Abstract
Propofol, an intravenous general anesthetic, produces many of its anesthetic effects in vivo by potentiating the responses of GABA type A receptors (GABAAR), members of the superfamily of pentameric ligand-gated ion channels (pLGICs) that contain anion-selective channels. Propofol also inhibits pLGICs containing cation-selective channels, including nicotinic acetylcholine receptors and GLIC, a prokaryotic proton-gated homolog from Gloeobacter violaceus. In the structure of GLIC co-crystallized with propofol at pH 4 (presumed open/desensitized states), propofol was localized to an intrasubunit pocket at the extracellular end of the transmembrane domain within the bundle of transmembrane α-helices [Nury, H, et. al. (2011) Nature 469, 428–431]. To identify propofol binding sites in GLIC in solution, we used a recently developed photoreactive propofol analog (2-isopropyl-5-[3-(trifluoromethyl)-3H-diazirin-3-yl]phenol or AziPm) which acts as an anesthetic in vivo and potentiates GABAAR in vitro. For GLIC expressed in Xenopus oocytes, propofol and AziPm inhibited current responses at pH 5.5 (EC20) with IC50s of 20 and 50 μM, respectively. When [3H]AziPm (7 μM) was used to photolabel detergent-solubilized, affinity-purified GLIC at pH 4.4, protein microsequencing identified propofol-inhibitable photolabeling of three residues in the GLIC transmembrane domain: Met-205, Tyr-254, and Asn-307 in the M1, M3, and M4 transmembrane helices, respectively. Thus, in GLIC in solution, propofol and AziPm bind competitively to a site in proximity to these residues, which in the GLIC crystal structure are in contact with the propofol bound in the intrasubunit pocket.
The primary target for many general anesthetics including propofol is the γ-aminobutyric acid type-A receptor (GABAAR), the main inhibitory receptor in the brain (1–3). Propofol potentiates GABAAR responses at anesthetic concentrations, while at higher concentrations it inhibits other members of the pentameric ligand-gated ion channel (pLGIC) superfamily that contain cation-selective channels, including nicotinic acetylcholine receptors (nAChR) (4–6) and the prokaryotic pLGIC from Gloeobacter violaceus (GLIC), a proton-gated ion channel (7). Identification of propofol binding sites in pLGICs is necessary to determine whether it binds to equivalent or distinct sites when it acts as a positive or a negative allosteric pLGIC modulator. Each pLGIC subunit is composed of an N-terminal extracellular domain, comprised primarily of β strands, and a transmembrane domain consisting of a loose bundle of 4 α-helices, designated M1–M4 (8,9). When 5 subunits assemble to form a pLGIC, the M2 helices from each subunit combine to line the ion channel and are protected from the lipid by the M1, M3, and M4 helices.
Crystal structures of a GABAAR are not as yet available. However, several structures of prokaryotic pLGICs homologous with the GABAAR have been solved with general anesthetics bound, including GLIC with an intrasubunit binding site in the transmembrane domain (TMD) for propofol (or desflurane) (10) and an intersubunit site for bromoform (11), as well as a binding site for ketamine in the extracellular domain (12). In ELIC, a pLGIC from Erwinia chrysanthemi, binding sites were identified for bromoform in the TMD in the ion channel and at an interface between adjacent subunits, and in the extracellular domain (13). When propofol was co-crystallized with GLIC, propofol bound to a site near the extracellular end of the TMD in a pocket formed by the 4 transmembrane helices, and site-directed mutations of residues lining this intrasubunit pocket affected the inhibitory action of propofol (10). In contrast, in GLIC expressed in Xenopus oocytes, a substituted Cys within this intrasubunit pocket was more susceptible to modification in the presence of propofol, while a substituted Cys at a position predicted to be in a pocket between helices of adjacent subunits (an intersubunit pocket) was protected from modification by propofol (14), which suggests that propofol binds to pockets between adjacent subunits.
Photoaffinity labeling allows the identification of amino acids in a protein that contribute to a drug binding site without any assumptions about the points of drug contact in a protein (reviewed in (15)), and photoreactive analogs of etomidate and mephobarbital have been used recently to identify two classes of intersubunit general anesthetic binding sites in the GABAAR transmembrane domain (16–18). A photoreactive propofol analog, 2-isopropyl-5-[3-(trifluoromethyl)-3H-diazirin-3-yl]phenol or AziPm, was developed recently that acts as an anesthetic in vivo and potentiates GABAAR responses in vitro (19). AziPm, which contains a photoreactive trifluoromethyl diazirine that can react with most amino acid side chains, also is a Torpedo (muscle-type) nicotinic acetylcholine receptor (nAChR) negative allosteric modulator (20). In the Torpedo nAChR in native membranes, there is propofol-inhibitable photolabeling by [3H]AziPm of amino acids in binding sites in (i) the ion channel and (ii) in the δ subunit helix bundle pocket, a pocket homologous to the propofol binding site identified in GLIC crystals (20). [3H]AziPm also photolabeled an amino acid in the transmembrane domain in the pocket between the γ and α subunits, but that photolabeling was enhanced rather than inhibited by propofol. In this work we demonstrate that AziPm inhibits GLIC currents in oocytes with an IC50 and Hill coefficient similar to propofol. Photolabeling detergent-solubilized, affinity-purified GLIC with [3H]AziPm at pH 4.4, a pH stabilizing the open or desensitized state (21), identified propofol-inhibitable labeling of residues in M1, M3, and M4 consistent with both AziPm and propofol occupying the propofol binding site identified by X-ray crystallography.
EXPERIMENTAL PROCEDURES
Materials
Milligram quantities of GLIC were obtained by expression in E. coli, detergent solubilization, and affinity purification as described (22). Purified GLIC was stored at 3 °C in 10 mM K2HPO4/KH2PO4 (pH 8.0), 150 mM NaCl, 0.075 % dodecyl maltoside. Non-radioactive 2-isopropyl-5-[3-(trifluoromethyl)-3H-diazirin-3-yl]phenol (AziPm) was synthesized as described (19) and [3H]AziPm (10 Ci/mmol) was prepared by custom tritiation at AmBios (Newington, CT). Propofol and 3-bromo-3-methyl-2-(2-nitrophenylthio)-3H-indole (BNPS-skatole) were from Sigma, o-phthalaldehyde (OPA) was from Alfa Aesar, and Lysobacter enzymogenes endoproteinase Lys-C (EndoLys-C) was from Roche Applied Science.
Two-electrode voltage clamp
Capped complementary RNA expressing GLIC was synthesized with the mMessage mMachine SP6 kit (Ambion), purified with the RNeasy kit (Qiagen), and 25 ng injected into Xenopus laevis stage 5–6 oocytes. Oocytes were maintained at 18° C in modified Barth’s saline. Two-electrode voltage clamp experiments were performed at room temperature 16–48 h after injection with a model OC-725C amplifier (Warner Instruments), and a 20 μl oocyte recording chamber (Automate Scientific). Oocytes were clamped to a holding potential of −60 mV. Currents were recorded in ND-96 supplemented with 10 mM MES and adjusted to the indicated pH. Propofol and AziPm were added at the concentrations indicated. Data were collected and processed using Clampex 10 (Molecular Devices). The currents were normalized to the current measured without drug and the data were fit to the equation:
where f(x) is the fraction of current remaining at drug concentration x, IC50 is the drug concentration at which 50 % inhibition occurs, and n is the Hill coefficient.
Photoaffinity labeling of GLIC with [3H]AziPm
Affinity purified GLIC (400 μg in 2.3 ml of 10 mM K2HPO4/KH2PO4 (pH 8.0), 150 mM NaCl, 0.075 % DDM) was adjusted to pH 4.4 by slow titration and gentle vortexing with 550 μl of 250 mM Na Acetate (pH 4), and [3H]AziPm (10 Ci/mmol) was added to 7 μM. The sample was divided and 300 μM propofol was added to half, resulting in a final GLIC concentration of 0.14 mg protein/ml, i.e. ~4 μM binding sites for a ligand binding to a single site in each subunit. After 30 min on ice, samples were irradiated at 365 nm for 30 min, denatured in sample buffer, and GLIC subunit was resolved by SDS-PAGE. The Coomassie blue stained band of GLIC at ~37 kDa was excised and eluted for 5 days in 100 mM NH4HCO3, 2.5 mM dithiothreitol, and 0.2 % SDS, pH 8.4 at 20 °C with gentle agitation. The eluates containing the labeled GLIC protein were filtered, concentrated, acetone precipitated, and resuspended in 100 μl of 15 mM Tris and 0.1% SDS, pH 8.5.
Enzymatic/chemical digestion, reversed-phase HPLC, and automated Edman degradation
Aliquots of GLIC were digested with 1.5 U of EndoLys-C for 3 weeks at 20 °C. Additional aliquots were loaded directly onto PVDF filters for BNPS-skatole chemical cleavage as described (23,17). Reversed-phase HPLC was performed on an Agilent 1100 series HPLC. Separations were achieved at 40 °C on a Brownlee Aquapore Bu-300 7 μM 100 mm column with aqueous solvent 0.08 % trifluoroacetic acid, organic solvent 60 % acetonitrile, 40 % isopropanol, and 0.05 % trifluoroacetic acid, and a gradient beginning at 5% organic and reaching 100 % organic in 75 min. Flow rate was 0.2 ml/min and fractions were collected every 2.5 min. Sequence analysis was performed on a Procise 492 (ABI) protein sequencer using 2/3 of each cycle for amino acid detection and 1/3 for 3H counting (measured 3H & pmol data are shown in the 3H release plots). Digests were loaded onto Prosorb (ABI) PVDF membranes for sequencing. To chemically isolate specific peptides during sequence analysis, the sequencer was paused prior to an expected proline and the filter treated with o-phthalaldehyde (OPA) before proceeding. OPA covalently blocks Edman degradation at all free amino-termini residues except prolines (24,25).
Modeling and computational docking
The CHARMm-based molecular dynamics simulated annealing program CDOCKER (Accelrys Discovery Studio) was used to dock propofol or AziPm into the propofol pocket in one subunit (C) of the GLIC crystal structure (PDB:3P50). A binding site sphere (10 Å radius) was centered on the propofol molecule of the PDB structure, and then all non-protein molecules were removed from the structure. Beginning with 13 (propofol) or 26 (AziPm) randomly oriented molecules, each of which had 35 random computer reorientations and 35 MD-driven molecular alterations (i.e. 1,225 starting points for each), the 100 lowest energy solutions for each starting molecule were collected (1,300 solutions for propofol; 2,600 for AziPm).
To compare GLIC and nAChR photolabeling results, a homology model of the Torpedo californica nAChR based on the GLIC structure (PDB:3P50) was made using the Discovery Studio. To align the TMD sequences of GLIC and Torpedo nAChR subunits, a secondary/tertiary structural alignment was made between GLIC and neuronal β2 nAChR TMD NMR structure (PDB: 2LM2) (26) using the Superimpose Proteins function, and the resulting sequence alignment was combined with a sequence alignment between Torpedo nAChR subunit TMD sequences and neuronal β2 nAChR TMD which share > 50% identity. The final TMD alignment between GLIC and Torpedo nAChR subunits necessitated one residue inserts in the M1–M2 (δPro-250) and M2–M3 (δLeu-283) loops. The level of the intrasubunit helix bundle pocket and the composition of the M2–M3 loop in this GLIC-based model differs from that of the cryoelectron microscopy structure of the nAChR (PDB:2BG9) (8) which was used previously to locate the amino acids photolabeled by nAChR inhibitors (20, 27– 29). In the GLIC-based structure, the photolabeled amino acids contribute to a better defined common binding site situated among the M1, M2, and M3 helices. Comparison of the structures of the two models of the nAChR δ subunit transmembrane domain, including the locations of photolabeled amino acids, are shown in the Supporting Information in stereo representation in Supplemental Figures S1 and S2 and in Supplemental Table 1.
RESULTS
Propofol and AziPm inhibit GLIC
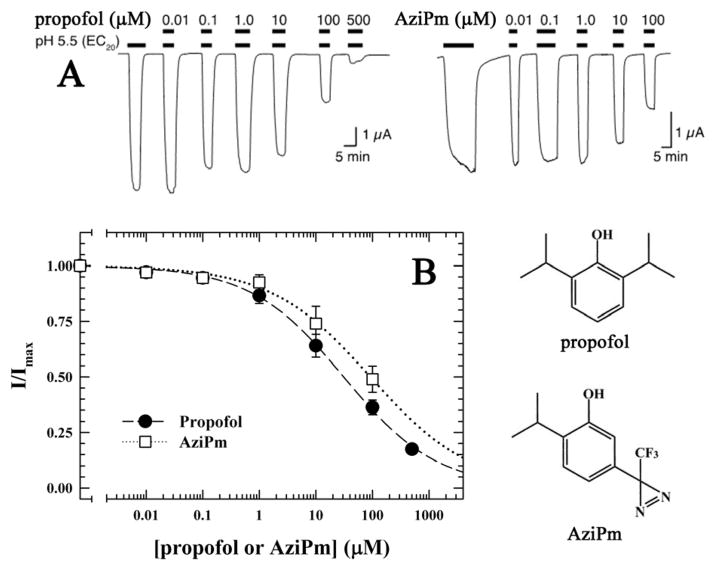
GLIC expressed in Xenopus was activated by a decrease in pH from 7.4 to 5.5 for ~20 seconds, a shift in pH that produced ~20 % of the maximal current response elicited by a shift to pH 3.5. Co-application of propofol with the decrease in pH produced a concentration-dependent inhibition of current responses with an IC50 of 21.4 ± 1.2 μM and a Hill coefficient of 0.67 ± 0.05 (Fig. 1). Similarly, co-application of AziPm with the decrease in pH inhibited GLIC currents with an IC50 of 51.4 ± 5.3 μM and a Hill coefficient of 0.63 ± 0.11.
Figure 1. Propofol and AziPm both inhibit H+ activated GLIC currents.
A) Representative current traces of oocyte-expressed GLIC exposed to pH 5.5 corresponding to the EC20 in the presence of various concentrations of propofol or AziPm as indicated with black bars over the traces. Limited solubility of AziPm prevented analysis at higher concentrations. B) Inhibition curves for propofol and AziPm at the pH corresponding to the EC20 (pH 5.5). Response is expressed as the fraction of current induced in the presence of the indicated concentrations of propofol (●) or AziPm (□) relative to that in their absence. The data (n=7) were fit to Hill equations with IC50s of 21.4 ± 1.2 and 51.4 ± 5.3 μM, respectively, and had Hill coefficients of −0.67 ± 0.05 and −0.63 ± 0.11, respectively. Error bars represent SEM. Also included are the chemical structures of propofol and AziPm.
Characterization of [3H]AziPm photolabeling of GLIC
Affinity-purified GLIC in detergent solution was equilibrated with [3H]AziPm (7 μM) for 30 min on ice at pH 4.4, in the absence or presence of 300 μM propofol. After irradiation for 30 min at 365 nm, [3H]AziPm-labeled GLIC subunit was isolated by SDS-PAGE. Sequence analysis of the intact subunit revealed the presence of 3 N-terminal residues (GPM) before the published sequence (GPMGQDM…)2. No 3H labeled residues were encountered in the first 20 cycles of Edman degradation, and 3H appeared to be stably incorporated in GLIC.
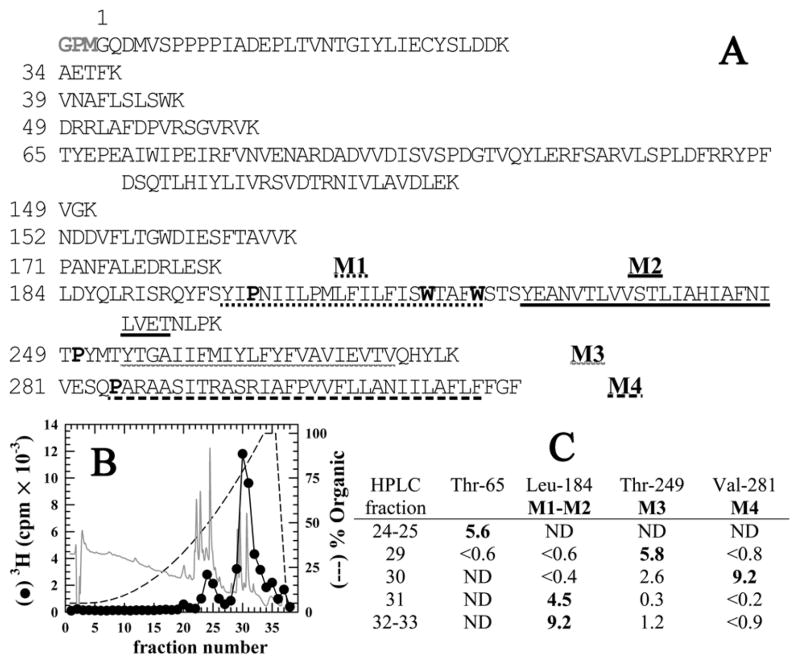
To initially characterize regions of [3H]AziPm photoincorporation, photolabeled GLIC was digested with EndoLys-C which cleaves after Lys residues and is predicted to fractionate GLIC into eleven peptides, three of which contain the four hydrophobic transmembrane helices M1–M4 (Fig 2A). When the digests were fractionated by reversed-phase HPLC, 80 % of the recovered 3H eluted at > 60 % organic (Fig 2B), and sequence analysis of those fractions identified only the 3 peptides containing the transmembrane segments (Fig 2C). An additional 15 % of 3H eluted in a hydrophilic peak that contained an 84 amino acid GLIC fragment beginning at Thr-65. Therefore, the majority of [3H]AziPm labeling was limited to the transmembrane region of GLIC.
Figure 2. Reversed-phase HPLC fractionation of EndoLys-C digests of GLIC: [3H]AziPm primarily photoincorporate into the hydrophobic, transmembrane domain of GLIC.
A) Shown are the sequences of the GLIC fragments produced by enzymatic cleavage with EndoLys-C (specific for Lys). The residue numbering is that used in the crystal structure (PDB:3P50 (10)) and does not include the 3 additional N-terminal residues (GPM) identified by sequence analysis of intact, expressed GLIC. The transmembrane helices M1–M4 are underlined. The Pro residues used to chemically isolate M1, M3, & M4 during sequencing are bolded as are the Trp residues subjected to chemical cleavage to sequence M2. B) Reversed-phase HPLC fractionation of an EndoLys-C digest of [3H]AziPm labeled GLIC. 82% of the recovered 3H eluted in hydrophobic fractions (>75 % organic, fractions 28–38). In gray is the absorbance profile at 215 nm. C) Selected fractions containing 3H were sequenced, with the peptides detected quantitated in picomoles. In fractions 29–33, the only GLIC fragments detected were those containing the transmembrane helices. ND, not detected.
Identification of propofol-inhibitable labeling in M3, M4, and M1
In the amino acid sequence of the GLIC fragment produced by EndoLys-C that begins at Leu-184 and contains the M1 and M2 helices, Pro-199, located near the N-terminus of M1, would occur in the 16th cycle of Edman degradation, while the fragments containing M3 and M4 contain prolines in cycles 2 (Pro-250) and 5 (Pro-285), respectively. This allowed us to characterize [3H]AziPm photoincorporation in each of the fragments by sequencing aliquots of the total digests and chemically isolating the fragment of interest by treatment of the sample at the cycle of Edman degradation containing the proline of interest with o-phthalaldehyde (OPA), which block the N-terminus of all peptides not containing a Pro in that cycle (24,25).
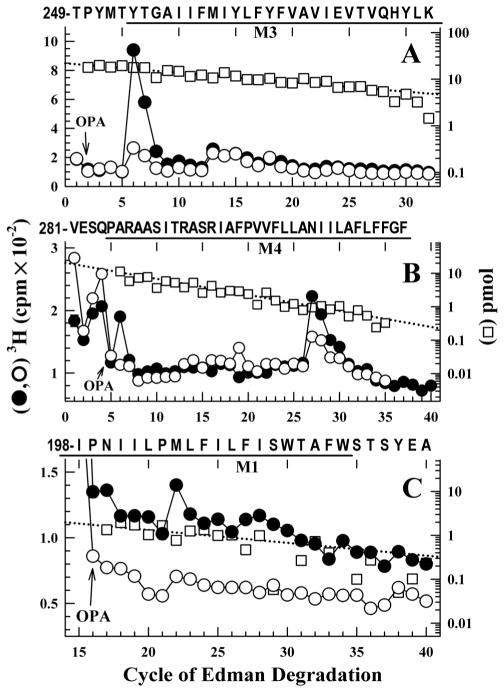
For analysis of [3H]AziPm photolabeling in M3, sequencing samples were treated with OPA before cycle 2 (Fig. 3A). After the OPA treatment, the only 2 fragments detected were the fragment beginning at Thr-249 and the N-terminal peptide (which also contains a Pro in cycle 2). Since sequence analysis of intact [3H]AziPm-labeled GLIC labeled had not identified any peaks of 3H release in 20 cycles of Edman degradation, the peak of 3H release in cycle 6 indicated photolabeling of Tyr-254 (115 cpm/pmol) which was inhibited ~80% by 300 μM propofol. The minor peaks of 3H release in cycles 13, 15, and 18 indicated that there may also be propofol-insensitive photolabeling in M3 of Met-261, Tyr-263, and Tyr-266 at <20% the level of labeling of Tyr-254.
Figure 3. Propofol-inhibitable [3H]AziPm labeling identified in M3, M4, and M1.
3H (●,○) and PTH-amino acids (□) released when equal aliquots of EndoLys-C digests of GLIC photolabeled with [3H]AziPm in the absence (●) or presence (○,□) of 300 μM propofol were sequenced with OPA treatment in cycles 2 (A), 5 (B), or 16 (C). A) After OPA treatment in cycle 2, the primary sequence began at Thr-249 before M3 (I0 = 20 pmol, both conditions). The GLIC amino-terminal fragment, the only other peptide from an EndoLys-C digest of GLIC with a proline in cycle 2, was present (11 pmol, both conditions). The peak of 3H release in cycle 6 indicated of photolabeling of Tyr-254 at an efficiency of 115 cpm/pmol, which 300 μM propofol inhibited by 83%. The small peak of 3H release in cycle 13 indicated propofol-insensitive photoabeling of Met-261 at ~20 cpm/pmol. B) After OPA treatment in cycle 5, the primary sequence began at Val-281 before M4 (-PPF, I0 = 15 pmol; +PPF (□), I0 = 21 pmol). The only other peptide present was lag from the fragment beginning Thr-65 (13 pmol), which contains Pro-68 in cycle 4 that would also be present in cycle 5 as a consequence of the ~90% repetitive yield of Edman degradation. The peak of 3H release in cycle 27 indicated photolabeling of Asn-307 in M4 at 300 cpm/pmol which 300 μM propofol inhibited by 70%. Since treatment with OPA blocks ~90 % of the free amino termini of peptides not containing a Pro, the peak of 3H release in cycle 6 after the OPA treatment is consistent with photolabeling of Tyr-254 if the M3 fragment beginning Thr-249 is present at ~7% the level seen in Panel A. C) After OPA treatment in cycle 16, the primary sequence detected originally began at Leu-184 (-PPF, I0 = 9 pmol; +PPF (□) I0 = 5 pmol). The peak of 3H release in cycle 22 indicated photolabeling of Met-205 at ~40 cpm/pmol, which propofol inhibited by ~40 %.
For analysis of photolabeling in M4, sequencing samples were treated with OPA before cycle 5 (Fig. 3B). After the OPA treatment, the primary sequence began at Val-281 before M4 and the peak of 3H in cycle 27 indicated labeling of Asn-307 (300 cpm/pmol) within the M4 helix which propofol inhibited by 70 %3. For analysis of photolabeling in M1, sequencing samples were treated with OPA before cycle 16 (Fig. 3C). After the OPA treatment, the only sequence detected originally began at Leu-184. Release of 3H in cycle 22 indicated photolabeling of Met-205 (41 cpm/pmol) in M1 which propofol inhibited by ~40 %.
[3H]AziPm photolabeling within M2
To characterize photoincorporation in M2, we took advantage of the distribution of Trp residues within GLIC primary structure and the ability of BNPS-skatole to selectively cleave peptide bonds C-terminal to tryptophan producing fragment with free amino termini susceptible to Edman degradation (23,17). Of the 5 Trp residues in GLIC (residues 47, 72, 160, 213, and 217), only Trp-213 & Trp-217 are in the transmembrane region, near the end of the M1 helix (Fig 2A). When photolabeled GLIC was sequenced for 30 cycles after treatment with BNPS-skatole, all 6 expected peptides were identified, with the fragment beginning at Ser-218 before M2 present at ~ 10 pmol. Any photolabeling of residues in M2, if it occurred, was at <10% the efficiency of photolabeling of Asn-307 in M4 and insensitive to propofol.
AziPm docking to the propofol binding site in GLIC
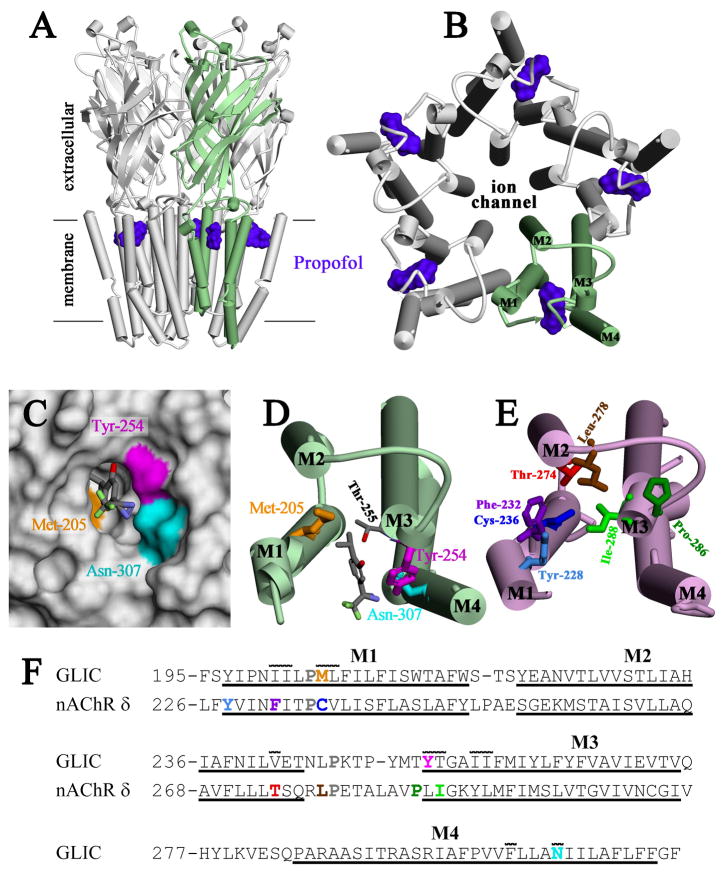
Propofol docking to one of the propofol binding sites in GLIC produced 1,300 lowest energy solutions, each reproducing the orientation of propofol in the PDB structure and differing only by rotations of < 50 ° of the external isopropyl group. CDocker interaction energies between GLIC and the propofol ranged between −24.7 and −26.3 kCal/mol. For AziPm docking to the same propofol binding site, 2,600 lowest energy solutions were collected with CDocker interaction energies between −24.5 and −26.3 kCal/mol. For all solutions, the diazirine nitrogens were positioned between the M1 and M4 helices, oriented towards lipid. In the lowest energy orientation, depicted in Figs. 4C & 4D and repeated in 2,212 of 2,600 solutions, the diazirine nitrogens were within ~3 Å of the hydroxyl of photolabeled Tyr-254 in M3 and the side chain amide nitrogen of photolabeled Asn-307 in M4. The hydroxyl oxygen of AziPm was 3.4 Å from the hydroxyl of Thr-255 (Fig. 4D), suggestive of a hydrogen bond as postulated by Nury et al. for desflurane (10).
Figure 4. Location of [3H]AziPm-photolabeled residues in the propofol binding site in GLIC and in the equivalent binding site in the Torpedo nAChR δ subunit.
Shown in panels A–D are views of the crystallographic model of GLIC co-crystalized with propofol (PDB:3P50) and in panel E a view of the nAChR δ subunit transmembrane domain (TMD) from an nAChR homology model derived from the GLIC structure (see Methods and Supporting Information Supplemental Figures S1 and S2). α-Helices are shown as cylinders and β-sheets as ribbons. A) A view of GLIC from the side and B) a view of the GLIC TMD from the base of the extracellular domain, with one subunit colored green. The positions of propofol within the structure are shown as Connolly surfaces (purple). C) Connolly surface representation of the propofol binding pocket, viewed from the lipid, with the surfaces contributed by the photolabeled residues color-coded: Tyr-254 (magenta), Asn-307 (cyan), and Met-205 (gold). The lowest energy docking solution for AziPm is included in stick format, color-coded by atom type: gray, carbon; red, oxygen; blue, nitrogen; light green, fluorine. D) A view of a single subunit’s TMD from the same perspective as in B, illustrating the position of the docked AziPm solution and the [3H]AziPm-photolabeled residues in stick format, color-coded as in C. The propofol/AziPm binding pocket resides between the M1, M3 and M4 helices. E) A view of the nAChR δ subunit TMD, oriented similar to GLIC in D. Residues in the δ subunit helix bundle pocket photolabeled by various photoreactive nAChR inhibitors are shown in stick format: [3H]AziPm Phe-232, Cys-236, & Thr-274 (20); [14C]halothane δTyr-228 (33); [125I]TID Ile-288, Phe-232, Cys-236, Thr-274, & Leu-278 (27,34,35); [3H]benzophenone Pro-286, Ile-288, & Phe-232 (28); [3H]azietomidate Cys-236 (41); and [3H]TFD-etomidate Phe-232 & Cys-236 (29). In contrast to the propofol/AziPm site in GLIC, the drug binding pocket in the nAChR δ subunit resides between the M1, M2, & M3 helices. F) The subunit primary structure alignment in the M1–M3 region used to make the nAChR homology model from the GLIC structure. The conserved prolines in all pLGICs are shown in gray. Also included is the M4 sequence from GLIC. The extent of the transmembrane helices is denoted by underlining. Photolabeled residues in GLIC and the Torpedo nAChR δ subunit are color-coded as in D & E. Residues in GLIC in contact with propofol in the crystal structure (10) have a line over them, illustrating the similarities between the intrasubunit pockets of GLIC and Torpedo nAChR δ subunit.
DISCUSSION
In this study we use AziPm, a photoreactive analog of propofol that inhibits pH-gated GLIC responses with a potency similar to that of propofol, to identify AziPm and propofol binding sites in GLIC in detergent solution. When GLIC was photolabeled with [3H]AziPm at pH 4.4, a condition that stabilizes open or desensitized states (30,31), we identified propofol-inhibitable photolabeling of three GLIC residues: Tyr-254 in M3, Asn-307 in M4, and Met-205 in M1. In the structure of GLIC co-crystallized with propofol (Fig. 4A–D), these three residues line the propofol binding pocket, with Tyr-254 (M3) and Asn-307 (M4) in contact with each other. AziPm docks to this same pocket (Figs. 4C & 4D) with similar interaction energies as propofol and in a preferred orientation with the diazirine ~3 Å from Tyr-254 and Asn-307, the most prominently photolabeled residues. Thus, our photolabeling results establish that AziPm and propofol bind to GLIC in solution at pH 4.4 at the same site that propofol binds to in the crystal structure. When in the native homopentameric form, GLIC has potentially five equivalent binding sites for propofol or AziPm. In a recent study (32), we used molecular dynamics simulations to compare the conformational transitions of GLIC under various states of propofol occupancy of these equivalent sites. The work concluded that asymmetric binding underlies the propofol functional inhibition of GLIC.
When [3H]AziPm was used to photolabel another pLGIC, the Torpedo nAChR in native membranes (20), propofol-inhibitable labeling was identified of amino acids contributing to two distinct binding sites in the TMD: (i) a site within the ion channel, which was photolabeled in the resting, closed channel state (−agonist) but not in the desensitized state (+agonist); and (ii) an intrasubunit site within the δ subunit helix bundle, homologous to the propofol binding site in GLIC, which was photolabeled in the nAChR desensitized state (+agonist), but not in the resting state. The inhibition of ion channel photolabeling may be allosteric, since propofol desensitizes the Torpedo nAChR in the absence of agonist. In the presence of agonist, however, propofol inhibition of photolabeling in the δ subunit helix bundle pocket (δPhe-232, δCys-236 (M1) and δThr-274 (M2), Figure 4E) appears competitive, indicating that both propofol (volume, 180 Å3) as well as AziPm (volume, 178 Å3) bind in the desensitized state to this intrasubunit site. Photoaffinity labeling studies have established that this pocket within the δ subunit helix bundle is a binding site for other small, hydrophobic nAChR inhibitors, including the volatile general anesthetic [14C]halothane (volume, 85 Å3) (33) and a drug related in structure to AziPm, [125I]TID (3-(trifluoromethyl)-3-(m-iodophenyl)diazirine (volume, 150 Å3)). Similar to AziPm, [125]TID photolabeled this site in the equilibrium desensitized state but not in the resting state (34). While GLIC and Torpedo nAChR have in common an intrasubunit helix bundle binding site for small hydrophobic inhibitors, the site in GLIC is located predominantly between M1, M3, & M4 helices whereas the site in the nAChR is between the M1, M2, and M3 helices (Fig 4D & 4E).
While we also found evidence of [3H]AziPm photolabeling at lower efficiency of amino acids in M3 (Tyr-263 and Tyr-266) that contribute to the pocket at the interface between subunits, that photolabeling was not inhibited by propofol at 300 μM. Thus, we found no evidence in our photolabeling study that propofol binds to an intersubunit site in GLIC in detergent at pH 4.4 equivalent to the intersubunit GLIC propofol binding site predicted based upon propofol’s protection at pH 8 of modification of the GLIC L241C mutant expressed in Xenopus oocytes (14). Since in that study it was also shown that propofol enhanced the rate of modification of a Cys substituted for Thr-255, which projects within the intrasubunit propofol binding pocket in the crystal structure adjacent to our photolabeled Tyr-254 in M3 (Fig. 4D), further studies are required to determine whether propofol binds preferentially in GLIC in detergent to an intrasubunit site at pH 4.4 (open/desensitized states) but to an intersubunit site at pH 8 (resting, closed channel state). Strong state-dependence has been seen for drug binding to the homologous intrasubunit binding site in the δ subunit of the Torpedo nAChR in native membranes, which was not photolabeled by [125I]TID in the resting state, and was photolabeled in the transient open/ fast desensitized states at >20-fold higher efficiency than in the equilibrium desensitized state (27,35). However, recent electron spin resonance studies found evidence of a pH-dependent change of structure of purified GLIC reincorporated into liposomes but not in detergent solution (36).
While propofol and AziPm, inhibitors of GLIC and the nAChR, bind to equivalent intrasubunit binding sites in these pLGICs, these anesthetics are unlikely to bind to equivalent sites in GABAARs when they potentiate GABA responses. Based upon the locations in early GABAAR structural models of the GABAAR amino acids in the β subunit M1 and M3 helices that were identified by mutational analyses as propofol and etomidate sensitivity determinants, these anesthetics were proposed to bind to an intrasubunit site within the β subunit that would be analogous to the propofol binding site in GLIC (reviewed in (37)). However, in more robust GABAAR structural models that can now be derived from the structures of GLIC or GluCl, these β subunit anesthetic sensitivity determinants are located within an intersubunit binding site at the interface between β and α subunits (17,38). Identification of GABAAR general anesthetic binding sites by use of photoreactive etomidate and barbiturate analogs has established that there are two classes of intersubunit anesthetic binding sites in the αβγ GABAAR transmembrane domain, one at the β+−α− subunit interfaces that contain the GABA binding site in the extracellular domain, and the second at the α+ −β− and γ+−β− interfaces (16,18). While etomidate binds selectively to the β+α−; sites and some barbiturates bind selectively to the α+/γ+−β− sites, propofol binds to both sites with similar affinity, based upon the concentration dependence of propofol inhibition of photolabeling (18). Direct photolabeling of GABAARs with [3H]AziPm or other recently developed photoreactive propofol analogs (39,40) will be necessary to determine whether propofol also binds to intrasubunit sites in GABAAR equivalent to the propofol site in GLIC.
Supplementary Material
The abbreviations used are
- pLGIC
pentameric ligand-gated ion channel
- GLIC
Gloeobacter violaceus ligand-gated ion channel
- AziPm
2-isopropyl-5-[3-(trifluoromethyl)-3H-diazirin-3-yl]phenol
- nAChR
nicotinic acetylcholine receptor
- GABAAR
γ-aminobutyric acid type-A receptor
- PPF
propofol
- OPA
o-phthalaldehyde
- EndoLys-C
Lysobacter enzymogenes endoproteinase Lys-C
- TMD
transmembrane domain
Footnotes
This research was supported in part by USPHS grants GM-06358 (PT), GM-58448 (JBC), and GM-55876 (RGE).
In this report we use the numbering of amino acids in GLIC primary structure that is used in the GLIC crystal structure (10), with the first 3 residues (GPM) in our subunit omitted.
After treatment with OPA in cycle 5, there was also a secondary sequence of the Thr-65 fragment which contains Pro-68 in cycle 4 that was present at low level in cycle 5 due to the finite repetitive yield of Edman degradation. The 3H release in cycle 27 was not associated with Val-90 from this peptide, since no 3H release was detected in cycle 26 when the digest was sequenced without OPA (data not shown).
CONFLICT OF INTEREST DISCLOSURE: The authors declare no competing financial interest.
SUPPORTING INFORMATION: Two, 3-panel stereo images of pLGIC subunit domains are included that show the structures of GLIC (PDB:3P50) and the nAChR δ subunit in a GLIC-derived homology model compared with the nAChR cryoelectron microscopy structure (PDB:2BG9). The AziPm photolabeled residues in GLIC and the nAChR δ subunit are indicated, as well as additional nAChR δ subunit residues photolabeled by nicotinic antagonists in this helix bundle binding site. One table of representative distances between photolabeled residues in the two nAChR structures is included. These comparisons indicate that a GLIC-derived homology model of the nAChR is more consistent with the photolabeling results than the nAChR cryoelectron microscopy structure.
This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Krasowski MD, Jenkins A, Flood P, Kung AY, Hopfinger AJ, Harrison NL. General anesthetic potencies of a series of propofol analogs correlate with potency for potentiation of gamma-aminobutyric acid (GABA) current at the GABA(A) receptor but not with lipid solubility. J Pharmacol Exp Ther. 2001;297:338–351. [PubMed] [Google Scholar]
- 2.Rudolph U, Antkowiak B. Molecular and neuronal substrates for general anaesthetics. Nat Rev Neurosci. 2004;5:709–720. doi: 10.1038/nrn1496. [DOI] [PubMed] [Google Scholar]
- 3.Franks NP. General anaesthesia: from molecular targets to neuronal pathways of sleep and arousal. Nat Rev Neurosci. 2008;9:370–386. doi: 10.1038/nrn2372. [DOI] [PubMed] [Google Scholar]
- 4.Dilger JP, Vidal AM, Mody HI, Liu Y. Evidence for direct actions of general anesthetics on an ion channel protein - A new look at a unified mechanism of action. Anesthesiology. 1994;81:431–442. doi: 10.1097/00000542-199408000-00022. [DOI] [PubMed] [Google Scholar]
- 5.Violet JM, Downie DL, Nakisa RC, Lieb WR, Franks NP. Differential sensitivities of mammalian neuronal and muscle nicotinic acetylcholine receptors to general anesthetics. Anesthesiology. 1997;86:866–874. doi: 10.1097/00000542-199704000-00017. [DOI] [PubMed] [Google Scholar]
- 6.Flood P, Ramirezlatorre J, Role L. α4β2 neuronal nicotinic acetylcholine receptors in the central nervous system are inhibited by isoflurane and propofol, but α7-type nicotinic acetylcholine receptors are unaffected. Anesthesiology. 1997;86:859–865. doi: 10.1097/00000542-199704000-00016. [DOI] [PubMed] [Google Scholar]
- 7.Weng Y, Yang LY, Corringer PJ, Sonner JM. Anesthetic Sensitivity of the Gloeobacter violaceus Proton-Gated Ion Channel. Anesth Analg. 2010;110:59–63. doi: 10.1213/ANE.0b013e3181c4bc69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4 Å resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 9.Baenziger JE, Corringer PJ. 3D structure and allosteric modulation of the transmembrane domain of pentameric ligand-gated ion channels. Neuropharmacology. 2011;60:116–125. doi: 10.1016/j.neuropharm.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Nury H, Van Renterghem C, Weng Y, Tran A, Baaden M, Dufresne V, Changeux JP, Sonner JM, Delarue M, Corringer PJ. X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel. Nature. 2011;469:428–431. doi: 10.1038/nature09647. [DOI] [PubMed] [Google Scholar]
- 11.Sauguet L, Howard RJ, Malherbe L, Lee US, Corringer PJ, Adron Harris R, Delarue M. Structural basis for potentiation by alcohols and anaesthetics in a ligand-gated ion channel. Nat Commun. 2013;4:1697. doi: 10.1038/ncomms2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan JJ, Chen Q, Willenbring D, Mowrey D, Kong XP, Cohen A, Divito CB, Xu Y, Tang P. Structure of the Pentameric Ligand-Gated Ion Channel GLIC Bound with Anesthetic Ketamine. Structure. 2012;20:1463–1469. doi: 10.1016/j.str.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spurny R, Billen B, Howard RJ, Brams M, Debaveye S, Price KL, Weston DA, Strelkov SV, Tytgat J, Bertrand S, Bertrand D, Lummis SCR, Ulens C. Multisite Binding of a General Anesthetic to the Prokaryotic Pentameric Erwinia chrysanthemi Ligand-gated Ion Channel (ELIC) J Biol Chem. 2013;288:8355–8364. doi: 10.1074/jbc.M112.424507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh B, Satyshur KA, Czajkowski C. Propofol Binding to the Resting State of the Gloeobacter violaceus Ligand-gated Ion Channel (GLIC) Induces Structural Changes in the Inter- and Intrasubunit Transmembrane Domain (TMD) Cavities. J Biol Chem. 2013;288:17420–17431. doi: 10.1074/jbc.M113.464040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vodovozova EL. Photoaffinity labeling and its application in structural biology. Biochemistry-Moscow. 2007;72:1–20. doi: 10.1134/s0006297907010014. [DOI] [PubMed] [Google Scholar]
- 16.Li G-D, Chiara DC, Sawyer GW, Husain SS, Olsen RW, Cohen JB. Identification of a GABAA receptor anesthetic binding site at subunit interfaces by photolabeling with an etomidate analog. J Neurosci. 2006;26:11599–11605. doi: 10.1523/JNEUROSCI.3467-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiara DC, Dostalova Z, Jayakar SS, Zhou X, Miller KW, Cohen JB. Mapping General Anesthetic Binding Site(s) in Human α1β3 γ-Aminobutyric Acid Type A Receptors with [3H]TDBzl-Etomidate, a Photoreactive Etomidate Analogue. Biochemistry. 2012;51:836–847. doi: 10.1021/bi201772m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiara DC, Jayakar SS, Zhou X, Zhang X, Savechenkov PY, Bruzik KS, Miller KW, Cohen JB. Specificity of Intersubunit General Anesthetic-binding Sites in the Transmembrane Domain of the Human α1β3γ2 γ-Aminobutyric Acid Type A (GABAA) Receptor. J Biol Chem. 2013;288:19343–19357. doi: 10.1074/jbc.M113.479725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall MA, Xi J, Lor C, Dai S, Pearce R, Dailey WP, Eckenhoff RG. m-Azipropofol (AziPm) a Photoactive Analogue of the Intravenous General Anesthetic Propofol. J Med Chem. 2010;53:5667–5675. doi: 10.1021/jm1004072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jayakar SS, Dailey WP, Eckenhoff RG, Cohen JB. Identification of Propofol Binding Sites in a Nicotinic Acetylcholine Receptor with a Photoreactive Propofol Analog. J Biol Chem. 2013;288:6178–6189. doi: 10.1074/jbc.M112.435909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bocquet N, de Carvalho LP, Cartaud J, Neyton J, Le Poupon C, Taly A, Grutter T, Changeux J–P, Corringer P–J. A prokaryotic proton-gated ion channel from the nicotinic acetylcholine receptor family. Nature. 2007 Jan 4;445:116–119. doi: 10.1038/nature05371. [DOI] [PubMed] [Google Scholar]
- 22.Chen Q, Cheng MH, Xu Y, Tang P. Anesthetic Binding in a Pentameric Ligand-Gated Ion Channel: GLIC. Biophys J. 2010;99:1801–1809. doi: 10.1016/j.bpj.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crimmins DL, McCourt DW, Thoma RS, Scott MG, Macke K, Schwartz BD. In situ chemical cleavage of proteins immobilized to glass-fiber and polyvinylidenedifluoride membranes: Cleavage at tryptophan residues with 2-(2′-nitrophenylsulfenyl)-3-methyl-3′-bromoindolenine to obtain internal amino acid sequence. Anal Biochem. 1990;187:27–38. doi: 10.1016/0003-2697(90)90412-3. [DOI] [PubMed] [Google Scholar]
- 24.Brauer AW, Oman CL, Margolies MN. Use of o-phthalaldehyde to reduce background during automated Edman degradation. Anal Biochem. 1984;137:134–142. doi: 10.1016/0003-2697(84)90359-2. [DOI] [PubMed] [Google Scholar]
- 25.Middleton RE, Cohen JB. Mapping of the acetylcholine binding site of the nicotinic acetylcholine receptor: [3H]-nicotine as an agonist photoaffinity label. Biochemistry. 1991;30:6987–6997. doi: 10.1021/bi00242a026. [DOI] [PubMed] [Google Scholar]
- 26.Bondarenko V, Mowrey D, Tillman T, Cui T, Liu LT, Xu Y, Tang P. NMR structures of the transmembrane domains of the α4β2 nAChR. Biochim Biophys Acta. 2012;1818:1261–1268. doi: 10.1016/j.bbamem.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arevalo E, Chiara DC, Forman SA, Cohen JB, Miller KW. Gating-enhanced accessibility of hydrophobic sites within the transmembrane region of the nicotinic acetylcholine receptor’s δ-subunit - A time-resolved photolabeling study. J Biol Chem. 2005;280:13631–13640. doi: 10.1074/jbc.M413911200. [DOI] [PubMed] [Google Scholar]
- 28.Garcia G, III, Chiara DC, Nirthanan S, Hamouda AK, Stewart DS, Cohen JB. [3H]Benzophenone photolabeling identifies state-dependent changes in nicotinic acetylcholine receptor structure. Biochemistry. 2007;46:10296–10307. doi: 10.1021/bi7008163. [DOI] [PubMed] [Google Scholar]
- 29.Hamouda AK, Stewart DS, Husain SS, Cohen JB. Multiple Transmembrane Binding Sites for p-Trifluoromethyldiazirinyl-etomidate, a Photoreactive Torpedo Nicotinic Acetylcholine Receptor Allosteric Inhibitor. J Biol Chem. 2011;286:20466–20477. doi: 10.1074/jbc.M111.219071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Velisetty P, Chakrapani S. Desensitization Mechanism in Prokaryotic Ligand-gated Ion Channel. J Biol Chem. 2012;287:18467–18477. doi: 10.1074/jbc.M112.348045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velisetty P, Chalamalasetti SV, Chakrapani S. Conformational Transitions Underlying Pore Opening and Desensitization in Membrane-embedded Gloeobacter violaceus Ligand-gated Ion Channel (GLIC) J Biol Chem. 2012;287:36864–36872. doi: 10.1074/jbc.M112.401067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mowrey D, Cheng MH, Liu LT, Willenbring D, Lu XH, Wymore T, Xu Y, Tang P. Asymmetric Ligand Binding Facilitates Conformational Transitions in Pentameric Ligand-Gated Ion Channels. JACS. 2013;135:2172–2180. doi: 10.1021/ja307275v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiara DC, Dangott LJ, Eckenhoff RG, Cohen JB. Identification of nicotinic acetylcholine receptor amino acids photolabeled by the volatile anesthetic halothane. Biochemistry. 2003;42:13457–13467. doi: 10.1021/bi0351561. [DOI] [PubMed] [Google Scholar]
- 34.Hamouda AK, Chiara DC, Blanton MP, Cohen JB. Probing the structure of the affinity-purified and lipid-reconstituted Torpedo nicotinic acetylcholine receptor. Biochemistry. 2008;47:12787–12794. doi: 10.1021/bi801476j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamodo IH, Chiara DC, Cohen JB, Miller KW. Conformational changes in the nicotinic acetylcholine receptor during gating and desensitization. Biochemistry. 2010;49:156–165. doi: 10.1021/bi901550p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dellisanti CD, Ghosh B, Hanson SM, Raspanti JM, Grant VA, Diarra GM, Schuh AM, Satyshur K, Klug CS, Czajkowski C. Site-directed spin labeling reveals pentameric ligand-gated ion channel gating motions. PLOS Biology. 2013;11:e1001714. doi: 10.1371/journal.pbio.1001714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hemmings HC, Akabas MH, Goldstein PA, Trudell JR, Orser BA, Harrison NL. Emerging molecular mechanisms of general anesthetic action. Trends Pharmacol Sci. 2005;26:503–510. doi: 10.1016/j.tips.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Bertaccini EJ, Yoluk O, Lindahl ER, Trudell JR. Assessment of Homology Templates and an Anesthetic Binding Site within the γ-Aminobutyric Acid Receptor. Anesthesiology. 2013;119:1087–1095. doi: 10.1097/ALN.0b013e31829e47e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart DS, Savechenkov PY, Dostalova, Chiara DC, Ge R, Raines DE, Cohen JB, Forman SA, Bruzik KS, Miller KW. p-(4-Azipentyl)propofol: A Potent Photoreactive General Anesthetic Derivative of Propofol. J Med Chem. 2011;54:8124–8135. doi: 10.1021/jm200943f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yip GMS, Chen ZW, Edge CJ, Smith EH, Dickinson R, Hohenester E, Townsend RR, Fuchs K, Sieghart W, Evers AS, Franks NP. A propofol binding site on mammalian GABAA receptors identified by photolabeling. Nat Chem Biol. 2013;9:715–720. doi: 10.1038/nchembio.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiara DC, Hong FH, Arevalo E, Husain SS, Miller KW, Forman SA, Cohen JB. Time-resolved photolabeling of the nicotinic acetylcholine receptor by [3H]Azietomidate, an open-state inhibitor. Mol Pharmacol. 2009;75:1084–1095. doi: 10.1124/mol.108.054353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.