Abstract
Polymorphisms in APOL1 are associated with CKD, including HIV-related CKD, in individuals of African ancestry. The apolipoprotein L1 (APOL1) protein circulates and is localized in kidney cells, but the contribution of APOL1 location to CKD pathogenesis is unclear. We examined associations of plasma APOL1 levels with plasma cytokine levels, dyslipidemia, and APOL1 genotype in a nested case-control study (n=270) of HIV-infected African Americans enrolled in a multicenter prospective observational study. Patients were designated as having CKD when estimated GFR (eGFR) decreased to <60 ml/min per 1.73 m2 (eGFR<60 cohort) or protein-to-creatinine ratios became >3.5 g/g (nephrotic proteinuria cohort). Circulating APOL1 levels did not associate with APOL1 genotype, CKD status, or levels of proinflammatory cytokines, but did correlate with fasting cholesterol, LDL cholesterol, and triglyceride levels. At ascertainment, CKD-associated polymorphisms (risk variants) in APOL1 associated with the eGFR<60 cohort, but not the nephrotic-range proteinuria cohort. Of note, in both the eGFR<60 and nephrotic proteinuria cohorts, CKD cases with two APOL1 risk variants had significant declines in eGFR over a median of 4 years compared with individuals with one or no risk variants. APOL1 risk genotype was not associated with changes in proteinuria. Higher circulating proinflammatory cytokine levels were independently associated with CKD but not APOL1 genotype. In conclusion, the function of variant APOL1 proteins derived from circulation or synthesized in the kidney, but not the level of circulating APOL1, probably mediates APOL1-associated kidney disease in HIV-infected African Americans.
Nondiabetic CKD in individuals of African ancestry have been linked to polymorphisms in the gene for apolipoprotein L1 (APOL1),1–5 a protein component of HDL particles with a known function in the immune clearance of Trypanosoma brucei infections.6 CKD is associated with two coding variants of the APOL1 gene known as G1 and G2, both of higher allele frequency in African and African descendent populations compared with white populations where they are almost absent. Evidence suggests that the prevalence of the G1 and G2 variants may have increased in African populations because of a selective advantage from their ability to kill a broader range of Trypanosoma species.1,2,7 Individuals carrying at least one G1 or G2 allele have additional protection from trypanosomiasis; however, individuals with two G1 or G2 alleles are at increased risk for nondiabetic CKD.2,4,5
The pathogenic mechanisms responsible for CKD associated with APOL1 risk variants are unknown. We recently showed that, in addition to being secreted and circulated in the blood,8 APOL1 is localized in podocytes, proximal tubular epithelial cells, and small-artery endothelium in normal kidney.9 Thus, the contribution of circulating versus kidney-localized variant APOL1s to CKD pathogenesis is unknown. In kidney transplantation, two studies suggest that graft loss is associated with the APOL1 genotype of the allograft, not the recipient.10,11 However, the association of APOL1 plasma levels with CKD phenotypes or APOL1 genotype has not been studied.
To address these issues, we examined circulating APOL1 levels with APOL1 genotype and renal function in HIV-infected African Americans in the AIDS Clinical Trials Group (ACTG) Longitudinal Linked Randomized Trials (ALLRT) cohort because the occurrence of HIV-associated nephropathy (HIVAN) and renal outcomes in HIV-infected patients are strongly associated with APOL1 risk alleles.12–14 In addition, we examined the relationship between circulating APOL1 levels and proinflammatory cytokines known to induce APOL1 expression and previously associated with CKD and HIV/AIDS progression.15,16 Additional analyses examined associations of APOL1 levels with dyslipidemia and the role of APOL1 genotype on CKD progression using longitudinal data.
Results
Study Population
Established in 2000, ALLRT is a multicenter, observational cohort study that coenrolled participants from ongoing ACTG randomized clinical trials of antiretroviral therapy or other treatment strategies for longitudinal follow-up.17,18 Serum creatinine concentrations were measured every 16 weeks for estimated GFRs (eGFRs), and protein-to-creatinine (UP/Cr) ratios were determined on random urine samples every 48 weeks. However, renal biopsies were not performed in ALLRT. CKD cases were selected from self-identified African Americans (Table 1) using two criteria: eGFR<60 ml/min per 1.73 m2 or nephrotic-range proteinuria (UP/Cr > 3.5 g/g). Only one patient fulfilled both CKD criteria and was included in the eGFR<60 cohort. Control African American participants had eGFR≥60 ml/min per 1.73 m2 and normal protein excretion throughout follow-up (see Concise Methods for details and control matching criteria).
Table 1.
Demographic baseline characteristics of participants (all cases and controls were HIV-infected African Americans)
| Characteristic | eGFR<60 Cohort | Nephrotic Proteinuria Cohort | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases (n=46) | IQR | Controls (n=92) | IQR | P Value | Cases (n=44) | IQR | Controls (n=88) | IQR | P Value | |
| Median age (yr) | 50 | 44, 58 | 42 | 33, 49 | <0.001 | 45 | 37, 52 | 40 | 33,48 | 0.05 |
| Men, n (%) | 23 (50) | 68 (74) | <0.01 | 34 (77) | 75 (85) | 0.24 | ||||
| Median eGFR (ml/min per 1.73 m2) | 54.2 | 49.3, 58.4 | 105.8 | 94.9, 125.4 | <0.001a | 100.7 | 78.4, 114.9 | 105.8 | 93.6, 118.6 | 0.08 |
| Median UP/Cr (g/g) | 0.20 | 0.1, 0.77 | 0.10 | 0.06, 0.149 | <0.01 | 6.37 | 5.17, 13.00 | 0.05 | 0.04, 0.07 | <0.001a |
| Median CD4 count (cells/mm3) | 284 | 165, 607 | 361 | 144, 608 | 0.81 | 419 | 255, 554 | 410 | 244, 673 | 0.61 |
| Viral load<50 copies/ml, n (%) | 26 (58) | 54 (59) | 0.30 | 33 (77) | 66 (75) | 0.69 | ||||
| Median log10 viral load | 1.77 | 1.69, 3.37 | 1.69 | 1.69, 4.14 | 0.05 | 1.69 | 1.69, 2.76 | 1.69 | 1.69, 2.27 | 0.46 |
| Diabetes, n (%)b | 7 (15) | 10 (11) | 0.45 | 9 (21) | 1 (1) | <0.001a | ||||
| Median fasting blood glucose (mg/dl) | 91 | 83, 97 | 89 | 83, 98 | 0.46 | 91 | 76, 108 | 87 | 79, 97 | 0.04a |
| Hypertension, n (%)b | 33 (72) | 33 (36) | <0.001 | 24 (55) | 27 (31) | <0.01 | ||||
| HCV infection, n (%)b | 13 (28) | 14 (15) | 0.09 | 10 (23) | 5 (6) | <0.01 | ||||
| Median fasting cholesterol (mg/dl) | 189 | 156, 224 | 173 | 150, 202 | 0.07 | 189 | 173, 230 | 180 | 164, 201 | 0.26 |
| Median fasting HDL cholesterol (mg/dl) | 43 | 37, 54 | 48 | 38, 56 | 0.76 | 46 | 35, 65 | 50 | 41, 61 | 0.54 |
| Median fasting LDL cholesterol (mg/dl) | 110 | 91, 135 | 102 | 77, 123 | 0.20 | 105 | 67, 140 | 108 | 90, 125 | 0.78 |
| Median fasting triglycerides (mg/dl) | 146 | 101, 256 | 105 | 75, 150 | 0.02 | 161 | 114, 228 | 100 | 74, 138 | 0.02 |
IQR, interquartile range.
Determined by Wilcoxon or Fisher exact tests because logistic regression models did not converge.
Diagnosis before case/control assignment.
APOL1 Risk Variants and CKD Phenotypes: Univariate Models
APOL1 allele and genotype frequencies (Table 2) did not deviate from Hardy-Weinberg equilibrium (P>0.05). Cases in the eGFR<60 cohort had higher G1 and G2 risk allele frequencies than their control group. However, APOL1 risk alleles in this cohort were less prevalent in cases than previously reported for patients with biopsy-proven HIVAN13,19 but were similar to allele frequencies reported in an HIV-infected CKD cohort that excluded patients with HIVAN (Supplemental Table 1).12 G1 and G2 allele frequencies among cases and controls in the nephrotic proteinuria cohort were similar to each other and to published estimates from African American control populations.2,13,20,21 Exclusion of diabetic patients (n=27) from the analysis did not substantively change APOL1 allele frequencies (Supplemental Table 2).
Table 2.
APOL1 risk allele genotype and allele frequency in CKD and control groups
| Variable | Genotype, n (%) | Allele Frequency | ||||
|---|---|---|---|---|---|---|
| 0 Risk Alleles | 1 Risk Allele | 2 Risk Alleles | G0a | G1 | G2 | |
| eGFR<60 cohort | ||||||
| Cases (n=44b) | 10 (23) | 27 (61) | 7 (16) | 0.53 | 0.31 | 0.16 |
| Controls (n=91b) | 40 (44) | 46 (50) | 5 (6) | 0.69 | 0.19 | 0.12 |
| Nephrotic proteinuria cohort | ||||||
| Cases (n=39b) | 16 (41) | 20 (51) | 3 (8) | 0.67 | 0.18 | 0.15 |
| Controls (n=88) | 43 (49) | 34 (39) | 11 (12) | 0.68 | 0.22 | 0.10 |
G0 is the common allele not associated with CKD and corresponds to reference sequence NP_003652 (G1 refers to two nonsynonymous single nucleotide polymorphisms rs73885319 and rs60910145, and G2 refers to a six base pair in-frame deletion rs71785313).
Numbers are different from those in Table 1 because genotyping was not possible in eight participants; see Concise Methods.
At ascertainment, APOL1 risk variants significantly associated with CKD in adjusted analyses comparing cases in the eGFR<60 cohort to their control group (Table 3). An additive pattern of inheritance best accounted for the association (P=0.01 for partial F-test comparing additive versus recessive inheritance patterns). Individuals with one and two APOL1 risk variant (G1 or G2) had increased odds for eGFR<60 (odds ratio, 2.8 and 6.2, respectively) compared with patients with no risk variants. APOL1 risk variants were not associated with cases in the nephrotic proteinuria cohort under any pattern of inheritance. Of note, these individuals with nephrotic proteinuria had preserved eGFRs (median eGFR, 100.7 ml/min per 1.73 m2), which were similar to those in controls (median eGFR, 105.8 ml/min per 1.73 m2).
Table 3.
Unadjusted odds ratios for the association of APOL1 risk alleles with CKD phenotypes
| Variable | Additive | Recessive | Dominant | |||||
|---|---|---|---|---|---|---|---|---|
| 1 versus 0 Risk Alleles | 2 versus 0 Risk Alleles | 2 versus 0 or 1 Risk Alleles | 1 or 2 versus 0 Risk Alleles | |||||
| OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | |
| eGFR<60 cohort | 2.80 (1.13 to 6.95) | 0.03 | 6.18 (1.55 to 24.59) | 0.01 | 2.80 (0.89 to 8.82) | 0.08 | 3.05 (1.25 to 7.47) | 0.01 |
| Nephrotic proteinuria cohort | 1.89 (0.75 to 4.78) | 0.18 | 0.68 (0.17 to 2.63) | 0.57 | 0.52 (0.14 to 1.95) | 0.33 | 1.40 (0.62 to 3.16) | 0.42 |
OR, odds ratio; 95% CI, 95% confidence interval.
APOL1 Levels and CKD Phenotypes
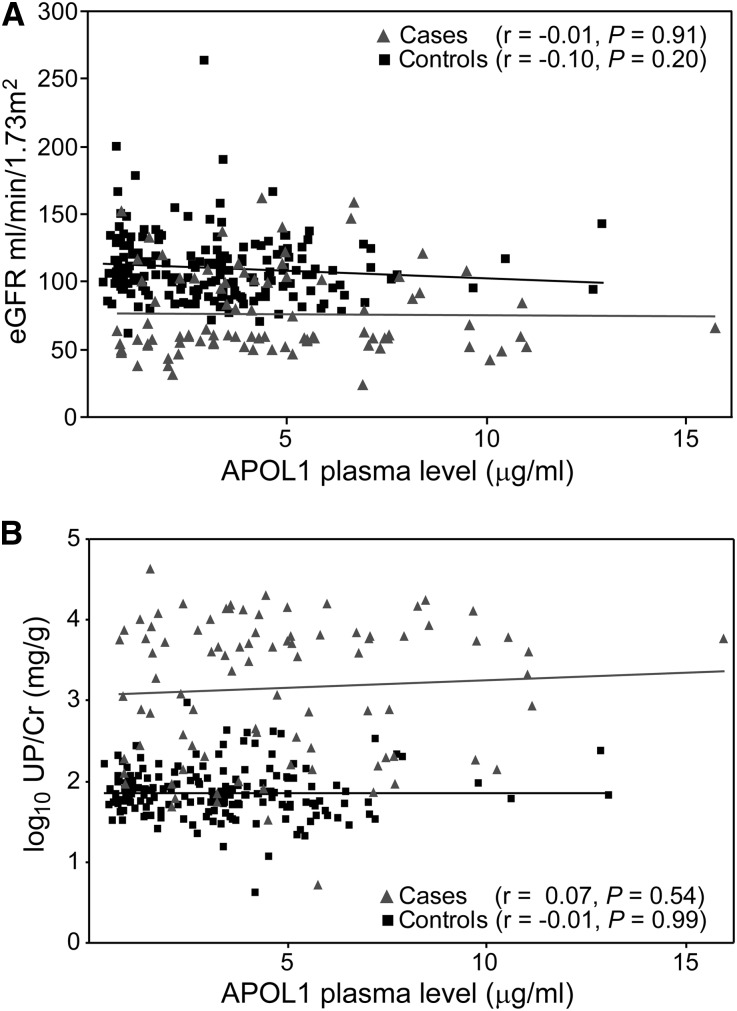
Plasma APOL1 protein levels were quantified from samples obtained at the ALLRT study visit when patients were classified as a case or control (baseline). Circulating APOL1 protein levels ranged from 386 to 15,743 ng/ml and were similar to previously reported levels in normolipidemic patients.22,23 APOL1 plasma levels did not correlate with eGFR (Figure 1A) or proteinuria (Figure 1B) in cases or controls. When we examined the eGFR<60 cohort alone, we found a trend toward higher APOL1 plasma levels in cases (median, 3953 ng/ml in cases versus 3241ng/ml in controls, P=0.07), but plasma APOL1 levels did not correlate with eGFR in cases or controls (r=−0.01 [P=0.91] for cases and r=−0.10 [P=0.19] for controls). In the nephrotic proteinuria cohort, APOL1 levels were significantly higher in cases than controls (median, 4308 ng/ml in cases versus 2984 ng/ml in controls, P<0.001). However, APOL1 plasma levels also did not correlate with proteinuria among cases or controls in this cohort (r=−0.02 [P=0.81] for cases and r=0.06 [P=0.46] for controls).
Figure 1.
APOL1 plasma levels did not correlate with eGFR or proteinuria. Scatter plots of APOL1 plasma levels relative to (A) eGFR or (B) UP/Cr in all participants (cases and controls combined). Linear regression trend lines are shown; Pearson correlation coefficients (r) and P values are presented in key.
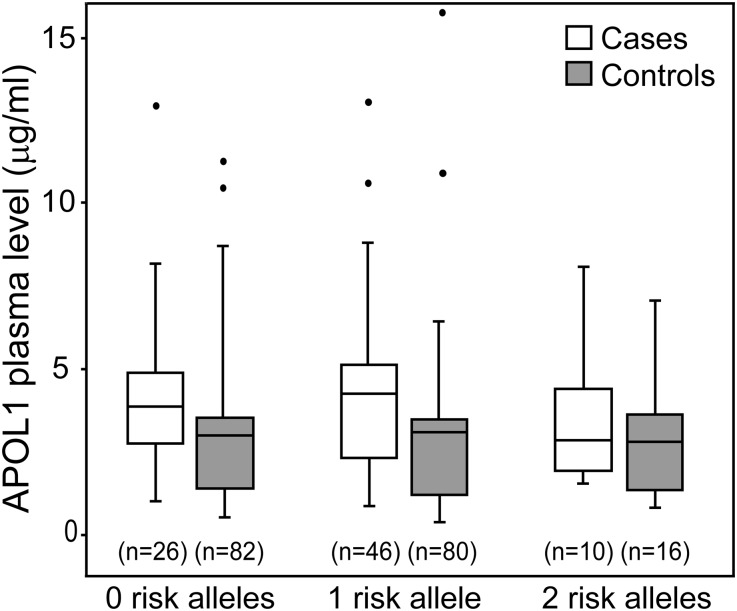
APOL1 levels also did not differ significantly according to the number of APOL1 risk alleles in cases or controls (Figure 2, Supplemental Table 3). When eGFR<60 and nephrotic proteinuria cohorts were analyzed separately (data not shown), again no significant association was seen between APOL1 level and the number of risk alleles and no significant interaction was seen between APOL1 levels and the number of APOL1 risk alleles in conditional logistic regression models (P=0.64 and P=0.45 for the interaction term for eGFR<60 and nephrotic proteinuria cohorts, respectively). Thus, APOL1 plasma levels were not associated with APOL1 risk variant status and did not correlate with CKD phenotypes.
Figure 2.
APOL1 plasma levels did not associate with APOL1 genotype. Box whisker plot associating APOL1 genotype with APOL1 plasma levels in cases and controls stratified by number of risk alleles. Boxes denote interquartile range with a median line; outliers are indicated with filled circles outside range whiskers. Statistical analyses for genotype comparisons are presented in Supplemental Table 3.
APOL1 Levels and Dyslipidemia
In both CKD cohorts, cases had suboptimal lipid profiles compared with controls (Table 1). Cases in the eGFR<60 and nephrotic proteinuria cohorts had significantly higher median fasting triglyceride levels compared with controls, and median fasting total cholesterol levels also were significantly higher in the eGFR<60 cohort. Plasma APOL1 levels weakly correlated with fasting cholesterol, fasting triglycerides, and fasting LDL cholesterol, but APOL1 levels did not correlate with fasting HDL cholesterol levels (Table 4). This is consistent with prior studies that found cholesterol, LDL cholesterol, and triglyceride levels to be independently associated with APOL1 levels, with triglycerides the strongest correlate of APOL1 level in dyslipidemic patients.22,24 We found no correlation between APOL1 genotype and levels of any lipid class (data not shown); however, the available lipid data did not include quantification of HDL subfractions. A prior report has demonstrated an inverse correlation between the number of APOL1 risk alleles and medium-size HDL particle concentration.25
Table 4.
Correlations of APOL1 plasma levels with fasting lipid and plasma cytokine levels
| Variable | All Controls | All Cases | ||||
|---|---|---|---|---|---|---|
| Patients (n) | Spearman ρ | P Value | Patients (n) | Spearman ρ | P Value | |
| Total cholesterol | 147 | 0.27 | <0.001 | 65 | 0.12 | 0.35 |
| Triglycerides | 147 | 0.09 | 0.28 | 65 | 0.25 | 0.04 |
| LDL cholesterol | 143 | 0.20 | 0.02 | 61 | −0.05 | 0.70 |
| HDL cholesterol | 146 | 0.10 | 0.21 | 63 | 0.13 | 0.32 |
| C-reactive protein | 179 | 0.03 | 0.72 | 89 | 0.01 | 0.90 |
| β2-Microglobulin | 179 | −0.11 | 0.14 | 89 | 0.13 | 0.24 |
| sTNFR1 | 179 | −0.08 | 0.30 | 89 | 0.05 | 0.65 |
| sTNFR2 | 179 | −0.17 | 0.02 | 89 | 0.00 | 1 |
| IL-6 | 179 | 0.11 | 0.13 | 89 | −0.02 | 0.85 |
| CCL2 | 179 | −0.06 | 0.45 | 89 | 0.06 | 0.56 |
| CCL5 | 179 | 0.16 | 0.03 | 89 | 0.19 | 0.07 |
| IFN-α | 179 | −0.06 | 0.40 | 89 | 0.04 | 0.74 |
Complete fasting lipid data were not available for all patients at the case definition time point (baseline) when APOL1 plasma levels were assayed. Correlation values between |0.2 and 0.39| are considered weak.
APOL1 Levels and Inflammatory Cytokines
To determine whether APOL1 plasma levels were associated with an inflammatory process, we quantified several proinflammatory cytokines that have been previously associated with poorer outcomes in HIV-1 infection15,26,27 or CKD progression.16 At the point when cases and controls were defined (baseline), cases in both the eGFR<60 and nephrotic proteinuria cohorts had significantly higher levels of soluble TNF receptor (sTNFR)1, sTNFR2, IL-6, chemokine (C-C motif) ligand (CCL)2, and β2-microglobulin compared with controls; in the nephrotic proteinuria cohort, CCL5 and IFN-α were also higher, but not C-reactive protein levels (Supplemental Figure 1). Cytokine levels, however, did not correlate with APOL1 levels (sTNFR2 is shown as a representative cytokine in Figure 3; all other cytokine correlations are in Table 4). Additionally, cytokine levels did not differ significantly according to the number of APOL1 risk variants in cases or controls (Supplemental Table 3).
Figure 3.
APOL1 plasma levels did not correlate with sTNFR2 levels. Scatter plot of APOL1 plasma levels relative to sTNFR2 levels in all cases and controls. See Table 4 for correlations with other cytokines. Linear regression trend lines are shown; Pearson correlation coefficients (r) and P values are presented in key.
APOL1 Risk Variants and CKD Phenotypes: Multivariable Models
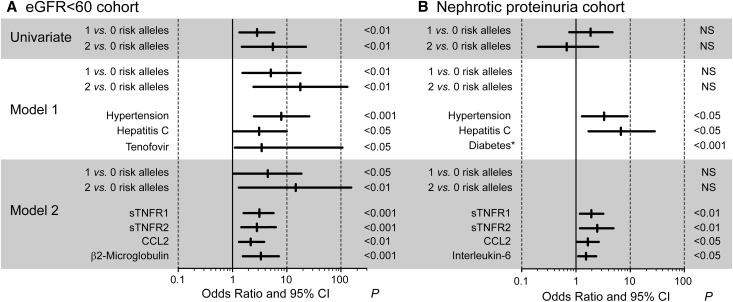
Multivariable conditional logistic regression models tested associations between APOL1 risk alleles and CKD after adjusting for risk factors associated with CKD (including hypertension, hepatitis C virus (HCV) infection status, and tenofovir treatment) in the eGFR<60 cohort (model 1; Figure 4A, Supplemental Table 4) or nephrotic range proteinuria cohort (model 1; Figure 4B, Supplemental Table 5). In addition to these CKD risk factors, model 2 also included cytokine levels as described in the Concise Methods. CKD cases and controls were matched for HIV viral loads and CD4 counts so these variables formed matched sets for the model and did not yield risk estimates. In the eGFR<60 cohort, the association of CKD with one and two APOL1 risk alleles remained significant after adjustment for other risk factors, and the effect size (odds ratios) for CKD was greater for two risk variants compared with one risk variant. However, APOL1 risk alleles were not associated with CKD in patients with nephrotic proteinuria.
Figure 4.
APOL1 risk alleles associated with CKD in the eGFR<60 cohort but not the nephrotic range proteinuria cohort. Forest plots of risk factors for CKD. Odds ratios, 95% confidence intervals (95% CIs), and P values for the associations between CKD in the (A) eGFR<60 cohort and (B) nephrotic proteinuria cohort. The associations with APOL1 genotype and CKD is depicted in a univariate conditional logistic regression model (univariate), or with comorbid conditions and tenofovir exposure estimated from a single multivariable conditional logistic regression model that included each of these covariates (model 1). These covariates also were included in separate multivariable models that tested associations with the individual cytokines (model 2). Odds ratios for risk alleles shown in model 2 are for sTNFR2 (see Supplemental Table 4 for odds ratios with other cytokines). Only cytokines with significant associations are shown. The complete results of all univariate and multivariate analyses are presented in Supplemental Tables 4 and 5. *Because of the small number of patients with diabetes (n=9 cases and n=1 control), association was determined by Fisher exact test and diabetes was not included in the model. NS, not significant.
In addition to APOL1 risk variant status, hypertension, hepatitis C virus infection, and tenofovir exposure were independently associated in model 1 with the eGFR<60 cases compared with the controls. However, after adjustment for cytokine levels, HCV infection status and tenofovir exposure were no longer significantly associated with an eGFR<60 ml/min per 1.73 m2. In model 1, hypertension and HCV also were significantly associated with nephrotic-range proteinuria. As expected, diabetes was associated with nephrotic proteinuria, but the sample size was small (present in nine cases and one control; P=0.001 by Fisher exact test).
Increased plasma levels of sTNFR1, sTNFR2, and CCL2 were each associated with significantly higher odds ratios for cases in both the eGFR<60 and nephrotic proteinuria cohorts versus controls in both univariate and multivariable model 2 analyses (Supplemental Tables 4 and 5). Because cases and controls were matched on viral load and CD4 count, the association of CKD with cytokine levels is probably not related to differences in the control of plasma HIV-RNA by antiretroviral therapy. This suggests there were significant contributions by cytokines to increased CKD risk in HIV-1 infection that was independent of contributions by APOL1 risk genotype status and other CKD risk factors.
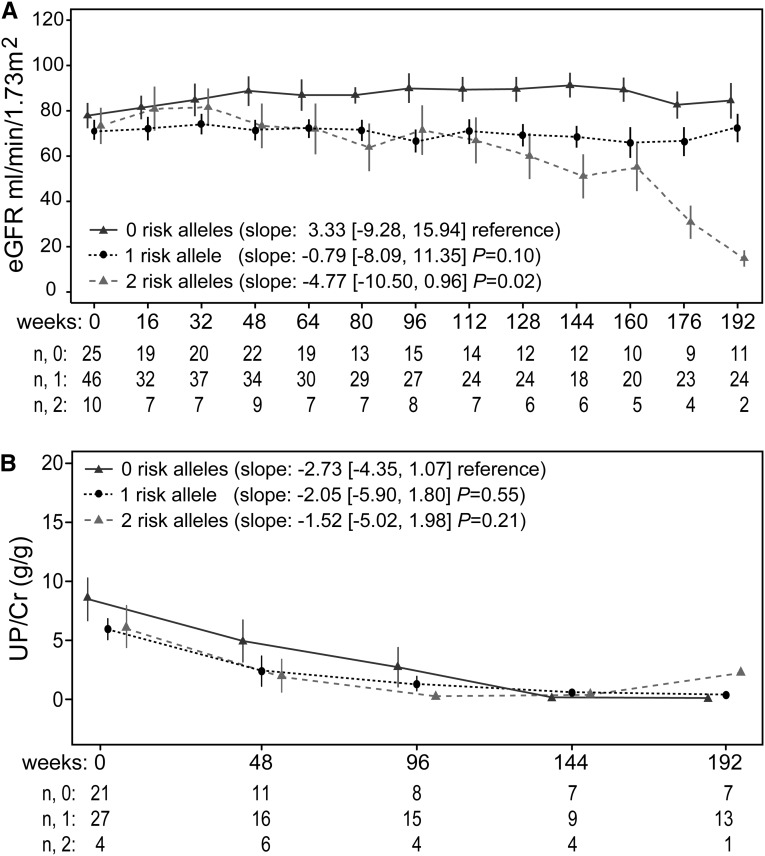
Longitudinal Analysis of Kidney Function
Rates of eGFR decline were analyzed in patients with no, one, or two APOL1 risk alleles in a combined CKD cohort (including patients with both case definitions; Figure 5A) or with the eGFR<60 and nephrotic proteinuria cohorts analyzed separately (Supplemental Figure 2, A and C). In both the individual and combined cohort analyses, cases with two APOL1 risk variants had a significantly faster rate of eGFR decline than did cases with no APOL1 risk alleles in adjusted longitudinal models. In the combined CKD cohort analysis, cases with one APOL1 risk variant had a trend toward more rapid eGFR decline compared with cases with no risk variants (P=0.10) (Figure 5A). The eGFR slopes did not significantly differ according to APOL1 risk alleles among controls from either CKD cohort because this was a control selection criterion.
Figure 5.
APOL1 risk alleles associated with eGFR decline but not with changes in proteinuria. (A) Longitudinal changes in eGFR in all cases (combined eGFR<60 and nephrotic proteinuria cohorts); slopes in ml/min per year. (B) Longitudinal changes in UP/Cr ratios in all cases; slopes in g/g per year. Controls did not change over time (see matching criteria in Concise Methods section). Graphical data are time point means with SEM bars. Slopes were determined for each participant and are presented in the legend as a mean slope (interquartile range) and P values versus no risk alleles. Below each graph are the numbers of participants with available data (n) per each time point for no, one, or two risk alleles.
Proteinuria declined substantially and significantly in the combined CKD cohort (Figure 5B) and in patients with nephrotic-range proteinuria (Supplemental Figure 2D), but APOL1 risk allele status was not associated with this change. However, this analysis is limited by missing proteinuria data at some time points subsequent to the baseline visit. In patients with eGFR<60 ml/min (Supplemental Figure 2B), proteinuria did not change significantly over time.
Discussion
The primary objective of our study was to ascertain whether APOL1 circulating levels were associated with CKD phenotypes in HIV-infected patients. Our results provided several novel observations regarding APOL1 circulating levels but did not reveal an obvious link with CKD. First, there was no association between APOL1 plasma levels and APOL1 genotype, indicating the changes in the APOL1 protein caused by the G1 and G2 polymorphisms did not alter their production or secretion into the circulation. Second, APOL1 plasma levels did not correlate with eGFR or proteinuria. Thus, circulating APOL1 protein levels do not appear to directly contribute to CKD. However, our study does not exclude a causal role for circulating APOL1 proteins in CKD due to functional changes caused by the G1 and G2 polymorphisms. APOL1 G1 and G2 proteins differ from G0 in their ability to bind the trypanosome serum response associated protein and kill trypanosomes.2 Consequently, the potential for the G1 and G2 variants, either from the circulation or expressed in renal cells, to affect additional biologic functions of APOL1 seems likely.
APOL1 plasma levels also did not correlate with the level of the inflammatory cytokines examined. This observation was unexpected because several prior studies have shown APOL1 expression is inducible in endothelial cells in vitro by TNF, IFN-α, and IFN-γ.9,23,28–30 APOL1 is a component of a subfraction of HDL particles,8 but the source of circulating APOL1 remains unknown. Numerous tissues express APOL1, and a prior study suggests the vascular endothelium is the primary source of circulating APOL1;29 however, the HDL particles that contain APOL1 are produced by the liver. In our study, sTNFR1 and sTNFR2 (markers of TNF synthesis) and IFN-α levels did not correlate with plasma APOL1 levels, suggesting that the major source of circulating APOL1 was not induced by these cytokines.
An additional unexpected finding was a lower than anticipated risk allele frequency. The intent of our study design was to examine a population with a high APOL1 risk allele frequency, such as seen in HIVAN.13,19 The ALLRT cohort is representative of the population living with HIV who are receiving care in the United States,31 and most participants in our study achieved excellent viral suppression. Because HIVAN is effectively prevented or treated with antiretroviral therapy,32 CKD in our cohort was probably not HIVAN. In the absence of biopsies, we can only speculate on the causes of CKD in the ALLRT cohort. Aside from HIVAN, other frequent CKDs seen in the setting of HIV infection include FSGS, hypertensive nephrosclerosis (both also associated with APOL1 risk alleles), drug toxicities, diabetic nephropathy, and various immune-mediated nephropathies, some attributable to hepatitis B virus or HCV coinfection.33–36 In our nephrotic proteinuria cohort, diabetes and HCV infection were more prevalent and significant risk factors for CKD, but CKD in this group was not associated with APOL1 risk alleles. However, APOL1 risk alleles were a significant risk factor for CKD in the eGFR<60 cohort, in which >70% of the patients were hypertensive. This cohort appears phenotypically similar to patients in the African American Study of Kidney Disease and Hypertension trial in which APOL1 risk alleles associated with a hypertension-related CKD without heavy proteinuria.37 Even in the absence of biopsy-proven diagnoses, our results corroborated several prior studies emphasizing the significant contributions of non-HIV risk factors for CKD, especially in persons receiving antiretroviral therapy.38–41 Thus, we suspect the cause of CKD in the eGFR<60 cohort is not HIVAN but are other forms of APOL1-associated kidney disease, while the nephrotic proteinuria cohort does not represent APOL1-associated kidney disease. Regardless, APOL1 levels did not correlate with either CKD phenotype, supporting our conclusion that circulating levels of APOL1 are not a mediator of injury in APOL1-associated CKD.
In addition, the association of APOL1 risk variants with CKD in our eGFR<60 cohort best fit an additive inheritance model. In prior publications, the recessive model best described the association of APOL1 risk variants and advanced CKD. However, several studies have suggested an association of a single-copy APOL1 risk variant in patients with both categorical and time-to-event renal phenotypes.2,13,19,42,43 These prior studies support our observation that a single risk variant may contribute to disease pathogenesis, and a better understanding of CKD phenotypes or biologic mechanisms may clarify additive versus recessive effects. Ultimately, the biologic mechanism of APOL1-associated CKD is currently unknown and cannot be presumed on the basis of statistical genetic analyses; rather, it must be determined empirically.
Other significant CKD risk factors included several cytokines, most notably sTNFR1 and sTNFR2, which have been previously linked with HIVAN44 and other CKDs.45–50 Although the cytokine levels were not associated with APOL1 genotype, several cytokines were independent risk factors for both CKD phenotypes. Although it is unclear how and why inflammatory cytokines are elevated in the setting of CKD, it is possible these cytokines may represent an additional pathogenic trigger that initiates or exacerbates CKD in individuals with APOL1 risk genotypes.
In the longitudinal analysis, progressive eGFR decline was significantly faster in cases with two versus no APOL1 risk variants and corroborates recent studies that found two APOL1 risk alleles predict CKD progression to ESRD.12,13,21 Consistent with a previous ALLRT study in which proteinuria improved in a subset of patients with heavy proteinuria,51 we also observed substantial improvements in proteinuria in patients with nephrotic-range proteinuria, but these improvements did not differ according to APOL1 risk variants. These observations also concur with a recent study that examined a cohort of HIV-infected patients with biopsy-proven disease that did not include HIVAN.12 In this group, time to ESRD but not proteinuria was associated with two APOL1 risk variants. Alternatively, a recent report from the Women Interagency HIV Study found a significant association between two APOL1 risk alleles and proteinuria,14 although the association was lost in patients with a history of AIDS.
In summary, circulating APOL1 levels were not associated with APOL1 genotype, inflammatory cytokine levels, eGFR < 60 ml/min per 1.73 m2 or nephrotic-range proteinuria in HIV-infected African Americans. Although the absence of renal biopsies is a limitation of our study, it does reflect routine patient care, where a definitive tissue diagnosis is often unavailable to guide management decisions. These observations may provide a foundation to develop diagnostic criteria and less invasive methods to better identify which HIV-1–infected African Americans may be at risk for progression to renal failure. In addition, the absence of any clear association of APOL1 plasma levels with CKD suggests that the function of variant APOL1 protein—either from the circulation or synthesized by kidney cells—may be a more likely mediator of kidney disease pathogenesis. Identifying the normal functions of APOL1 in kidney cells and how risk variants change these functions will be valuable future studies.
Concise Methods
Participants and Study Design
We used a 1:2 case-control study design to assess the relationship of APOL1 genotype and APOL1 plasma levels with CKD in HIV-infected self-reported African Americans (non-Hispanic black). CKD was defined by an eGFR<60 ml/min per 1.73 m2 for at least 3 months using the Modification of Diet in Renal Disease equation or by nephrotic-range proteinuria with a UP/Cr >3.5 g/g on at least one measurement. For proteinuria cases, matching controls had a UP/Cr <0.1 g/g at all time points and had no more than one eGFR measurement<60 ml/min per 1.73m2, whereas controls matched to cases defined by eGFR<60 ml/min per 1.73 m2 had no more than one eGFR measurement<60 ml/min per 1.73 m2 and UP/Cr<1.0 g/g at all time points during follow-up. Cases and controls were matched on CD4 cell counts (<200 versus≥200 cells/µl) and plasma HIV-1 RNA (<200 versus≥200 copies/ml). The total number of patients in the study was 270, with 46 cases and 92 controls in the eGFR<60 cohort and 44 cases and 88 controls in the nephrotic proteinuria cohort. Because of missing data, some comparisons had fewer patients; deviations from the above numbers are noted in the respective figures or tables. All participants provided written informed consent through the ACTG, and this study was conducted with oversight from the ACTG and the MetroHealth Medical Center Institutional Review Board and adhered to the Declaration of Helsinki.
APOL1 Genotyping
Archived DNA isolated from peripheral blood mononuclear cells was obtained from the ACTG repository. Genotypes were determined by direct sequencing of PCR-amplified DNA using a previously described method.9 Of the 270 identified cases and controls, APOL1 genotype was not obtained in eight patients because DNA was not available (n=6), DNA did not amplify (n=1), and one participant withdrew consent. A Pearson chi-square test was used to determine whether APOL1 risk variants were in Hardy–Weinberg equilibrium.
APOL1 ELISA and Cytokine Analysis
Archived frozen plasma was obtained from the ACTG repository representing the time point when the patient presented to a follow-up clinic visit and the laboratory examination demonstrated an eGFR<60 ml/min per 1.73 m2 or nephrotic proteinuria (baseline sample). APOL1 levels were quantified using a human specific sandwich ELISA (USCN Life Sciences, Inc., Houston, TX). Plasma was diluted 1:200 and assayed in duplicate according to the manufacturer's instructions. Plasma cytokine levels were assayed in duplicate using multiplexing kits and analyzer from Meso Scale Discovery (Sector Imager 2400; Gaithersburg, MD) according to the manufacturer's protocols and specifications. APOL1 and cytokine plasma levels were not obtained in two participants because plasma samples were not available at time of assay. All plasma assays were conducted at one of Case Western Reserve University's Clinical Laboratory Improvement Amendments–certified Clinical Research Units.
Statistical Analyses
Baseline differences between cases and controls were evaluated using conditional logistic regression; where these models did not converge, continuous variables were compared using the Wilcoxon rank-sum test and categorical variables were compared using the Fisher exact test. Spearman rank correlations examined associations between APOL1 levels and other continuous variables; because cases and controls were matched, these analyses were stratified by case or control status. The multivariable conditional logistic regression was the primary analysis of baseline associations; additional baseline correlations that were performed were not adjusted for multiple testing and the longitudinal analyses were exploratory.
For the primary analysis conditional logistic regression models (which incorporated the matching factors CD4 count and HIV-1 RNA level) tested associations between APOL1 genotypes (as no, one, or two risk alleles) and each CKD phenotype, using a dominant (one or two versus no risk alleles), recessive (two versus no or one risk allele) or additive (one and two versus no risk alleles) model of inheritance for APOL1 genotypes.52 A partial F-test was used to select between the models that detected significant associations with APOL1 genotypes. Multivariable models for each CKD outcome included APOL1 genotypes (always in the model) and additional risk factors or potential confounders, including HCV infection (defined as antibody positive or a hepatitis C diagnosis before case or control assignment), hypertension or diabetes status, fasting plasma total cholesterol at the time of case or control designation, and tenofovir exposure, defined by whether tenofovir was prescribed when patients were designated as a case or control, and tested interactions between APOL1 genotype and APOL1 plasma levels. Because age and sex are included in the Modification of Diet in Renal Disease equation, these variables were only included in longitudinal models of proteinuria. Covariates that were associated with the outcome at a significance of P<0.20 by univariate association were added in the order of highest to lowest significance, using the forward selection method, and they were retained in the final model if significant at P<0.10.
Additional multivariable models tested the independent contributions of proinflammatory cytokines (C-reactive protein, IL-6, sTNFR1, sTNFR2, β2-microglobulin, IFN-α, CCL2, and CCL5 as the average of duplicate measurements, all after log10 transformation) by adding each cytokine level separately to the previously described multivariable model. To minimize contributions by outliers, cytokine averages that were above or below the 95th percentile for the study population were set to the upper and lower 95th percentile values, respectively. To facilitate comparisons between markers, each level was subtracted from the mean for the study population of that cytokine, and divided by its SD (Z-score).
Associations between APOL1 genotype and changes in eGFR or UP/Cr over time were examined using separate mixed-effects linear models among cases and controls that were adjusted for HCV infection; pharmacologically treated hypertension and diabetes; and time-varying CD4 cell counts, HIV-1 RNA concentrations, and fasting plasma total cholesterol concentrations. Age and sex were included in models of UP/Cr but not eGFR. These models assumed an autoregressive covariance structure and included random intercepts and random slopes. Variability of slopes was summarized by the interquartile range of the empirical Bayes estimates.
Disclosures
S.G. received unrestricted research grants from Merck & Co., Janssen/Tibotec Therapeutics, and Gilead Sciences; compensation for one lecture in 2012 to Merck & Co. pertaining to HIV-related renal disease; and travel support from Gilead Sciences in 2011 to present data at the International AIDS Conference for a tenofovir-related study. M.R.P. as filed a patent in relation to this topic. J.R.S. is a member of CKD Advisory Board for Eli Lilly and Company. R.C.K. received grant support from Gilead Sciences.
Supplementary Material
Acknowledgments
We thank Drs. Tyler Miller, Michael Lederman, and Jeffrey Schelling for suggestions and critical review of the manuscript, the staff of the Clinical Research Units at MetroHealth Medical Center and University Hospitals for technical support, and the ACTG site staff and ALLRT participants for their time and effort.
This study was supported by the Leonard C. Rosenberg Renal Research Foundation of the Cleveland Centers for Dialysis Care, MetroHealth Medical Center institutional funds, the National Institutes of Health ACTG supported by AI68636, AI68634, AI38858, AI69501, and AI38855, and the Clinical and Translational Science Collaborative of Cleveland supported by the National Center for Advancing Translational Sciences grant UL1TR000439. L.A.B. and R.C.K. are members of the Case Western Reserve University Center for AIDS Research, supported by National Institutes of Health grant AI036219. R.C.K. is also supported by the Department of Veterans Affairs, VISN10 Geriatric Research Educational and Clinical Centers, Louis Stokes Cleveland Veterans Affairs Medical Center.
This work was presented in preliminary form at the 9th International Podocyte Symposium in Miami, FL, on April 22–25, 2012, and the International AIDS Conference 2012 in Washington, DC, on July 22–27, 2012.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2013070700/-/DCSupplemental.
References
- 1.Genovese G, Tonna SJ, Knob AU, Appel GB, Katz A, Bernhardy AJ, Needham AW, Lazarus R, Pollak MR: A risk allele for focal segmental glomerulosclerosis in African Americans is located within a region containing APOL1 and MYH9. Kidney Int 78: 698–704, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR: Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329: 841–845, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Genovese G, Friedman DJ, Pollak MR: APOL1 variants and kidney disease in people of recent African ancestry. Nat Rev Nephrol 9: 240–244, 2013 [DOI] [PubMed] [Google Scholar]
- 4.Tzur S, Rosset S, Shemer R, Yudkovsky G, Selig S, Tarekegn A, Bekele E, Bradman N, Wasser WG, Behar DM, Skorecki K: Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet 128: 345–350, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ulasi II, Tzur S, Wasser WG, Shemer R, Kruzel E, Feigin E, Ijoma CK, Onodugo OD, Okoye JU, Arodiwe EB, Ifebunandu NA, Chukwuka CJ, Onyedum CC, Ijoma UN, Nna E, Onuigbo M, Rosset S, Skorecki K: High population frequencies of APOL1 risk variants are associated with increased prevalence of non-diabetic chronic kidney disease in the Igbo people from South-Eastern Nigeria. Nephron Clin Pract 123: 123–128, 2013 [DOI] [PubMed] [Google Scholar]
- 6.Pays E, Vanhollebeke B: Human innate immunity against African trypanosomes. Curr Opin Immunol 21: 493–498, 2009 [DOI] [PubMed] [Google Scholar]
- 7.Lecordier L, Vanhollebeke B, Poelvoorde P, Tebabi P, Paturiaux-Hanocq F, Andris F, Lins L, Pays E: C-terminal mutants of apolipoprotein L-I efficiently kill both Trypanosoma brucei brucei and Trypanosoma brucei rhodesiense. PLoS Pathog 5: e1000685, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duchateau PN, Pullinger CR, Orellana RE, Kunitake ST, Naya-Vigne J, O’Connor PM, Malloy MJ, Kane JP: Apolipoprotein L, a new human high density lipoprotein apolipoprotein expressed by the pancreas. Identification, cloning, characterization, and plasma distribution of apolipoprotein L. J Biol Chem 272: 25576–25582, 1997 [DOI] [PubMed] [Google Scholar]
- 9.Madhavan SM, O’Toole JF, Konieczkowski M, Ganesan S, Bruggeman LA, Sedor JR: APOL1 localization in normal kidney and nondiabetic kidney disease. J Am Soc Nephrol 22: 2119–2128, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee BT, Kumar V, Williams TA, Abdi R, Bernhardy A, Dyer C, Conte S, Genovese G, Ross MD, Friedman DJ, Gaston R, Milford E, Pollak MR, Chandraker A: The APOL1 genotype of African American kidney transplant recipients does not impact 5-year allograft survival. Am J Transplant 12: 1924–1928, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeves-Daniel AM, DePalma JA, Bleyer AJ, Rocco MV, Murea M, Adams PL, Langefeld CD, Bowden DW, Hicks PJ, Stratta RJ, Lin JJ, Kiger DF, Gautreaux MD, Divers J, Freedman BI: The APOL1 gene and allograft survival after kidney transplantation. Am J Transplant 11: 1025–1030, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fine DM, Wasser WG, Estrella MM, Atta MG, Kuperman M, Shemer R, Rajasekaran A, Tzur S, Racusen LC, Skorecki K: APOL1 risk variants predict histopathology and progression to ESRD in HIV-related kidney disease. J Am Soc Nephrol 23: 343–350, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopp JB, Nelson GW, Sampath K, Johnson RC, Genovese G, An P, Friedman D, Briggs W, Dart R, Korbet S, Mokrzycki MH, Kimmel PL, Limou S, Ahuja TS, Berns JS, Fryc J, Simon EE, Smith MC, Trachtman H, Michel DM, Schelling JR, Vlahov D, Pollak M, Winkler CA: APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol 22: 2129–2137, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estrella MM, Wyatt CM, Pearce CL, Li M, Shlipak MG, Aouizerat BE, Gustafson D, Cohen MH, Gange SJ, Kao WH, Parekh RS: Host APOL1 genotype is independently associated with proteinuria in HIV infection. Kidney Int 84: 834–840, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Appay V, Sauce D: Immune activation and inflammation in HIV-1 infection: causes and consequences. J Pathol 214: 231–241, 2008 [DOI] [PubMed] [Google Scholar]
- 16.Carrero JJ, Stenvinkel P: Inflammation in end-stage renal disease—what have we learned in 10 years? Semin Dial 23: 498–509, 2010 [DOI] [PubMed] [Google Scholar]
- 17.Krishnan S, Wu K, Smurzynski M, Bosch RJ, Benson CA, Collier AC, Klebert MK, Feinberg J, Koletar SL, ALLRT/A5001 Team : Incidence rate of and factors associated with loss to follow-up in a longitudinal cohort of antiretroviral-treated HIV-infected persons: An AIDS Clinical Trials Group (ACTG) Longitudinal Linked Randomized Trials (ALLRT) analysis. HIV Clin Trials 12: 190–200, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smurzynski M, Collier AC, Koletar SL, Bosch RJ, Wu K, Bastow B, Benson CA: AIDS clinical trials group longitudinal linked randomized trials (ALLRT): Rationale, design, and baseline characteristics. HIV Clin Trials 9: 269–282, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papeta N, Kiryluk K, Patel A, Sterken R, Kacak N, Snyder HJ, Imus PH, Mhatre AN, Lawani AK, Julian BA, Wyatt RJ, Novak J, Wyatt CM, Ross MJ, Winston JA, Klotman ME, Cohen DJ, Appel GB, D’Agati VD, Klotman PE, Gharavi AG: APOL1 variants increase risk for FSGS and HIVAN but not IgA nephropathy. J Am Soc Nephrol 22: 1991–1996, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman DJ, Kozlitina J, Genovese G, Jog P, Pollak MR: Population-based risk assessment of APOL1 on renal disease. J Am Soc Nephrol 22: 2098–2105, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster MC, Coresh J, Fornage M, Astor BC, Grams M, Franceschini N, Boerwinkle E, Parekh RS, Kao WH: APOL1 variants associate with increased risk of CKD among African Americans. J Am Soc Nephrol 24: 1484–1491, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duchateau PN, Movsesyan I, Yamashita S, Sakai N, Hirano K, Schoenhaus SA, O’Connor-Kearns PM, Spencer SJ, Jaffe RB, Redberg RF, Ishida BY, Matsuzawa Y, Kane JP, Malloy MJ: Plasma apolipoprotein L concentrations correlate with plasma triglycerides and cholesterol levels in normolipidemic, hyperlipidemic, and diabetic subjects. J Lipid Res 41: 1231–1236, 2000 [PubMed] [Google Scholar]
- 23.Page NM, Olano-Martin E, Lanaway C, Turner R, Minihane AM: Polymorphisms in the Apolipoprotein L1 gene and their effects on blood lipid and glucose levels in middle age males. Genes Nutr 1: 133–135, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albert TS, Duchateau PN, Deeb SS, Pullinger CR, Cho MH, Heilbron DC, Malloy MJ, Kane JP, Brown BG: Apolipoprotein L-I is positively associated with hyperglycemia and plasma triglycerides in CAD patients with low HDL. J Lipid Res 46: 469–474, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Freedman BI, Langefeld CD, Murea M, Ma L, Otvos JD, Turner J, Antinozzi PA, Divers J, Hicks PJ, Bowden DW, Rocco MV, Parks JS: Apolipoprotein L1 nephropathy risk variants associate with HDL subfraction concentration in African Americans. Nephrol Dial Transplant 26: 3805–3810, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalayjian RC, Machekano RN, Rizk N, Robbins GK, Gandhi RT, Rodriguez BA, Pollard RB, Lederman MM, Landay A: Pretreatment levels of soluble cellular receptors and interleukin-6 are associated with HIV disease progression in subjects treated with highly active antiretroviral therapy. J Infect Dis 201: 1796–1805, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuller LH, Tracy R, Belloso W, De Wit S, Drummond F, Lane HC, Ledergerber B, Lundgren J, Neuhaus J, Nixon D, Paton NI, Neaton JD, INSIGHT SMART Study Group : Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PLoS Med 5: e203, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duchateau PN, Pullinger CR, Cho MH, Eng C, Kane JP: Apolipoprotein L gene family: tissue-specific expression, splicing, promoter regions; discovery of a new gene. J Lipid Res 42: 620–630, 2001 [PubMed] [Google Scholar]
- 29.Monajemi H, Fontijn RD, Pannekoek H, Horrevoets AJ: The apolipoprotein L gene cluster has emerged recently in evolution and is expressed in human vascular tissue. Genomics 79: 539–546, 2002 [DOI] [PubMed] [Google Scholar]
- 30.Zhaorigetu S, Wan G, Kaini R, Jiang Z, Hu CA: ApoL1, a BH3-only lipid-binding protein, induces autophagic cell death. Autophagy 4: 1079–1082, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Althoff KN, Buchacz K, Hall HI, Zhang J, Hanna DB, Rebeiro P, Gange SJ, Moore RD, Kitahata MM, Gebo KA, Martin J, Justice AC, Horberg MA, Hogg RS, Sterling TR, Cescon A, Klein MB, Thorne JE, Crane HM, Mugavero MJ, Napravnik S, Kirk GD, Jacobson LP, Brooks JT, North American AIDS Cohort Collaboration on Research and Design : U.S. trends in antiretroviral therapy use, HIV RNA plasma viral loads, and CD4 T-lymphocyte cell counts among HIV-infected persons, 2000 to 2008. Ann Intern Med 157: 325–335, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucas GM, Eustace JA, Sozio S, Mentari EK, Appiah KA, Moore RD: Highly active antiretroviral therapy and the incidence of HIV-1-associated nephropathy: A 12-year cohort study. AIDS 18: 541–546, 2004 [DOI] [PubMed] [Google Scholar]
- 33.Fine DM, Fogo AB, Alpers CE: Thrombotic microangiopathy and other glomerular disorders in the HIV-infected patient. Semin Nephrol 28: 545–555, 2008 [DOI] [PubMed] [Google Scholar]
- 34.Kalayjian RC, Lau B, Mechekano RN, Crane HM, Rodriguez B, Salata RA, Krishnasami Z, Willig JH, Martin JN, Moore RD, Eron JJ, Kitahata MM: Risk factors for chronic kidney disease in a large cohort of HIV-1 infected individuals initiating antiretroviral therapy in routine care. AIDS 26: 1907–1915, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scherzer R, Estrella M, Li Y, Choi AI, Deeks SG, Grunfeld C, Shlipak MG: Association of tenofovir exposure with kidney disease risk in HIV infection. AIDS 26: 867–875, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wyatt CM, Morgello S, Katz-Malamed R, Wei C, Klotman ME, Klotman PE, D’Agati VD: The spectrum of kidney disease in patients with AIDS in the era of antiretroviral therapy. Kidney Int 75: 428–434, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lipkowitz MS, Freedman BI, Langefeld CD, Comeau ME, Bowden DW, Kao WH, Astor BC, Bottinger EP, Iyengar SK, Klotman PE, Freedman RG, Zhang W, Parekh RS, Choi MJ, Nelson GW, Winkler CA, Kopp JB, SK Investigators : Apolipoprotein L1 gene variants associate with hypertension-attributed nephropathy and the rate of kidney function decline in African Americans. Kidney Int 83: 114–120, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganesan A, Krantz EM, Huppler Hullsiek K, Riddle MS, Weintrob AC, Lalani T, Okulicz JF, Landrum M, Agan B, Whitman TJ, Ross MJ, Crum-Cianflone NF, Infectious Disease Clinical Research Program HIV/STI Working Group : Determinants of incident chronic kidney disease and progression in a cohort of HIV-infected persons with unrestricted access to health care. HIV Med 14: 65–76, 2013 [DOI] [PubMed] [Google Scholar]
- 39.Tsui J, Vittinghoff E, Anastos K, Augenbraun M, Young M, Nowicki M, Cohen MH, Peters MG, Golub ET, Szczech L: Hepatitis C seropositivity and kidney function decline among women with HIV: data from the Women’s Interagency HIV Study. Am J Kidney Dis 54: 43–50, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi AI, Shlipak MG, Hunt PW, Martin JN, Deeks SG: HIV-infected persons continue to lose kidney function despite successful antiretroviral therapy. AIDS 23: 2143–2149, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jotwani V, Li Y, Grunfeld C, Choi AI, Shlipak MG: Risk factors for ESRD in HIV-infected individuals: traditional and HIV-related factors. Am J Kidney Dis 59: 628–635, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tzur S, Rosset S, Skorecki K, Wasser WG: APOL1 allelic variants are associated with lower age of dialysis initiation and thereby increased dialysis vintage in African and Hispanic Americans with non-diabetic end-stage kidney disease. Nephrol Dial Transplant 27: 1498–1505, 2012 [DOI] [PubMed] [Google Scholar]
- 43.Kanji Z, Powe CE, Wenger JB, Huang C, Ankers E, Sullivan DA, Collerone G, Powe NR, Tonelli M, Bhan I, Bernhardy AJ, Dibartolo S, Friedman D, Genovese G, Pollak MR, Thadhani R: Genetic variation in APOL1 associates with younger age at hemodialysis initiation. J Am Soc Nephrol 22: 2091–2097, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruggeman LA, Drawz PE, Kahoud N, Lin K, Barisoni L, Nelson PJ: TNFR2 interposes the proliferative and NF-κB-mediated inflammatory response by podocytes to TNF-α. Lab Invest 91: 413–425, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vielhauer V, Stavrakis G, Mayadas TN: Renal cell-expressed TNF receptor 2, not receptor 1, is essential for the development of glomerulonephritis. J Clin Invest 115: 1199–1209, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niewczas MA, Ficociello LH, Johnson AC, Walker W, Rosolowsky ET, Roshan B, Warram JH, Krolewski AS: Serum concentrations of markers of TNFalpha and Fas-mediated pathways and renal function in nonproteinuric patients with type 1 diabetes. Clin J Am Soc Nephrol 4: 62–70, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernández-Real JM, Vendrell J, García I, Ricart W, Vallès M: Structural damage in diabetic nephropathy is associated with TNF-α system activity. Acta Diabetol 49: 301–305, 2012 [DOI] [PubMed] [Google Scholar]
- 48.Keller C, Katz R, Cushman M, Fried LF, Shlipak M: Association of kidney function with inflammatory and procoagulant markers in a diverse cohort: A cross-sectional analysis from the Multi-Ethnic Study of Atherosclerosis (MESA). BMC Nephrol 9: 9, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gohda T, Niewczas MA, Ficociello LH, Walker WH, Skupien J, Rosetti F, Cullere X, Johnson AC, Crabtree G, Smiles AM, Mayadas TN, Warram JH, Krolewski AS: Circulating TNF receptors 1 and 2 predict stage 3 CKD in type 1 diabetes. J Am Soc Nephrol 23: 516–524, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niewczas MA, Gohda T, Skupien J, Smiles AM, Walker WH, Rosetti F, Cullere X, Eckfeldt JH, Doria A, Mayadas TN, Warram JH, Krolewski AS: Circulating TNF receptors 1 and 2 predict ESRD in type 2 diabetes. J Am Soc Nephrol 23: 507–515, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta SK, Smurzynski M, Franceschini N, Bosch RJ, Szczech LA, Kalayjian RC, AIDS Clinical Trials Group Longitudinal Linked Randomized Trials Study Team : The effects of HIV type-1 viral suppression and non-viral factors on quantitative proteinuria in the highly active antiretroviral therapy era. Antivir Ther 14: 543–549, 2009 [PMC free article] [PubMed] [Google Scholar]
- 52.Lewis CM: Genetic association studies: Design, analysis and interpretation. Brief Bioinform 3: 146–153, 2002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.