Summary
FRUITFULL is uncovered as a modulator of the function of flowering time integrators, highlighting the importance of protein–protein complexes in the fine-tuning of the flowering time response.
Key words: Flowering, FUL, SVP, SOC1, FLC, MADS-box factors.
Abstract
The role in flowering time of the MADS-box transcription factor FRUITFULL (FUL) has been proposed in many works. FUL has been connected to several flowering pathways as a target of the photoperiod, ambient temperature, and age pathways and it is has been shown to promote flowering in a partially redundant manner with SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1). However, the position of FUL in these genetic networks, as well as the functional output of FUL activity during floral transition, remains unclear. In this work, a genetic approach has been undertaken to understand better the functional hierarchies involving FUL and other MADS-box factors with well established roles as floral integrators such as SOC1, SHORT VEGETATIVE PHASE (SVP) or FLOWERING LOCUS C (FLC). Our results suggest a prominent role of FUL in promoting reproductive transition when photoinductive signalling is suppressed by short-day conditions or by high levels of FLC expression, as in non-vernalized winter ecotypes. A model is proposed where the sequential formation of FUL–SVP and FUL–SOC1 heterodimers may mediate the vegetative and meristem identity transitions, counteracting the repressive effect of FLC and SVP on flowering.
Introduction
Arabidopsis thaliana adult life cycle comprises three major phase transitions that are mainly characterized by the identity of the lateral structures produced by the shoot apical meristem (SAM). The vegetative phase transition marks the change from the production of juvenile leaves to the production of adult leaves. Both types of leaves form a rosette through the period of vegetative growth of the plant and, then, triggered by both environmental and endogenous cues, the SAM undergoes two subsequent phase transitions leading to reproductive development: the reproductive transition that causes bolting of the primary inflorescence and the production of cauline leaves subtending secondary inflorescences, and the meristem identity transition, after which the SAM will produce floral meristems directly (Araki, 2001; Yamaguchi et al., 2009; Huijser and Schmid, 2011).
Both reproductive and meristem identity transitions, that are collectively named as floral transition, are highly controlled by developmental and environmental signals. Six promoting pathways have been proposed to regulate this process (reviewed in Fornara et al., 2010; Srikanth and Schmid, 2011): the photoperiod, vernalization, ambient temperature, age, autonomous, and gibberellin pathways. The first three pathways respond to environmental signals such as daylength and seasonal or day growth temperature, while the age and autonomous patways respond to endogenous signals, and the gibberellin pathway responds to both environmental and endogenous clues. All these pathways converge at the level of a few genes, named floral transition integrators.
Within this group of floral transition integrators, several members of the MADS-box family have major roles: the expression of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) is activated by the photoperiod, age and gibberellin pathways to promote floral transition (Borner et al., 2000; Lee et al., 2000; Samach et al., 2000; Lee and Lee, 2010) which is, in part, mediated by the activation of the floral identity gene LEAFY (LFY) (Lee et al., 2008; Liu et al., 2008). Conversely, FLOWERING LOCUS C (FLC) and SHORT VEGETATIVE PHASE (SVP) act as floral transition repressors (Hartmann et al., 2000; Michaels and Amasino, 1999; Sheldon et al., 1999). High levels of FLC expression compete the inductive floral signals at the SAM, and thus, flowering is promoted when the vernalization and autonomous pathways repress FLC expression (Michaels and Amasino, 1999; Lee et al., 2000; Sheldon et al., 1999, 2000; Hepworth et al., 2002; Michaels et al., 2004; Kim et al., 2009). Likewise, the expression of the flowering repressor SVP is controlled by the autonomous, thermosensory, and gibberellin pathways (Lee et al., 2007; Li et al., 2008). FLC and SVP are able to form heterodimers that directly bind to the SOC1 promoter to down-regulate SOC1 expression, as well as to other floral transition integrators such as FLOWERING LOCUS T (FT) (Lee et al., 2007; Fujiwara et al., 2008; Li et al., 2008).
The MADS-box transcription factor FRUITFULL (FUL), a closely related gene to the flower meristem identity genes APETALA1 (AP1) and CAULIFLOWER, has been associated with several developmental processes. In addition to its well-known function during fruit development, FUL roles in floral meristem identity specification, shoot maturation, and the control of floral transition have also been described (Hempel et al., 1997; Gu et al., 1998; Ferrándiz et al., 2000a , b; Melzer et al., 2008; Shikata et al., 2009; Wang et al., 2009).
FUL is partially redundant with SOC1 in flowering promotion. Although the ful mutants are only slightly late flowering under long-day growth conditions (Ferrándiz et al., 2000a ), the double ful soc1 mutants show a strong delay in floral transition (Melzer et al., 2008). As SOC1, FUL is one of the earliest responsive genes to photoinductive signals (Hempel et al., 1997; Schmid et al., 2003) being a target of the FT–FD dimer (Schmid et al., 2003; Teper-Bamnolker and Samach, 2005; Torti et al., 2012). FUL also responds to signals derived from the age pathway, being one of the most responsive genes to the SQUAMOSA PROMOTER BINDING LIKE (SPL) proteins (Shikata et al., 2009; Wang et al., 2009; Yamaguchi et al., 2009). A recent study also places FUL in the promotion of flowering in response to ambient temperature through the action of miR156/SPL3 and FT (Kim et al., 2012).
In spite of mounting evidence linking FUL to the main flowering pathways, the importance of FUL in controlling these processes, as well as its position, downstream effectors, and mode of action in these pathways are still unclear. In this study, genetic analyses have been used to understand better the regulatory hierarchies involving FUL and other floral integrators of the MADS-box family such as SOC1, SVP, and FLC in the control of floral transition in Arabidopsis. Our results show that FUL is able to act both upstream and co-operatively with SOC1, forming a heterodimer and binding directly to the LFY promoter. In addition, it is shown that the promotive effect of FUL on floral transition depends of the presence of a functional allele of SVP and that FUL is able to counteract the repressive effect of FLC on flowering both affecting FLC expression levels and probably competing with FLC for common targets. Taking all these data together, a dynamic model is proposed for the role of FUL during floral transition, where the progressive formation of different heterodimers of FUL and other MADS transcription factors may act as a molecular switch between the vegetative and reproductive states.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana plants were grown in cabinets at 21 °C under LD (16h light) or SD (8h light) conditions, illuminated by cool-white fluorescent lamps (150 µE m–2 s–1), in a 1:1:1 by vol. mixture of sphagnum:perlite:vermiculite. To promote germination, seeds were stratified on soil at 4 °C for 3 d in the dark. The Arabidopsis plants used in this work were in the Col-0 background, except ful-1 and 35S::SOC1, that were in Ler. Mutant alleles and transgenic lines have been previously described: soc1-2 (Lee et al., 2000), ful-1 (Gu et al., 1998), ful-2 (Ferrándiz et al., 2000a ), svp-32 (Lee et al., 2007), FRI FLC (Lee and Amasino, 1995), 35S::SOC1, (Lee et al., 2000), 35S::FUL (Ferrándiz et al., 2000b ), 35S::SVP (Masiero et al., 2004), 35S::FLC (Michaels and Amasino, 1999), LFY:GUS (Blázquez et al., 1997) and FLC:GUS (Sheldon et al., 2002).
35S::FUL::GFP was generated by cloning the FUL CDS into the pEarley103 vector (Earley et al., 2006). Agrobacterium strain C58 pM090 was used to transform Arabidopsis using the floral dip protocol (Clough and Bent, 1998), and transgenic lines carrying a single transgene insertion and with similar phenotypes to the reference 35S::FUL line were selected.
Flowering time measurements
Flowering time was scored as number of leaves at bolting. The number of rosette and cauline leaves was counted when the bolting shoot had produced the first open flower. At least 15 genetically identical plants were used to score flowering time of each genotype. The Student’s t-test was used to test the significance of flowering time differences.
Chromatin immunoprecipitation (ChIP)
35S::FUL and 35S::FUL::GFP seeds were grown for 15 d in soil and inflorescences were collected for analysis. The ChIP experiments were performed as previously described by Sorefan et al. (2009) with minor modifications using an anti-GFP antibody (Abcam, Ab290). Q-PCR was performed using the SYBR®Green PCR Master Mix (Applied Biosystems) in a ABIPRISM 7700 sequence detection system (Applied Biosystems). The values correspond to the ratios between the pull-down DNA with the GFP antibody from 35S::FUL and 35S::FUL:GFP lines and between a 10% fraction of the input genomic DNA from both samples, all of them initially normalized by ACT7 or UBQ10 genomic region. The primers used for this study are described in Supplementary Table S1 at JXB online.
Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from whole plants with the RNeasy Plant Mini kit (Qiagen). 2 µg of total RNA were used for cDNA synthesis performed with the First-Strand cDNA Synthesis kit (Invitrogen) and the qPCR master mix was prepared using the iQTM SYBR Green Supermix (Bio-Rad). Results were normalized to the expression of the TIP41-like reference gene. The PCR reactions were run and analysed using the ABI PRISM 7700 Sequence detection system (Applied Biosystems). Three technical and two biological replicates were performed for each sample. See Supplementary Table S1 at JXB online for the primer sequences.
β-Glucuronidase (GUS) staining and activity measurements
For GUS histochemical detection, samples were treated for 15min in 90% ice-cold acetone and then washed for 5min with washing buffer (25mM sodium phosphate, 5mM ferrocyanide, 5mM ferricyanide, and 1% Triton X-100) and incubated from 4–16h at 37 °C with staining buffer (washing buffer+1mM X-Gluc). Following staining, plant material was fixed, cleared in chloral hydrate, and mounted to be viewed under bright-field microscopy.
For quantitative measurements, the protocol described in Blazquez et al. (1997) was followed. Briefly, apices were incubated at 37 °C for 16h in 1mM MUG assay solution (1mM 4-methyl umbelliferyl glucuronide, 50mM sodium phosphate buffer pH 7, 10mM EDTA, 0.1% SDS, 0.1% Triton X-100), in individual wells of a microtitre plate. After the reaction had been stopped by the addition of 0.3M Na2CO3, fluorescence at 430nm was measured on a luminescence spectrophotometer equipped with an ELISA plate reader (Perkin Elmer, model LS50B).
Bimolecular Fluorescence Complementation (BiFC)
Open reading frames of full-length FUL, SOC1, and SVP CDS were cloned into vectors pYFPN43 and pYFPC43 (http://www.ibmcp.upv.es/FerrandoLabVectors.php), and BiFC was performed as previously described by Belda-Palazon et al. (2012).
Confocal microscopy
Confocal microscopy was performed using a Leica TCS SL (Leica Microsystems GmbH, Heidelberg, Germany) equipped with an Argon krypton laser (Leica).
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: FUL (AT5G60910), SOC1 (AT2G45660), SVP (AT2G22540), FLC (AT5G10140), FRI (AT4G00650), LFY (AT5G61850), UBQ10 (AT4G05320), act7 (AT5G09810), and tip41-like (AT4G34270).
Results
Genetic interactions of FUL and SOC1
The timing of both reproductive and meristem phase transitions were compared by the quantification of rosette and cauline leaves of wild-type, ful, and 35S::FUL plants. As previously reported, it was observed that the loss of FUL function caused a small delay in flowering time both in long-day (LD) and short-day (SD) conditions, while the over-expression of FUL caused a strong early flowering phenotype (Table 1) (Ferrándiz et al., 2000a ; Melzer et al., 2008). The late flowering phenotype of ful mutants mainly affected the onset of the meristem identity transition, since the number of rosette leaves did not significantly differ from the wild type, while the number of cauline leaves was increased in both LD and SD conditions (Table 1). In addition, when grown in SD, the axillary meristems of cauline leaves of single ful-2 mutants formed aerial rosettes (see Supplementary Fig. S1 at JXB online), and flowers were subtended by bracts (see Supplementary Fig. S1 at JXB online).
Table 1.
Genetic interaction of FUL and SOC1: effect on flowering
| Long day | Short day | |||
|---|---|---|---|---|
| Rosette leaves | Cauline leaves | Rosette leaves | Cauline leaves | |
| Columbia-0 | 10.2±1.0 | 3.2±0.4 | 55.1±3.4 | 9.3±0.7 |
| ful-2 | 10.7±0.8 | 4.4±0.5a | 59.9±3.8a | 23.7±3.2a |
| soc1-2 | 19.3±0.9a | 4.2±0.5a | 75.0±4.2a | 15.2±0.5a |
| ful-2 soc1-2 | 24.5±0.8a,b,c | 9.7±1.9a.b,c | 75.1±3.5a,b, | 28.1±1.7a,b,c |
| 35S::FUL | 3.5±0.5a | 1.7±0.7a | 10.6±0.9a | 3.6±0.7a |
| 35S::FUL soc1-2 | 9.0±1.1d | 2.2±0.7d | 44.6±12.8d | 7.2±4.5d |
| Landsberg er | 7.3±0.5 | 1.8±0.4 | nd | nd |
| ful-1 | 8.4±0.5e | 2.5±0.5e | nd | nd |
| 35S::SOC1 | 4.0±0.0e | 0.4±0.5e | nd | nd |
| 35S::SOC1 ful-1 | 4.0±0.0f | 0.7±0.5f,g | nd | nd |
| 35S::FUL 35S::SOC1 | 2.0±0.0g | 0.2±0.4g | nd | nd |
Flowering time is expressed as the mean of rosette and cauline leaves produced in long- and short-day conditions. Errors are represented as the standard deviation. Superscript letters indicate a significant difference (P <0.05) from (a) Col, (b) ful-2, (c) soc1-2, (d) 35S::FUL, (e) Ler, (f) ful-1, and (g) 35S::SOC1 controls, respectively, according to Student’s t-test; nd=not determined.
It has been described that FUL and SOC1 have similar roles and probably promote flowering redundantly (Melzer et al., 2008). However, it is still unclear how precisely these two factors interact genetically and how each of them contributes to the reproductive or the meristem identity transitions. To understand better the genetic relationship of FUL and SOC1, the effect on flowering time of different combinations of FUL and SOC1 loss- and gain-of-function alleles was compared.
In LD conditions, the ful-2 soc1-2 double mutant showed a synergistic late-flowering phenotype, in agreement with previously reported data (Melzer et al., 2008), producing more rosette leaves than the soc1-2 single mutant and more cauline leaves than both ful-2 and soc1-2 single mutants (Table 1). Additional phenotypes were observed such as the production of small leaves subtending flowers, the development of aerial rosettes at the cauline leaf axils, and frequent SAM reversion (see Supplementary Fig. S1B at JXB online), similar to what was observed in ful-2 single mutants grown in SD and in other studies (Torti et al., 2012).
The soc1-2 mutant grown in SD showed a dramatic increase in rosette leaf number, and also a delay in meristem identity transition, although not as important as the delay produced by ful-2 (Table 1). The ful-2 soc1-2 double mutants grown in SD produced a similar number of rosette leaves than the soc1-2 mutant, indicating that, in the absence of photoperiodic stimulus, the promoting role of FUL on the reproductive transition could depend on the presence of SOC1. On the other hand, the number of cauline leaves produced by ful-2 soc1-2 was only moderately higher than in ful-2 single mutants, suggesting that FUL would have a predominant effect in the control of meristem identity transition (Table 1).
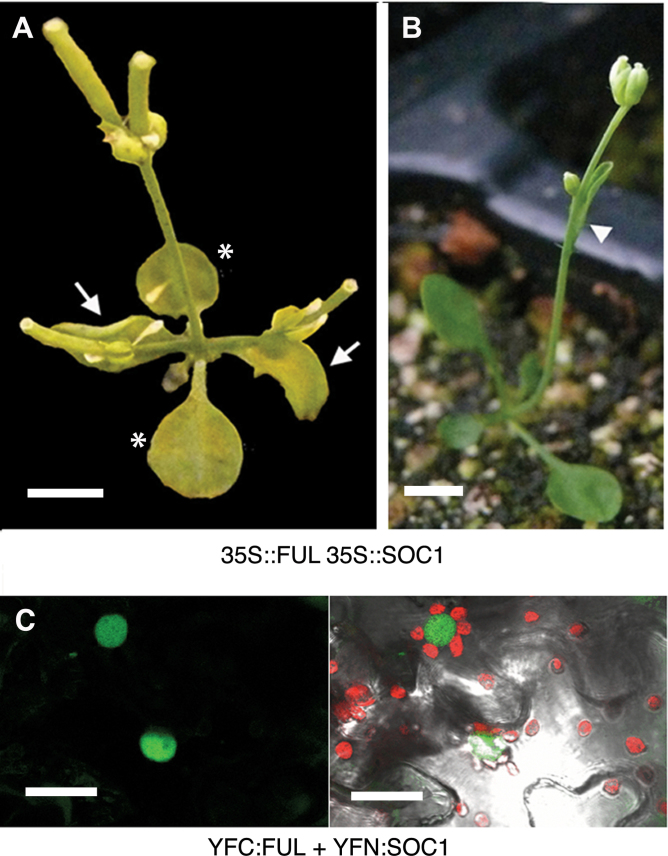
35S::FUL soc1-2 plants flowered earlier than the wild type, but significantly later than 35S::FUL lines (Table 1) supporting the idea that the flowering-promoting role of FUL was partially dependent on the presence of an active allele of SOC1. In contrast, 35S::SOC1 ful-1 plants were identical to 35S::SOC1 plants in rosette leaf number, while the absence of FUL only slightly increased the number of cauline leaves produced in the 35S::SOC1 background (Table 1). Finally, lines that over-expressed both genes simultaneously flowered extremely early, producing only two rosette leaves before the SAM directly differentiated into one or two flowers, although occasionally one cauline leaf with an axillary flower was formed (Table 1;Fig. 1A, B). Moreover, the axillary meristems from rosette leaves were also converted into flowers (Fig. 1A). This strong synergistic effect, together with the partial dependence of FUL on the presence of SOC1 to promote flowering, was compatible with FUL acting in part as an upstream regulator of SOC1, together with a subsequent co-operative action of both proteins in the regulation of putative common targets, although it did not exclude other possible scenarios.
Fig. 1.
Interaction of FUL with SOC1. (A, B) Phenotypes of 35S::FUL 35S::SOC1 double over-expression lines. Only two rosette leaves are produced (arrows in A) and occasionally one cauline leaf (arrowhead in B). All axillary meristems are determinate, directly producing flowers. Asterisks mark the cotyledons in (A). (C) Bimolecular Fluorescence Complementation in tobacco epidermal leaf cells between transiently expressed FUL and SOC1 fusions to the C- and N-terminal fragments of YFP, respectively. The left panel shows reconstituted YFP fluorescence (green) and the right panel is an overlay with a bright field image of the same sector where chlorophyll is shown in red. Negative controls for BiFC experiments are shown in Supplementary Fig. S3 at JXB online. Scale bars: 500mm (A, B), 40 µm (C).
SOC1 and LFY are FUL direct targets
It has been described that FUL and SOC1 are able to interact in yeast two-hybrid experiments as homo- and heterodimers (de Folter et al., 2005; Immink et al., 2012). To confirm this interaction in planta, a Bimolecular Fluorescence Complementation (BiFC) experiment was performed through transient expression on Nicotiana benthamiana leaves, observing FUL-SOC1 dimerization in the nuclei of the cells (Fig. 1C).
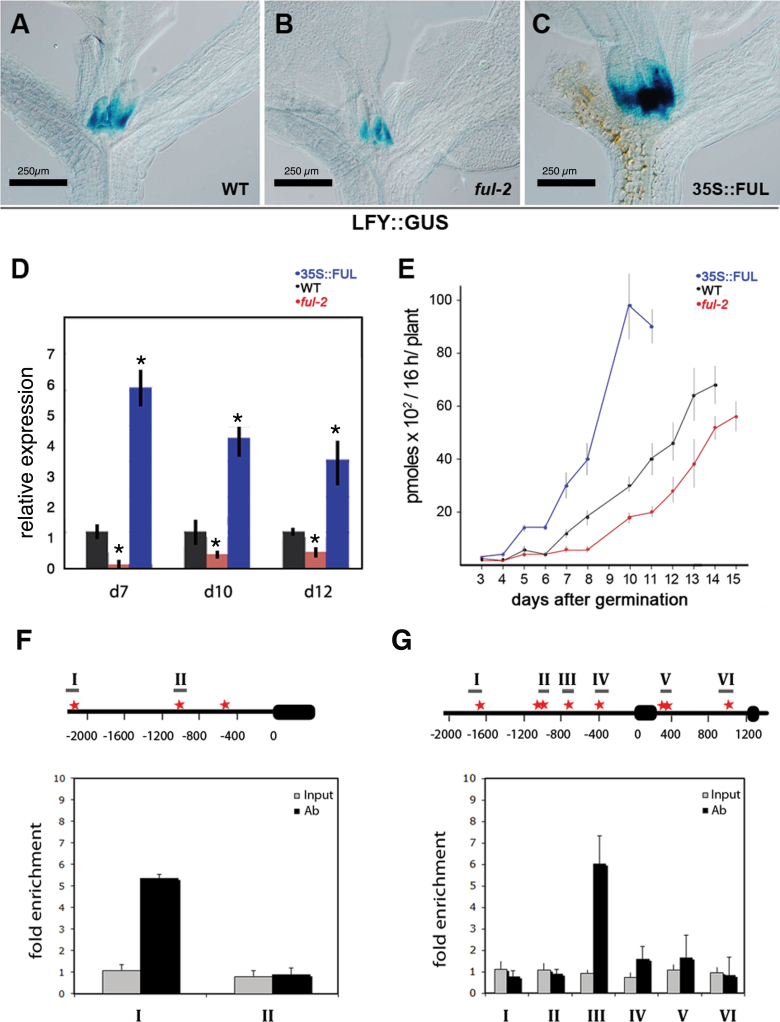
The floral identity gene LFY has been identified as a bona fide SOC1 direct target (Lee et al., 2008). In addition, FUL has been also suggested to up-regulate LFY (Ferrándiz et al., 2000a ). To confirm this suggestion, the expression of a LFY::GUS reporter line was analysed in the ful-2 and 35S::FUL backgrounds, and it was observed that the level of LFY expression was dependent on FUL, being lower in the ful-2 mutant and higher in the 35S::FUL line than in WT plants (Fig. 2A–C). These relative levels of expression were also confirmed by quantitative RT-PCR of LFY expression in apices at 7, 10, and 12 d after germination (Fig. 2D). In addition, GUS activity was also quantitatively determined in individual dissected apices, using the substrate 4-methyl umbelliferyl glucuronide (MUG), which is converted by GUS into the fluorescent product 4-MU. A time-course per-apex quantification was performed on the three genetic backgrounds, observing that LFY::GUS activity was consistently higher in 35S::FUL plants and lower in ful-2 plants than in the WT (Fig 2E). Chromatin immunoprecipitations (ChIP) experiments using a 35S::FUL::GFP line (see Supplementary Fig. S2 at JXB online) revealed that FUL was able to bind a region 2.2kb upstream to the ATG codon of the LFY gene (Fig. 2F), overlapping with a previously identified region also bound by SOC1 (Lee et al., 2008).
Fig. 2.
FUL regulates key genes in the floral transition process binding directly to SOC1 and LFY promoters. (A–C) Histochemical detection of LFY::GUS activity in the apices of 6-d-old wild type (A), ful-2 (B) or 35S::FUL (C) plants. Scale bars, 250 µm. (D) Relative expression of LFY analysed by qRT-PCR in WT, ful-2, and 35S::FUL plants at 7, 10, and 12 d after germination. The error bars depict the s.e. based on two biological replicates. Asterisks (*) indicate a significant difference (P <0.05) from the WT control according to Student’s t-test. (E) Quantification of LFY:GUS activity in WT, ful-2, and 35S::FUL backgrounds. Plants were grown on plates under long days (LD). At each time point, GUS activity was measured in at least 12 individual apices, and the means ±s.e are given. (F) (Top) Schematic diagram of the LFY upstream promoter region. First exon is represented by a black box, while the upstream genomic region is represented by a black line. The red stars indicate the sites containing either single mismatch or perfect match with the consensus binding sequence (CArG box) of MADS-domain proteins. Amplicons spanning these sites used in the ChIP analyses are represented by grey lines and marked by roman numbers. (Bottom) ChIP enrichment tests showing the binding of FUL-GFP to the LFY-I region. Bars represent the ratio of amplified DNA (35S::FUL:GFP/35S::FUL) in the starting genomic DNA (input) or in the immunoprecipitated DNA with the GFP antibody (Ab). (G) (Top) Schematic diagram of the SOC1 genomic region, including upstream promoter, exons 1 and 2 and the first intron. Exons are represented by black boxes, upstream genomic region and intron by a black line. The red stars mark CArG boxes. Amplicons spanning these sites used in the ChIP analyses are represented by grey lines and marked by roman numbers. (Bottom) ChIP enrichment tests showing the binding of FUL-GFP to the SOC1-III region. Bars represent the ratio of amplified DNA (35S::FUL:GFP/35S::FUL) in the starting genomic DNA (input) or in the immunoprecipitated DNA with the GFP antibody (Ab).
Moreover, FUL–GFP was also found to bind the SOC1 promoter, around 800bp upstream of the ATG codon (Fig. 2G). Again, this region bound by FUL overlaps with a region bound by SOC1 itself, which confirms in planta the Y1H experiment reported previously, which shows a FUL–SOC1 heterodimer binding to this fragment of the SOC1 promoter (Immink et al., 2012). Taken together, these results strongly support the hypothesis of SOC1 and FUL binding as heterodimers to the promoters of their target genes and could explain the genetic interactions observed.
Genetic interactions of FUL and SVP
SVP has been shown to repress SOC1 directly, in part by binding to the SOC1 promoter as a heterodimer with FLC, a potent repressor of flowering involved in the vernalization and autonomous pathways (Michaels and Amasino, 1999; Sheldon et al., 2002; Helliwell et al., 2006). Our results indicated that FUL could also act as an upstream regulator of SOC1, binding directly the SOC1 promoter. To explore whether FUL could interact with SVP to regulate SOC1, the effect on flowering time of different combinations of FUL and SVP loss- and gain-of-function alleles was characterized.
The svp-32 mutant showed a clear early-flowering phenotype both in LD and SD conditions, reducing the number of rosette leaves produced when compared with the WT control, as previously described by Lee et al. (2007) (Table 2). ful-2 svp-32 flowered with a similar number of leaves as the svp-32 single mutant (Table 2) (Torti et al., 2012), suggesting that SVP represses additional targets that can promote flowering in the absence of FUL, as has already been proposed by Torti et al. (2012). If this was true, we could expect plants over-expressing FUL in a svp background to flower earlier or at least like 35S::FUL plants. However, 35S::FUL svp-32 plants also flowered similarly to svp-32, both in LD and SD, (Table 2) suggesting an alternative scenario where FUL over-expression was not able to promote flower transition in the absence of an active SVP protein. Thus, the epistatic effect of svp mutation on both FUL loss- or gain-of-function may suggest that FUL required SVP to regulate its targets, and this could be mediated by the physical interaction of both factors.
Table 2.
Genetic interaction of FUL and SVP: effect on flowering
| Long day | Short day | |||
|---|---|---|---|---|
| Rosette leaves | Cauline leaves | Rosette leaves | Cauline leaves | |
| Columbia-0 | 12.4±1.7 | 2.5±0.4 | 64.4±6.0 | 8.6±0.8 |
| ful-2 | 12.9±0.9 | 3.8±0.6a | 70.2±7.0a | 20.8±3.8a |
| svp-32 | 5.6±0.5a | 2.8±0.4 | 16.4±2.1 | 4.6±1.0 |
| ful-2 svp-32 | 5.3±0.5b | 3.3±0.5 | 16.1±2.5 | 7.1±1.6 |
| 35S::FUL | 4.0±0.0a | 1.4±0.5a | 8.3±1.8a | 3.5±0.8a |
| 35S::FUL svp-32 | 5.8±0.4 | 2.5±0.5 | 14.9±2.1c,d | 3.4±1.2c |
| 35S::SVP | 27.5±1.7a | 7.3±1.0a | nd | nd |
| 35S::FUL 35S::SVP | 5.8±1.2e | 2.7±0.8d,e | nd | nd |
Flowering time is expressed as the mean of rosette and cauline leaves produced in long- and short-day conditions. Errors are represented as the standard deviation. Superscript letters indicate a significant difference (P <0.05) from (a) Col, (b) ful-2, (c) svp-32, (d) 35S::FUL, and (e) 35S::SVP controls, respectively, according to Student’s t-test; nd=not determined.
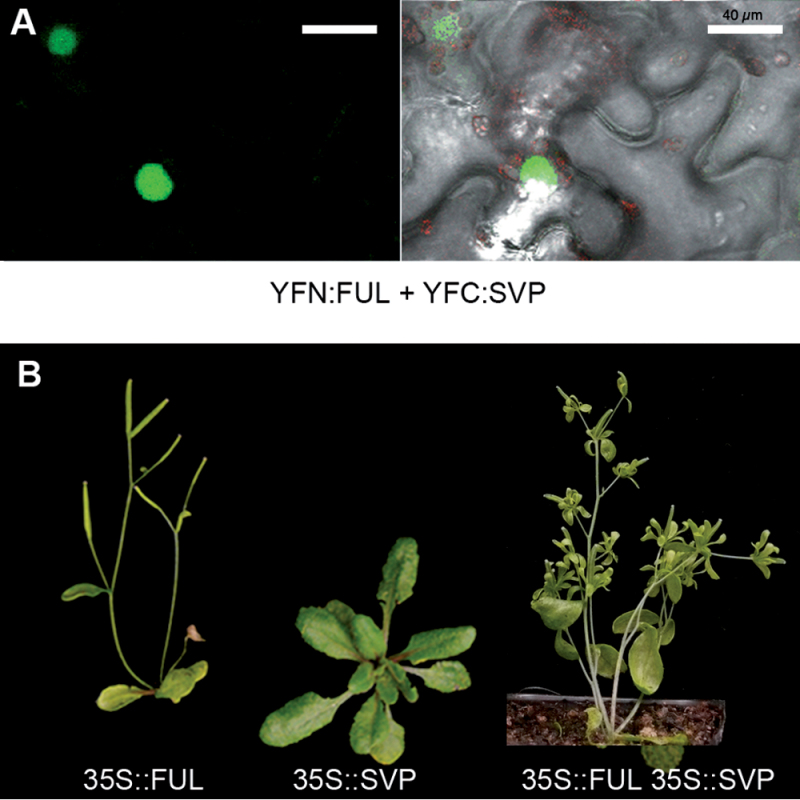
Interaction of FUL and SVP proteins has already been reported in yeast-two-hybrid experiments (de Folter et al., 2005; Immink et al., 2012). To test if this heterodimer also occurred in planta, a BiFC experiment was performed that confirmed such interaction (Fig 3A). If FUL required interaction with SVP to promote floral transition, it could be expected that simultaneous over-expression of FUL and SVP would result in early flowering, overcoming the late-flowering phenotype caused by SVP over-expression. A 35S::SVP 35S::FUL line was then generated and flowering time quantified in this double transgenic line. As described above, 35S::FUL flowered early, while 35S::SVP flowered very late, as expected for a potent repressor of flowering transition (Table 2; Fig. 3B). The line harbouring both the 35S::FUL and the 35S::SVP transgenes flowered early, similarly to 35S::FUL or 35S::FUL svp plants (Fig. 3B; Table 2). This phenotype indicated that SVP was not able to repress floral transition when both high levels of SVP and FUL were present, suggesting that the FUL–SVP dimer could suppress the repressor effect of SVP on flowering or even act as a flowering promoting factor.
Fig. 3.
Interaction of FUL with SVP. (A) BiFC experiments in tobacco leaf cells between transiently expressed FUL and SOC1 fusions to the C- and N-terminal fragments of YFP, respectively. The left panel shows YFP reconstituted fluorescence (green) and the right panel is an overlay with a bright field image of the same sector where chlorophyll is shown in red. Negative controls for BiFC experiments are shown in Supplementary Fig. S3 at JXB online. Scale bars: 40 µm. (B) Phenotypes of the 35S::FUL, 35S::SVP, and 35S::FUL 35S::SVP double over-expression lines. FUL over-expression reverts the late flowering phenotype of 35S::SVP, although inflorescence development is partially restored respect to the 35S::FUL plants.
Genetic interactions of FUL and FLC
Because the repressor effect of SVP in flowering transition is partially mediated by the formation of a heterodimer with FLC (Lee et al., 2007; Fujiwara et al., 2008; Li et al., 2008), the genetic relationship of FUL and FLC was studied.
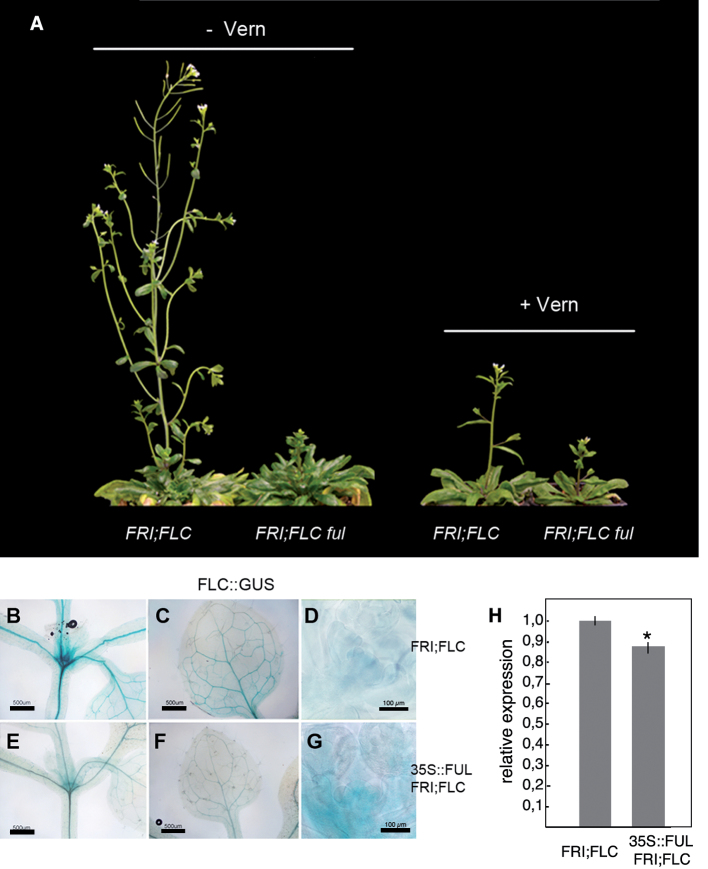
Much of the natural variation in flowering time in Arabidopsis depends on the allelic variation of FLC and its positive regulator FRI (Amasino, 2010). Late-flowering accessions usually bear functional alleles of both FLC and FRI, while most rapid-cycling accessions typically possess loss-of-function alleles of either gene. ful-2 mutants are in the Col-0 genetic background, which has a fri;FLC genotype and, therefore, an early-flowering habit (Sheldon et al., 1999; Johanson et al., 2000; Michaels, 2009). To study the effect of ful mutations in the presence of FLC, the ful-2 allele was introduced in a FRI;FLC genetic background derived from the introgression of a FRI functional allele into Col-0 (Lee and Amasino, 1995). FRI;FLC plants flower very late in all growing conditions, and are strongly responsive to vernalization treatment to induce flowering (Lee and Amasino, 1995). In LD conditions and without vernalization, the ful-2 mutation greatly enhanced the late-flowering phenotype of FRI;FLC plants, as FRI;FLC ful-2 produced many more rosette and cauline leaves than FRI;FLC plants (Table 3; Fig. 4A). Vernalization of both FRI;FLC and FRI;FLC ful-2 significantly accelerated the reproductive transition, and both lines flowered with a similar number of rosette leaves although FRI;FLC ful-2 still produced more cauline leaves (Table 3; Fig. 4A). Thus, vernalization significantly suppressed the effect of ful-2 on the floral transition of FRI;FLC plants, suggesting that, in the presence of high levels of FLC (such as in non-vernalized FRI;FLC plants), FUL was required to promote flowering and that this promotion could either be mediated by negative regulation of FLC or by counteracting the repressor effect of FLC on flowering.
Table 3.
Effect of vernalization in flowering time of ful mutants
| Long day | ||||
|---|---|---|---|---|
| –Vernalization | +Vernalization | |||
| Rosette leaves | Cauline leaves | Rosette leaves | Cauline leaves | |
| FRI FLC | 57.6±8.0 | 9.5±2.2 | 24.4±2.1 | 5.9±1.0 |
| FRI FLC ful-2 | 73.9±6.2** | 19.8±0.9** | 23.2±2.9 | 8.6±0.8 |
Flowering time is expressed as the mean of rosette and cauline leaves produced in long-day conditions. Errors are represented as the standard deviation. Asterisks (*) indicate a significant difference (P <0.05) from the FRI FLC control according to Student’s t-test.
Fig. 4.
FUL over-expression suppresses the effects of high levels of FLC. (A) Vernalization response of FRI;FLC and FRI;FLC ful-2 in LD. The ful-2 mutation greatly enhances the late flowering phenotype of FRI;FLC unvernalized plants (left), while a vernalization treatment causes both genotypes to flower similarly earlier (right). (B–G) Histochemical detection of FLC::GUS activity in FRI;FLC (B–D) and FRI;FLC 35S::FUL (E–G) plants. Apices of 10-d-old plants are compared in (B) and (E), the first rosette leaf in (C) and (F), and inflorescence apices of plants at bolting in (D) and (G). All plants were heterozygous for the FLC::GUS reporter and for the wild-type dominant alleles of FRI or FLC. 35S::FUL in (E–G) was also heterozygous. Scale bars: 500 µm (B, C, E, F) or 100 µm (D, G). (H) Relative expression of FLC analysed by qRT-PCR in FRI;FLC and FRI;FLC 35S::FUL plants 10 d after germination. The error bars depict the s.e. based on two biological replicates. An asterisk (*) indicates a significant difference (P <0.05) from the WT control according to Student’s t-test.
Flowering time was also analysed in plants resulting from crossing 35S::FUL to FRI;FLC and to 35S::FLC lines, thus generating F1 plants heterozygous for the FRI allele and hemizygous for the 35S::FUL transgene or hemizygous for both the 35S::FLC and the 35S::FUL transgenes. The results were compared with the flowering time of the corresponding F1s from crosses between FRI;FLC or 35S::FLC to the Col-0 wild type. Constitutive expression of FUL caused early flowering in FRI;FLC plants and was also able to promote flowering in the 35S::FLC background, although to a lesser extent than when FLC expression was controlled by its own regulatory sequences (Table 4). The activity of a FLC::GUS reporter in rosettes of 35S::FUL FRI;FLC plants was checked and it was found to be lower than in a FRI;FLC background (Fig. 4B, C, E, F). Quantitative RT-PCR showed that this reduction was modest, but significant (Fig. 4H), supporting that FUL could, at least partially, repress FLC expression. Moreover, while FRI;FLC plants only flowered when FLC levels were almost undetectable in the inflorescence, the 35S::FUL FRI;FLC plants flowered when FLC was still detected, indicating that FUL could also overcome the FLC repressive effect on flowering (Fig. 4D, G). Taking all these data together, it appeared that FUL was both repressing FLC expression and counteracting the negative effect of FLC on flowering, since plants were able to flower even in the presence of significant levels of FLC.
Table 4.
Genetic interaction of FUL and FLC: effect on flowering
| Long day | ||
|---|---|---|
| Rosette leaves | Cauline leaves | |
| FRI/+ | 56.5±1.7 | 12.0±1.4 |
| 35S::FUL/+ | 7.0±2.3 | 2.2±0.4 |
| 35S::FLC/+ | >80 | nd |
| 35S::FUL/+ FRI/+ | 9.7±1.1a,b | 2.3±0.8a |
| 35S::FUL/+ 35S::FLC/+ | 34.3±7.7b,c | 13.8±1.9b |
Flowering time is expressed as the mean of rosette and cauline leaves produced in long-day conditions. Errors are represented as the standard deviation. Superscript letters indicate a significant difference (P <0.05) from (a) FRI/+, (b) 35S::FUL/+, and (c) 35S::FLC/+ controls, respectively, according to Student’s t-test; nd=not determined.
Discussion
The results presented in this study show that FUL participates in both reproductive and meristem identity transitions modulating the activity of MADS-box factors with major regulatory roles in these phase changes. The role of FUL in promoting meristem identity transition is co-operative and partly dependent on SOC1, while the role of FUL in reproductive transition may be mediated both by interfering with the FLC–SVP dimer and/or changing the activity of SVP from a repressor to an activator of flowering. Taking together our genetic analyses and the results from BiFC dimerization experiments, it is proposed that these regulatory interactions are probably mediated by the sequential participation of FUL in heterodimers with SVP and SOC1 (Fig. 5).
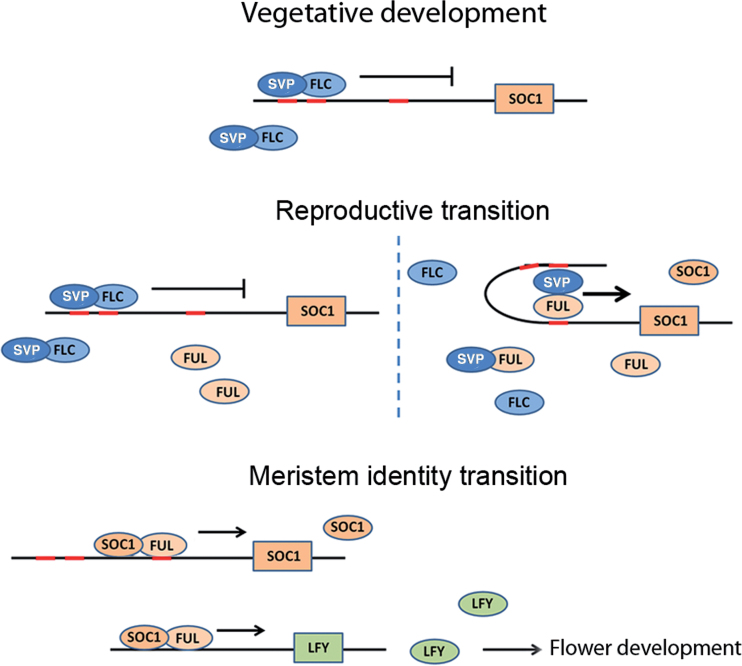
Fig. 5.
A proposed mechanistic model for the role of FUL during floral transition through interaction with SVP and SOC1 factors. During vegetative growth FLC and SVP repress the expression of SOC1 and other flowering promoting factors. Upon FUL accumulation, probably mediated by the age SPL-dependent pathway, FUL–SVP dimerization occurs. The FUL–SVP dimer could compete with the FLC–SVP dimer for binding sites in the SOC1 promoter and/or directly interfering with the FLC–SVP dimer formation. Lower repressive activity of the FLC-SVP dimer on SOC1 or even direct activation of SOC1 by FUL-SVP would lead to SOC1 accumulation, the dimerization of FUL-SOC1 and the activation of both SOC1 and LFY promoters, thus triggering flower initiation.
FUL promotes flower initiation together with SOC1
Previous studies indicate that FUL and SOC1 are able to act redundantly to promote floral transition. FUL and SOC1 share common upstream regulators, as they are both activated by the FT–FD complex and repressed by SVP (Lee et al., 2007; Li et al., 2008; Torti and Fornara, 2012). However, they also respond differently to other flowering pathways, FUL being more responsive to the age pathway and SOC1 to the gibberellin pathway (Wang et al., 2009; Yamaguchi et al., 2009; Porri et al., 2012). Moreover, recent work has also shown how SOC1 and FUL respond differently to the signals from the photoperiodic pathway, where the maintenance of SOC1 expression in the SAM depends more strongly on a continuous photoperiodic stimulus than that of FUL (Torti et al., 2012). These differences in regulation could partly explain the phenotypic effects that were observed in ful and soc1 mutants. When grown in SD, ful mutants show little effect in reproductive transition, while strongly delaying flower production, indicating that when other photoperiod-responsive genes like SOC1 are down-regulated, FUL plays an important role in promoting floral meristem initiation. Moreover, the presence of binding sites for FUL in the SOC1 promoter, the similar timing of reproductive transition in soc1 and ful soc1 mutants grown in SD, and the significant suppression of the early-flowering phenotype of 35S::FUL lines in the soc1 background, probably places FUL upstream of SOC1, suggesting that, in the absence of a photoperiodic stimulus, FUL could directly mediate the activation of SOC1. Moreover, previous reports on SOC1 binding to its own promoter (Immink et al., 2012) and our experiments showing binding of FUL to the same region of the SOC1 promoter also suggest that, once both factors are present, they could act in a positive feedback loop to maintain high levels of SOC1 expression. This positive feedback loop could also explain why a ful mutant grown in SD, where SOC1 expression is down-regulated, shows meristem reversion and bracts subtending flowers. On the other hand, no binding sites for SOC1 on the FUL promoter have been identified in a recent ChIP-seq experiment (Tao et al., 2012), and loss of FUL function does not modify the 35S::SOC1 early flowering phenotype, suggesting that FUL is not a target of SOC1 regulation and, therefore, of this feedback loop.
Our results also show that FUL and SOC1 appear to act co-operatively in promoting a sharp meristem identity transition through the activation of LFY. A similar model has been proposed for the interaction of SOC1 and AGL24, another MADS factor with a flowering promoting role (Michaels et al., 2003). SOC1 has been described as a cytoplasmic protein able to dimerize with AGL24, and to translocate to the nucleus to up-regulate LFY expression (Lee et al., 2008; Li et al., 2008). A similar mechanism appears to be working for FUL and SOC1, as it has been observed that FUL and SOC1 are able to dimerize in the nucleus, and that both SOC1 and FUL bind to the same region of the LFY promoter. Thus SOC1, AGL24 and FUL could be forming redundant dimers or a higher order molecular complex to ensure the initiation of floral meristems through LFY activation.
SVP behaviour as a repressor of flowering is probably suppressed by its interaction with FUL
Because svp mutations largely suppress the late-flowering phenotype of soc1 and ful mutants, it has been proposed that SVP represses additional flowering-promoting factors that would act in parallel to FUL and SOC1 and, therefore, even in the absence of FUL and SOC1 functions, the derepression of these factors would still cause early flowering (Torti et al., 2012). Our results, showing that FUL over-expression suppresses the strong late-flowering phenotype of SVP over-expression and that SVP and FUL are able to dimerize, may suggest a different interpretation. A possibility would be that FUL over-expression could overcome the down-regulation of these additional flowering-promoting factors repressed by SVP. However, this is in contradiction to our data showing that soc1 mutations only partially suppress 35S::FUL early-flowering phenotypes and by the phenotype of 35S::SVP 35S::SOC1 plants, which flower earlier than 35S::SVP plants but later than 35S::SVP 35S::FUL plants (Li et al., 2008). We can then speculate about the role of the SVP–FUL putative dimers. Our data are compatible with a model where SVP is inactivated as a flowering repressor upon interaction with FUL. This situation would parallel the switch in SVP activity triggered by SVP dimerization with different MADS transcription factors. Thus, it has been proposed that SVP represses flowering during vegetative development, but upon up-regulation of the flowering promoting factor AGL24 in the SAM, a SVP–AGL24 dimer is formed which is able to activate the expression of AP1 in early stages of flower development. This model also proposes that once AP1 is present, SVP would be displaced from the interaction with AGL24 to form a complex with AP1 which, in turn, represses the expression of floral organ identity genes, thus ensuring the proper development of floral meristems (Gregis et al., 2006, 2008, 2009).
It is then proposed that SVP would be repressing flowering until other pathways allow the accumulation of SVP interactors such as AGL24 or FUL which, in turn, would form protein complexes with SVP to switch off SVP activity as a flowering repressor.
The interaction of FUL and FLC appears to take place at two levels
Our work suggests a major role of FUL in promoting flowering on winter ecotypes, as revealed by the enhanced late-flowering phenotype produced by the ful-2 mutation in the FRI;FLC background. Again, this effect is different from that caused by mutations in SOC1, since it has been described that soc1 does not affect the number of rosette leaves of FRI;FLC plants or other mutants in the autonomous pathway (Moon et al., 2005). These different effects of ful and soc1 mutations in the FRI;FLC background are consistent with the described role of FLC in the repression of the photoperiodic stimuli, and the prominent role of FUL on flowering promotion under short days. Accordingly, FUL loss-of-function delays flowering in the soc1 and FRI;FLC backgrounds. While FT and SOC1 are bona fide targets of FLC negative regulation, no evidence in the literature has been found of FLC regulating FUL and, in agreement with that, no binding of FLC on the FUL promoter has been detected in ChIP-seq experiments (Deng et al., 2011). Thus, in non-vernalized winter ecotypes, the expression of FT and SOC1 should be repressed by FLC, but FUL expression would be regulated independently of FLC, most likely through signals from the age pathway mediated by miR156-targets of the SPL family (Wang et al., 2009; Wu et al., 2009; Yamaguchi et al., 2009).
It has also been observed that FUL over-expression was able both to reduce FLC expression in the FRI;FLC background and to counteract the FLC repressive effect on flowering independently of FLC regulation, as revealed by the partial suppression of the 35S::FLC extreme late-flowering phenotype by FUL over-expression. These results indicate that FUL could be antagonizing FLC at two different levels: by repressing its expression and by competing with FLC activity on its targets. FLC repression by FUL might not be direct, as FUL binding on the CArG boxes of the FLC promoter could not be detected in ChIP experiments, but it is shown by the observed reduction of FLC::GUS reporter activity in the vegetative tissues of 35S::FUL lines. On the other hand, FUL could also be competing with FLC for SVP dimerization, and thus reduce the repressive effect of FLC–SVP on targets such as FT or SOC1.
A model for FUL activity as a modulator of reproductive and meristem identity transitions
With our results on the observed protein–protein interactions as well as the genetic analyses of the FUL/SVP/SOC1 relationship, we can speculate on a possible mechanism of FUL action to regulate flowering transition in Arabidopsis (Fig. 5). During the vegetative phase, both FLC and SVP are able to repress SOC1 by binding as a heterodimer to the SOC1 promoter. When FLC and SVP levels are high, as for example in the FRI;FLC unvernalized plants, the photoperiodic pathway would be repressed even under long-day conditions. FUL expression would increase, gradually responding to signalling from the age pathway. FUL accumulation could then interfere with the FLC–SVP dimer activity, perhaps by displacing SVP from the complex to form an alternative SVP–FUL heterodimer, and thus releasing SOC1 repression, and/or leading to SOC1 activation. Upon subsequent SOC1 accumulation, a FUL–SOC1 dimer would form, driving SOC1 protein to the nucleus to maintain its own expression and to activate LFY expression and flower initiation, in a possibly redundant manner with AGL24–SOC1 heterodimers.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Inflorescence phenotypes of ful, soc1, and the ful soc1 double mutant.
Supplementary Fig. S2. Plants used in the ChIP experiments.
Supplementary Fig. S3. Negative controls for BiFC experiments.
Supplementary Table S1. Primers used in this study.
Acknowledgements
We thank Francisco Madueño and Jose Antonio Jarillo for critical reading of the manuscript and their helpful comments and suggestions. Our work was supported by grants BIO2009-09920 from the Spanish Ministerio de Ciencia e Innovación and BIO2012-32902 from the Spanish Ministerio de Economia y Competitividad to CF. VB was supported by a doctoral fellowship of the Generalitat Valenciana.
References
- Amasino R. 2010. Seasonal and developmental timing of flowering. The Plant Journal 61, 1001–1013 [DOI] [PubMed] [Google Scholar]
- Araki T. 2001. Transition from vegetative to reproductive phase. Current Opinion in Plant Biology 4, 63–68 [DOI] [PubMed] [Google Scholar]
- Belda-Palazon B, Ruiz L, Marti E, Tarraga S, Tiburcio AF, Culianez F, Farras R, Carrasco P, Ferrando A. 2012. Aminopropyltransferases involved in polyamine biosynthesis localize preferentially in the nucleus of plant cells. PloS One 7, e46907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez MA, Soowal L, Lee I, Weigel D. 1997. LEAFY expression and flower initiation in Arabidopsis . Development 124, 3835–3844 [DOI] [PubMed] [Google Scholar]
- Borner R, Kampmann G, Chandler J, Gleissner R, Wisman E, Apel K, Melzer S. 2000. A MADS domain gene involved in the transition to flowering in Arabidopsis. The Plant Journal 24, 591–599 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . The Plant Journal 16, 735–743 [DOI] [PubMed] [Google Scholar]
- de Folter S, Immink RG, Kieffer M, et al. 2005. Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. The Plant Cell 17, 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Ying H, Helliwell CA, Taylor JM, Peacock WJ, Dennis ES. 2011. FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proceedings of the National Academy of Sciences, USA 108, 6680–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. 2006. Gateway-compatible vectors for plant functional genomics and proteomics. The Plant Journal 45, 616–629 [DOI] [PubMed] [Google Scholar]
- Ferrándiz C, Gu Q, Martienssen R, Yanofsky M. 2000a. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1, and CAULIFLOWER. Development 127, 725–734 [DOI] [PubMed] [Google Scholar]
- Ferrándiz C, Liljegren S, Yanofsky M. 2000b. FRUITFULL negatively regulates the SHATTERPROOF genes during Arabidopsis fruit development. Science 289, 436–438 [DOI] [PubMed] [Google Scholar]
- Fornara F, de Montaigu A, Coupland G. 2010. SnapShot: control of flowering in Arabidopsis. Cell 141, 550, 550e1–2 [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Oda A, Yoshida R, et al. 2008. Circadian clock proteins LHY and CCA1 regulate SVP protein accumulation to control flowering in Arabidopsis. The Plant Cell 20, 2960–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Colombo L, Kater MM. 2006. AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. The Plant Cell 18, 1373–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Colombo L, Kater MM. 2008. AGAMOUS-LIKE24 and SHORT VEGETATIVE PHASE determine floral meristem identity in Arabidopsis. The Plant Journal 56, 891–902 [DOI] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Dorca-Fornell C, Kater MM. 2009. The Arabidopsis floral meristem identity genes AP1, AGL24, and SVP directly repress class B and C floral homeotic genes. The Plant Journal 60, 626–637 [DOI] [PubMed] [Google Scholar]
- Gu Q, Ferrandiz C, Yanofsky MF, Martienssen R. 1998. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125, 1509–1517 [DOI] [PubMed] [Google Scholar]
- Hartmann U, Höhmann S, Nettesheim K, Wisman E, Saedler H, Huijser P. 2000. Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. The Plant Journal 12, 351–360 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES. 2006. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. The Plant Journal 46, 183–192 [DOI] [PubMed] [Google Scholar]
- Hempel FD, Weigel D, Mandel MA, Ditta G, Zambryski P, Feldman LJ, Yanofsky MF. 1997. Floral determination and expression of floral regulatory genes in Arabidopsis . Development 124, 3845–3853 [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G. 2002. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO Journal 21, 4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P, Schmid M. 2011. The control of developmental phase transitions in plants. Development 138, 4117–4129 [DOI] [PubMed] [Google Scholar]
- Immink R, Pose D, Ferrario S, et al. 2012. Characterisation of SOC1’s central role in flowering by the identification of its up- and downstream regulators. Plant Physiology 160, 433–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347 [DOI] [PubMed] [Google Scholar]
- Kim DH, Doyle MR, Sung S, Amasino RM. 2009. Vernalization: winter and the timing of flowering in plants. Annual Reviews in Cell and Developmental Biology 25, 277–299 [DOI] [PubMed] [Google Scholar]
- Kim JJ, Lee JH, Kim W, Jung HS, Huijser P, Ahn JH. 2012. The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiology 159, 461–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I. 2000. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes and Development 14, 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Amasino RM. 1995. Effect of vernalization, photoperiod, and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiology 108, 157–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lee I. 2010. Regulation and function of SOC1, a flowering pathway integrator. Journal of Experimental Botany 61, 2247–2254 [DOI] [PubMed] [Google Scholar]
- Lee J, Oh M, Park H, Lee I. 2008. SOC1 translocated to the nucleus by interaction with AGL24 directly regulates leafy. The Plant Journal 55, 832–843 [DOI] [PubMed] [Google Scholar]
- Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH. 2007. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes and Development 21, 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H. 2008. A repressor complex governs the integration of flowering signals in Arabidopsis. Developmental Cell 15, 110–120 [DOI] [PubMed] [Google Scholar]
- Liu C, Chen H, Er HL, Soo HM, Kumar PP, Han JH, Liou YC, Yu H. 2008. Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135, 1481–1491 [DOI] [PubMed] [Google Scholar]
- Masiero S, Li MA, Will I, Hartmann U, Saedler H, Huijser P, Schwarz-Sommer Z, Sommer H. 2004. INCOMPOSITA: a MADS-box gene controlling prophyll development and floral meristem identity in Antirrhinum . Development 131, 5981–5990 [DOI] [PubMed] [Google Scholar]
- Melzer S, Lens F, Gennen J, Vanneste S, Rohde A, Beeckman T. 2008. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana . Nature Genetics 40, 1489–1492 [DOI] [PubMed] [Google Scholar]
- Michaels SD. 2009. Flowering time regulation produces much fruit. Current Opinion in Plant Biology 12, 75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. 1999. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. The Plant Cell 11, 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Bezerra IC, Amasino RM. 2004. FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proceedings of the National Academy of Sciences, USA 101, 3281–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Ditta G, Gustafson-Brown C, Pelaz S, Yanofsky M, Amasino RM. 2003. AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. The Plant Journal 33, 867–874 [DOI] [PubMed] [Google Scholar]
- Moon J, Lee H, Kim M, Lee I. 2005. Analysis of flowering pathway integrators in Arabidopsis. Plant and Cell Physiology 46, 292–299 [DOI] [PubMed] [Google Scholar]
- Porri A, Torti S, Romera-Branchat M, Coupland G. 2012. Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139, 2198–2209 [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. 2000. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis . Science 288, 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU. 2003. Dissection of floral induction pathways using global expression analysis. Development 130, 6001–6012 [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES. 1999. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. The Plant Cell 11, 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Conn AB, Dennis ES, Peacock WJ. 2002. Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. The Plant Cell 14, 2527–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. 2000. The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proceedings of the National Academy of Sciences, USA 97, 3753–3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata M, Koyama T, Mitsuda N, Ohme-Takagi M. 2009. Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant and Cell Physiology 50, 2133–2145 [DOI] [PubMed] [Google Scholar]
- Sorefan K, Girin T, Liljegren SJ, Ljung K, Robles P, Galvan-Ampudia CS, Offringa R, Friml J, Yanofsky MF, Ostergaard L. 2009. A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 459, 583–586 [DOI] [PubMed] [Google Scholar]
- Srikanth A, Schmid M. 2011. Regulation of flowering time: all roads lead to Rome. Cellular and Molecular Life Sciences 68, 2013–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Shen L, Liu C, Liu L, Yan Y, Yu H. 2012. Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. The Plant Journal 70, 549–561 [DOI] [PubMed] [Google Scholar]
- Teper-Bamnolker P, Samach A. 2005. The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. The Plant Cell 17, 2661–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torti S, Fornara F. 2012. AGL24 acts in concert with SOC1 and FUL during Arabidopsis floral transition. Plant Signaling and Behavior 7, 1251–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torti S, Fornara F, Vincent C, Andres F, Nordstrom K, Gobel U, Knoll D, Schoof H, Coupland G. 2012. Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. The Plant Cell 24, 444–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D. 2009. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana . Cell 138, 738–749 [DOI] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. 2009. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D. 2009. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Developmental Cell 17, 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.