Summary
This study characterized novel functions of APUM23, a PUF family protein, in Arabidopsis development. APUM23 functions in rRNA biosynthesis and has pleiotropic roles, influencing cell division and adaxial–abaxial polarity.
Key words: APUM23, Arabidopsis, KANADI, leaf development, polarity, PUF.
Abstract
The normal biological function of leaves, such as intercepting light and exchanging gasses, relies on proper differentiation of adaxial and abaxial polarity. KANADI (KAN) genes, members of the GARP family, are key regulators of abaxial identity in leaf morphogenesis. This study identified a mutant allele (apum23-3) of APUM23, which encodes a Pumilio/PUF domain protein and acts as an enhancer of the kan mutant. Arabidopsis APUM23 has been shown to function in pre-rRNA processing and play pleiotropic roles in plant development. The apum23-3 mutant also synergistically interacts with other leaf polarity mutants, affects proliferation of division-competent cells, and alters the expression of important leaf polarity genes. These phenotypes show that APUM23 has critical functions in plant development, particularly in polarity formation. The PUF gene family is conserved across kingdoms yet it has not been well characterized in plants. These results illuminating the functions of APUM23 suggest a novel role for PUF genes in Arabidopsis leaf development.
Introduction
The leaves of many plant species exhibit differences between their adaxial (dorsal) and abaxial (ventral) surfaces (Telfer and Poethig, 1994). Proper specification of adaxial and abaxial identity is required for the formation and function of leaves. In Arabidopsis thaliana, adaxial identity is specified by the class III homeodomain-leucine zipper (HD-ZIPIII) genes (McConnell et al., 2001; Emery et al., 2003; Prigge et al., 2005), Myb and LOB domain transcription factors ASYMMETRIC LEAVES1 (AS1) and AS2 (Lin et al., 2003; Xu et al., 2003), and trans-acting short-interfering RNA (ta-siRNA) (Allen et al., 2005; Williams et al., 2005; Hunter et al., 2006), whereas on the abaxial side, KANADI genes (Eshed et al., 2001, 2004; Kerstetter et al., 2001), AUXIN RESPONSE FACTORS (ARF) ETTIN (ETT/ARF3) and ARF4 (Pekker et al., 2005), YABBY genes (Sawa et al., 1999; Siegfried et al., 1999; Eshed et al., 2004), LITTLE ZIPPER (ZPR) genes (Wenkel et al., 2007), and microRNA165/166 (Bao et al., 2004; Kidner and Martienssen, 2004; Mallory et al., 2004) play important roles.
The HD-ZIPIII gene family consists of members such as REVOLUTA (REV), PHABULOSA (PHB), and PHAVOLUTA (PHV). These genes encode proteins with partially redundant functions. Gain-of-function of one of these genes or simultaneous downregulation of all three genes results in radial cotyledons and leaves (McConnell and Barton, 1998; Emery et al., 2003). Similarly to the HD-ZIPIII gene family, KANADI genes also have overlapping functions. Mutations in any single KAN gene have relatively mild defects in leaf polarity (Eshed et al., 2001; Kerstetter et al., 2001; McAbee et al., 2006). However, if several of these genes are non-functional, the resulting plants exhibit strong defects associated with the loss of abaxial identity. For instance, the kan1 kan2 double mutant has reduced leaf blade expansion and develops ectopic outgrowths on the abaxial side of the leaf. More dramatically, in the kan1 kan2 kan3 triple mutant, leaves are almost fully radialized and adaxialized (Eshed et al., 2004).
HD-ZIPIII and KANADI are thought to act antagonistically (Kidner and Timmermans, 2007). The HD-ZIPIII gene PHB is expressed abaxially in kan1 kan2 kan3 triple mutants and its adaxial expression is reduced when KAN2 is expressed throughout the leaf primordia (Eshed et al., 2004). In addition, KAN1 also represses AS2 by directly interacting with its promoter and regulating its transcription (Wu et al., 2008). These results demonstrate that the interactions of key adaxial–abaxial genes are critical for leaf polarity establishment and subsequent blade expansion.
This study identified a new regulator of leaf polarity, APUM23, which interacts with major polarity genes including KANADI, AS2, and REV. APUM23 encodes a protein belonging to the PUF RNA-binding protein family (Abbasi et al., 2010). Drosophila Pumilio is a founding member of this family and is required for the establishment of anterior–posterior polarity (Murata and Wharton, 1995) and stem cell maintenance (Lin and Spradling, 1997; Forbes and Lehmann, 1998) through translation inhibition. PUF proteins specifically bind to nanos response element sequences in the 3′-untranscribed region of target mRNAs (Zamore et al., 1999; Wang et al., 2001; White et al., 2001) and usually function in a complex with other RNA-binding proteins such as Nanos (Sonoda and Wharton, 1999) and Brat (Sonoda and Wharton, 2001). In Arabidopsis, more than 20 putative PUF genes (APUMs) have been predicted by various studies (Francischini and Quaggio, 2009; Abbasi et al., 2010; Tam et al., 2010). Biochemical experiments have shown that several Arabidopsis PUF proteins are able to bind to Drosophila nanos response element sequences and the 3′-untranscribed region of mRNAs from Arabidopsis genes involved in shoot stem cell maintenance, such as WUSCHEL and CLAVATA1 (Francischini and Quaggio, 2009), suggesting a evolutionarily conserved mechanism of PUF protein action. However, in contrast to the products of other APUM genes, APUM23 was found to be localized to the nucleolus and involved in pre-rRNA processing and rRNA maturation (Abbasi et al., 2010). This distinct function of APUM23 is probably due to its unique structure in the PUF RNA-binding domains (Tam et al., 2010).
APUM23 has been shown to play important roles in various aspects of plant growth (Abbasi et al., 2010). The current work shows that APUM23 has a previously undescribed role in regulating the activity of division-competent cells and the establishment of organ polarity. The results suggest that APUM23 is important for organ growth and pattern formation in Arabidopsis.
Materials and methods
Plant materials
The apum23-3, kan1-11, and kan2-5 plants used in this study were in the Columbia (Col) background. as1-1 (CS3774, in Col background), as2-2 (CS3118, in ER background), and rev-1 (CS3826, in No background) were obtained from the Arabidopsis Biology Resource Center (ABRC). The kan1-11 and kan2-5 alleles have been described previously (Wu et al., 2008). All plants were grown at 22 °C under long-day conditions (16/8 light/dark). Double mutants apum23-3 as1-1, apum23-3 as2-2, and apum23-3 rev-1 were generated by initially crossing two parental lines followed by backcrossing the F1 with both parents. The progeny of the backcross was selected for parental phenotypes. The selected plants were self-pollinated and the double mutants were identified from the next generation. F2 progeny from the self-pollinated F1 plants was also examined. The double mutants were found at the ratio of 1/16 or less, and no enhancement of double mutant phenotypes (or other novel phenotypes) was identified in the F2 plants, ruling out background effects on the phenotypes of the double mutants. In order to make the apum23-3 kan1-11 kan2-5 triple mutant, apum23-3 was first pollinated with kan1-11 kan2-5. The F1 then was backcrossed to kan1-11 kan2-5. In the progeny of the backcross, plants homozygous for kan1-11 and apum23-3 and heterozygous for kan2-5 were maintained, and the triple mutants were selected in their progeny by genotyping. The names of the double and triple mutants are abbreviated as kan1 kan2, apum23 as1, apum23 as2, apum23 rev, and apum23 kan1 kan2.
Anatomical analysis
Petioles of 21-d-old plants were fixed in 2% glutaraldehyde in 25mM sodium phosphate buffer (pH 6.8) with 0.1% Triton X-100 overnight at 4°C. Samples were washed in the same sodium phosphate buffer, dehydrated through an ethanol series, and embedded in wax blocks. Sections (8 μm) were made on a Jung Biocut microtome and stained with 0.1% toluidine blue.
Molecular identification of the enhancer of kan1 kan2
The enhancer of kan1 kan2 mutant was identified by a map-based approach. The single mutants were first crossed with L er plants. The F2 generation segregating the enhancer phenotype was used as the mapping population, in which 929 mutant plants were identified. Genomic DNA was isolated from these mutants and utilized for mapping by using InDel and cleaved-amplified polymorphic sequence (CAPS) markers based on the Cereon L er/Col SNP database (TAIR; http://www.arabidopsis.org). These molecular markers localized the mutation at the bottom of chromosome I, in an approximate 17-kb region containing 13 genes. Sequencing six candidate genes revealed a G→A change that disrupts the intron splicing in At1g72320 (APUM23). In order to confirm the molecular identity of the enhancer as an allele of APUM23, a 6.7-kb genomic DNA fragment containing the entire At1g72320 gene was released from the BAC T10D10 by restriction digestion with SalI and Acc65I (Fermentas, Hanover, MD, USA) and cloned in the binary vector pCAMBIA1300 and transformed into the enhancer mutant using the floral dip method (Clough and Bent, 1998). A control transformation was also performed with the empty pCAMBIA1300 vector. The T1 transgenic plants were selected on 1/2 MS medium containing 30mg l–1 hygromycin. In addition, a T-DNA line (SAIL_757_B08; apum23-1; Abbasi et al., 2010) was also obtained from the ABRC. The T-DNA insertion in this line was confirmed by sequencing.
Genetic complementation in yeast
Yeast Magic Marker nop9 strain (BY4743) was obtained as a heterozygous diploid knock out (open biosystems, Huntsville, AL, USA). APUM23 full-length cDNA was amplified from the BAC clone U09300 (obtained from ABRC) with primers GATEARW1N.f (5′-ATGGTTTCTGTTGGTTCTAAATCATTG-3′) and GATEA RW1N.r (5′-ACGTCTCAAATTCTCATTTTATTTGAATGCCG-3′). The PCR product was cloned into the pCR8 vector (Invitrogen, Eugene, OR, USA) and subcloned into the yeast expression vector BG1805 (a gift from Beth Grayhack) containing the GAL1 promoter and URA3 marker by GATEWAY cloning strategy (Invitrogen). The resulting plasmid (BG1805-APUM23), as well as the wild-type NOP9 in the same vector (BG1805-NOP9), which was purchased (open biosystems), was transformed into the nop9/+ diploid strain. An empty BG1805 vector was also transformed separately as a control. The transformed cells were sporulated and spread on Magic Medium (Pan et al., 2004) to select haploid cells that survived in the absence of uracil. The transformants were further confirmed by several sets of primers: NOP9A (5′-GTTTTCAAACTTGCTAGGCTGTATC-3′) and NOP9B (5′-CCAGTTCTTCTCTATCTAGCACACC-3′) were used to confirm the knock out of NOP9 gene in the nop9 genome, whereas NOP9A and kanB (5′-CTGCAGCGAGGAGCCGTAAT-3′) were used to detect the kanMX insertion that knocks out NOP9. Primers BNOP9.f (5′-CAAACCTTCAAATGAACGAATCAA-3′) and BNOP9.r (5′-agactcttcttgctggatgtgctc-3′) confirmed the presence of BG1805-NOP9 in the haploid cells and BARW1.f (5′-GCGAAGCGATGATTTTTGATCTAT-3′) and BARW1.r (5′-CTTCCTACGCATTCCCTTATTCCT-3′) detected BG1805-APUM23 in the haploid cells.
Haploid cells containing either NOP9 or APUM23 were inoculated in the liquid Magic Medium and grown in 30 °C until OD600 reached 0.6. The culture was serially diluted in sterile water and spotted on solid Magic Medium. The plates were incubated at 30 °C for 2 d for analysis.
Examination of β-glucuronidase (GUS) reporter activities
CYCB1:db:GUS (Harrar et al., 2003), pKAN1:GUS, and pAS2:GUS (Wu et al., 2008) were introduced in apum23-3 via crossing. GUS staining was performed according to Senecoff et al. (1996) with modifications. Plants were fixed in 80% acetone at –20 °C for 20min, then stained with 2mM X-Gluc in GUS-staining buffer (9mM potassium ferrocyanide and potassium ferricyanide) for 1h at 37 °C. After removing the chlorophyll with an ethanol series, the young leaves or roots were dissected, mounted in 10% glycerol, and observed under microscope.
Reverse-transcription PCR
Total RNA (2 μg) extracted from 10-d-old seedlings using TRIzol Reagent (Invitrogen) was treated with DNase I (Fermentas) and reverse transcribed with Superscript III (Invitrogen), and 1 μl of a 10-fold dilution was used as template for PCR. To quantify the unprocessed pre-rRNA, the cleavage site of 35S pre-RNA was specifically amplified by primers U1 (5′-CGTAACGAAGATGTTCTTGGC-3′) and U2 (5′-ATGCGTCCCTTCCATAAGTC-3′). The primer pair UBC.f (5′-TCAAGAGGTTGCAGCAAGA-3′) and UBC.r (5′-CTTTGCTCAACAACATCACG-3′) was employed to amplify a ubiquitin conjugating enzyme gene as a control. The PCR conditions were 94 °C for 15 s, 52 °C for 15 s, and 72 °C for 30 s, for 33 cycles. The DNA band intensity was measured by using a KODAK Molecular Imaging Software 4.0 (Eastman KODAK Company, Rochester, NY, USA). Bands were normalized using Gaussian curve with background subtraction. Mean intensities and standard error of the mean were calculated from three independent biological samples.
Quantitative real-time PCR
Young leaf primordia of Col and apum23-3 were dissected from seedlings that were grown on half-strength MS plates for 2 d after the emergence of the first two leaves. Total RNA was extracted from the primordia and reverse transcribed as already described. Quantitative real-time PCR (qRT-PCR) utilized Power SYBR Green PCR Master Mix (Applied Biosystems) for amplification. Primers used in qRT-PCR are listed in Supplementary Table S1 available at JXB online. Changes in gene expression were calculated from three biological replicates using the 2–ΔΔCt method (Livak and Schmittgen, 2001). The relative mRNA levels were normalized to the expression of GAPC2 (Husar et al., 2011; Mafra et al., 2012).
Results
Identification of an enhancer of the kan1 kan2 leaf polarity phenotype
An EMS mutagenesis screen was carried out to identify enhancers of the kan1-11 kan2-5 (abbreviated as kan1 kan2) double mutant. kan1 kan2 plants have upward-curling leaves with ectopic outgrowths on the abaxial side (Eshed et al., 2001; Fig. 1C–I). One enhancer mutation was identified that enhanced kan1 kan2 by having rosette leaves with reduced blade expansion (Fig. 1D–J). This defect became more severe as leaf number increased. For instance, leaf 9 of the triple mutant was almost completely radialized indicating a dramatic loss of leaf polarity (Fig. 1J). In addition, the abaxial outgrowths of the kan1 kan2 mutant were also reduced in the triple mutant.
Fig. 1.
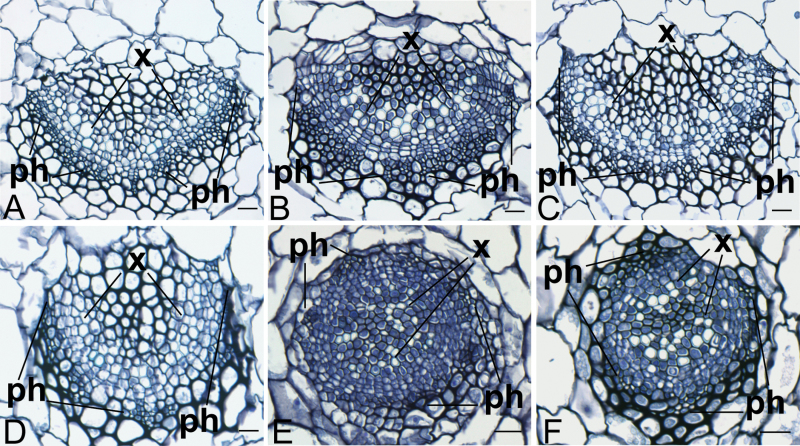
apum23-3 enhances the kan1 kan2 double mutant phenotype. (A–D) 10-d-old wild type (Col and apum23-3) (A), 21-d-old Col and apum23-3 (B), 30-d-old kan1 kan2 (C), and apum23 kan1 kan2 (D). (E–J) Individual leaves 5, 7, and 9 of kan1 kan2 (E, G, I) compared with those of apum23 kan1 kan2 (F, H, J): note that blade expansion on kan1 kan2 is reduced in the triple mutant. (K–O) Sections of petioles to examine the vascular structures of Col (K), apum23-3 (L), kan1 kan2 (M), and apum23 kan1 kan2 (N, O) plants: O is a higher magnification of the vascular tissue in the black box in N, which shows the dramatic adaxialization in the triple mutant indicated by the xylem-surrounding-phloem phenotype. ph, phloem; x, xylem. Bars, 5mm (E–J) and 20 μm (K–O) (this figure is available in colour at JXB online).
In order to further characterize the mutant phenotypes, the internal structure of the leaf petioles was analyzed. Distinct from the relatively normal midveins of kan1 kan2 double mutant (Fig. 1M), transverse sections showed that the triple mutant had a disorganized vascular pattern consisting of very few phloem cells. These phloem cells were surrounded by adjacent xylem tissues suggesting a strong loss of abaxial identity (Fig. 1N, O). This enhancer is a recessive mutation and the single mutant had elliptical, flat and serrated leaves with a greyish green adaxial side (Fig. 1A, B; see also Fig. 6A). In contrast to the phenotype seen in the triple mutant, the enhancer mutant had very subtle vascular defects with a midvein structure similar to that observed in the wild type Col (Fig. 1K, L).
Fig. 6.
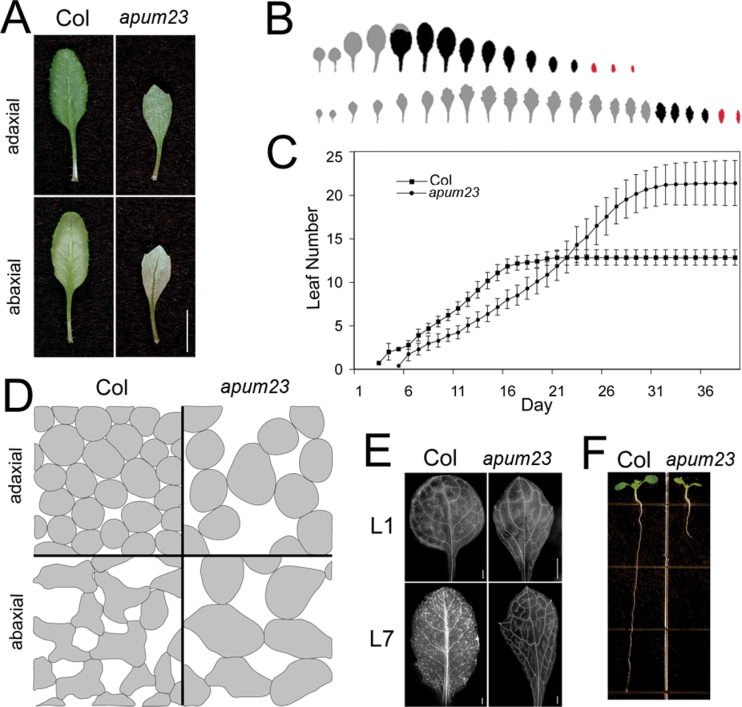
APUM23 plays pleotropic roles in plant development. (A) Adaxial and abaxial surfaces of leaf 5 of 21-d-old Col and apum23-3 plants. (B) Outline of leaves from Col (upper) and apum23-3 (lower), indicating the delayed transition from juvenile to adult stage: light grey indicates leaves lacking abaxial trichomes; black indicates leaves with abaxial trichomes; red indicates bracts. (C) The rate of leaf initiation in Col and apum23-3: the first visible leaf appears 2 d later in apum23-3 but leaf production persists longer, resulting in more leaves (n = 20). (D) Camera-lucida drawings of the adaxial and abaxial mesophyll layers of leaf 5 in Col and apum23-3. (E) Chloral hydrate-cleared leaves of Col and apum23-3 show reduced complexity of vascular strands in apum23-3. (F) Root growth of Col and apum23-3 on 1/2 MS media. Bar, 1cm (A) and 1mm (E) (this figure is available in colour at JXB online).
The kan1 kan2 enhancer is a mutant allele of APUM23
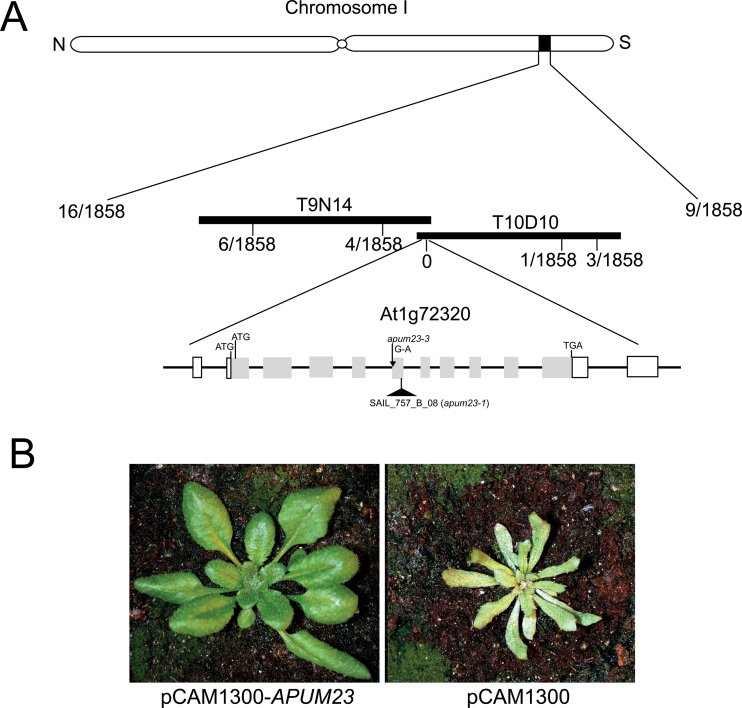
The enhancer mutation was mapped to a region containing 13 genes at the bottom of chromosome I, corresponding to the overlap of bacterial artificial chromosomes T10D10 and T9N14. Sequencing of six genes in this region in the enhancer mutant background revealed a single nucleotide mutation (G→A) in APUM23 (At1g72320; Abbasi et al., 2010) that disrupted the 3′-splice site of intron 5 in this gene (Fig. 2A). This suggested that this lesion may account for the enhancer phenotype. The molecular identity of the enhancer was verified by transgene complementation. A 6.7-kb genomic DNA fragment including the wild-type APUM23 was transformed into the enhancer mutant line and resulted in transgenic plants with a wild-type phenotype, while plants carrying the empty vector did not show complementation (Fig. 2B). Moreover, T-DNA insertion alleles of APUM23 also showed similar phenotypes to those of the enhancer (Abbasi et al., 2010; Supplementary Fig. S1 available at JXB online), which further confirmed that the kan1 kan2 enhancer mutation corresponds to a new allele of APUM23, named apum23-3.
Fig. 2.
Molecular identification of apum23-3. (A) Positional cloning and gene structure of APUM23: solid black lines represent two bacterial artificial chromosomes (T9N14 and T10D10) containing APUM23. The recombination frequency at each molecular marker is shown as number of recombinations/number of chromosomes analysed. The genomic structure of APUM23 (At1g72320) is illustrated with grey solid bars representing exons and black lines representing introns. APUM23 has two non-coding exons at the 5′- and 3′-ends, which are represented by hollow bars. The lesion in apum23-3 and the SAIL T-DNA line (apum23-1) are also shown. (B) Transgenic plant containing the APUM23 genomic sequence transformed into the apum23-3 mutant background shows complete rescue of the apum23 mutant phenotype (left), whereas transgenic plants containing the empty vector pCAMBIA1300 still show the mutant phenotype (right) (this figure is available in colour at JXB online).
apum23-3 interacts synergistically with mutants of genes specifying adxaxial identity
In addition to enhancing the kan1 kan2 phenotype, it was also found that apum23-3 showed synergistic interactions with mutants affecting adaxial fate, such as revoluta-1 (rev-1), asymmetric leaves1-1 (as1-1), and asymmetric leaves2-2 (as2-2). REV encodes a member of the HD-ZIP III gene family of transcription factors that redundantly promote adaxial identity (Emery et al., 2003). Due to the overlapping roles of these HD-ZIP III genes, the rev single mutant did not show strong defects in the vegetative stage (Fig. 3A). However, the apum23 rev double mutant had pin-shaped outgrowths in the centre of the rosette that arose after the emergence of several apum23-like, but more elongated leaves (Fig. 3B, C). Petiole sections of the expanded leaves of apum23 rev double mutants displayed abaxialization of vascular tissues with phloem partially surrounding xylem (Fig. 4D), although the vascular pattern of rev-1 was similar to that of the wild type (Fig. 4A). Furthermore, apum23-3 also enhanced the inflorescence defects of rev. The rev mutant inflorescences contained filamentous organs due to the failure of normal floral organ formation (Talbert et al., 1995; Otsuga et al., 2001; Fig 5A, D). The apum23-3 mutant strongly enhanced this defect, such that the double mutant displayed a completely sterile inflorescence with filamentous organs (Fig. 5C, F, G).
Fig. 3.
apum23-3 interacts with adaxial polarity mutants. Phenotypes of 21-d-old rev-1 (A), as1-1 (D), as2-2 (G), apum23 rev (B), apum23 as1 (E), and apum23 as2 (H). C, F, and I are higher magnifications of B, E, and H, respectively. White arrowheads in E and H indicate the trumpet-shaped leaves in apum23 as1 and apum23 as2; black arrowheads in C, F, and I show the pin-shaped structures in the double mutants; black arrows in I show the branched radialized structures in apum23 as2. Bars, 1mm (C, F, I) and 1cm in all other panels (this figure is available in colour at JXB online).
Fig. 4.
Vascular phenotypes of leaf petioles of adaxial polarity mutants and double mutants with apum23-3. Transverse sections of leaf petioles showing vascular organization in 21-d-old rev-1 (A), as1-1 (B), as2-2 (C), apum23 rev (D), apum23 as1 (E), and apum23 as2 (F) plants. Synergistic interactions between apum23 and the adaxial mutants are shown by the partial or complete phloem-surrounding-xylem structure in the double mutants. ph, phloem; x, xylem. Bar, 20 μm (this figure is available in colour at JXB online).
Fig. 5.
apum23-3 enhances the inflorescence phenotype of rev-1. (A–C) 30-d-old rev-1 (A), apum23-3 (B), and apum23 rev (C) plants. (D–G) The inflorescences of plants shown in A, B, and C, respectively; G is a higher magnification of F. White arrowhead in D indicates the degenerate flowers in rev; white arrowheads in G indicate several of the sterile and filamentous structures in apum23 rev. Bars, 1cm (A–C), 1mm (D–F), and 100 μm (G) (this figure is available in colour at JXB online).
As well as the HD-ZIPIII genes, AS1 and AS2 are also key regulators of adaxial fate. AS1 encodes a Myb domain transcription factor closely related to the PHANTASTICA (PHAN) gene product that plays a role in adaxial determination in Antirrhinum (Waites and Hudson, 1995; Waites et al., 1998). AS2 interacts with AS1 (Xu et al., 2006) and participates in establishing leaf polarity by antagonizing KAN genes (Wu et al., 2008). as1 and as2 both produce asymmetric, rumpled leaves with ectopic leaflet-like structures on petioles (Tsukaya and Uchimiya, 1997; Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001; Sun et al., 2002; Fig. 3 D, G). apum23-3 displayed similar interactions with as1 and as2. In the apum23 as1 and apum23 as2 double mutants, only the first four or five leaves were expanded and the subsequent leaves were replaced by pin-shaped or branched structures (Fig. 3F, I). The expanded leaves were also distinct from those of single mutants in that they occasionally produced a ‘trumpet’ shape (Fig. 3E, H). The vasculature of the petioles was also analysed in these mutants. The as1 and as2 mutants often have a slightly abaxialized vein pattern (Ha et al., 2007; Fig. 4B, C). However, in double mutants with apum23, the abaxialization was strikingly enhanced. The midveins of these plants were radialized with phloem surrounding xylem (Fig. 4E, F), which is typical of vascular structures completely lacking adaxial identity.
apum23-3 displays pleotropic defects in development
These strong genetic interactions with key leaf polarity genes imply that APUM23 may play an important role in leaf development. To explore this role, the apum23-3 single-mutant phenotypes were further characterized. The apum23-3 homozygous mutant displayed a variety of developmental defects including delayed leaf formation (2 d later than the wild type, Fig. 6C), delayed phase transition from juvenile to adult as measured by the appearance of the first abaxial trichomes (on leaf 17 in apum23-3 versus leaf 5 in Col; Fig. 6B), and reduced leaf venation complexity (Fig. 6E). The morphology of the subepidermal mesophyll in apum23-3 was also examined as compared to the wild type (Fig. 6D). In wild-type leaves, adaxial mesophyll cells were round and densely packed, whereas abaxial mesophyll consisted of irregular cells with large air spaces. In apum23-3, both adaxial and abaxial mesophyll cells were fewer but slightly bigger than the wild type, and the abaxial cells were more regular in shape. Thus, apum23 affected the differentiation of both adaxial and abaxial cells in the leaf blade. In addition, apum23-3 also had shorter roots (0.9±0.1cm) as compared to Col (4.1±0.1cm, Fig. 6F).
apum23-3 affects the expression of leaf polarity genes
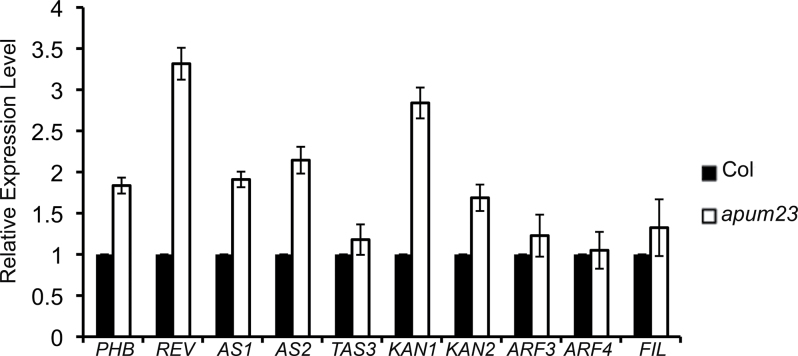
APUM23 has been found to be expressed ubiquitously in a variety of organs in Arabidopsis (Abbasi et al., 2010), which implies that it is unlikely to have tissue specific functions in leaf polarity. In order to further examine the effect of APUM23 on leaf polarity genes, the expression of KAN1 and AS2 was assayed in the wild type and apum23-3 mutants using promoter:GUS reporter lines. In wild-type seedlings, pKAN1:GUS was expressed in the meristem and the abaxial side of young leaf primordia (Supplementary Fig. S2A available at JXB online), while pAS2:GUS showed a complementary pattern with GUS present only in the adaxial side (Supplementary Fig. S2C available at JXB online). The spatial expression patterns of both GUS reporters were not obviously different in the apum23-3 background (Supplementary Fig. S2B, D available at JXB online), indicating that the spatial expression domains of KAN1 and AS2 are not affected by apum23-3. qRT-PCR was then utilized to measure the mRNA levels of several key leaf polarity genes in the young leaf primordia in the wild type and apum23-3. PHB, REV, AS1, AS2, KAN1, and KAN2 were all expressed at higher level in apum23-3 than in the wild type; however, the expression of TAS3 (ta-siRNA), ARF3, ARF4, and FILAMENTOUS FLOWER (FIL, a major member in the YABBY gene family) were not significantly altered in apum23-3 as compared to those in the wild type (Fig. 7). These results indicated that loss of APUM23 can affect the transcript levels of both adaxial and abaxial regulatory genes, which may be associated with the synergistic phenotypes observed in these double and triple mutant analyses.
Fig. 7.
Expression of leaf polarity genes in apum23-3. Results of qRT-PCR show the expression of major leaf polarity genes (PHB, REV, AS1, AS2, TAS3, KAN1, KAN2, ARF3, ARF4, and FIL), in young leaves of Col (black) and apum23-3 (white). The relative expression level of genes was normalized by the expression of GAPC2. Error bars represent the standard error of the mean.
apum23-3 displays defective cell division patterns in both leaves and roots
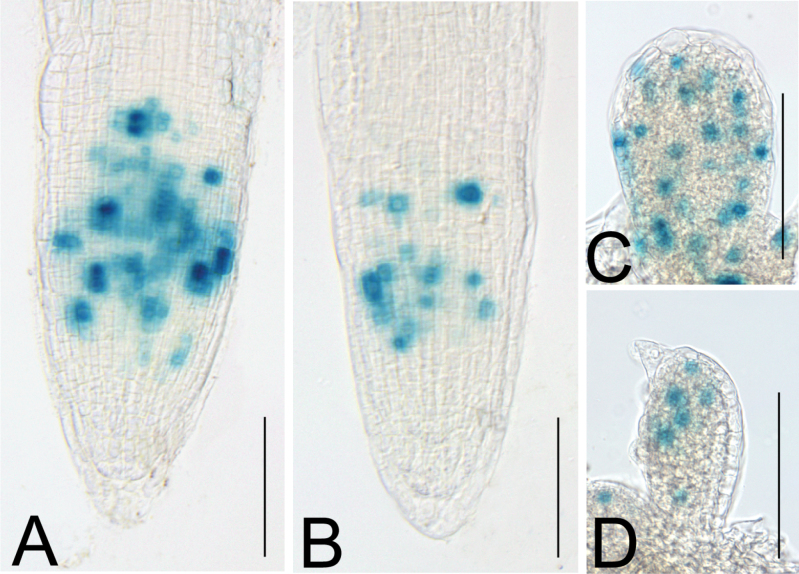
The apum23-3 mutant had fewer cells in the mesophyll and shorter roots, suggesting a reduction in cell proliferation. To test this idea, the pattern of cell division was examined in both the wild type and apum23-3 using the CYCB1:db:GUS reporter gene (Harrar et al., 2003). GUS staining showed that the number of division-competent cells was reduced in both leaves and roots of apum23-3 as compared to the wild type (Fig. 8). The putative function of APUM23 in promoting cell division is consistent with its strong expression in developmentally active tissues (Abbasi et al., 2010) and is likely correlated with the growth defects seen in apum23 mutants (Fig. 6).
Fig. 8.
Cell division patterns in Col and apum23-3. CYCB1:db:GUS expression in roots of Col (A) and apum23-3(B) and young leaves of Col (C) and apum23-3(D). Bars, 100 μm (this figure is available in colour at JXB online).
APUM23 functions in pre-rRNA processing
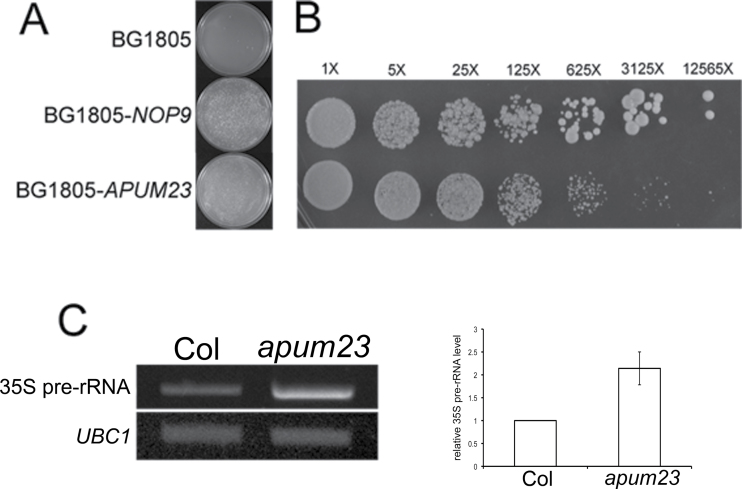
APUM23 belongs to the Pumilio/PUF gene family that is evolutionarily conserved across kingdoms (Spassov and Jurecic, 2003; Abbasi et al., 2010). The APUM23 protein has been found to play a specific role in the cleavage of 35S pre-rRNA, a critical step for 18S rRNA biosynthesis (Abbasi et al., 2010). This function is similar to that of NOP9, a PUF gene in Saccharomyces cerevisiae (Thomson et al., 2007; Abbasi et al., 2010). In order to further confirm the functional homology of APUM23 and NOP9, a complementation experiment was performed in yeast cells. Because the null nop9 mutant is lethal, a heterozygous diploid strain, nop9/+, was transformed with NOP9 or APUM23 full-length cDNA, and Magic Marker technology (Pan et al., 2004) was utilized to select the nop9 haploid cells carrying NOP9 or APUM23 after sporulation. Growth tests showed that APUM23 could partially rescue nop9 defects (Fig. 9A, B), which suggested some level of functional similarity between these two proteins. Because the cleavage of 35S pre-rRNA is conserved in eukaryotes (Venema and Tollervey, 1999) and the cleavage site in Arabidopsis has been identified in previous studies (Saez-Vasquez et al., 2004; Shi et al., 2005), the accumulation of the unprocessed 35S pre-RNA was examined in the wild type and apum23-3 using established methods (Petricka and Nelson, 2007). As expected, the amount of unprocessed 35S rRNA was 2.1-fold higher in apum23-3 as compared to the wild type (Fig. 9C). These results further support and extend the observation that APUM23 plays a critical role in 35S pre-rRNA processing (Abbasi et al., 2010).
Fig. 9.
APUM23 functions in 18S rRNA biosynthesis. (A) Colonies of the haploid nop9 mutant transformed with empty vector BG1805, NOP9, and APUM23, as indicated. (B) Comparisons of a dilution series of haploid nop9 mutant cells transformed with NOP9 (upper) and APUM23 (lower), plated on Magic Medium. Cells with APUM23 form smaller colonies than those with NOP9. Fold-dilution is indicated above each column. (C) Enrichment of unprocessed 35S pre-RNA in apum23-3 is revealed by semiquantitative reverse-transcription PCR. Error bar represents the standard error of the mean. UBC1 serves as the loading control.
Discussion
APUM23 is a unique PUF protein involved in rRNA biosynthesis and functions in regulating cell division in Arabidopsis
The Arabidopsis genome encodes more than 20 putative PUF proteins (Francischini and Quaggio, 2009; Abbasi et al., 2010; Tam et al., 2010). Among all these PUF family members, APUM23 is the only one that possesses PUF repeats outside the conserved C-terminal PUM-HD region (Tam et al., 2010). Consistent with this unusual structural feature, the major molecular function of APUM23 in regulating 35S pre-rRNA processing (Abbasi et al., 2010; this study) is also distinct from those of other PUF genes in Arabidopsis (Francischini and Quaggio, 2009).
Other Arabidopsis mutants disrupting rRNA synthesis also have morphological and developmental defects similar to the phenotypes of apum23 alleles (Abbasi et al., 2010; this study), such as narrow and pointed leaves, defective vein patterning, and reduced root growth and leaf initiation (Shi et al., 2005; Kojima et al., 2007; Petricka and Nelson, 2007). These growth defects have been proposed to be associated with impaired cell division in plants (Petricka and Nelson, 2007). In the current study, the cell division patterns in apum23 mutants were characterized using CYCB1:db:GUS and the results showed that cell division activity is indeed reduced in both roots and leaves of apum23-3 (Fig. 8). This reduction in cell division is likely associated with the lower number of mesophyll cells in the leaf blade, which results in the pale green colour of the mutant leaves (Fig. 6). These mesophyll cells are also slightly bigger than those of the wild type probably due to ‘compensation’ mechanisms that coordinate cell number and cell size in an organ (Ferjani et al., 2007; Fig. 6). Reduced cell division is also possibly related to the narrow and flat leaves in apum23-3, because growth repression results in leaves with reduced curvature and blade expansion (White, 2006). In addition, the short root phenotype is also likely attributable to the reduced activity of cell division zone (Fig. 6). Defective cell division seems to be tightly correlated with the major growth abnormalities observed in the apum23-3 mutant, so clarifying the genetic mechanisms that account for the reduction in cell division may help lead to a better understanding of the regulation of organ growth by APUM23.
Role of APUM23 in the regulation of leaf polarity
The apum23-3 mutant on its own displayed a mild leaf polarity defect. There was a modestly decreased distinction between the adaxial and abaxial mesophyll (Fig. 6), but vascular structures did not differ noticeably from the wild type (Fig. 1). However, mRNAs of a number of polarity genes (PHB, REV, AS1, AS2, KAN1, KAN2) accumulated to a higher level in the apum23 mutants than in the wild type during early stages of leaf development (Fig. 7), which suggests that APUM23 does influence the expression of these critical leaf polarity regulators. Normal leaf polarity is highly dependent on the balance between antagonistic adaxial- and abaxial-specifying genes. In the apum23 mutant, this balance may not be strongly altered because both types of key regulators are simultaneously upregulated, so the defects in leaf polarity are relatively mild. However, if one or more of these key genes are also mutated, the balance will be altered and proper polarity formation may be disrupted to an even greater degree, which may explain the strong polarity defects observed in double and triple mutants with apum23.
The major molecular function of APUM23 is to regulate rRNA biosynthesis, which acts primarily on the translational machinery (Abbasi et al., 2010; this study), but how APUM23 controls the transcription of leaf polarity genes is still not understood. One plausible mechanism is that APUM23 may act indirectly on leaf polarity genes through translational regulation of genes that, in turn, control the transcription of leaf polarity genes. Alternatively, it may be possible that APUM23 also regulates the biogenesis of other RNA species, such as small RNAs, required for leaf polarity. It has been reported that genes involved in rRNA processing are also required for miRNA biogenesis (Fukuda et al., 2007). In leaf polarity, miRNA play important roles to posttranscriptionally regulate critical genes that APUM23 interacts with, such as HD-ZIP III genes (Bao et al., 2004; Juarez et al., 2004; Kidner and Martienssen, 2004; Mallory et al., 2004). Interestingly, HD-ZIP III genes also have similar synergistic interactions with genes encoding subunits of ribosome proteins that interact with rRNA to constitute the ribosome (Pinon et al., 2008; Yao et al., 2008). It has been hypothesized that these ribosomal genes may interact with the small RNA silencing complex RISC to mediate miRNA or siRNA functions, which in turn affect the function of leaf polarity genes (Pinon et al., 2008). Given that the phenotypes of the single mutants of these ribosomal genes and their double mutants with leaf polarity genes both resemble those of apum23 (Pinon et al., 2008; Yao et al., 2008), it is possible that these two essential components of the ribosome may act in related posttranscriptional pathways to control the expression of leaf polarity genes. Future studies to identify the proteins that interact with APUM23 will hopefully provide more insights into this possibility and help to unravel further the function of this unique type of PUF protein in Arabidopsis.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. 1. Phenotype of the confirmed T-DNA insertion line of APUM23 SAIL_757_B08 (apum23-1).
Supplementary Fig. 2. Spatial expression patterns of KAN1 and AS2 in Col and apum23-3.
Supplementary Table S1. Primers used in qRT-PCR.
Acknowledgements
The authors thank Beth Grayhack for providing the yeast expression vector BG1805 and Yaël Harrar for the seeds of CYCB1:db:GUS. They also thank Jeon Hong for helping with the initial mapping of apum23-3 mutant. This work was supported by the National Science Foundation (IBN-0343179 to R.A.K) and the Charles and Johanna Busch Foundation (to R.A.K and T.H.).
References
- Abbasi N, Kim HB, Park NI, Kim HS, Kim YK, Park YI, Choi SB. 2010. APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis . The Plant Journal 64, 960–976 [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221 [DOI] [PubMed] [Google Scholar]
- Bao N, Lye KW, Barton MK. 2004. microRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Developmental Cell 7, 653–662 [DOI] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. 2000. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis . Nature 408, 967–971 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743 [DOI] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. 2003. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Current Biology 13, 1768–1774 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL. 2001. Establishment of polarity in lateral organs of plants. Current Biology 11, 1251–1260 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Izhaki A, Baum SF, Floyd SK, Bowman JL. 2004. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131, 2997–3006 [DOI] [PubMed] [Google Scholar]
- Ferjani A, Horiguchi G, Yano S, Tsukaya H. 2007. Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiology 144, 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes A, Lehmann R. 1998. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 125, 679–690 [DOI] [PubMed] [Google Scholar]
- Francischini CW, Quaggio RB. 2009. Molecular characterization of Arabidopsis thaliana PUF proteins—binding specificity and target candidates. FEBS Journal 276, 5456–5470 [DOI] [PubMed] [Google Scholar]
- Fukuda T, Yamagata K, Fujiyama S, et al. 2007. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nature Cell Biology 9, 604–611 [DOI] [PubMed] [Google Scholar]
- Ha CM, Jun JH, Nam HG, Fletcher JC. 2007. BLADE-ON-PETIOLE 1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial–abaxial polarity genes. The Plant Cell 19, 1809–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrar Y, Bellec Y, Bellini C, Faure JD. 2003. Hormonal control of cell proliferation requires PASTICCINO genes. Plant Physiology 132, 1217–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutierrez-Nava M, Poethig SR. 2006. trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis . Development 133, 2973–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husar S, Berthiller F, Fujioka S, et al. 2011. Overexpression of the UGT73C6 alters brassinosteroid glucoside formation in Arabidopsis thaliana . BMC Plant Biology 11, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MC. 2004. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428, 84–88 [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS. 2001. KANADI regulates organ polarity in Arabidopsis . Nature 411, 706–709 [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. 2004. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428, 81–84 [DOI] [PubMed] [Google Scholar]
- Kidner CA, Timmermans MC. 2007. Mixing and matching pathways in leaf polarity. Current Opinion in Plant Biology 10, 13–20 [DOI] [PubMed] [Google Scholar]
- Kojima H, Suzuki T, Kato T, et al. 2007. Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. The Plant Journal 49, 1053–1063 [DOI] [PubMed] [Google Scholar]
- Lin H, Spradling AC. 1997. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124, 2463–2476 [DOI] [PubMed] [Google Scholar]
- Lin WC, Shuai B, Springer PS. 2003. The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial–abaxial patterning. The Plant Cell 15, 2241–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- Mafra V, Kubo KS, Alves-Ferreira M, Ribeiro-Alves M, Stuart RM, Boava LP, Rodrigues CM, Machado MA. 2012. Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PLoS One 7, e31263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. 2004. microRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO Journal 23, 3356–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAbee JM, Hill TA, Skinner DJ, Izhaki A, Hauser BA, Meister RJ, Venugopala Reddy G, Meyerowitz EM, Bowman JL, Gasser CS. 2006. ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. The Plant Journal 46, 522–531 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Barton MK. 1998. Leaf polarity and meristem formation in Arabidopsis . Development 125, 2935–2942 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. 2001. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713 [DOI] [PubMed] [Google Scholar]
- Murata Y, Wharton RP. 1995. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell 80, 747–756 [DOI] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. 2000. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127, 5523–5532 [DOI] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE. 2001. REVOLUTA regulates meristem initiation at lateral positions. The Plant Journal 25, 223–236 [DOI] [PubMed] [Google Scholar]
- Pan X, Yuan DS, Xiang D, Wang X, Sookhai-Mahadeo S, Bader JS, Hieter P, Spencer F, Boeke JD. 2004. A robust toolkit for functional profiling of the yeast genome. Molecular Cell 16, 487–496 [DOI] [PubMed] [Google Scholar]
- Pekker I, Alvarez JP, Eshed Y. 2005. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. The Plant Cell 17, 2899–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Nelson TM. 2007. Arabidopsis nucleolin affects plant development and patterning. Plant Physiology 144, 173–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon V, Etchells JP, Rossignol P, Collier SA, Arroyo JM, Martienssen RA, Byrne ME. 2008. Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 135, 1315–1324 [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. 2005. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. The Plant Cell 17, 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Vasquez J, Caparros-Ruiz D, Barneche F, Echeverria M. 2004. A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro . Molecular Cell Biology 24, 7284–7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S, Watanabe K, Goto K, Kanaya E, Morita EH, Okada K. 1999. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes and Development 13, 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y. 2001. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128, 1771–1783 [DOI] [PubMed] [Google Scholar]
- Senecoff JF, McKinney EC, Meagher RB. 1996. De novo purine synthesis in Arabidopsis thaliana. II. The PUR7 gene encoding 5′-phosphoribosyl-4-(N-succinocarboxamide)-5-aminoimidazole synthetase is expressed in rapidly dividing tissues. Plant Physiology 112, 905–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi DQ, Liu J, Xiang YH, Ye D, Sundaresan V, Yang WC. 2005. SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. The Plant Cell 17, 2340–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL. 1999. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis . Development 126, 4117–4128 [DOI] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP. 1999. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes and Development 13, 2704–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP. 2001. Drosophila Brain Tumor is a translational repressor. Genes and Development 15, 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassov DS, Jurecic R. 2003. The PUF family of RNA-binding proteins: does evolutionarily conserved structure equal conserved function? IUBMB Life 55, 359–366 [DOI] [PubMed] [Google Scholar]
- Sun Y, Zhou Q, Zhang W, Fu Y, Huang H. 2002. ASYMMETRIC LEAVES1, an Arabidopsis gene that is involved in the control of cell differentiation in leaves. Planta 214, 694–702 [DOI] [PubMed] [Google Scholar]
- Talbert PB, Adler HT, Parks DW, Comai L. 1995. The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana . Development 121, 2723–2735 [DOI] [PubMed] [Google Scholar]
- Tam PP, Barrette-Ng IH, Simon DM, Tam MW, Ang AL, Muench DG. 2010. The Puf family of RNA-binding proteins in plants: phylogeny, structural modeling, activity and subcellular localization. BMC Plant Biology 10, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A, Poethig RS. 1994. Development Leaf in Arabidopsis . In: Meyerowitz EM, Somerville CR, editors, Arabidopsis. Plainview: Cold Spring Harbor Laboratory Press, pp 379–401 [Google Scholar]
- Thomson E, Rappsilber J, Tollervey D. 2007. Nop9 is an RNA binding protein present in pre-40S ribosomes and required for 18S rRNA synthesis in yeast. RNA 13, 2165–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H, Uchimiya H. 1997. Genetic analyses of the formation of the serrated margin of leaf blades in Arabidopsis: combination of a mutational analysis of leaf morphogenesis with the characterization of a specific marker gene expressed in hydathodes and stipules. Molecular and General Genetics 256, 231–238 [DOI] [PubMed] [Google Scholar]
- Venema J, Tollervey D. 1999. Ribosome synthesis in Saccharomyces cerevisiae . Annual Review of Genetics 33, 261–311 [DOI] [PubMed] [Google Scholar]
- Waites R, Hudson A. 1995. phantastica: a gene required for dorsoventrality in leaves of Antirrhinum majus . Development 121, 2143–2154 [Google Scholar]
- Waites R, Selvadurai HR, Oliver IR, Hudson A. 1998. The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum . Cell 93, 779–789 [DOI] [PubMed] [Google Scholar]
- Wang X, Zamore PD, Hall TM. 2001. Crystal structure of a Pumilio homology domain. Molecular Cell 7, 855–865 [DOI] [PubMed] [Google Scholar]
- Wenkel S, Emery J, Hou BH, Evans MM, Barton MK. 2007. A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. The Plant Cell 19, 3379–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DW. 2006. PEAPOD regulates lamina size and curvature in Arabidopsis . Proceedings of the National Academy of Sciences, USA 103, 13238–13243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White EK, Moore-Jarrett T, Ruley HE. 2001. PUM2, a novel murine puf protein, and its consensus RNA-binding site. RNA 7, 1855–1866 [PMC free article] [PubMed] [Google Scholar]
- Williams L, Carles CC, Osmont KS, Fletcher JC. 2005. A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proceedings of the National Academy of Sciences, USA 102, 9703–9708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Lin WC, Huang T, Poethig RS, Springer PS, Kerstetter RA. 2008. KANADI1 regulates adaxial–abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2 . Proceedings of the National Academy of Sciences, USA 105, 16392–16397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Xu Y, Dong A, Sun Y, Pi L, Xu Y, Huang H. 2003. Novel as1 and as2 defects in leaf adaxial–abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130, 4097–4107 [DOI] [PubMed] [Google Scholar]
- Xu L, Yang L, Pi L, Liu Q, Ling Q, Wang H, Poethig RS, Huang H. 2006. Genetic interaction between the AS1–AS2 and RDR6–SGS3–AGO7 pathways for leaf morphogenesis. Plant Cell Physiology 47, 853–863 [DOI] [PubMed] [Google Scholar]
- Yao Y, Ling Q, Wang H, Huang H. 2008. Ribosomal proteins promote leaf adaxial identity. Development 135, 1325–1334 [DOI] [PubMed] [Google Scholar]
- Zamore PD, Bartel DP, Lehmann R, Williamson JR. 1999. The PUMILIO-RNA interaction: a single RNA-binding domain monomer recognizes a bipartite target sequence. Biochemistry 38, 596–604 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.