. 1A). The slow-greening phenotype was apparent in other newly formed organs of sg1, including the stems, inflorescences, and siliques. The young inflorescences and siliques of sg1 were white or pale green, and became green as they matured (Fig. 1B, C). These observations are consistent with a pigment deficiency in sg1, and therefore the levels of chlorophyll a and chlorophyll b were measured at different growth stages of leaf development. The sixth leaves of 3-, 4-, and 5-week-old sg1 mutant plants (exhibiting albino, pale-green, and green leaves, respectively) and the corresponding Col leaves were used. Consistent with their phenotypes, the chlorophyll contents increased as the leaves turned green; when the sixth leaves of 5-week-old sg1 mutants became green, the chlorophyll contents were markedly higher than those of the albino leaves, but lower than those of the WT (see Supplementary Table S1 available at JXB online). Over time, the chlorophyll contents of the mutant and wild-type leaves became comparable. At all growth stages, the mutant plants were smaller than the WT (Fig. 1A).
Fig. 1.
The phenotypes of the sg1 mutant. (A) Seedlings of the sg1 mutant and WT (Col) grown for 1–5 weeks in soil. (B, C) Inflorescences and young siliques of the Col and sg1 mutant. d, days after germination. Bars=1cm in (A); 0.5cm in (B, C).
Many chloroplast-development mutants of Arabidopsis can grow well when supplied with sucrose as a carbon source (Koch, 1996; Chi et al., 2008; Yu et al., 2009); further, they may show abnormal embryo development (Uwer et al., 1998; Apuya et al., 2001; Kobayashi et al., 2007). To investigate whether SG1 is involved in chloroplast development, sg1 seedlings were grown on MS medium without or with 2% sucrose, and the sg1 embryogenesis of sg1 homozygotes was observed. It was determined that sucrose partially alleviated the albino phenotype of sg1, and also that embryogenesis of sg1 in heterozygote plants was delayed (see Supplementary Fig. S1 available at JXB online). The results suggest that sg1 affects chloroplast development during the early stages of seedling growth.
Differentiation of proplastids into chloroplasts
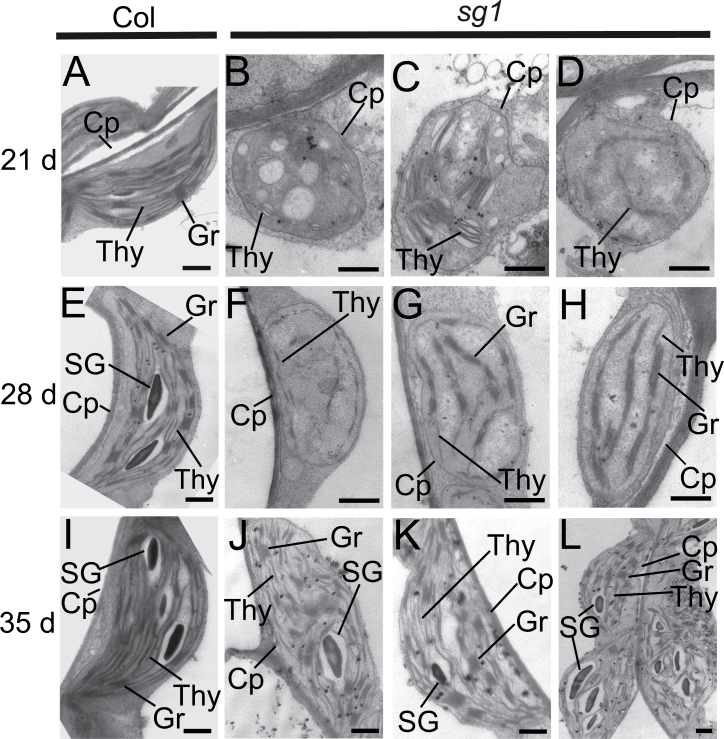
The delayed-greening phenotype of sg1 implies defective chloroplast development. Transmission electron microscopy was used to examine the chloroplast ultrastructures of the sixth leaves of 3-, 4-, and 5-week-old mutant plants (exhibiting albino, pale-green, and green leaves, respectively). In wild-type plants grown under normal conditions, the proplastid to chloroplast transition occurs at the shoot apical meristem during the early stages of development (Charuvi et al., 2012). Thus, in the present study, the chloroplasts of WT leaves at the same developmental stages were already differentiated and crescent-shaped, and contained well-developed thylakoid membranes with grana stacks (Fig. 2A, E, I). Starch grains were lacking in the chloroplasts of 3-week-old WT plants (Fig. 2A), but were present in the chloroplasts of 4- and 5-week-old plants (Fig. 2E, I). The albino leaves of 3-week-old sg1 seedlings contained few well-developed crescent-shaped chloroplasts, but many smaller, abnormal, irregularly shaped chloroplasts, similar to proplastids (Fig. 2B–D). These abnormal chloroplasts could be classified into three types according to their morphologies. The first type was rounded and highly vacuolated, with almost no thylakoid membrane, and appeared undifferentiated (Fig. 2B). The second type had fewer vacuoles and easily observable thylakoid membranes (Fig. 2C), possibly representing an intermediate form between the proplastid and chloroplast. The third type had discontinuous thylakoid membranes, resembling chloroplasts at an early stage of development (Fig. 2D). The pale-green leaves of 4-week-old sg1 seedlings contained differentiated chloroplasts that were smaller and had fewer thylakoid membranes than did chloroplasts of WT seedlings at the same growth stage (Fig. 2F–H); these chloroplasts resembled WT chloroplasts at an early stage of development. Some of the thylakoid membranes were discontinuous and some were arranged as grana stacks (Fig. 2F–H). Similar to the WT, the green leaves of 5-week-old sg1 plants contained chloroplasts with well-developed thylakoid membranes and grana stacks, and well-developed starch grains (Fig. 2J–L). The conversion of proplastids to chloroplasts in sg1 occurred over a period of ~3 weeks. These results suggest the delayed transition of proplastids to chloroplasts in sg1, consistent with a slow-greening phenotype.
Fig. 2.
Transmission electron micrographs of chloroplasts from the sixth leaves of 3- to 5-week-old Col and sg1 plants. (A, E, I) Chloroplast structures from the wild-type plants. (B–D), (F–H), and (J–L) Chloroplast structures from the sg1 mutant plants. Cp, chloroplast; SG, starch grain; Thy, thylakoid; Gr, grana thylakoids. Bars=1 μm.
Gene cloning and complementation of sg1
To verify whether sg1 is a nuclear recessive mutant, the M3 generation families of sg1 were crossed reciprocally with wild-type plants. Plants in the F1 generations of sg1 (♂)×Col (♀) and Col (♂)×sg1 (♀) were as green as the WT. The offspring of F1 plants from both of the crosses segregated in a 3:1 ratio (see Supplementary Table S2 available at JXB online). These results suggest that sg1 is a single recessive gene mutation with nuclear inheritance.
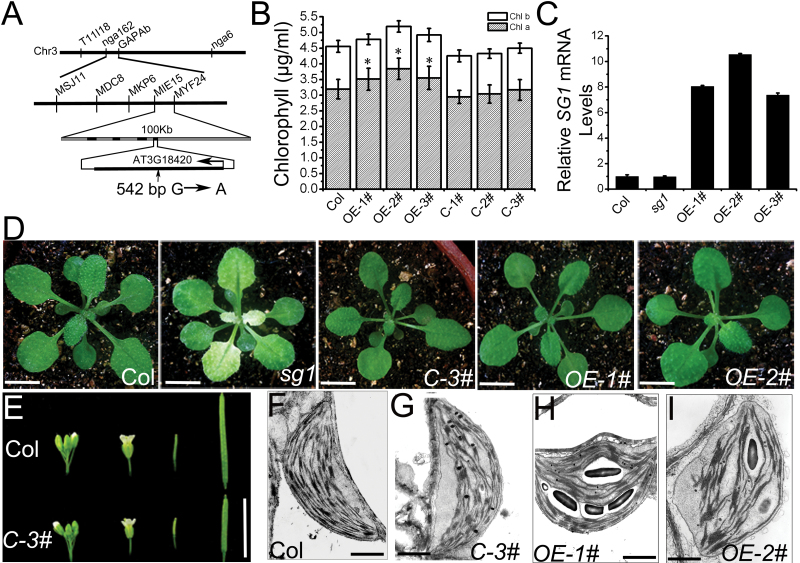
A map-based cloning approach was used to identify the mutated gene, by crossing sg1 with the Landsberg erecta (Ler) ecotype of Arabidopsis, to generate an F2 mapping population. SSLP markers (see Supplementary Table S3 available at JXB online) were selected for primary determination of the linkage group. On the basis of 21 F2 plants, it was concluded that SG1 was located on the upper arm of chromosome 3, in the interval between the markers NGA162 (7.24%) and GAPAb (16.24%) (Fig. 3A). Additional InDel markers and a CAPS marker (Fig. 3A; Supplementary Table S4 available at JXB online) were used to refine the position of SG1. On the basis of 146 F2 plants, the location of SG1 was narrowed down to an ~110kb region between the markers MIE15 and MYF24. In this region, only four genes encoding proteins predicted to be involved in chloroplast localization were identified, namely AT3G18230, AT3G18270, AT3G18390, and AT3G18420. Sequence analysis of the open reading frames of these four candidate genes revealed the existence of a single G to A mutation in base pair 542 from the start codon ATG of AT3G18420 genomic DNA; this mutation caused a conversion of arginine to lysine in amino acid 181 of SG1 protein (Fig. 3A). The expression level of SG1 in the sg1 mutant was determined, and it was shown that the G to A substitution did not affect mRNA accumulation (Fig. 3C). To confirm that the SG1 gene is AT3G18420, the sg1 mutant was genetically complemented with the full-length AT3G18420 cDNA under control of the promoter (1500bp upstream of the open reading frame) of AT3G18420. A total of 22 T1 transgenic plants were screened for pSG1::SG1 with a sg1 background. Subsequent phenotypic observations confirmed that the complemented mutants had WT traits (Fig. 3D). Further, the chlorophyll contents of 28-day-old rescued transgenic plants were similar to those of the WT (Fig. 3B), and ultrastructural examination of the chloroplasts from the sixth leaves of these plants revealed that they were well developed and similar to those of the WT (Fig. 3F). These results indicate that AT3G18420 can complement the chloroplast differentiation defects in the sg1 mutant, thereby further suggesting that AT3G18420 is responsible for the sg1 phenotypes. Two T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center. The T-DNA insertion harboured 296bp upstream of the ATG translation start codon in the SALK_046229C line, and 74bp downstream of the TAA translation stop codon in the SALK_026339 line. The results of RT–PCR indicated that the SG1 gene could be expressed in both T-DNA lines, without apparent effects on the chloroplasts.
Fig. 3.
Identification of the SG1 gene. (A) Map-based cloning, with the locations of molecular markers and mutation site indicated. (B) Chlorophyll concentration in mature rosette leaves of 4-week-old Col and overexpressing and complemented plants. (C) qRT–PCR analysis of SG1 transcription levels in sg1 and differently overexpressing transgenic lines with a Col background. (D) Seedlings of sg1 mutants, complemented (C-3#), and overexpressing transgenic plants (OE-1# and OE-2#) grown for 4 weeks in soil. (E) Inflorescences and siliques of complemented plants (C-3#). (F–I) Transmission electron micrographs of chloroplasts in 4-week-old leaves from Col, the complemented (C-3#), and overexpressing transgenic plants (OE-1# and OE-2#). Bars=0.5cm in (D, E); 1 μm in (F–I). Values represent the mean±SD of three independent experiments. Asterisks denote significant differences (P < 0.05).
To investigate further the function of SG1 in chloroplast development, a plasmid containing the full-length cDNA of AT3G18420, under control of the Cauliflower mosaic virus 35S promoter (CaMV 35S), was constructed, which was transformed to the WT. Thirty-one T1 overexpressing (OE) transgenic plants were screened, and no visible phenotypic effects were observed (Fig. 3C, D). The chlorophyll contents were measured and the ultrastructures of three OE lines were examined; it was determined that the content of chlorophyll a was slightly higher than that of WT plants (Fig. 3B). The chloroplasts of the OE lines were somewhat irregularly shaped, and had a slightly higher proportion of thylakoid membranes, especially in the grana; these characteristics may be responsible for the higher chlorophyll a content (Fig. 3B, H, I). The results indicate that SG1 plays an important role in chloroplast development.
Encoding of a conserved, widely expressed, chloroplast-localized TPR-containing protein by SG1
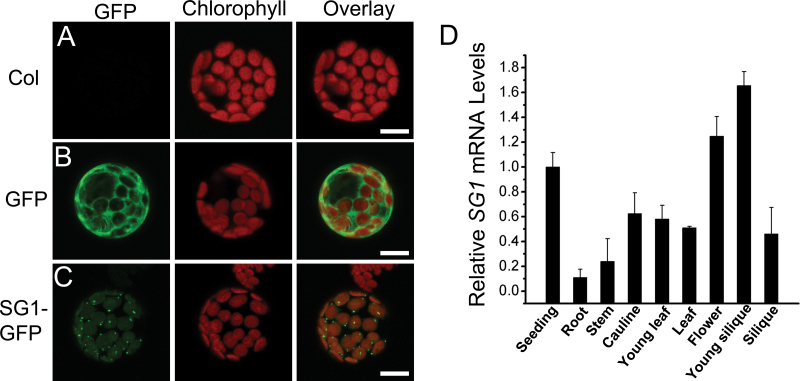
Analysis of the complete Arabidopsis sequence by using BLAST revealed that the nuclear genome contains a single copy of the SG1 gene. The results of phylogenetic analysis and protein alignments indicated that SG1 was conserved during the evolutionary process; further, it shares significant identity with the (hypothetical) Arabidopsis proteins—AT2G37400, AT3G53560, AT5G02590, and AT3G09490—which are chloroplast lumen common family proteins (see Supplementary Fig. S2 available at JXB online). SG1 encodes a putative polypeptide of 316 amino acids, with four TPR motifs (see Supplementary Fig. S2B available at JXB online). To confirm the subcellular localization of SG1, the transient expression of the p35S-SG1-GFP plasmid was examined in living Arabidopsis protoplasts. As a control, Arabidopsis protoplasts were transformed with a plasmid containing only GFP, under control of the 35S promoter. The GFP signals were observed by using confocal laser-scanning microscopy. In the absence of transformation, only chlorophyll autofluorescence was detected (Fig. 4A). After transformation with the control vector, GFP signals accumulated ubiquitously in the cytosol (Fig. 4B). In contrast, in the p35S-SG1-GFP-transformed protoplasts, GFP signals were coincident with chlorophyll autofluorescence; further, some SG1–GFP fusion protein was observed accumulated as spots in the chloroplast, which may be important for its function (Fig. 4C).
Fig. 4.
Subcellular localization and expression profiles of SG1. (A) Protoplast from a wild-type plant. (B, C) Protoplast transformed with p35S::GFP and p35S::SG1::GFP. Left to right: fluorescent image of GFP, chlorophyll autofluorescence, and merged image of GFP and chlorophyll autofluorescence. (D) Expression profiles of the SG1 gene based on qRT–PCR analysis of SG1 transcripts in various organs. Seedling, 10-day-old seedlings; root, roots from 10-day-old seedlings; stem, stem only, with all leaves and inflorescences removed; cauline, cauline leaves; young leaf, the sixth leaves of 5-week-old plants; leaf, mature rosette leaves of 5-week-old plants; flower, flower clusters; young silique, siliques at 1 week after fertilization; silique, green siliques at >1 week after fertilization. Bars=10 μm. Values represent the mean±SD of three independent experiments.
Next, the expression profiles of SG1 was examined. The mRNAs were isolated from different tissues of WT plants, and the SG1 expression level was detected by using qRT–PCR. The housekeeping gene β-tubulin, the expression levels of which remain similar across different tissues, was used to normalize different samples. The expression level of SG1 in 10-day-old seedlings was arbitrarily set to 1. It was determined that SG1 was widely expressed in all Arabidopsis tissues (Fig. 4D). The highest expression levels were observed in young siliques and flower clusters, while the lowest expression level was observed in roots (Fig. 4D). SG1 was ubiquitously expressed throughout the plant; however, its expression was preferentially associated with green tissues, particularly newly formed tissues. These results indicate that SG1 localizes to the chloroplast and exhibits ubiquitous expression.
Disrupted expression levels of genes associated with chloroplast development, photosynthesis, or chlorophyll biosynthesis
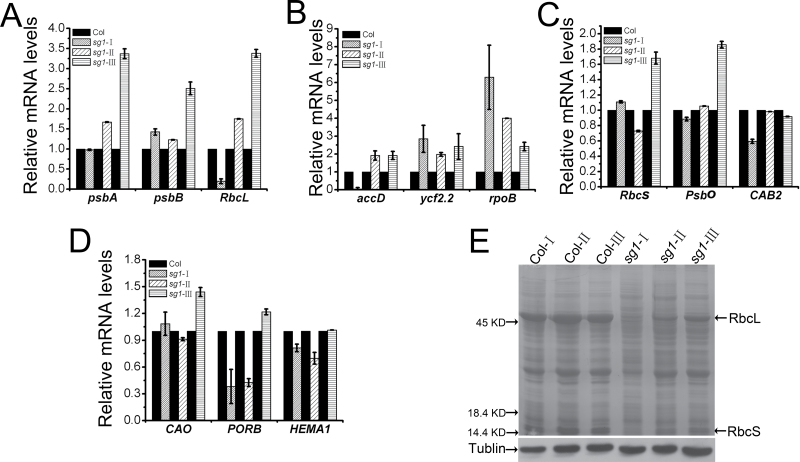
The TPR or TPR-related motif-pentatricopeptide repeat (PPR) proteins are reported to be involved in chloroplast gene expression (Pfalz et al., 2006; Chi et al., 2008; Su et al., 2012). Since abnormal chloroplast development was observed in sg1, the effect of the loss of SG1 on the expression of chloroplast-related genes was investigated using qRT–PCR. The transcription levels of plastid-encoded polymerases (PEPs) and nucleus-encoded polymerases (NEPs), which transcribe chloroplast genes, nuclear-encoded chloroplast genes, and chlorophyll biosynthesis genes, were examined in the albino, pale-green, and green leaves of sg1 plants, and also in the corresponding WT leaves (Fig. 5A–D). Three PEP genes, namely those encoding two members of photosystem II complexes (psbA and psbB) and a RuBisCO large subunit (RbcL), were selected (Fig. 5A). Three NEP genes were also selected, namely an accD, which encodes a carboxytransferase β subunit of the acetyl-CoA carboxylase (ACCase) complex, ycf2.2, which encodes a predicted chloroplast-localized ATP-binding protein, and rpoB, which encodes a chloroplast DNA-dependent RNA polymerase B subunit (Fig. 5B). Three nuclear-encoded chloroplast genes: RuBisCO small subunit (RbcS), light-harvesting chlorophyll a/b-binding protein (CAB2/LHCB1.1), and oxygen evolving polypeptide 1 (PsbO) were detected (Fig. 5C). Genes that are crucial to chlorophyll biosynthesis, namely CAO (encoding chlorophyllide-a oxygenase), HEMA1 (encoding glutamyl-tRNA reductase 1), and PORB (encoding protochlorophyllide oxidoreductase B), were also selected for detection (Fig. 5D). The expression levels of psbA, RbcL, accD, PsbO, and PORB showed very similar tendencies. The expression levels of RbcL, accD, and PORB (especially RbcL and accD) were markedly lower in the albino leaves of mutant plants than in the WT; however, the expression levels of all five genes gradually increased as the leaves became green, to reach higher levels than those of the WT (Fig. 5A–D). Nevertheless, the expression of the rpoB gene showed the opposite expression pattern. Its expression level was markedly higher in albino leaves than in the leaves of the WT; further, the expression level gradually decreased as the leaves grew, but remained slightly higher in green leaves than in the leaves of the WT (Fig. 5B). The expression of psbB, ycf2.2, RbcS, and CAO was a little higher in sg1 albino leaves than in the WT. These four genes and HEMA1 (lower in albino leaves) decreased in pale-green leaves while they increased when the leaves turned green (Fig. 5A–D). The expression level of CAB2 was lower in albino leaves than in the leaves of the WT; however, in pale-green and green leaves, it increased to approximately the same level as in the leaves of the WT (Fig. 5C). Thus, the expression levels of all four types of genes were affected in sg1. In addition, the soluble proteins were profiled in leaves of sg1 mutant plants at different growth stages. It was determined that albino leaves of mutant plants contained significantly lower amounts of RbcL and RbcS (Fig. 5E). As the leaves became green, the amounts of these two proteins gradually increased (Fig. 5E). The increase in RbcL was in accordance with the transcript data. On the other hand, the expression level of RbcS did not change in the albino leaves, despite a significant decrease in the level of encoded protein. Furthermore, in the green leaves of sg1, the expression levels of RbcL and RbcS were higher than those in the corresponding WT leaves; however, the protein levels were lower than were those of the corresponding WT leaves. Therefore, SG1 may also be involved in chloroplast protein biosynthesis and/or degradation.
Fig. 5.
Transcript and protein analysis from leaves of sg1 plants at different stages. (A–D) The expression levels of plastid-encoded polymerase (PEP) transcribed chloroplast genes, nucleus-encoded polymerase (NEP) transcribed chloroplast genes, nuclear-encoded chloroplast genes, and chlorophyll biosynthesis genes. (E) Total protein resolved by SDS–PAGE from leaves of sg1 and corresponding wild-type plants at different growth stages. The terms sg1-I, sg1-II, and sg1-III refer to the sixth leaves that exhibited albino, pale-green, and green leaves in 3- to 5-week-old sg1 mutants, respectively; Col-I, Col-II, and Col-III refer to the corresponding leaves in Col plants. Western blotting of β-tubulin was used to confirm equal loading.
Genetic interaction of GUN1 and GUN4 with SG1
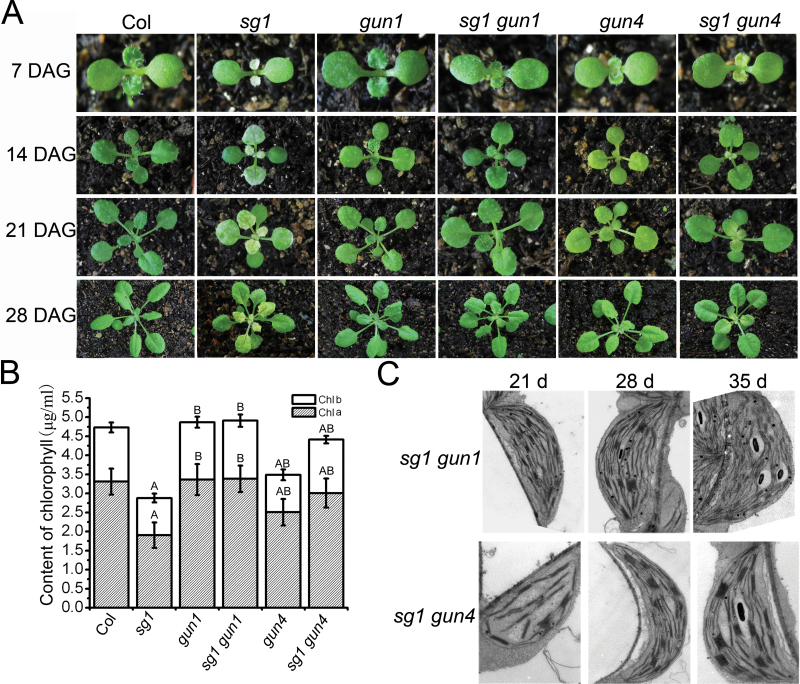
To investigate further the putative pathways in which SG1 may be involved in chloroplast development, sg1 gun1 and sg1 gun4 double mutants were constructed by crossing sg1 with gun1-1 and gun4-1 mutants. Interestingly, both sg1 gun1 and sg1 gun4 double mutants alleviated the delayed-greening phenotype of sg1 (Fig. 6A). The leaves of the sg1 gun1 double mutant were of a similar green colour to the leaves of the WT plants (Fig. 6A). Further, this double mutant had no albino leaves, but the young buds and basal parts of the second and third inner leaves were slightly pale green (Fig. 6A). Similarly, few or no albino leaves were observed in sg1 gun4, and its mature leaves were completely green, unlike the pale-green phenotype of gun4-1 (Mochizuki et al., 2001; Larkin et al., 2003) (Fig. 6A). In both of the double mutants, the newly formed inflorescences and siliques were of a similar green colour to those of WT plants. The chlorophyll contents of 2-week-old Col, sg1, gun1, sg1 gun1, gun4, and sg1 gun4 seedlings were compared after removal of the cotyledons. It was determined that both of the double mutants had higher chlorophyll contents than did the sg1 mutant (Fig. 6B). The gun4 mutant showed chlorophyll defects; however, the chlorophyll contents of the sg1 gun4 double mutant were considerably higher than were those of the sg1 and gun4 single mutants while they were lower than those of the WT. The chlorophyll contents of the sixth leaves (at different stages) of these plants were also measured, and concurring results were obtained (see Supplementary Table S1 available at JXB online).
Fig. 6.
The gun1 and gun4 mutants alleviated the delayed greening phenotype of sg1. (A) Seedlings of Col, sg1, gun1, sg1 gun1, gun4, and sg1 gun4 grown for 1–4 weeks in soil. (B) Chlorophyll concentrations of 2-week-old Col, sg1, gun1, sg1 gun1, gun4, and sg1 gun4 seedlings with cotyledons removed. (C) Chloroplast structures of the sixth leaves from 3- to 5-week-old sg1 gun1 and sg1 gun4 plants. Bars=0.5cm in A; 1 μm in B. Values represent the mean±SD of three independent experiments. aSignificantly different from Col, P < 0.05; ASignificantly different from Col, P < 0.01; BSignificantly different from sg1, P < 0.01.
The development of chloroplasts in the sixth leaves of 3-, 4-, and 5-week-old double mutants was observed. As expected, the transition from proplastid to chloroplast in sg1 gun1 and sg1 gun4 double mutants was significantly advanced relative to that in sg1 plants, but slightly delayed in comparison with the WT (Figs 2A, E, I, 6C). In 3- to 5-week-old sg1 gun1 double mutants, the development of chloroplasts in the sixth leaves was the same as in WT plants (Fig. 2A, E, I; Fig. 6C, upper panels). In the 3-week-old sg1 gun4 double mutant, prophase chloroplasts formed, but the abundance of thylakoid membranes was lower than that in the corresponding WT plants (Fig. 2A, E, I; Fig. 6C, lower panels). Further, these chloroplasts were similar to those present in the sixth leaves of 4-week-old sg1 plants (Fig. 2G, H; Fig. 6C, lower panels). Mature chloroplasts were present in the sixth leaves of 4-week-old and 5-week-old sg1 gun4 plants (Fig. 6C, lower panels).
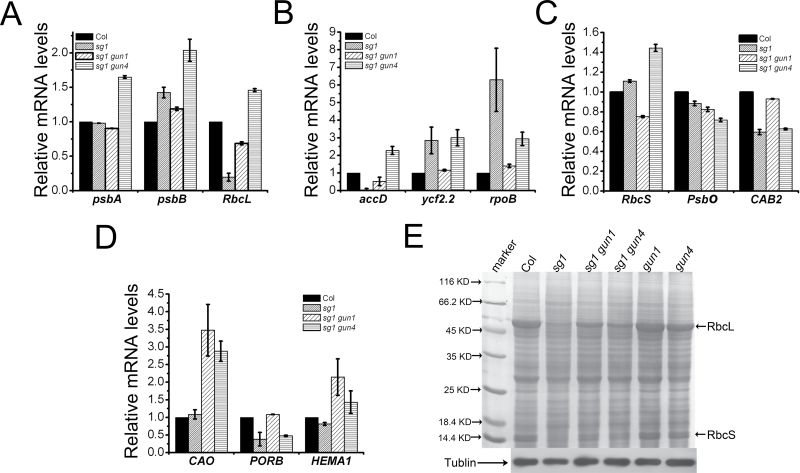
The expression levels of chloroplast-related genes were also quantified in 2-week-old Col, sg1, sg1 gun1, and sg1 gun4 plants. To eliminate the influence of green cotyledons, these were removed before RNA isolation was performed. The results showed that both gun1 and gun4 mutations in the sg1 background affected the expression of chloroplast-related genes compared with those in sg1. For example, both double mutants increased the expression levels of RbcL and accD (expressed at very low levels in sg1), and decreased the expression level of rpoB (expressed at much higher levels in sg1) (Fig. 7A–D). Also, the chlorophyll biosynthesis genes CAO and HEMA1 were increased in both double mutants compared with the WT and sg1 mutant (Fig. 7A–D). The levels of soluble proteins were profiled in 2-week-old Col, sg1, gun1, sg1 gun1, gun4, and sg1 gun4 plants (with cotyledons removed). Consistent with the observations of the alleviated phenotypes, the amounts of RbcL and RbcS in the double mutant plants were significantly higher than were those in sg1, but lower than were those in the WT (Fig. 7E). Subsequently the sg1 gun5 double mutant was constructed, and it was revealed that the mutation of GUN5 (with a function similar to that of GUN1 and GUN4 in retrograde signalling) did not alleviate the sg1 phenotype. On the contrary, it slightly enhanced the albino phenotypes of sg1 (see Supplementary Fig. S3 available at JXB online). Taken together, the present results indicate that the gun1 and gun4 mutations in sg1 may partially restore the disordered expression patterns of chloroplast relative genes, thereby alleviating the sg1 phenotypes.
Fig. 7.
Transcript and protein analysis of 2-week-old Col, sg1, sg1 gun1, and sg1 gun4. (A–D) The expression levels of plastid-encoded polymerase (PEP) transcribed chloroplast genes, nucleus-encoded polymerase (NEP) transcribed chloroplast genes, nuclear-encoded chloroplast genes, and chlorophyll biosynthesis genes. (E) Total proteins resolved by SDS–PAGE from different genotype seedlings. The mRNA and total proteins were isolated from 2-week-old plants with cotyledons removed.
Discussion
TPR-containing proteins comprise a common group of proteins that participate in protein–protein interactions or assembly of multiprotein complexes. The TPR domain consists of a degenerate, 34 amino acid sequence, which is present in tandem arrays of 3–16 motifs (D’Andrea and Regan, 2003; Whitfield and Mainprize, 2010). The TPR proteins have been found to be involved in many diverse processes within eukaryotic cells, including synaptic vesicle fusion (Young et al., 2003), peroxisomal targeting and import (Brocard and Hartig, 2006; Fransen et al., 2008), and mitochondrial and chloroplast import (Baker et al., 2007; Mirus et al., 2009). In the present study, a novel chloroplast-localized TPR-containing protein, SG1, was identified which is required for chloroplast development.
Influence of TPR protein SG1 mutation on normal chloroplast development
A large number of TPR-containing proteins are predicted to target to either mitochondria or chloroplasts (Small and Peeters, 2000). Consistent with their localization, many TPR proteins, for example pTAC2, Nac2, Toc64, Pyg7, and LAP1, have been reported to be involved in chloroplast development (Boudreau et al., 2000; Sohrt and Soll, 2000; Peng et al., 2006; Pfalz et al., 2006; Stockel et al., 2006; Kalanon and McFadden, 2008). These TPR proteins regulate chloroplast development in many ways, including gene expression (pTAC2), mRNA processing (Nac2), and protein transport and assembly (Toc64, Pyg7, and LAP1). SG1 is also required for chloroplast development; however, the mechanism of regulation differs from those of previously reported TPR proteins. First, and most importantly, SG1 is only required for the early stage of chloroplast development; once the plant has grown, the chloroplast becomes normal, and the seedling can grow photoautotrophically. In contrast, many previously reported TPR mutations, such as ptac2 and pyg7, are lethal (Pfalz et al., 2006; Stockel et al., 2006). Secondly, previously reported TPR protein regulation of chloroplast gene expression showed high specificity; for example, pTAC2 directly regulates the expression of PEP genes (Pfalz et al., 2006). In contrast, the sg1 mutant showed disrupted expression of PEP genes, NEP genes, nuclear-encoded chloroplast genes, and chlorophyll biosynthesis genes. It could not be determined whether SG1 was involved in mRNA processing, protein transport, or assembly. However, the inconsistency between the mRNA levels and protein levels of RbcL and RbcS in the sg1 mutant suggests that SG1 may be involved in protein biosynthesis or degradation in chloroplast development.
The present protein alignment data indicated that the mutation site of SG1 is located within the first TPR motif; this site is not a conserved site in the motif, but is conserved in its homologous sequences in Arabidopsis (see Supplementary Fig. S2B available at JXB online). The mutation may not affect the protein–protein interaction scaffolds formed by the TPR motif, but affect the proper function of SG1 in chloroplast development. The gene analysis showed that the expression of chloroplast-related genes was changed in the process of the sg1 albino leaf becoming green; for example, RbcL and accD which were decreased in albino leaf were up-regulated, while rpoB which was increased in albino leaf was down-regulated (Fig. 5A–D). These changes may re-establish a new balance among the chloroplast-related genes in the sg1 mutant and result in the leaf turning green. Further studies should be conducted to clarify the mechanisms by which SG1 regulates chloroplast development.
Mutation of GUN1 and GUN4 ameliorating sg1 phenotypes
The chloroplast developmental status has been shown to control a set of nuclear genes that encode chloroplast-localized proteins via a process known as retrograde signalling (Surpin et al., 2002). GUN genes (including GUN1–GUN6) are important for sending signals to the nucleus, to regulate nuclear-encoded chloroplast gene expression (Susek et al., 1993). In the present study, the gun1 and gun4 mutations ameliorated the slow-greening phenotypes of sg1 (Fig. 6A), and the leaves of sg1 gun1 and sg1 gun4 double mutants showed higher chlorophyll contents than did the leaves of sg1 mutant plants, at all growth stages (see Supplementary Table S1 available at JXB online). These results indicate that GUN1 and GUN4 genetically interact with SG1. GUN1 and GUN4 are both important factors in the retrograde signalling pathway of chloroplast development. Therefore, to investigate whether other components of retrograde signalling interact with SG1, the sg1 gun5 double mutant was constructed, and it was revealed that sg1 gun5 did not alleviate the sg1 phenotypes (see Supplementary Fig. S3 available at JXB online). Possible explanations are that GUNs play very important but differing roles in retrograde signalling; SG1 genetically interacts with GUN1 and GUN4 through their differing roles from GUN5 in retrograde signalling; or that the mutation of GUN1 and GUN4 ameliorates sg1 phenotypes through other roles in chloroplast development, apart from their involvement in the retrograde signalling pathway.
The present gene expression data revealed that gun1 and gun4 mutation in plants with the sg1 background altered the disturbed expression pattern of chloroplast-related genes in sg1, which may partially restore the imbalanced expression of chloroplast-related genes caused by sg1 mutation. The expression changes brought by gun1 and gun4 are not identical, and were therefore capable of ameliorating sg1 phenotypes to different degrees. Further, the results of protein analysis showed that the contents of RbcL and RbcS in sg1 gun1 and sg1 gun4 double mutants were significantly higher than were those in the sg1 mutant. The abundance of RbcL and RbcS was higher in sg1 gun1 than in sg1 gun4 (Fig. 7E), possibly indicating the different degrees to which gun1 and gun4 restore the phenotypes of sg1. Taken together, the results indicate that gun1 and gun4 can partially restore the imbalance of chloroplast-related genes in sg1, thereby alleviating the defective phenotypes.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Early growth of sg1 favoured by sucrose, and delayed embryogenesis during sg1 seed development.
Figure S2. Phylogenetic analysis and amino acid sequence alignment of SG1 TPR domains.
Figure S3. The phenotypes of the sg1 gun5 double mutant.
Table S1. Chlorophyll contents of leaves from different genotypes at different growth stages.
Table S2. The segregated ratio of different phenotypic seedlings in the F1 offspring of a reciprocal cross between Col and sg1.
Table S3. Primers for markers used in first mapping.
Table S4. Primers for markers used for fine mapping.
Table S5. Primers for qRT–PCR.
Acknowledgements
We are grateful to Professor Enrique Lopez-Juez (University of London, UK) for providing gun1-1, gun4-1, and gun5-1 seeds. We thank Professor Scott D. Russell (University of Oklahoma) and Professor Xiaoping Gou (Lanzhou University) for suggestions on the manuscript, and Dr Sodmergen (Peking University) for advice on the experimental methods. This study was supported by the Ministry of Agriculture of the People’s Republic of China (Grant NO. 2013ZX08009-003-002), the National Natural Science Foundation of China (NSFC) (Grant NO. 31070247, 91017002, and 31271460), and the Open Fund of the National Key Laboratory of Plant Molecular Genetics, Institute of Plant Physiology and Ecology, CAS.
References
- Abdallah F, Salamini F, Leister D. 2000. A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis . Trends in Plant Science 5, 141–142 [DOI] [PubMed] [Google Scholar]
- Albrecht V, Ingenfeld A, Apel K. 2006. Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: the impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Molecular Biology 60, 507–518 [DOI] [PubMed] [Google Scholar]
- Albrecht V, Ingenfeld A, Apel K. 2008. Snowy cotyledon 2: the identification of a zinc finger domain protein essential for chloroplast development in cotyledons but not in true leaves. Plant Molecular Biology 66, 599–608 [DOI] [PubMed] [Google Scholar]
- Albrecht V, Simkova K, Carrie C, Delannoy E, Giraud E, Whelan J, Small ID, Apel K, Badger MR, Pogson BJ. 2010. The cytoskeleton and the peroxisomal-targeted snowy cotyledon3 protein are required for chloroplast development in Arabidopsis . The Plant Cell 22, 3423–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluru MR, Yu F, Fu A, Rodermel S. 2006. Arabidopsis variegation mutants: new insights into chloroplast biogenesis. Journal of Experimental Botany 57, 1871–1881 [DOI] [PubMed] [Google Scholar]
- Apuya NR, Yadegari R, Fischer RL, Harada JJ, Zimmerman JL, Goldberg RB. 2001. The Arabidopsis embryo mutant schlepperless has a defect in the chaperonin-60 alpha gene. Plant Physiology 126, 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Hirohashi T, Kikuchi S, Belcher S, Osborne E, Yano S, Terashima I, Barkan A, Nakai M. 2004. Maize mutants lacking chloroplast FtsY exhibit pleiotropic defects in the biogenesis of thylakoid membranes. The Plant Cell 16, 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MJ, Frazier AE, Gulbis JM, Ryan MT. 2007. Mitochondrial protein-import machinery: correlating structure with function. Trends in Cell Biology 17, 456–464 [DOI] [PubMed] [Google Scholar]
- Boudreau E, Nickelsen J, Lemaire SD, Ossenbuhl F, Rochaix JD. 2000. The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO Journal 19, 3366–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard C, Hartig A. 2006. Peroxisome targeting signal 1: is it really a simple tripeptide? Biochimica et Biophysica Acta 1763, 1565–1573 [DOI] [PubMed] [Google Scholar]
- Carol P, Kuntz M. 2001. A plastid terminal oxidase comes to light: implications for carotenoid biosynthesis and chlororespiration. Trends in Plant Science 6, 31–36 [DOI] [PubMed] [Google Scholar]
- Charuvi D, Kiss V, Nevo R, Shimoni E, Adam Z, Reich Z. 2012. Gain and loss of photosynthetic membranes during plastid differentiation in the shoot apex of Arabidopsis . The Plant Cell 24, 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, Ma J, Zhang D, Guo J, Chen F, Lu C, Zhang L. 2008. The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis . Plant Physiology 147, 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743 [DOI] [PubMed] [Google Scholar]
- Cui L. 2006. Chloroplast DB: the chloroplast genome database. Nucleic Acids Research 34, D692–D696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea LD, Regan L. 2003. TPR proteins: the versatile helix. Trends in Biochemical Science 28, 655–662 [DOI] [PubMed] [Google Scholar]
- Dyall SD. 2004. Ancient invasions: from endosymbionts to organelles. Science 304, 253–257 [DOI] [PubMed] [Google Scholar]
- Fransen M, Amery L, Hartig A, Brees C, Rabijns A, Mannaerts GP, Van Veldhoven PP. 2008. Comparison of the PTS1- and Rab8b-binding properties of Pex5p and Pex5Rp/TRIP8b. Biochimica et Biophysica Acta 1783, 864–873 [DOI] [PubMed] [Google Scholar]
- Han CD, Coe EH, Jr, Martienssen RA. 1992. Molecular cloning and characterization of iojap (ij), a pattern striping gene of maize. EMBO Journal 11, 4037–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizaki T, Matsumura H, Nakayama K, Che FS, Terauchi R, Inaba T. 2009. Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiology 151, 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalanon M, McFadden GI. 2008. The chloroplast protein translocation complexes of Chlamydomonas reinhardtii: a bioinformatic comparison of Toc and Tic components in plants, green algae and red algae. Genetics 179, 95–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Miura E, Matsushima R, Sakamoto W. 2007. White leaf sectors in yellow variegated2 are formed by viable cells with undifferentiated plastids. Plant Physiology 144, 952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Lee KP, Baruah A, Nater M, Gobel C, Feussner I, Apel K. 2009. (1) O2-mediated retrograde signaling during late embryogenesis predetermines plastid differentiation in seedlings by recruiting abscisic acid. Proceedings of the National Academy of Sciences, USA 106, 9920–9924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Somers DE. 2010. Rapid assessment of gene function in the circadian clock using artificial microRNA in Arabidopsis mesophyll protoplasts. Plant Physiology 154, 611–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Kondo M, Fukuda H, Nishimura M, Ohta H. 2007. Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proceedings of the National Academy of Sciences, USA 104, 17216–17221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch KE. 1996. Carbohydrate-modulated gene expression in plants. Annual Review of Plant Physiology and Plant Molecular Biology 47, 509–540 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. 1986. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Molecular and General Genetics 204, 383–396 [Google Scholar]
- Kosambi DD. 1943. The estimation of map distances from recombination values. Annals of Human Genetics 12, 172–175 [Google Scholar]
- Larkin RM, Alonso JM, Ecker JR, Chory J. 2003. GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299, 902–906 [DOI] [PubMed] [Google Scholar]
- Leon P, Arroyo A, Mackenzie S. 1998. Nuclear control of plastid and mitochondrial development in higher plants. Annual Review of Plant Physiology and Plant Molecular Biology 49, 453–480 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Wellburn AR. 1983. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochemical Society Transactions 11, 591–592 [Google Scholar]
- Liu X, Yu F, Rodermel S. 2010. Arabidopsis chloroplast FtsH, var2 and suppressors of var2 leaf variegation: a review. Journal of Integrative Plant Biology 52, 750–761 [DOI] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG. 1996. The dynamics of seedling and cotyledon cell development in Arabidopsis thaliana during reserve mobilization. International Journal of Plant Sciences 157, 280–295 [Google Scholar]
- Michaels SD, Amasino RM. 1998. A robust method for detecting single-nucleotide changes as polymorphic markers by PCR. The Plant Journal 14, 381–385 [DOI] [PubMed] [Google Scholar]
- Mirus O, Bionda T, von Haeseler A, Schleiff E. 2009. Evolutionarily evolved discriminators in the 3-TPR domain of the Toc64 family involved in protein translocation at the outer membrane of chloroplasts and mitochondria. Journal of Molecular Modeling 15, 971–982 [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. 2001. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proceedings of the National Academy of Sciences, USA 98, 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J. 2006. Plastid-to-nucleus retrograde signaling. Annual Review of Plant Biology 57, 739–759 [DOI] [PubMed] [Google Scholar]
- Peng L, Ma J, Chi W, Guo J, Zhu S, Lu Q, Lu C, Zhang L. 2006. LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana . The Plant Cell 18, 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmuller R. 2006. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. The Plant Cell 18, 176–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Albrecht V. 2011. Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiology 155, 1545–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K. 1997. The genetic control of plastid division in higher plants. American Journal of Botany 84, 1017. [PubMed] [Google Scholar]
- Richly E, Leister D. 2004. An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene 329, 11–16 [DOI] [PubMed] [Google Scholar]
- Robertson D, Laetsch WM. 1974. Structure and function of developing barley plastids. Plant Physiology 54, 148–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W, Uno Y, Zhang Q, Miura E, Kato Y, Sodmergen 2009. Arrested differentiation of proplastids into chloroplasts in variegated leaves characterized by plastid ultrastructure and nucleoid morphology. Plant and Cell Physiology 50, 2069–2083 [DOI] [PubMed] [Google Scholar]
- Shimada H, Mochizuki M, Ogura K, Froehlich JE, Osteryoung KW, Shirano Y, Shibata D, Masuda S, Mori K, Takamiya K. 2007. Arabidopsis cotyledon-specific chloroplast biogenesis factor CYO1 is a protein disulfide isomerase. The Plant Cell 19, 3157–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small ID, Peeters N. 2000. The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends in Biochemical Science 25, 46–47 [DOI] [PubMed] [Google Scholar]
- Sohrt K, Soll J. 2000. Toc64, a new component of the protein translocon of chloroplasts. Journal of Cell Biology 148, 1213–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin LA, Newcomb EH. 2000. Membrane structure and membranous organelles. In: Buchanan RB, Gruissem W, Jones RL, eds. Biochemistry and molecular biology of plants. Rockville, MD: American Society of Plant Biology, 2–50 [Google Scholar]
- Stockel J, Bennewitz S, Hein P, Oelmuller R. 2006. The evolutionarily conserved tetratrico peptide repeat protein pale yellow green7 is required for photosystem I accumulation in Arabidopsis and copurifies with the complex. Plant Physiology 141, 870–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su N, Hu ML, Wu DX, et al. 2012. Disruption of a rice pentatricopeptide repeat protein causes a seedling-specific albino phenotype and its utilization to enhance seed purity in hybrid rice production. Plant Physiology 159, 227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpin M, Larkin RM, Chory J. 2002. Signal transduction between the chloroplast and the nucleus. The Plant Cell 14 Suppl, S327–S338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J. 1993. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74, 787–799 [DOI] [PubMed] [Google Scholar]
- Uwer U, Willmitzer L, Altmann T. 1998. Inactivation of a glycyl-tRNA synthetase leads to an arrest in plant embryo development. The Plant Cell 10, 1277–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C, Mainprize IL. 2010. TPR motifs: hallmarks of a new polysaccharide export scaffold. Structure 18, 151–153 [DOI] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Chory J. 2011. Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Current Biology 21, 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Barral JM, Ulrich Hartl F. 2003. More than folding: localized functions of cytosolic chaperones. Trends in Biochemical Science 28, 541–547 [DOI] [PubMed] [Google Scholar]
- Yu QB, Jiang Y, Chong K, Yang ZN. 2009. AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana . The Plant Journal 59, 1011–1023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.