Abstract
Mutations in PCBD1 are causative for transient neonatal hyperphenylalaninemia and primapterinuria (HPABH4D). Until now, HPABH4D has been regarded as a transient and benign neonatal syndrome without complications in adulthood. In our study of three adult patients with homozygous mutations in the PCBD1 gene, two patients were diagnosed with hypomagnesemia and renal Mg2+ loss, and two patients developed diabetes with characteristics of maturity onset diabetes of the young (MODY), regardless of serum Mg2+ levels. Our results suggest that these clinical findings are related to the function of PCBD1 as a dimerization cofactor for the transcription factor HNF1B. Mutations in the HNF1B gene have been shown to cause renal malformations, hypomagnesemia, and MODY. Gene expression studies combined with immunohistochemical analysis in the kidney showed that Pcbd1 is expressed in the distal convoluted tubule (DCT), where Pcbd1 transcript levels are upregulated by a low Mg2+-containing diet. Overexpression in a human kidney cell line showed that wild-type PCBD1 binds HNF1B to costimulate the FXYD2 promoter, the activity of which is instrumental in Mg2+ reabsorption in the DCT. Of seven PCBD1 mutations previously reported in HPABH4D patients, five mutations caused proteolytic instability, leading to reduced FXYD2 promoter activity. Furthermore, cytosolic localization of PCBD1 increased when coexpressed with HNF1B mutants. Overall, our findings establish PCBD1 as a coactivator of the HNF1B-mediated transcription necessary for fine tuning FXYD2 transcription in the DCT and suggest that patients with HPABH4D should be monitored for previously unrecognized late complications, such as hypomagnesemia and MODY diabetes.
Hypomagnesemia is a common clinical manifestation in patients with mutations in the transcription factor hepatocyte nuclear factor 1 homeobox B (HNF1B [Mendelian Inheritance in Man (MIM) 189907]).1 Mutations in HNF1B are associated with an autosomal dominant syndrome characterized by renal malformations with or without cysts, liver and genital tract abnormalities, gout, and maturity-onset diabetes of the young type 5 (MODY5; renal cysts and diabetes syndrome [MIM 137920]).1,2 Hypomagnesemia (plasma Mg2+ levels<0.7 mmol/L) with hypermagnesuria affects up to 50% of the HNF1B patients.1,3
In adult kidney, HNF1B is expressed in epithelial cells along all segments of the nephron. The role of HNF1B in renal Mg2+ handling was, however, pinpointed to the distal convoluted tubule (DCT), where the final urinary Mg2+ excretion is determined.1,4 In DCT, the Na+/K+-ATPase provides the necessary driving force for active Mg2+ reabsorption from the prourine into the blood.4 Heterozygous mutations in the FXYD2 gene, encoding the γ-subunit of the Na+/K+-ATPase, result in an autosomal dominant renal hypomagnesemia with hypocalciuria (MIM 154020).5 More recently, functional HNF1B binding sites were identified in the promoter region of FXYD2, suggesting that impaired transcription of FXYD2 by HNF1B results in renal Mg2+ wasting.1,6 Additional HNF1B target genes in kidney include renal cystic genes7–9 as well as genes involved in tubular electrolyte transport.10,11
HNF1B forms heterotetrameric complexes with the protein pterin-4α-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 homeobox A (PCBD1 [MIM 126090]).12 PCBD1 is a protein of 12 kDa with two distinct biologic functions: transcriptional coactivation of HNF1A-mediated transcription within the nucleus and PCBD1 (EC 4.2.1.96) in the cell cytosol.12,13 The enzymatic activity of PCBD1, together with dihydropteridine reductase, regenerates tetrahydrobiopterin (BH4), which is the cofactor for phenylalanine hydroxylase and other aromatic amino acid hydrolases.14 The crystal structure of PCBD1 revealed that the protein forms a tetramer of identical subunits comprising two saddle-like shaped dimers.15,16 HNF1 binding sites are located at the same surface that mediates interaction of the PCBD1 homodimers on the opposite side of the catalytic domain.17 PCBD1 knockout mice display hyperphenylalaninemia, predisposition to cataracts, and mild glucose intolerance.18 Homozygous or compound heterozygous PCBD1 mutations in humans are associated with transient neonatal hyperphenyalaninemia and high urinary levels of primapterin (HPABH4D; or primapterinuria [MIM 264070]).19–21 To date, there have been no reports of late complications or possible phenotypic consequences of impaired stimulation of the HNF1 transcription factors.
In our study, the occurrence of hypomagnesemia and MODY diabetes was investigated in three patients carrying PCBD1 mutations. We evaluated whether PCBD1 plays a role in renal Mg2+ reabsorption by directly affecting HNF1B-regulated FXYD2 transcription to gain new insight into the molecular basis of the PCBD1–HNF1B interaction.
Results
Homozygous PCBD1 Mutations Are Associated with Hypomagnesemia and Renal Mg2+ Wasting
We diagnosed hypomagnesemia and hypermagnesuria in two patients carrying mutations on both alleles in the PCBD1 gene (Table 1). In patient 1, hypomagnesemia was partially corrected with oral Mg2+ supplements at a dose of 500 mg/d (0.64–0.76 mmol/L after first supplementation, 0.66–0.69 mmol/L after second supplementation), although at the expense of increased magnesuria (fractional excretion of Mg2+ [FEMg]; 4.6%–7.8%, N<4%). He had nonspecific symptoms that might be related to hypomagnesemia, including fatigue, muscular pain, weakness and cramps in arms, numbness, difficulty with memory, chest pains, and blurred vision, with some improvement after Mg2+ supplementation. An abdominal ultrasound showed slightly increased echogenicity of both liver and kidney of uncertain cause; renal size was normal, with no cysts present (Supplemental Figure 2, A and B). Renal function was normal (GFR=128 ml/min per 1.73 m2, N≥90). Laboratory investigations of patient 2 revealed a relatively high 24-hour urinary Mg2+ excretion (5.25 mmol/24 h, N=2–8) (Table 1) in the presence of hypomagnesemia (0.65 mmol/L), with no secondary symptoms. Serum and 24-hour urinary Ca2+ excretion were within the normal range as was creatinine clearance (126 ml/min, N=89–143). Ultrasound examinations excluded the presence of renal hypoplasia or cysts (Supplemental Figure 2C). Patient 3 showed borderline serum Mg2+ levels (0.74 mmol/L) but displayed normal urinary Mg2+ excretion (FEMg=2.5%).
Table 1.
Pertinent laboratory investigations for three patients with PCBD1 mutations
| Parameter | Patient 1 (BIODEF 272) | Patient 2 (BIODEF 329) Baseline | Patient 3 (BIODEF 319) Baseline | Reference Range | |
|---|---|---|---|---|---|
| Baseline | 500 mg/d Mg2+ | ||||
| Serum indices | |||||
| Na+ (mmol/L) | 141 | — | 138 | 139 | 135–145 |
| K+ (mmol/L) | 4.0 | — | 4.5 | 4.0 | 3.5–5.0 |
| Mg2+ (mmol/L) | 0.64a | 0.76 | 0.65a | 0.74 | 0.7–1.1 |
| Ca2+ (mmol/L) | 2.41 | 2.55 | 2.25 | 2.43 | 2.20–2.65 |
| Pi (mmol/L) | 1.13 | 1.16 | — | 1.15 | 0.8–1.4 |
| Creatinine (mmol/L) | 0.069 | 0.066 | 0.062 | 0.056 | 0.045–0.110 |
| HbA1c (%) | 7.2a | — | 4.95 | 6.5a | 4.3–6.1 |
| Uric acid (μmol/L) | 310 | — | — | — | 135–510 |
| Urinary indices | |||||
| Mg2+ (mmol/L) | 6.16 | 3.96 | — | 2.6 | — |
| Mg2+ (mmol/24 h) | — | — | 5.25 | — | 2–8 |
| FEMg (%) | 6.6a | 11.1a | — | 3.5 | <4 |
| Ca2+ (mmol/L) | 4.47 | 3.04 | — | 2.99 | — |
| Ca2+ (mmol/24 h) | — | — | 5.61 | — | <7.5 |
| FeCa (%) | 0.9 | 1.8 | — | 0.86 | >1 |
| Pi (mmol/L) | 40.29 | 13.61 | — | 18.2 | — |
| FePi (%) | 17 | 17.6 | — | — | 5–20 |
| Creatinine (mmol/L) | 14.29 | 4.40 | — | 8.0 | — |
| Creatinine (mmol/24 h) | — | — | 14 | — | 9–18 |
| GFR (ml/min per 1.73 m2) | 128 | 135 | — | 124 | ≥90 |
| CCr (ml/min) | — | — | 126 | — | 89–143 |
FEMg was calculated using the following formula: FEMg=(UMg×PCr)/[(0.7×PMg)×UCr×100]. GFR was calculated by the Modification of Diet in Renal Disease formula. FeCa, fractional excretion of Ca2+; FePi, fractional excretion of phosphate (Pi); CCr, creatinine clearance.
aAbnormal value.
MODY in Two Patients with PCBD1 Mutations
In addition to hypomagnesemia, patient 1 was diagnosed with diabetes. Type 1 autoimmune diabetes was unlikely, because the patient lacked islet cell antibodies and showed normal serum C-peptide levels (0.69 nmol/L, N=0.3–1.32 nmol/L). Because MODY patients are generally highly sensitive to sulphonylureas,22 he was treated with Gliclazide 80 mg two times per day, to which he responded well, allowing his previous insulin treatment of 20–30 units/d to be discontinued. Additional extrarenal manifestations in HNF1B disease include liver test abnormalities.23,24 Liver function tests in patient 1 revealed that the plasma levels of high-sensitivity C-reactive protein (hs-CRP) were significantly low at <0.1 mg/L (N<8 mg/L) (Table 2). IgG was slightly low at 5.66 g/L (N=6.94–16.18 g/L), whereas the remaining liver function tests were all within the normal range. Based on his glycosylated hemoglobin (HbA1c) levels, we concluded that patient 2 did not develop diabetes (HbA1c=4.95%, N=4.3%–6.1%) (Table 1). Abdominal ultrasound analysis showed that the pancreas and liver were normal (Supplemental Figure 2, D and E). Patient 3 was also diagnosed with diabetes of no autoimmune origin because of the absence of islet cell antibodies. HbA1c levels were 6.5% (N=4.3%–6.1%), and glucose was 6.4 mmol/L (N=4.4–6.1 mmol/L). MODY types 1, 2, and 3 caused by mutations in HNF4A, GCK, and HNF1A, respectively, were excluded by DNA analysis. Presently, the patient is not receiving medications or insulin treatment.
Table 2.
Liver function tests in patient 1 (BIODEF 272)
| Parameter | Patient 1 (BIODEF 272) | Reference Range |
|---|---|---|
| Albumin (g/L) | 41 | 38–50 |
| Prealbumin (g/L) | 0.381 | 0.1–0.4 |
| hs-CRP (mg/L) | <0.1a | <8 |
| α-1 antitrypsin (g/L) | 1.4 | 0.9–2.6 |
| C3 (g/L) | 1.17 | 0.8–2.1 |
| C4 (g/L) | 0.18 | 0.15–0.5 |
| IgG (g/L) | 5.66a | 6.94–16.18 |
| IgM (g/L) | 0.75 | 0.6–3.0 |
| IgA (g/L) | 1.99 | 0.7–4.0 |
| ALT (U/L) | 20a | 21–72 |
| AST (U/L) | 26 | 15–46 |
| ALP (U/L) | 108 | 30–130 |
C3, complement component 3; C4, complement component 4; ALT, alanine transaminase; AST, aspartate transaminase; ALP, alkaline phosphatase.
aAbnormal value.
Pcbd1 Expression in DCT Is Modulated by Dietary Mg2+ Content
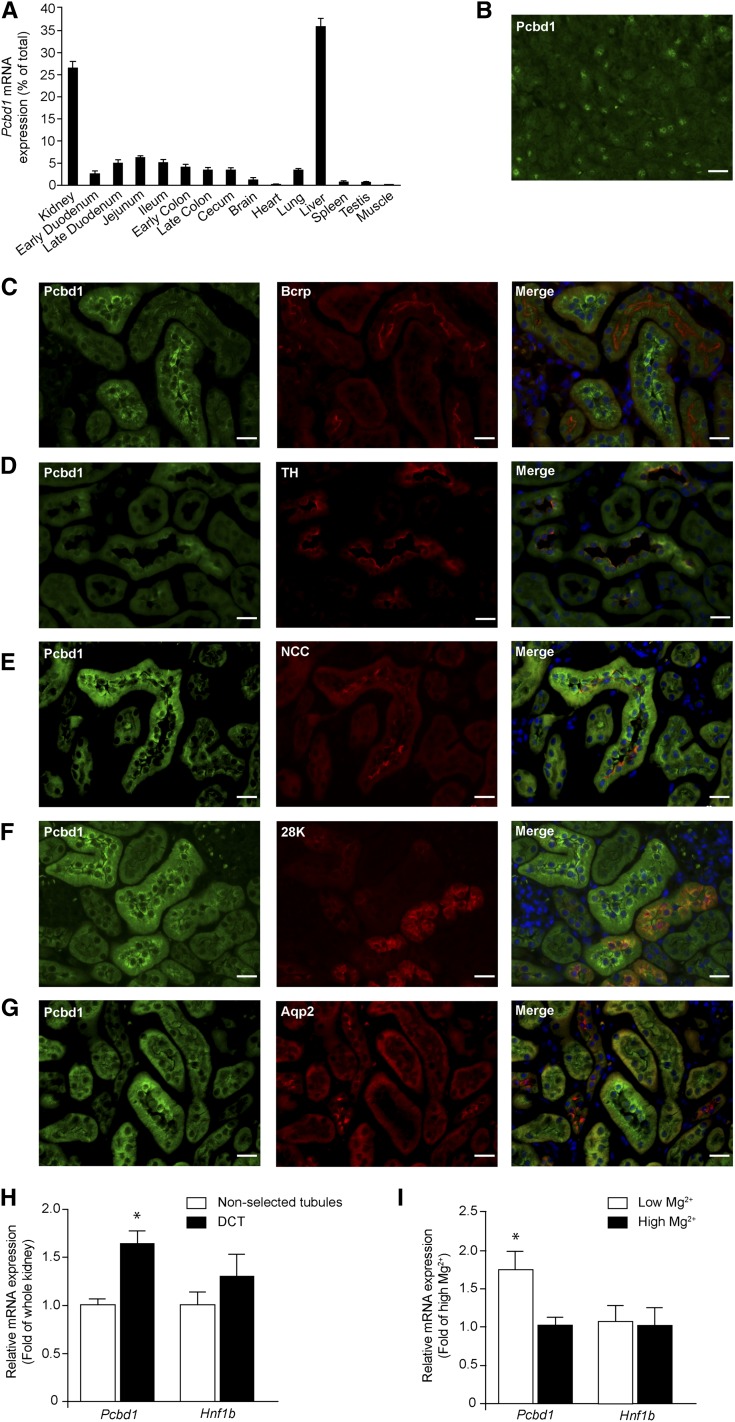
We examined Pcbd1 mRNA expression levels in a mouse tissue panel using real-time RT-PCR. Highest expression was measured in kidney and liver (Figure 1A). Furthermore, immunohistochemical analysis of mouse pancreas sections revealed that Pcbd1 is expressed in pancreatic cells (Figure 1B). Mouse kidney sections were stained for Pcbd1 to evaluate whether Pcbd1 locates to the sites of renal Mg2+ reabsorption (Figure 1, C–G). Pcbd1 staining colocalized with Na+/Cl− cotransporter (NCC) in DCT and in part, Tamm Horsfall and calbindin-D28K expression in the cortical thick ascending limb of Henle’s loop (TAL) and connecting tubule, respectively. No colocalization of Pcbd1 was detected with markers of the proximal tubule or collecting duct, which are Bcrp and Aqp2, respectively. The expression of Pcbd1 in DCT was confirmed in isolated DCT fragments. Pcbd1 expression was significantly higher in DCT compared with whole kidney (Figure 1H). Hnf1b transcript was not enriched in the DCT fraction. Interestingly, Pcbd1 expression in the DCT was significantly upregulated when mice were fed a low Mg2+-containing diet compared with a high Mg2+-containing diet (Figure 1I). Mice on a Mg2+-deficient diet displayed a significantly lower 24-hour Mg2+ excretion (Supplemental Figure 1B) in response to the hypomagnesemia (1.1 versus 2.2 mmol/L, N=1.2–1.5 mmol/L25) (Supplemental Figure 1A). Differences were not observed in the serum and urinary levels of phenylalanine between the experimental groups (Supplemental Figure 1, C and D).
Figure 1.
Pcbd1 is expressed in the DCT of the kidney. (A) Tissue expression pattern of the Pcbd1 transcript. Pcbd1 mRNA expression level was measured in a panel of mouse tissues by quantitative RT-PCR and normalized for Gapdh1 expression. Data represent the mean of three individual experiments±SEM and are expressed as the percentage of the total tissue expression. (B) Immunohistochemical analysis of Pcbd1 in mouse pancreas tissue. Scale bar, 20 μm. (C–G) Mouse kidney sections were costained for Pcdb1 (green) and (C) Bcrp (red), (D) TH (red), (E) NCC (red), (F) 28K (red), or (G) Aqp2 (red). Nuclei are shown by 4′,6-diamidino-2-phenylindole staining (blue). TH, Tamm Horsfall; 28K, calbindin-D28K. Scale bar, 20 μm. (H) Kidney expression pattern of Pcbd1 shows the highest expression in DCT. The mRNA expression levels of Pcbd1 and Hnf1b in (black bars) COPAS-selected mouse DCT and (white bars) control (nonselected) kidneys were measured by quantitative RT-PCR and normalized for Gapdh expression. Data represent the mean of three individual experiments±SEM and are expressed as fold difference compared with the expression in nonselected tubules. *P<0.05 versus total kidney. (I) DCT expression of Pcbd1 is regulated by dietary Mg2+ intake. The mRNA expression levels of Pcbd1 and Hnf1b in COPAS-selected mouse DCT kidney tubules from mice fed with (white bars) low and (black bars) high Mg2+-containing diets were measured by real-time RT-PCR and normalized for Gapdh expression. Data represent the mean of four individual experiments±SEM and are expressed as fold difference compared with the expression in high Mg2+-containing diets. *P<0.05 versus high Mg2+. Aqp2, aquaporin 2; Bcrp, breast cancer resistance protein.
PCBD1 Enhances FXYD2 and PKHD1 Promoter Activation by HNF1B
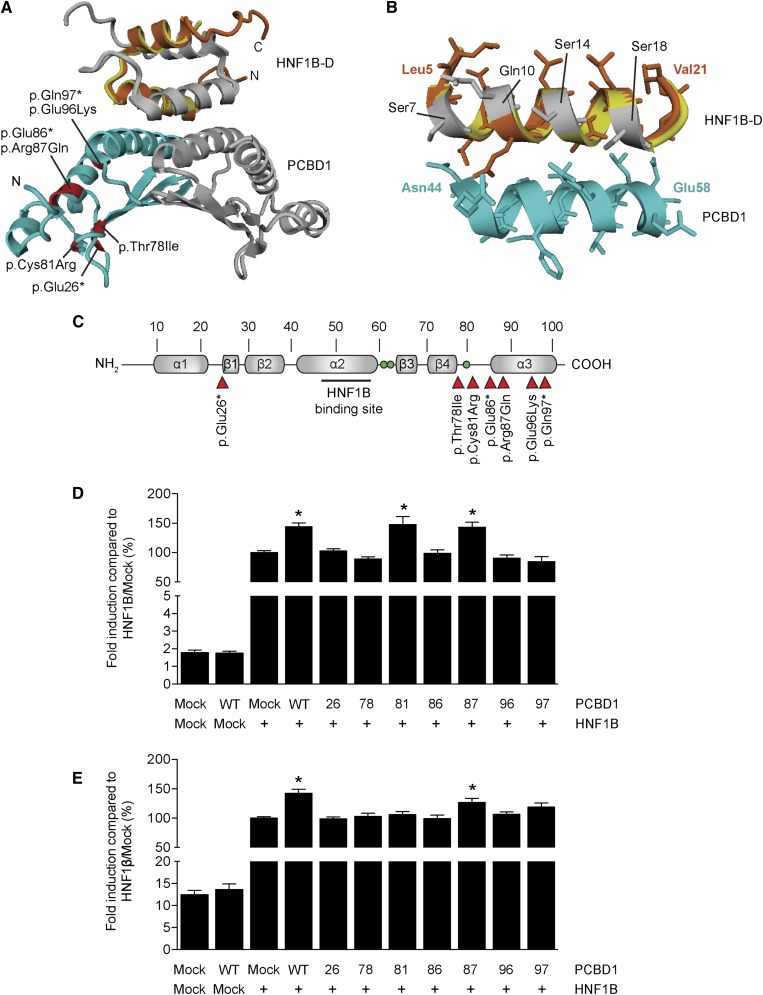
To investigate whether the PCBD1 p.Glu26*, p.Arg87Gln, p.Glu96Lys, and p.Gln97* mutations can lead to an impairment of the interaction with HNF1B, we generated a structural homology model of the PCBD1–HNF1B dimerization domain (HNF1B–D) complex using the structure of the PCBD1–HNF1A dimerization domain tetramer (Figure 2, A and B). The p.Glu26* mutation leads to a major protein truncation and complete loss of the interaction domain with HNF1B (Figure 2B). Although p.Gln97* does not directly affect the HNF1 binding domain of PCBD1, the mutation results in a truncation of the central α-helix of the protein. Consequently, the interaction domain might be destabilized, explaining the PCBD1 dysfunction. Similarly, the negatively charged Glu96 residue could be important for the stabilization of the interaction domain, which may be hampered by the p.Glu96Lys mutation.
Figure 2.
PCBD1 coactivates HNF1B-induced FXYD2 and PKHD1 promoter activity. (A) Homology model of the PCBD1–HNF1B dimerization domain (HNF1B–D) tetramer modeled using the structure of the PCBD1–HNF1A dimerization domain (HNF1A–D) complex (Protein Data Bank ID code 1F93). (Light blue and grey) The PCBD1 dimer binds the (orange and grey) HNF1B dimer through helix sequences. The HNF1A–D monomer is shown in yellow. Residues in the PCBD1 protein that were found mutates in patients affected by hyperphenylalaninemia are depicted in red. (B) Homology model of the interaction site within the PCBD1–HNF1B dimerization domain (HNF1A–D) complex. (Orange) The bound HNF1B monomer forms a helix bundle with the (light blue) PCBD1 monomer. The HNF1A–D monomer is shown in yellow. The residues that differ between HNF1B–D and HNF1A–D are visualized in grey. (C) Linear representation of the secondary structure elements of the human PCBD1 protein. Red arrowheads indicate the positions of the patient mutations described in the literature. Green balls indicate the histidine residues involved in the dehydratase active site (His61, His62, and His79). A luciferase assay was performed in HEK293 cells transiently cotransfected with a (D) Firefly luciferase FXYD2 and (E) PKHD1 promoter construct and HNF1B or mock DNA in the presence of wild-type or mutant PCBD1. A Renilla luciferase construct was cotransfected to correct for transfection efficiency. Firefly/Renilla luciferase ratios were determined as a measure of promoter activity. Results are depicted as percentage compared with HNF1B-Mock transfected cells. *P<0.05 versus HNF1B/Mock (n=0). Mock, mock DNA; WT, wild type; 26, p.Glu26+; 78, p.Thr78lle; 81, p.Cys81Arg; 86, p.Glu86*; 87, Arg87Gln; 96, p.Glu96Lys; 97, p.Gln97*.
To test the effects of the mutations on PCBD1 function, we studied all previously described patient mutations in their ability to coactivate FXYD2 transcription in a luciferase assay (Figure 2C).19–21 Expression of wild-type PCBD1 did not change the luciferase activity compared with mock-transfected cells, whereas HNF1B significantly enhanced the FXYD2 promoter activity (Figure 2D). Interestingly, coexpression of PCBD1 with HNF1B further increased FXYD2 promoter activation by ∼1.5 fold (Figure 2D). Of all PCBD1 mutants, only PCBD1 p.Arg87Gln and p.Cys81Arg maintained their coactivator properties (Figure 2D). The transcription of another established target gene of HNF1B in the kidney, PKHD1,8 was assessed in the presence of PCBD1. PKHD1 promoter activity showed a ∼1.5-fold increase expression levels when HNF1B and PCBD1 were coexpressed (Figure 2E). PCBD1 p.Arg87Gln, but not p.Cys81Arg, was the only PCBD1 mutant that retained its coactivator activity (Figure 2E).
Mutations Detected in HPABH4D Patients Cause Protein Degradation of PCBD1
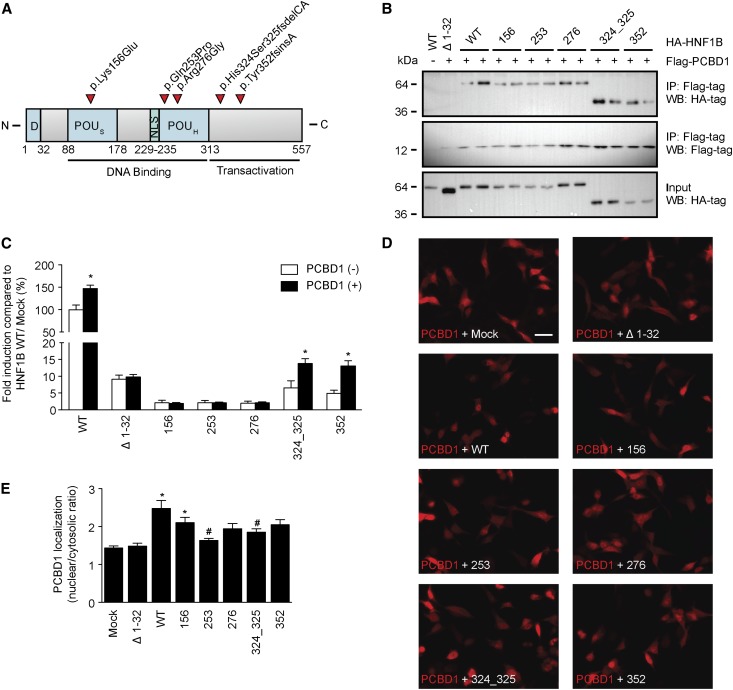
To explain why the PCBD1 mutants are not capable of enhancing HNF1B-induced transcription, we examined their subcellular localization and their capacity to bind HNF1B. Immunostaining for PCBD1 in transiently transfected human embryonic kidney (HEK293) cells revealed that wild-type PCBD1 translocates to the nucleus on coexpression with HNF1B compared with mock DNA (Figure 3A). Furthermore, PCBD1 p.Glu26*, p.Glu86*, p.Glu96Lys, and p.Gln97* were not expressed, whereas p.Thr78Ile and p.Cys81Arg were detected significantly less than the wild-type protein. PCBD1 p.Arg87Gln was the only mutant showing a comparable expression level with wild-type PCBD1 (Figure 3A). Coimmunoprecipitation studies confirmed that only PCBD1 p.Arg87Gln, p.Thr78Ile, and p.Cys81Arg were expressed and able to bind HNF1B (Figure 3B, top and bottom panels). Importantly HNF1B was equally expressed in all conditions (Figure 3B, middle panel). To examine whether mutated PCBD1 proteins are degraded by the proteasomal pathway for misfolded proteins, PCBD1-expressing HEK293 cells were treated for 24 hours with 10 nM proteasome inhibitor MG-132 (N-[(phenylmethoxy)carbonyl]-L-leucyl-N-[(1S)-1-formyl-3-methylbutyl]-L-leucinamide). Protein expression was restored for all of the PCBD1 mutants, with the only exception being p.Glu26* (Figure 3C).
Figure 3.
Several PCBD1 mutations lead to protein degradation HEK293 cells. (A) Immunocytochemistry analysis of the subcellular localization of HA-tagged PCDB1 wild type or HA-tagged PCDB1 mutants when coexpressed in a 1:1 ratio with FLAG-tagged HNF1B or mock DNA in HEK293 cells. Red signal represents immunodetected HA epitopes. Nuclei stained with 4′,6-diamidino-2-phenylindole are shown in blue. Scale bar, 10 μm. Representation immunocytochemical images are shown. (B) HA-tagged PCDB1 wild-type, PCDB1 mutant, or mock DNA were transiently expressed in HEK293 cells with or without FLAG-tagged HNF1B. Immunoprecipitations on nuclear extracts using an anti-FLAG antibody were separated by SDS-PAGE, and Western blots were probed with (top) anti-HA and (middle) anti-FLAG antibodies. (Bottom) HA-PCDB1 input (25%) expression was also included in the analysis. The immunoblots shown are representative of three independent experiments. (C) Western blot analysis of HA-tagged PCDB1 mutants expressed in HEK293 cells treated with (+) or without (−) 10 nM MG-132 for 24 hours. A representative immunoblot is shown. IP, immunoprecipitation; Mock, mock DNA; WB, Western blot; WT, wild-type; 26, p.Glu26+; 78, p.Thr78lle; 81, p.Cys81Arg; 86, p.Glu86*; 87, Arg87Gln; 96, p.Glu96Lys; 97, p.Gln97*.
HNF1B Mutations Affect the Subcellular Localization of PCBD1
We evaluated five HNF1B mutations (p.Lys156Glu, p.Gln253Pro, p.Arg276Gly, p.His324Ser325fsdelCA, and p.Tyr352fsinsA) for their ability to bind and functionally respond to PCBD1 (Figure 4A).1,3 All HNF1B mutants, except HNF1B Δ2–30, bound PCBD1 in coimmunoprecipitation studies in transiently transfected HEK293 cells (Figure 4B, top panel). PCBD1 and HNF1B were expressed in all conditions tested (Figure 4B, middle and bottom panels). FXYD2 promoter luciferase assays showed that only HNF1B p.His324Ser325fsdelCA and p.Tyr352fsinsA, which retained partial transcriptional activity, respond to the coactivation by PCBD1 to the same extent as HNF1B wild type (∼1.5-fold) (Figure 4C). Importantly, immunocytochemical analysis revealed that coexpression of PCBD1 with the HNF1B mutants p.Gln253Pro and p.His324Ser325fsdelCA causes a predominant cytosolic localization of PCBD1 compared with the nuclear translocation observed on coexpression with HNF1B wild type (Figure 4, D and E).
Figure 4.
HNF1B mutations result in cytosolic localization of PCBD1. Effect of HNF1B mutations on PCDB1 binding, transcription coactivation, and subcellular localization. (A) Linear representation of the human HNF1B protein. Red arrowheads indicate patient mutations that were tested in this study. D, dimerization domain; NLS, nuclear localization signal; POUH, atypical POU homeodomain; POUS, POU-specific domain. (B) HA-tagged HNF1B wild-type (WT), HNF1B mutants, or HNF1B lacking the extracts using an anti-FLAG antibody were separated by SDS-PAGE, and Western blots (WBs) were probed with (top) anti-HA or (middle) anti-FLAG antibodies. (Lower) HA-HNF1B input (25%) expression was also included in the analysis. The immunoblots shown are representative for three independent experiments. IP, immunoprecipitation. (C) A luciferase assay was performed in HEK293 cells transiently cotransfected with a Firefly luciferase FXYD2 promoter construct and each of the HNF1B variants (black bars) with and (white bars) without PCDB1. A Renilla luciferase construct was cotransfected to correct for transfection efficiency. Firefly/Renilla luciferase ratios were determined as a measure of promoter activity. Results are depicted as percentage compared with HNF1B/Mock transfected cells. *P<0.05 compared with the HNF1B/Mock condition (n=9). (D) Immunocytochemistry analysis of the subcellular localization of FLAG-tagged PCDB1 when coexpressed in a 1:1 ratio with HA-tagged HNF1B WTs, HA-tagged HNF1B mutants, or mock DNA HEK293 cells. Red signal represents immunodetected FLAG epitopes. Scale bar, 20 μm. The immunocytochemical images shown are representative for three independent experiments. (E) Quantification of the nuclear versus cytosolic localization of PCDB1 when coexpressed in a 1:1 ratio with mock DNA (n=24), HNF1B Δ1–32 (n=33), WT (n=41), 156 (n=38), 253 (n=35), 276 (n=33), 324_325 (n=37), or 352 (n=36). *P<0.05 compared with Mock. #P<0.05 compared with WT. Mock, mock DNA; Δ1–32, HNF1B lacking the dimerization domain; 156, p.Lys156Glu; 253, p.Gln253Pro; 276, p.Arg276Gly; 324_325, His324Ser325fsdelCA; 352, p.Tyr352fsinsA.
Discussion
Mutations in PCBD1 have been shown to cause a transient and benign form of neonatal HPABH4D.19–21 Here, we present the first follow-up study of HPABH4D patients reporting the onset of late complications linked to the deficient activity of PCBD1 as a transcriptional coactivator of HNF1B. Our results suggest that PCBD1 acts as an important transcriptional regulator of FXYD2, contributing to renal Mg2+ reabsorption in DCT. Our observations are based on the following results: (1) hypomagnesemia with renal Mg2+ wasting was reported in two of three patients carrying mutations in the PCBD1 gene; (2) two patients were diagnosed with MODY; (3) PCBD1 is present in the pancreas and kidney, predominantly in DCT, where its expression is sensitive to dietary Mg2+ content; (4) in vitro data showed that PCBD1 binds HNF1B to costimulate the FXYD2 promoter, and its activity contributes to Mg2+ reabsorption in DCT; and (5) PCBD1 mutations reported in HPABH4D patients caused proteolytic instability, leading to degradation through the proteasomal pathway.
To our knowledge, we are the first to report that PCBD1 mutations associate with hypomagnesemia because of renal Mg2+ wasting as well as MODY. In the kidney, the key sites for Mg2+ reabsorption are TAL and DCT. Although defects in the molecular pathway for Mg2+ handling in TAL lead to a concomitant waste of Ca2+, shortcomings in DCT cause hypermagnesuria associated with hypo- or normocalciuria. Based on the serum Ca2+ levels and urinary Ca2+ excretion values, it is likely that, in our patients, the DCT is primarily affected. Fluorescence-based sorting of DCT-enhanced green fluorescent protein (eGFP) tubules using a Complex Object Parameric Analyzer and Sorter (COPAS) coupled with real-time PCR analysis confirmed an enrichment of Pcbd1 expression in DCT tubules compared with other nephron segments. Pcbd1 expression in DCT was increased in mice fed with a low Mg2+ diet, suggesting that Pcbd1 is important for renal Mg2+ reabsorption in DCT. Serum and urinary phenylalanine levels were stable in the mice, and thus, they did not modulate Pcbd1 expression. Immunohistochemical analysis showed that Pcbd1 expression is not limited to DCT but partially extended to cortical TAL and connecting tubule. Overall, these findings confirm previous immunohistochemical studies showing Pcbd1 expression in the cortex of the kidney.26 Because Pcbd1 was present in the cytosol of renal cells, we hypothesized that the relative abundance of PCBD1 and HNF1B in the kidney may favor the cytosolic localization. Nevertheless, a nuclear localization of PCBD1 in the kidney could occur according to tissue-specific dynamics of the PCBD1–HNF1B complex.
Our results showed that PCBD1 enhances the FXYD2 promoter activation by HNF1B in vitro. The relevance of FXYD2 activity in Mg2+ handling in DCT has been suggested previously, because both patients with FXYD2 mutations and patients carrying HNF1B mutations may present with hypomagnesemia.1,5 It was shown that HNF1B binds the FXYD2 promoter to specifically regulate the transcription of the γ-subunit of the Na+/K+-ATPase isoform-α.1,6 γα-Subunit is mainly expressed in proximal tubule and medullary TAL, but it is also expressed in DCT (Supplemental Figure 3),6,27 where PCBD1 is expressed. To date, the exact molecular mechanism by which the γ-subunit regulates Mg2+ handling in DCT remains elusive.4
In the kidney, HNF1B is ubiquitously expressed, where it regulates the transcription of several cystic genes,9 like PKHD1. Mutations in PKHD1 are causative for autosomal recessive polycystic kidney disease (MIM 263200).8 Similar to the FXYD2 promoter, a PKHD1 reporter construct showed increased expression levels when HNF1B and PCBD1 were cotransfected in HEK293 cells. However, the renal expression of PKHD1 in vivo is restricted to the loops of Henle and collecting duct, where PCBD1 expression is negligible.28 Thus, under physiologic conditions, renal PKHD1 gene transcription seems to not be affected by PCBD1. This finding is supported by the absence of kidney abnormalities in patients with PCBD1 mutations.
Additional evidence for an impaired HNF1-mediated transcription in HPABH4D patients was provided by the diagnosis of MODY in patients 1 and 3. MODY is a monogenic form of autosomal dominant type 2 diabetes characterized by age of onset often below 25 years and negative pancreatic autoantibodies. Heterozygous mutations in HNF1B and its homolog HNF1A associate with MODY types 5 and 3, respectively.29 We showed that Pcbd1 is localized in the nuclei of pancreatic cells. Knowing that PCBD1 acts as transcriptional coactivator of both HNF1B and HNF1A, a dysregulation of HNF1B and/or HNF1A is potentially responsible for the MODY diagnosed in patients 1 and 3. Interestingly, the low plasma hs-CRP levels in patient 1 are in line with two recent studies showing lower hs-CRP levels in MODY3 patients than individuals with other forms of diabetes and nondiabetic controls.30,31 This evidence favors the diagnosis of MODY3-like diabetes in patient 1. Patient 2 was not diagnosed with diabetes, but he will be monitored for later onset.
In this study, we showed that the HPABH4D patient mutations reported in the literature lead to protein degradation of PCBD1 through the proteasome pathway. Our data, together with previous studies on proteolytic instability for the same mutants in both mammalian and bacterial expression systems, support the hypothesis that this degradation process may also occur in HPABH4D patients.19,20,32 Considering the inheritance of the disease, HNF1-mediated transcription seems to be sensitive to changes in PCBD1 quantity only when both PCBD1 alleles are affected, suggesting that PCBD1 probably belongs to an ancillary regulatory mechanism to which other HNF1B partners may participate.33 In vitro data in HEK293 cells revealed that PCBD1 p.Arg87Gln was the only mutant showing an expression level comparable with the wild-type protein. Nevertheless, in patient 2, p.Arg87Gln is homozygously present on both alleles, with a p.Glu26* mutation that leads to a significant decrease in the cellular content of PCBD1.20 PCBD1 p.Glu26* degradation could not be rescued by proteasome inhibition, suggesting that its transcript is degraded by mRNA surveillance mechanisms.34,35 PCBD1 p.Thr78Ile and p.Cys81Arg were significantly less expressed than the wild-type protein. Both mutants showed binding to HNF1B, but only PCBD1 p.Cys81Arg coactivated the FXYD2 promoter. PCBD1 p.Cys81Arg did not coactivate the PKHD1 promoter, suggesting insufficient expression to enhance the transcription of PKHD1. In the near future, it would be of interest to screen patients with PCBD1 p.Thr78Ile and p.Cys81Arg mutations for complications related to the HNF1B disease.
PCBD1 monomers are small molecules that can passively diffuse from the cytosol into the nucleus.36 It was suggested that, by assembling through the same interface, PCBD1 homotetramer and PCBD1–HNF1 complexes are mutually exclusive.17 In our immunocytochemical analysis, the HNF1B mutants p.Gln253Pro and p.His324Ser325fsdelCA significantly stimulated a cytosolic localization of PCBD1 compared with the nuclear PCBD1 localization observed in the presence of wild-type HNF1B. This finding suggests that HNF1B mutations may disturb the stability of the PCBD1–HNF1 complexes in the nucleus and therefore, favor the formation of PCBD1 homotetramers in the cytosol of the cell.1,3 The reduced nuclear localization of PCBD1 will indirectly result in a decreased coactivation of HNF1B and could contribute to the hypomagnesemia observed in some HNF1B patients. Because the presentation and development of HNF1B disease is diverse, variations in HNF1B interacting proteins may be responsible for the phenotypic heterogeneity. Screening of HNF1B patients for polymorphisms in PCBD1 should, therefore, be considered.
In conclusion, we identified PCBD1 as a new molecular player in renal Mg2+ handling. Our results suggest that PCBD1 regulates the HNF1B-mediated FXYD2 transcription, influencing active renal Mg2+ reabsorption in DCT. So far, HPABH4D caused by PCBD1 mutations has been considered a transient, benign condition, primarily related to impaired BH4 regeneration. To date, 23 patients with PCBD1 mutations linked to HPABH4D are listed in the International Database of Tetrahydrobiopterin Deficiencies (BIODEF database; http://www.biopku.org/biodef/).37 Here, we suggest that patients affected by HPABH4D should be monitored for late complications related to the interactions with HNF1 transcription factors, including hypomagnesemia and MODY.
Concise Methods
Patients
The three patients reported in this study were ascertained by contacting the authors of papers related to hyperphenylalaninemia, tetrahydrobiopterin-deficient HPABH4D (MIM 264070).19–21 Of 23 patients listed in the International Database of Tetrahydrobiopterin Deficiencies (BIODEF database; http://www.biopku.org/biodef/),37 BIODEF 272, 329, and 319 were assessed by P.F. (Canada), R.G. (Germany), and J.B.C.d.K. (The Netherlands), respectively. The results are included in this study. Informed consent to participate in this study was obtained, and the procedures followed were in accordance with the standards of the medical ethics committee of each participating institution. Patients were not treated with any medication that could affect serum levels of Mg2+ (e.g., diuretics, calcineurin inhibitors, and corticosteroids).
Patient 1 (BIODEF 272) was reported in detail previously.19 In summary, he was found to have borderline hyperphenylalaninemia on newborn screening, but this markedly increased to a peak of 2589 μmol/L at 3.5 weeks. The diagnosis of HPABH4D was suspected because of primapterinuria (7-biopterin) as well as a marked response to BH4. There was no parental consanguinity; molecular analysis confirmed the diagnosis, showing that he was homozygous for a c.312C>T (p.Gln97*) mutation in the PCBD1 gene. He was treated with phenylalanine restriction and BH4 until 4 months of age, at which time his phenylalanine levels normalized without additional treatment on a normal diet. Intermittent follow-up into adolescence revealed normal health, growth, and cognitive development. He was recontacted for this study at the age of 19 years; at that time, it was noticed that he had become an insulin-dependent diabetic about 6 months previously.
In patient 2 (BIODEF 329), initial neonatal screening showed hyperphenylalaninemia with high urinary levels of primapterin (7-biopterin),19,20 which resolved after daily treatment with BH4. There was parental consanguinity, and molecular studies showed that he was homozygote for two separate mutations in the PCBD1 gene: c.99G>T (p.Glu26*) and 283G>A (p.Arg87Gln). He was followed by pediatricians at the Klinik für Kinder und Jugendmedizin.
Patient 3 (BIODEF 319) is the first child of nonconsanguineous parents. A previous sibling of the patient died neonatally, whereas a younger brother was born healthy. The patient was diagnosed with hyperphenylalaninemia and primapterinuria at the age of 1 month.20 Molecular analysis revealed compound heterozygous c.309G>A/c.312C>T (p.Glu96Lys/p.Gln97*) mutations in the PCBD1 gene. She needed cofactor BH4 replacement therapy on a normal diet to maintain phenylalanine levels within the reference range. At the moment of recruitment for this study, she was 17 years old. At that time, she had stopped taking BH4 around 1 year earlier and had slightly elevated phenylalanine levels of 120 μmol/L (n<60 μmol/L). It was also noticed that, just before turning 16 years old, she was diagnosed with diabetes. MODY diabetes types 1, 2, and 3 were excluded by DNA analysis. The patient is presently healthy and developing normally.
Calculation of FEMg
The FEMg was calculated using the following formula: FEMg=(UMg×PCr)/[(0.7×PMg)×UCr×100]. U and P refer to the urine and serum concentrations, respectively of Mg2+ (Mg) and creatinine (Cr). Serum Mg2+ concentration was multiplied by 0.7, because only approximately 70% of the circulating Mg2+ was unbound by albumin and therefore, able to be filtered across the glomerulus.
Animal Study
All the experimental procedures are in compliance with the animal ethics board of the Radboud University Nijmegen. The transgenic parvalbumin-eGFP mice were a gift from Dr. Hannah Monyer.38 In the Mg2+ diet experiment, the mice were fed low (0.02% wt/wt) or high (0.48% wt/wt) Mg2+-containing diets for 15 days (SSNIFF Spezialdiäten GmbH, Soest, Germany). During the last 48 hours of the experiment, the mice were housed in metabolic cages for urine collection (24 hours of adaptation and 24 hours of sampling). Blood samples were taken at the start of the experiment and just before euthanasia. Parvalbumin-eGFP–positive tubules were isolated as described previously.39 In short, mice ages 4–6 weeks were anesthetized and perfused transcardially with ice cold buffer of 145 mmol/L NaCl, 5 mmol/L KCl, 1 mmol/L NaH2PO4, 2.5 mmol/L CaCl2, 1.8 mmol/L MgSO4, 10 mmol/L glucose, and 10 mmol/L Hepes/NaOH (pH 7.4). Kidneys were harvested and digested in buffer of 145 mmol/L NaCl, 5 mmol/L KCl, 1 mmol/L NaH2PO4, 2.5 mmol/L CaCl2, 1.8 mmol/L MgSO4, 10 mmol/L glucose, and 10 mmol/L Hepes/NaOH (pH 7.4) containing 1 mg/ml collagenase (Worthington, Lakewood, NJ) and 2000 units/ml hyaluronidase (Sigma-Aldrich, Houten, The Netherlands) for three cycles of a maximum of 15 minutes each at 37°C. Subsequently, the kidney tubules between 40 and 100 μm were collected by filtration. Tubules collected from the three digestions were ice cooled and sorted by the COPAS (Union Biometrica, Holliston, MA). Sorted tubules were directly collected in 1% (vol/vol) β-mercaptoethanol containing RNA extraction buffer that was supplied by the RNeasy RNA Extraction Kit (Qiagen, Venlo, The Netherlands). Per mouse, 4000 eGFP-fluorescent tubules were collected; 4000 tubules were pooled on a microcolumn for RNA extraction according to the manufacturer’s protocol. Subsequently, reverse transcription of the RNA by M-MLV reverse transcription (Invitrogen) was performed for 1 hour at 37°C. Gene expression levels were determined by quantitative real-time PCR on a BioRad Analyzer and normalized for Gapdh expression levels. Real-time PCR primers (Supplemental Table 1) were designed using the online computer NCBI/Primer-BLAST software.
Phenylalanine Measurements
Phenylalanine in plasma and urine samples was measured by liquid chromatography–tandem mass spectrometry (MS) by a standardized method, in which the MS was operated in positive mode. For these MS measurements, a Waters Premier triple quadrupole mass spectrometer (tandem MS) interfaced with an electrospray ionization source and equipped with an Alliance ultra-performance liquid chromatography (Waters, Etten-Leur, The Netherlands) was used. Briefly, to a 10-µl plasma or urine sample, 13C6-labeled phenylalanine was added as an internal control to correct for possible ion suppression. Samples were deproteinized with methanol, diluted with water, and injected on an RP C18 column (Waters Atlantis T3; 3 µm, 2.1×100 mm). Samples were eluted in 17.5% methanol/0.1 M formic acid. Positively charged [M-H]+ phenylalanine (m/z=166) and 13C6-phenylalanine (m/z=172) were selected as parent ions. After collision-induced dissociation, the losses of -NH3 and -COOH groups were chosen as quantifier ions (m/z=120 and =126, respectively), and the losses of -NH3, -COOH, and -OH2 groups (m/z=103 and 109, respectively) were chosen as qualifier ions. In Multiple Reaction Monitoring mode, the transitions m/z=166→120 and m/z=166→103 (for phenylalanine) and m/z=172→126 and m/z=172→109 (for 13C6-phenylalanine) were measured. Phenylalanine concentrations were calculated using a calibration curve.
DNA Constructs
Human HNF1B wild types and mutants were cloned into the pCINeo hemagglutinin (HA) internal ribosome entry site (IRES) GFP vector as described previously.6 HNF1B was FLAG-tagged at the NH2 terminal by Phusion PCR using NheI and AgeI restriction sites. To obtain an HNF1B lacking the dimerization domain spanning residues 2–30 (Δ2–30), HNF1B was amplified using a Phusion polymerase (Finnzymes, Vantaa, Finland) with specific primers covering from residue p.Glu31. The HNF1B-Δ2–30 was ligated into pCINeo IRES GFP using AscI/AgeI restriction sites. The open reading frame of human PCBD1 was amplified using Phusion polymerase from PCBD1 pCMV-SPORT6 (Genbank accession number BC006324; ImaGenes) and subcloned into the pCINeo HA IRES GFP vector using AgeI/EcoRI restriction sites. Wild-type PCBD1 was FLAG-tagged at the NH2 terminal by PCR using NheI/XhoI restriction sites. PCBD1 mutations were inserted in the construct using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer’s protocol. The luciferase reporter plasmid containing the FXYD2 promoter linked to the coding region of firefly luciferase was produced as previously described.6 Briefly, the human FXYD2 promoter region that controls transcription of the γ-subunit isoform-α (−3229/+91 bp from the transcription start site) was amplified from genomic DNA using Phusion polymerase (Finnzymes) and cloned into pGL3-Basic (Promega) using KpnI/BglII restriction sites. The human PKHD1 promoter region extending from exon 1 to 1.5 kb upstream to the transcription start site was amplified from genomic DNA and cloned into pGL3-Basic using KpnI/HindIII sites. The pRL-CMV vector encoding Renilla luciferase under control of a CMV promoter was commercially available (Promega, Fitchburg, WI). Primer sequences used for cloning or mutagenesis PCR are reported in Supplemental Table 2. All constructs were verified by sequence analysis.
Cell Culture
HEK293 cells were grown in DMEM (Bio Whittaker-Europe, Verviers, Belgium) containing 10% (vol/vol) FCS (Thermo Fisher HyClone), 10 μl/ml nonessential amino acids, and 2 mmol/L l-glutamine at 37°C in a humidity-controlled incubator with 5% (vol/vol) CO2. The cells were transiently transfected with the respective constructs using polyethylenimine cationic polymer (Polysciences, Inc.) at a 1:6 DNA to polyethylenimine cationic polymer ratio for 48 hours unless otherwise stated.
Western Blotting
HEK293 cells were transfected for 24 hours with HA-PCBD1 mutants and treated at the same time with 10 mmol/L MG-132. Protein lysates were denatured in Laemmli containing 100 mmol/L dithiothreitol for 30 minutes at 37°C and subsequently subjected to SDS-PAGE. Then, immunoblots were incubated with a mouse anti-HA (high-affinity 3F10, 1:5000; Roche) primary antibody and peroxidase-conjugated sheep anti-mouse secondary antibodies (1:10,000; Jackson Immunoresearch).
Immunocytochemistry
HEK293 cells were seeded in 12-well plates on glass coverslips and cotransfected with 400 ng PCBD1 constructs and 400 ng HNF1B constructs or empty pCINeo IRES GFP vector (mock DNA). After 48 hours, HEK293 cells were fixed with 4% (wt/vol) paraformaldehyde for 30 minutes at 4°C followed by the subsequent steps at room temperature: permeabilization for 15 minutes with 0.3% (vol/vol) Triton X-100 in PBS, incubation for 15 minutes with 50 mmol/L NH4Cl, and finally, incubation in blocking buffer (16% [vol/vol] goat serum, 0.3% [vol/vol] Triton X-100, and 0.3 M NaCl in PBS). After incubation overnight at 4°C with a rabbit anti-FLAG M2 (F7425, 1:100; Sigma-Aldrich) or a mouse anti-HA (6E2, 1:100; Cell Signaling Technology), cells were washed three times with Tris-buffered NaCl Tween-20 (150 mmol/L NaCl, 0.1 mol/L Tris/HCl [pH 7.5], and 0.05% [vol/vol] Tween-20) and subsequently incubated with a secondary goat anti-rabbit antibody (A4914, 1:300; Sigma-Aldrich) or a sheep anti-mouse (1:300; Jackson Immunoresearch) coupled to AlexaFluor 594 for 45 minutes at room temperature. After incubation with 4′,6-diamidino-2-phenylindole for 30 minutes at room temperature, cells were washed three times with Tris-buffered NaCl Tween-20 and finally mounted with Fluoromount-G (SouthernBiotech). Photographs were taken using a Zeiss Axio Imager 1 microscope (Oberkochen, Germany) equipped with an HXP120 Kubler Codix fluorescence lamp and a Zeiss Axiocam MRm digital camera. Images were analyzed by use of the software ImageJ.40
Immunohistochemistry
Staining was performed on 5-μm sections of PLP fixed frozen mouse kidney or pancreas samples. Subsequently, sections were washed three times with buffer of 0.15 mol/L NaCl and 0.1 mol/L Tris/HCl (pH 7.6). Antigen retrieval was done with 10 mmol/L sodium citrate (pH 6.0), and nonspecific binding was blocked by incubation in blocking buffer for 1 hour. Sections were stained overnight at 4°C with a polyclonal rabbit anti-PCBD1 antibody (F3862, 1:5000; kind gift from Prof. Beat Thöny).26 The following day, sections were washed and incubated with a goat anti-rabbit secondary antibody coupled to Alexa Fluor 488 (A11008, 1:300; Invitrogen) for 1 hour at room temperature. Kidney sections were costained for Bcrp, Tamm Horsfall, NCC, calbindin-D28k (D28K), or Aqp2. Briefly, sections were washed and incubated for 2 hours at room temperature with rat anti-Bcrp (Clone BXP-9, MC-980, 1:200; Kamya Biomedical Company), sheep anti-Tamm Horsfall (8595–0054, 1:1500; AbD Serotec), guinea pig anti-NCC (1:50; gift from Jan Loffing), mouse anti-28K (C-9848, 1:1500; Sigma-Aldrich), or guinea pig anti-Aqp2 (1:1000; gift from Peter Deen). Secondary antibodies were coupled with Alexa Fluor 495 (1:300; Invitrogen) and incubated for 45 minutes at room temperature. Subsequently, sections were washed, incubated for 10 minutes with 4′,6-diamidino-2-phenylindole, and mounted with Mowiol. Photographs were taken using a Zeiss Axio Imager 1 microscope equipped with an HXP120 Kubler Codix fluorescence lamp and a Zeiss Axiocam MRm digital camera.
Coimmunoprecipitation
HEK293 cells were seeded on 55-mm Petri dishes and cotransfected with 5 μg PCBD1 constructs and 5 μg HNF1B constructs or an empty pCINeo IRES GFP vector (mock DNA); 48 hours after transfection, nuclear and cytosolic fractions were prepared using the NE-PER Nuclear Protein Extraction Kit (Pierce). The next incubation steps were all done under rotary agitation at 4°C; 35 μl protein A-agarose beads (Santa Cruz Biotechnology) were previously incubated overnight at 4°C with 2.5 μg rabbit anti-FLAG antibody (F7425; Sigma) and washed with IPP500 (500 mmol/L NaCl, 10 mmol/L Tris adjusted to pH 8.0 with HCl, 0.1% [vol/vol] NP-40, 0.1% [vol/vol] Tween-20, and 0.1% [wt/vol] BSA, and protein inhibitors) four times. After sampling 60 μl as the input control, the remaining 180 μl lysate sample was added to the antibody beads mixture overnight at 4°C. Then, the beads were collected by centrifugation at 2000 rpm for 2 minutes at 4°C and washed four times with lysis buffer. The proteins were separated from the beads by incubation for 30 minutes at 37°C in 1× Laemmli sample buffer supplemented with 100 mmol/L dithiothreitol and detected by immunoblotting using a mouse anti-HA (high-affinity 3F10, 1:5000; Roche) or a mouse anti-FLAG M2 (F3165, 1:5000; Sigma-Aldrich) primary antibodies and peroxidase-conjugated sheep anti-mouse secondary antibodies (1:10,000; Jackson Immunoresearch).
Luciferase Reporter Assay
HEK293 cells were seeded on 12-well plates and transfected with the following DNA amounts: 700 ng FXYD2 promoter luciferase construct, 50 ng PCBD1 constructs, and 50 ng HNF1B constructs or empty pCINeo IRES GFP vector (mock DNA). To correct for transfection efficiency, 10 ng pRL-CMV was used as a reference. Firefly and Renilla luciferase activities were measured by use of a Dual-Luciferase Reporter Assay (Promega) 48 hours after transfection.
Homology Modeling
A homology model was built using the modeling script in the WHAT IF & YASARA Twinset with standard parameters.41,42 The structure of the PCBD1 dimer in complex with the HNF1A dimerization domain dimer was used as a template for modeling of HNF1B (Protein Data Bank ID code 1f93). Our model contains only the dimerization domain (31 amino acids) of HNF1B, and it has 68% sequence identity with the dimerization domain of HNF1A.
Statistical Analyses
All results are depicted as mean±SEM. Statistical analyses were conducted by unpaired t test when comparing two experimental conditions and one-way ANOVA with Bonferroni test when comparing more conditions. P values <0.05 were considered significant.
Disclosures
None.
Supplementary Material
Acknowledgments
The authors are grateful to the patients for their participation in this study. We appreciate the help of Drs. N. Blau and B. Thöny in the recruitment of the patients and provision of the PCBD1 antibody. We also thank Lonneke Duijkers and Annelies van Angelen for their excellent technical support.
This work was supported by The Netherlands Organization for Scientific Research (NWO) Grants ZonMw 9120.8026 and NWO ALW 818.02.001, a European Young Investigator Award 2006, and Vici Grant NWO-ZonMw 016.130.668 (to J.G.J.H.) as well as EURenOmics funding from the European Union Seventh Framework Programme (FP7/2007-2013, Agreement 305608).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2013040337/-/DCSupplemental.
References
- 1.Adalat S, Woolf AS, Johnstone KA, Wirsing A, Harries LW, Long DA, Hennekam RC, Ledermann SE, Rees L, van’t Hoff W, Marks SD, Trompeter RS, Tullus K, Winyard PJ, Cansick J, Mushtaq I, Dhillon HK, Bingham C, Edghill EL, Shroff R, Stanescu H, Ryffel GU, Ellard S, Bockenhauer D: HNF1B mutations associate with hypomagnesemia and renal magnesium wasting. J Am Soc Nephrol 20: 1123–1131, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edghill EL, Bingham C, Ellard S, Hattersley AT: Mutations in hepatocyte nuclear factor-1beta and their related phenotypes. J Med Genet 43: 84–90, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faguer S, Decramer S, Chassaing N, Bellanné-Chantelot C, Calvas P, Beaufils S, Bessenay L, Lengelé JP, Dahan K, Ronco P, Devuyst O, Chauveau D: Diagnosis, management, and prognosis of HNF1B nephropathy in adulthood. Kidney Int 80: 768–776, 2011 [DOI] [PubMed] [Google Scholar]
- 4.Glaudemans B, Knoers NV, Hoenderop JG, Bindels RJ: New molecular players facilitating Mg(2+) reabsorption in the distal convoluted tubule. Kidney Int 77: 17–22, 2010 [DOI] [PubMed] [Google Scholar]
- 5.Meij IC, Koenderink JB, van Bokhoven H, Assink KF, Groenestege WT, de Pont JJ, Bindels RJ, Monnens LA, van den Heuvel LP, Knoers NV: Dominant isolated renal magnesium loss is caused by misrouting of the Na(+),K(+)-ATPase gamma-subunit. Nat Genet 26: 265–266, 2000 [DOI] [PubMed] [Google Scholar]
- 6.Ferrè S, Veenstra GJ, Bouwmeester R, Hoenderop JG, Bindels RJ: HNF-1B specifically regulates the transcription of the γa-subunit of the Na+/K+-ATPase. Biochem Biophys Res Commun 404: 284–290, 2011 [DOI] [PubMed] [Google Scholar]
- 7.Gong Y, Ma Z, Patel V, Fischer E, Hiesberger T, Pontoglio M, Igarashi P: HNF-1beta regulates transcription of the PKD modifier gene Kif12. J Am Soc Nephrol 20: 41–47, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiesberger T, Bai Y, Shao X, McNally BT, Sinclair AM, Tian X, Somlo S, Igarashi P: Mutation of hepatocyte nuclear factor-1beta inhibits Pkhd1 gene expression and produces renal cysts in mice. J Clin Invest 113: 814–825, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gresh L, Fischer E, Reimann A, Tanguy M, Garbay S, Shao X, Hiesberger T, Fiette L, Igarashi P, Yaniv M, Pontoglio M: A transcriptional network in polycystic kidney disease. EMBO J 23: 1657–1668, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kikuchi R, Kusuhara H, Hattori N, Kim I, Shiota K, Gonzalez FJ, Sugiyama Y: Regulation of tissue-specific expression of the human and mouse urate transporter 1 gene by hepatocyte nuclear factor 1 alpha/beta and DNA methylation. Mol Pharmacol 72: 1619–1625, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Kikuchi R, Kusuhara H, Hattori N, Shiota K, Kim I, Gonzalez FJ, Sugiyama Y: Regulation of the expression of human organic anion transporter 3 by hepatocyte nuclear factor 1alpha/beta and DNA methylation. Mol Pharmacol 70: 887–896, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Mendel DB, Khavari PA, Conley PB, Graves MK, Hansen LP, Admon A, Crabtree GR: Characterization of a cofactor that regulates dimerization of a mammalian homeodomain protein. Science 254: 1762–1767, 1991 [DOI] [PubMed] [Google Scholar]
- 13.Citron BA, Davis MD, Milstien S, Gutierrez J, Mendel DB, Crabtree GR, Kaufman S: Identity of 4a-carbinolamine dehydratase, a component of the phenylalanine hydroxylation system, and DCoH, a transregulator of homeodomain proteins. Proc Natl Acad Sci U S A 89: 11891–11894, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werner ER, Blau N, Thöny B: Tetrahydrobiopterin: Biochemistry and pathophysiology. Biochem J 438: 397–414, 2011 [DOI] [PubMed] [Google Scholar]
- 15.Endrizzi JA, Cronk JD, Wang W, Crabtree GR, Alber T: Crystal structure of DCoH, a bifunctional, protein-binding transcriptional coactivator. Science 268: 556–559, 1995 [DOI] [PubMed] [Google Scholar]
- 16.Ficner R, Sauer UH, Stier G, Suck D: Three-dimensional structure of the bifunctional protein PCD/DCoH, a cytoplasmic enzyme interacting with transcription factor HNF1. EMBO J 14: 2034–2042, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose RB, Bayle JH, Endrizzi JA, Cronk JD, Crabtree GR, Alber T: Structural basis of dimerization, coactivator recognition and MODY3 mutations in HNF-1alpha. Nat Struct Biol 7: 744–748, 2000 [DOI] [PubMed] [Google Scholar]
- 18.Bayle JH, Randazzo F, Johnen G, Kaufman S, Nagy A, Rossant J, Crabtree GR: Hyperphenylalaninemia and impaired glucose tolerance in mice lacking the bifunctional DCoH gene. J Biol Chem 277: 28884–28891, 2002 [DOI] [PubMed] [Google Scholar]
- 19.Thöny B, Neuheiser F, Kierat L, Blaskovics M, Arn PH, Ferreira P, Rebrin I, Ayling J, Blau N: Hyperphenylalaninemia with high levels of 7-biopterin is associated with mutations in the PCBD gene encoding the bifunctional protein pterin-4a-carbinolamine dehydratase and transcriptional coactivator (DCoH). Am J Hum Genet 62: 1302–1311, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thöny B, Neuheiser F, Kierat L, Rolland MO, Guibaud P, Schlüter T, Germann R, Heidenreich RA, Duran M, de Klerk JB, Ayling JE, Blau N: Mutations in the pterin-4alpha-carbinolamine dehydratase (PCBD) gene cause a benign form of hyperphenylalaninemia. Hum Genet 103: 162–167, 1998 [DOI] [PubMed] [Google Scholar]
- 21.Citron BA, Kaufman S, Milstien S, Naylor EW, Greene CL, Davis MD: Mutation in the 4a-carbinolamine dehydratase gene leads to mild hyperphenylalaninemia with defective cofactor metabolism. Am J Hum Genet 53: 768–774, 1993 [PMC free article] [PubMed] [Google Scholar]
- 22.Thanabalasingham G, Owen KR: Diagnosis and management of maturity onset diabetes of the young (MODY). BMJ 343: d6044, 2011 [DOI] [PubMed] [Google Scholar]
- 23.Bellanné-Chantelot C, Chauveau D, Gautier JF, Dubois-Laforgue D, Clauin S, Beaufils S, Wilhelm JM, Boitard C, Noël LH, Velho G, Timsit J: Clinical spectrum associated with hepatocyte nuclear factor-1beta mutations. Ann Intern Med 140: 510–517, 2004 [DOI] [PubMed] [Google Scholar]
- 24.Bingham C, Hattersley AT: Renal cysts and diabetes syndrome resulting from mutations in hepatocyte nuclear factor-1beta. Nephrol Dial Transplant 19: 2703–2708, 2004 [DOI] [PubMed] [Google Scholar]
- 25.Chollet D, Franken P, Raffin Y, Malafosse A, Widmer J, Tafti M: Blood and brain magnesium in inbred mice and their correlation with sleep quality. Am J Physiol Regul Integr Comp Physiol 279: R2173–R2178, 2000 [DOI] [PubMed] [Google Scholar]
- 26.Résibois A, Cuvelier L, Svoboda M, Heizmann CW, Thöny B: Immunohistochemical localisation of pterin-4alpha-carbinolamine dehydratase in rat peripheral organs. Histochem Cell Biol 111: 381–390, 1999 [DOI] [PubMed] [Google Scholar]
- 27.Arystarkhova E, Wetzel RK, Sweadner KJ: Distribution and oligomeric association of splice forms of Na(+)-K(+)-ATPase regulatory gamma-subunit in rat kidney. Am J Physiol Renal Physiol 282: F393–F407, 2002 [DOI] [PubMed] [Google Scholar]
- 28.Ward CJ, Yuan D, Masyuk TV, Wang X, Punyashthiti R, Whelan S, Bacallao R, Torra R, LaRusso NF, Torres VE, Harris PC: Cellular and subcellular localization of the ARPKD protein; fibrocystin is expressed on primary cilia. Hum Mol Genet 12: 2703–2710, 2003 [DOI] [PubMed] [Google Scholar]
- 29.Maestro MA, Cardalda C, Boj SF, Luco RF, Servitja JM, Ferrer J: Distinct roles of HNF1beta, HNF1alpha, and HNF4alpha in regulating pancreas development, beta-cell function and growth. Endocr Dev 12: 33–45, 2007 [DOI] [PubMed] [Google Scholar]
- 30.McDonald TJ, Shields BM, Lawry J, Owen KR, Gloyn AL, Ellard S, Hattersley AT: High-sensitivity CRP discriminates HNF1A-MODY from other subtypes of diabetes. Diabetes Care 34: 1860–1862, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owen KR, Thanabalasingham G, James TJ, Karpe F, Farmer AJ, McCarthy MI, Gloyn AL: Assessment of high-sensitivity C-reactive protein levels as diagnostic discriminator of maturity-onset diabetes of the young due to HNF1A mutations. Diabetes Care 33: 1919–1924, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnen G, Kowlessur D, Citron BA, Kaufman S: Characterization of the wild-type form of 4a-carbinolamine dehydratase and two naturally occurring mutants associated with hyperphenylalaninemia. Proc Natl Acad Sci U S A 92: 12384–12388, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dudziak K, Mottalebi N, Senkel S, Edghill EL, Rosengarten S, Roose M, Bingham C, Ellard S, Ryffel GU: Transcription factor HNF1beta and novel partners affect nephrogenesis. Kidney Int 74: 210–217, 2008 [DOI] [PubMed] [Google Scholar]
- 34.Baker KE, Parker R: Nonsense-mediated mRNA decay: Terminating erroneous gene expression. Curr Opin Cell Biol 16: 293–299, 2004 [DOI] [PubMed] [Google Scholar]
- 35.Behm-Ansmant I, Izaurralde E: Quality control of gene expression: A stepwise assembly pathway for the surveillance complex that triggers nonsense-mediated mRNA decay. Genes Dev 20: 391–398, 2006 [DOI] [PubMed] [Google Scholar]
- 36.Sourdive DJ, Transy C, Garbay S, Yaniv M: The bifunctional DCOH protein binds to HNF1 independently of its 4-alpha-carbinolamine dehydratase activity. Nucleic Acids Res 25: 1476–1484, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blau N, Barnes I, Dhondt JL: International database of tetrahydrobiopterin deficiencies. J Inherit Metab Dis 19: 8–14, 1996 [DOI] [PubMed] [Google Scholar]
- 38.Meyer AH, Katona I, Blatow M, Rozov A, Monyer H: In vivo labeling of parvalbumin-positive interneurons and analysis of electrical coupling in identified neurons. J Neurosci 22: 7055–7064, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markadieu N, San-Cristobal P, Nair AV, Verkaart S, Lenssen E, Tudpor K, van Zeeland F, Loffing J, Bindels RJ, Hoenderop JG: A primary culture of distal convoluted tubules expressing functional thiazide-sensitive NaCl transport. Am J Physiol Renal Physiol 303: F886–F892, 2012 [DOI] [PubMed] [Google Scholar]
- 40.Schneider CA, Rasband WS, Eliceiri KW: NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vriend G: WHAT IF: A molecular modeling and drug design program. J Mol Graph 8: 52–56, 1990 [DOI] [PubMed] [Google Scholar]
- 42.Krieger E, Koraimann G, Vriend G: Increasing the precision of comparative models with YASARA NOVA—a self-parameterizing force field. Proteins 47: 393–402, 2002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.