Abstract
Objective
Our objective was to execute a prospective cohort study to determine relationships between plasma mtDNA DAMP levels and the occurrence of systemic inflammatory response syndrome (SIRS), multiple organ dysfunction syndrome (MODS), and mortality.
Background
Mitochondrial DNA damage-associated molecular patterns (DAMPs) accumulate in the circulation after severe injury. Observations in animal models demonstrate that mtDNA DAMPs contribute to organ dys-function; however, the link between plasma mtDNA DAMPs and outcome in severely injured human subjects has not been established.
Methods
DNA was isolated from plasma samples taken from severely injured patients at hospital days 0, 1, and 2. Real-time PCR was used to quantify selected ≈200 base pair sequences of mtDNA within the COX1, ND1, and ND6 genes, as well as from the D-Loop transcriptional regulatory region. MODS was defined as a Denver Multiple Organ Failure score of 4 or greater.
Results
MtDNA DAMPs were quantified as PCR threshold cycle number. Lower threshold cycles indicate increased mtDNA DAMP content. Patients with SIRS had significantly increased mtDNA DAMP levels in all 4 sequences examined (32.14 ± 0.90 vs 29.00 ± 1.15 for COX1, 31.90 ± 0.47 vs 30.16 ± 1.42 for ND1, 32.40 ± 0.61 vs 28.94 ± 1.13 for ND6, and 33.12 ± 0.83 vs 28.30 ± 1.14 for D-Loop). Patients who developed MODS also had elevated mtDNA DAMP levels compared with those who did not (32.57 ± 0.74 vs 27.12 ± 0.66 for COX1, 32.45 ± 0.65 vs 28.20 ± 0.73 for ND1, 32.52 ± 0.56 vs 27.60 ± 0.79 for ND6, and 32.85 ± 0.75 vs 27.86 ± 1.27 for D-Loop). Patients with above-median mtDNA DAMP levels had a significantly elevated relative risk for mortality. Four patients died secondary to severe MODS.
Conclusions
These findings comprise the first observational evidence that plasma mtDNA DAMPs is associated with the evolution of SIRS, MODS, and mortality in severely injured human subjects.
Keywords: damage-associated molecular patterns, DAMPs, multiple organ dysfunction syndrome, SIRS, systemic inflammatory response syndrome, trauma
Multiple organ dysfunction syndrome (MODS) is a common and potentially fatal consequence of severe injury. Although mitochondrial and bioenergetic dysfunction have been incriminated in the pathogenesis of MODS for many years,1–6 involvement of mitochondria-derived mediators may be more dynamic than previously appreciated. In this regard, fragments of the mitochondrial genome released into the circulation after injury—termed mtDNA damage-associated molecular patterns (DAMPs)—have been postulated to function as intercellular signals propagating damage from the initial site of injury to distant organs through Toll-like-receptor– mediated activation of inflammatory and resident cells.7,8 The evidence incriminating mtDNA DAMPs in MODS is derived from cell culture studies, isolated perfused organ, and intact animal models and from indirect observations in human subjects.7,9–11 In the latter context, although it is known that mtDNA DAMPs can be mobilized by long bone fracture or reparative operative procedures (eg, femoral reaming), and that isolated mtDNA DAMPs given to intact animals or to immune cells in vitro recapitulate many of the attributes of the systemic inflammatory response syndrome,7 the association between mtDNA DAMPs and human MODS remains largely speculative. Accordingly, to provide evidence linking mtDNA DAMPs to MODS we executed a prospective cohort study to determine the relationships between plasma mtDNA DAMP levels and the occurrence of MODS, SIRS, and mortality in severely injured human patients.
METHODS
This prospective cohort study approved by institutional review board enrolled 14 consecutive severely injured patients who were admitted to the surgical trauma intensive care unit (ICU) at the University of South Alabama Medical Center from August 2012 to October 2012 (ClinicalTrials.gov; NCT 01812941). Study enrollment criteria consisted of any patient admitted secondary to a traumatic mechanism of injury with an injury severity score (ISS) of greater than 15, age more than 18 years, and nonpregnant. DNA was isolated from 8 mL of peripheral blood, which was originally placed into BD Vacutainer CPT tubes with Sodium Citrate (Becton, Dickinson and Company, NJ). These blood samples were taken at hospital days 0 (within 8 hours of admission), 1, 2, and 6. Clinical variables were recorded at the time of blood draw; laboratory variables were obtained at the discretion of the attending clinicians within 4 hours of blood sampling for mtDNA DAMP analyses.
Samples were centrifuged within 1 hour of collection at 1200 RCF for 25 minutes at 21°C, and 200 μL of the plasma fraction was decanted and processed using the Qiagen DNEasy kit to isolate DNA (Qiagen, Inc, CA). Quantitative real time polymerase chain reaction (qPCR), using USB VeriQuest Fast SYBR Green qPRC Master Mix (Affymetrix, Inc., Santa Clara, CA) and the manufacturer's protocol, was applied to quantify selected ~200 bp sequences corresponding to the COX1, ND1, ND6, and D-Loop mitochondrial genomic regions as previously described by our group.12,13 Primers for qRT-PCR analyses of the indicated mtDNA sequences are listed in Table 1. The relative abundances of plasma mtDNA DAMPs were expressed as threshold cycles (Tc). Of note, higher Tc represent lower levels of mtDNA DAMPs. As negative controls, we verified that the selected mtDNA sequences were below detectable limits in solutions used in the assay and in plasma from control, noninjured human subjects, as previously reported.7
TABLE 1.
Primers for qPCR Detection of Plasma mtDNA Sequences
| Mt DNA Sequence | Forward (F) and Reverse (R) Primer |
|---|---|
| COX1 | F: TCA TCT GTA GGC TCA TTC |
| R: GCG ATC CAT ATA GTC ACT | |
| ND1 | F: GCT ACG ACC AAC TCA TAC |
| R: GAA TGC TGG AGA TTG TAA TG | |
| ND6 | F: CCA TCG CTG TAG TAT ATC CAA |
| R: TCG GGT GTG TTA TTA TTC TGA | |
| D-Loop | F: ATC AAC CTT CAA CTA TCA |
| R: ACT GTA ATG TGC TAT GTA |
Mt indicates mitochondrial.
The primary outcome variable was death secondary to MODS. Secondary outcomes included MODS and SIRS. MODS was defined as a Denver Postinjury Multiple Organ Failure Score of 4 or greater.14 This scoring system rates the dysfunction of 4 organ systems (pulmonary, renal, hepatic, and cardiovascular), which are evaluated daily throughout the patient's ICU stay. SIRS was defined by the American College of Chest Physicians (ACCP)/Society of Critical Care Medicine (SCCM) definition.15
The data were statistically evaluated using several strategies. The mtDNA DAMP levels, as a continuous variable, were compared to the presence of MODS, SIRS, and death as a categorical variable. A relative risk analysis was derived to determine the association between high mtDNA DAMPs (using the median Tc level) and MODS. Statistically significant differences (P < 0.05) were detected by nonparametric analyses.
RESULTS
There were 14 consecutive patients enrolled during the study period. One patient was excluded due to a moribund presentation and rapid determination of clinical brain death secondary to an isolated traumatic brain injury. Ten patients were enrolled after blunt trauma and 2 after severe burns. Seven enrolled patients were found to have a severe traumatic brain injury, of which 3 required initial craniotomy. Individual patient characteristics are noted in Table 2. Average age and ISS were 39.1 ± 5.1 and 21.1 ± 1.7, respectively. There were no statistically significant differences in age or ISS with regard to outcomes assessed in terms of MODS/death (P = 0.12 and 0.26, respectively) or SIRS (P = 0.72 and 0.22, respectively).
TABLE 2.
Individual Patient Characteristics
| Patient | Age, y | Gender | Mechanism | ISS | SIRS* | Highest Denver† | Mortality |
|---|---|---|---|---|---|---|---|
| 1 | 41 | M | MVC | 17 | Yes | 2 | No |
| 2 | 82 | F | 34% Burn | 16 | No | 4 | Yes |
| 3 | 39 | M | Assault | 38 | Yes | 8 | Yes |
| 4 | 39 | M | MVC | 22 | No | 2 | No |
| 5 | 59 | M | 50% Burn | 25 | Yes | 2 | No |
| 6 | 56 | M | MCC | 22 | No | 2 | No |
| 7 | 21 | M | MCC | 21 | No | 1 | No |
| 8 | 21 | M | MCC | 16 | No | 2 | No |
| 9 | 26 | F | MVC | 17 | No | 0 | No |
| 10 | 36 | M | MCC | 25 | No | 6 | Yes |
| 11 | 20 | M | Fall | 16 | No | 1 | No |
| 12 | 21 | M | MCC | 22 | Yes | 0 | No |
| 13 | 48 | M | MVC | 17 | Yes | 4 | Yes |
| 14‡ | 20 | M | GSW | 25 | Yes | 3 | Yes |
Meeting ≥2 SIRS criteria by ACCP/SCCM definitions.
A Denver MOF score ≥4 at any time during the study was defined as MODS.
Excluded from study due to a moribund presentation and rapid determination of clinical brain death.
GSW indicates gunshot wound; MCC, motorcycle collision; MVC, motor vehicle collision.
Mortality and Multiple Organ System Dysfunction
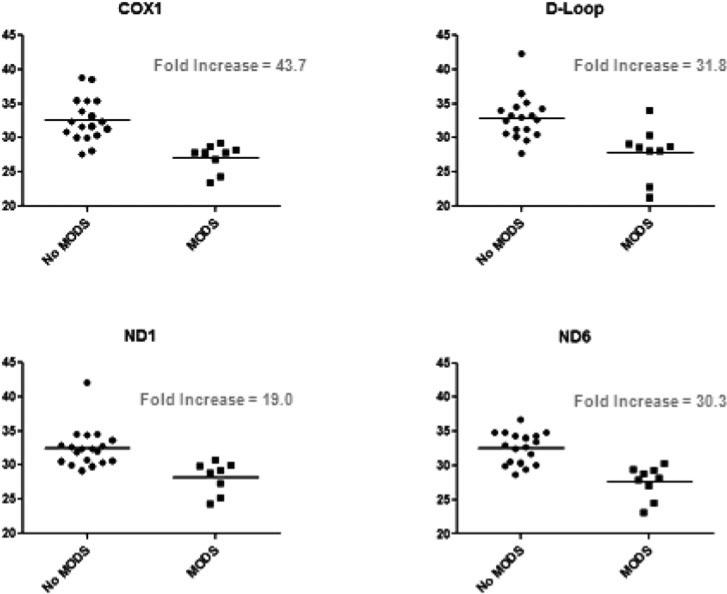
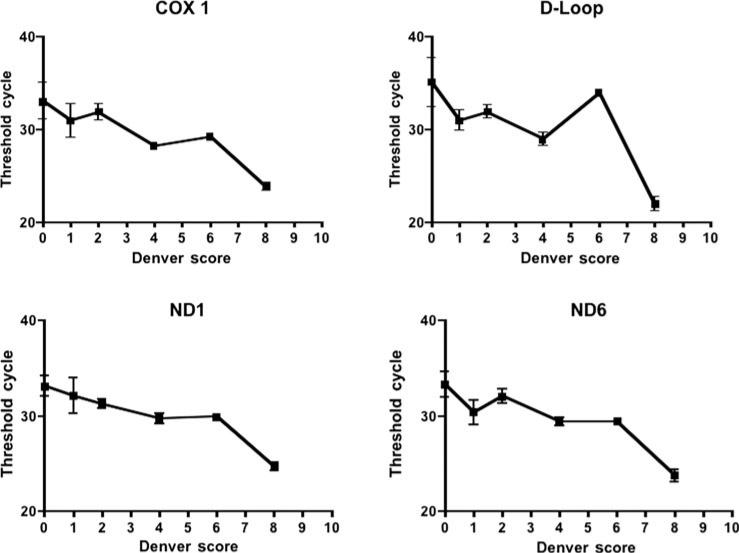
There were 4 patients in our cohort who developed MODS, and these 4 patients were the only nonsurvivors. Nonsurvivors had significantly higher mtDNA DAMPs than survivors (Table 3 and Fig. 1). The relative risk of MODS and death was significantly higher in patients displaying increases in any of the mtDNA sequences measured (Table 3). Furthermore, as shown in Figure 2, there were intriguing trends associating maximum Denver Multiple Organ Failure scores and mtDNA DAMP levels for all mtDNA sequences studied. Finally, survivors and nonsurvivors did not differ in terms of ISS, worst base deficit, blunt trauma, burn trauma, or number of transfused blood products (Table 4).
TABLE 3.
Associations of Plasma mtDNA DAMP Levels with SIRS, MODS, and Mortality*
| MtDNA DAMP | No SIRS Tc (n = 8) | SIRS Tc (n = 5) | Fold Increase | No MODS Tc (n = 9) | MODS Tc (n = 4) | Fold Increase | Mortality Relative Risk (95% CI) |
|---|---|---|---|---|---|---|---|
| COX1 | 32.1 ± 0.9 | 29.0 ± 1.2 | 8.8† | 32.6 ± 0.7 | 27.1 ± 0.7 | 43.7† | 20.4† (1.3–318) |
| D-Loop | 33.1 ± 0.8 | 28.3 ± 1.1 | 28.3† | 32.9 ± 0.8 | 27.9 ± 1.3 | 31.8† | 8.0† (1.16–55.2) |
| ND1 | 31.9 ± 0.5 | 30.2 ± 1.4 | 3.3† | 32.5 ± 0.7 | 28.2 ± 0.7 | 19.0† | 8.0† (1.15–55.8) |
| ND6 | 32.4 ± 0.6 | 28.9 ± 1.1 | 11.0† | 32.5 ± 0.5 | 27.6 ± 0.8 | 30.3† | 20.4† (1.3–318) |
MtDNA DAMP levels for 4 sequences of interest were examined, segregating patients by the presence of SIRS and MODS, with statistically significant differences in each sequence. As each PCR cycle represents a doubling of DNA quantity, fold increase thus describes the relative starting quantity of mtDNA DAMPs in patients with each syndrome versus those without. Mortality relative risk describes the increased risk of mortality for having a below-median Tc; in this cohort, the median Tc ≈ 30 for each mtDNA DAMP examined.
Day 6 data excluded to minimize survivor bias.
Statistically significant difference at P < 0.05 by nonparametric analyses.
FIGURE 1.
Associations of Plasma mtDNA DAMP Levels with MODS/Mortality. There were only 4 patients in our cohort who developed MODS, and these 4 patients were the only nonsurvivors. DAMP levels were significantly elevated in patients who developed MODS/Death when compared to survivors. All four sequences exhibited statistically significant differences at P < 0.05 by nonpara-metric analyses. Day 6 data were excluded to minimize survivor bias. The relative abundances of plasma mtDNA DAMPs were expressed as threshold cycles (Tc). Of note, higher Tc represents lower levels of mtDNA DAMPs.
FIGURE 2.
Serum mtDNA DAMP levels (quantified as the PCR Tc) expressed as a function of the Denver Organ Failure Score. For all 4 mtDNA sequences examined, in patients with greater Denver scores, Tc tended to be lower indicating greater amounts of circulating mtDNA DAMPs.
TABLE 4.
Group Characteristics Between Survivors and Nonsurvivors
| Survivor (n = 9) | Nonsurvivor (n = 4) | P | |
|---|---|---|---|
| Male | 8 | 3 | 1.0 |
| ISS | 19.8 ± 1.1 | 24 ± 5.1 | 0.27 |
| Worst base deficit* | 4.4 ± 0.64 | 8.49 ± 3.45 | 0.11 |
| Blunt trauma | 7 | 3 | 1.0 |
| Traumatic brain injury | 6 | 1 | 0.27 |
| Burn | 1 | 1 | 1.0 |
| Blood products* | 3.89 ± 2.3 | 18.75 ± 14.9 | 0.17 |
No statistically significant differences were noted in gender, injury severity, worst base deficit, mechanism, presence of traumatic brain injury, transfusion requirement, or presence of SIRS. Transfusion requirements are described as total units of transfused products, all types; no statistically significant differences were noted when analyzed by individual product types (not shown). P values determined by unpaired t test or Fisher exact test as appropriate for variable type.
Blood products include packed red blood cells, platelets, and fresh frozen plasma.
In first 24 hrs.
Systemic Inflammatory Response Syndrome
Five patients demonstrated SIRS by ACCP/SCCM definitions within 48 hours of presentation. Patients meeting SIRS criteria had significantly higher plasma levels of all mtDNA sequences measured than those who failed to meet SIRS criteria (Table 3). No relationship was noted between presence of SIRS and the development of MODS/death.
Serum mtDNA DAMP Levels and Kinetics
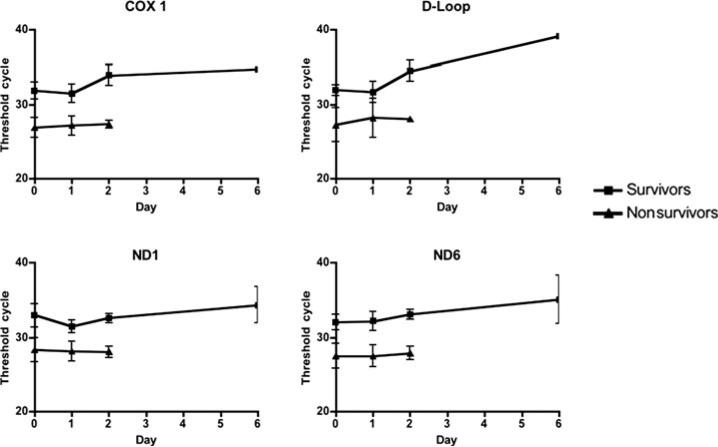
Plasma mtDNA DAMP levels were quantified as PCR Tc, with each cycle doubling DNA quantity until a detection threshold is achieved. When pooled according to outcome, plasma mtDNA DAMP levels did not significantly change over time in either survivors or nonsurvivors (Fig. 3). A Tc greater than 35 is considered below the limits of detection of template DNA; therefore, we did not statistically analyze changes above this level. Patients with average Tc greater than 30 after 24 hours did not develop MODS/Death. Likewise, patients with average Tc less than 30 beyond 24 hours all developed MODS and died. All 4 patients who developed MODS and died had less than 30 Tc for all time points.
FIGURE 3.
Serum mtDNA DAMP levels displayed as a function of time after hospital admission. The number of PCR Tc, and thus mtDNA DAMP levels, did not significantly change during patients’ hospital stays for both survivors and nonsurvivors. Tc greater than 35 are considered equivalent and indicative of negligible mtDNA content for sequences of interest. All mortality occurred before day 6 in the present cohort.
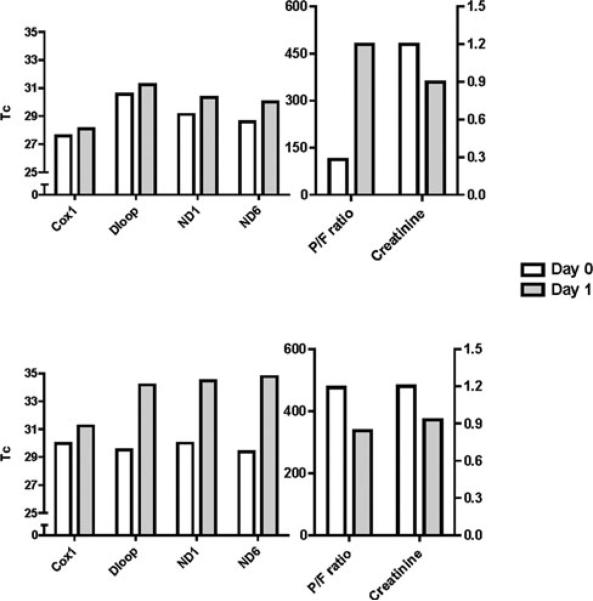
Despite the lack of time-dependent changes in levels of pooled mtDNA DAMP sequences, 2 individual patients displayed conspicuous time courses of mtDNA DAMP levels relative to clinical outcome. Both had initial Tc values very close to the median Tc for all patients enrolled in the study, Tc ≈ 30. In contrast to the majority of patients in which Tc values were constant over the clinical courses, Tc values for these 2 patients increased to more than 30 Tc within 24 hours; neither developed MODS nor died (Fig. 4). One of these patients displayed dramatic improvement in creatinine and P/F ratio whereas the other had improvement in serum creatinine but exhibited normal P/F ratios over the time period.
FIGURE 4.
Tc and clinical markers for 2 survivors with initially elevated serum mtDNA DAMPs. Two patients who presented with relatively greater plasma mtDNA DAMP levels among survivors improved clinically with concomitant decreases in mtDNA DAMPs; one patient (top) noted a modest normalization of serum creatinine and a dramatic improvement in pulmonary function, whereas a second patient (bottom) revealed a modest improvement in creatinine and maintenance of a normal P/F with subsequent uneventful removal from mechanical ventilation.
DISCUSSION
Rapidly emerging evidence suggests that DAMPs originating from damaged mitochondria and released into the circulation after traumatic injury contribute to the evolution of MODS.8 Included on the list of mitochondrial derived DAMPs are formylated peptides, the mtDNA-binding protein and transcriptional regulator mitochondrial transcription factor A (TFAM), and mtDNA itself.8 Mitochondrial DNA DAMPs are particularly interesting for several reasons. First, given the widespread recognition that oxidant stress is a key pathophysiologic component of severe trauma and illness, it is noteworthy that DNA in the mitochondrial genome is considerably more sensitive to oxidative damage than nuclear DNA.16–18 Oxidative damage to the sugar backbone of mtDNA leads to mtDNA “degradation,”19 for which the fate of degraded DNA is poorly understood. Second, Hauser and colleagues demonstrated in severely injured patients that plasma mtDNA sequences increased about 3500-fold in comparison with normal control subjects. When mtDNA fragments were administered to intact rats, the ensuing inflammatory response recapitulates many elements of SIRS in human subjects.7,20,21 Fragments of mtDNA also caused the TLR-9-dependent activation of neutrophils.7,8,11,22 Finally and most recently, it has been demonstrated that mtDNA DAMPs erode barrier integrity in cultured endothelial cells,9 thus mimicking another key pathophysiologic event in MODS. Although collectively these findings incriminate mtDNA DAMPs in trauma-related MODS, observational evidence for such an association in a severely injured human patient cohort has been lacking.
The current study presents an interim analysis of the relation between mtDNA DAMP levels and outcome of severe injury, defined as an ISS of more than 15, in a comparatively small number of patients. In light of the limitations associated with the small size of the cohort analyzed, we were surprised to find that plasma levels of 4 selected mtDNA sequences, corresponding to mtDNA-encoded COX1, ND1, ND6 proteins and the D-Loop transcriptional regulatory region, were elevated in severely injured patients with SIRS and MODS relative to patients who did not develop these syndromes. The relative risk of mortality also was greater in patients with elevated circulating mtDNA DAMPs. Perhaps even more striking, the disparity in mtDNA DAMP levels distinguishing survivors from those who ultimately developed MODS and died was evident in the first plasma sample taken after admission. These findings are the first to describe an association between circulating mtDNA DAMPs and outcome in severely injured human subjects, and support the concept that mtDNA DAMPs function to propagate injury from the initial site to distant organ dysfunction in severely injured patients.
One of the most interesting observations reported herein was that the first determination of mtDNA DAMPs levels, made within 8 hours of hospital admission, distinguished between survivors and nonsurvivors. These findings should be viewed in light of the fact that mtDNA DAMPs are one of an expanding number of biomarkers associated with outcome in trauma patients, and it remains to be determined how mtDNA DAMPs compare to other plasma markers.23,24 Related to this, expression profiling recently has been used to describe the clinical course of severely injured or ill patients,25,26 and here, too, the relative utility of plasma mtDNA DAMP measurements could be questioned. It is noteworthy, though, that determination of mtDNA DAMP abundance in plasma is less costly and laborious than expression profiling where the predictive value of the transcriptional abnormalities is evident only after a protracted time of observation.
Another interesting observation in this study pertained to the 2 patients who were admitted with plasma mtDNA DAMP levels near the median for the cohort, but whose mtDNA DAMP abundance declined early during the hospital course. The reduction in mtDNA DAMPs was associated in time with clinical improvement. Although factors controlling the disposition of mtDNA DAMPs are entirely unexplored, it is an appealing idea that, as inferred by Sursal and colleagues,10 pathways suppressing further mtDNA DAMP formation acting in concert with mechanisms of mtDNA DAMP degradation act to limit the persistence of mtDNA DAMPs in the circulation and thereby forestall propagation of MODS.
In recent reports by Seok et al25 and Xaio et al,26 it was noted that a multitude of genes encoding inflammatory mediators, pattern recognition receptors, antimicrobial function, antigen recognition, T-cell proliferation, and apoptosis, etc, were altered in a near-simultaneous manner by trauma, burn, or endotoxemia. The implication of these findings for design of new therapies seems clear; the challenge to develop single agents for treating the numerous immuno-logical complications after trauma will be daunting because of the vast number of mediators apparently involved. If, however, the proposal that circulating mtDNA DAMPs are important for initiating such global transcriptional and ensuing pathophysiologic events is valid, then determining the mechanisms controlling mtDNA DAMP formation and disposition might identify isolated pharmacologic targets for controlling the ensuing genomic storm or other adverse pathophysiologic events in severe injury or illness. In this context, rapidly accumulating evidence shows that oxidative stress-induced cytotoxicity in multiple cell types is triggered by via a pathway involving oxidative mtDNA damage.18,27–32 Particularly relevant are recent findings that fusion proteins directing DNA glycosylases (either Ogg1 or Endo III) to mitochondria protect against oxidant-induced endothelial barrier degradation in perfused lungs,33 ventilator-induced lung injury in intact mice,34 and hyperoxic dysmorphogenesis in fetal rat lung explants.35 In isolated mitochondria, oxidant stress has been shown to release mtDNA sequences in a fragment size-dependent manner that exit the organelle through the mitochondrial permeability transition pore.36 Viewed collectively, these observations support the idea that a pharmacologic strategy to prevent or reverse oxidative mtDNA damage could attenuate fragmentation of the mitochondrial genome, the resulting export of mtDNA fragments into the circulatory system, and ensuing propagation of injury to secondary sites. Obviously, additional studies will be required to verify key aspects of this concept.
CONCLUSIONS
These findings comprise the first evidence that plasma mtDNA DAMPs are linked to the evolution of MODS and mortality in severely injured human subjects.
DISCUSSANTS
T.R. Billiar (Pittsburgh, PA)
The cloning of the mammalian TLR4 gene and the description of its key roles in sensing bacterial endotoxin in 1997 and 1998 would not only lead to the awarding of a 2011 Nobel prize to Bruce Beutler and Jules Hoffmann, but it also ushered in a new era in the field of innate immunity by providing the first conclusive evidence that the immune system is programmed to detect microbes and their molecules through specific gene encoded pattern recognition receptors.
Within 2 years, it was evident that not only components of the microbial cell wall were detected through Toll-like receptors, but also DNA and RNA sequences. By 2001, data began to appear indicating that some of these same receptors could be activated by molecules of endogenous origin, providing support for the danger hypothesis of immune responsiveness originally proposed by Polly Matzinger in 1994. Experimental evidence has also mounted over the past decade that the immuno-inflammatory response in trauma models is driven in part by the recognition of endogenous molecules released by stressed or damaged tissues by pattern recognition receptors.
That mitochondria are a potentially important source of these endogenous molecules should come with very little surprise when considering the putative proteobacteria origins of mitochondria. What is truly remarkable about the findings presented here is that even with a relatively small cohort of trauma patients, Dr Richards and colleagues were able to identify a strong correlation between the early and sustained elevations in circulating mitochondrial DNA sequences and the development of SIRS, MODS, and death.
This observation does not prove that mitochondrial DNA drives the response, but it certainly suggests that further work is warranted and highly supports the seminal work of Hauser and colleagues published in 2010.
I have 3 specific questions: first, the mechanisms of mitochondrial DNA release are not necessarily evident, especially in view of the marked differences in mitochondrial DNA levels between patients with similar ISSs. With cell death a primary source, we would perhaps expect to see other markers of cell death go up, such as lactate dehydrogenase, uric acid, or even nuclear DNA. Did you measure any of these endpoints?
The second question, hypomethylated DNA, as found in bacteria and mitochondria, activate inflammatory signaling through DNA sensors such as Toll-like receptor 9. In addition to neutrophil activation, this should also lead to cytokine and chemokine production. Did you correlate mitochondrial DNA levels with well-established circulating markers of SIRS, such as IL6 or IP10?
Finally, we have shown in experimental studies that Toll-like 9 is engaged more avidly in the setting of hemorrhagic shock and oxidative stress. There was at least a suggestion that higher based deficit and a greater need for transfusion and, hence, shock might have been present in the patients who went on to have higher mitochondrial DNA levels. Could you comment further on the mechanisms leading to DNA release?
Response from W.O. Richards (Mobile, AL)
We are students of your work there at Pittsburgh and we agree that the immune-inflammatory response you have shown in trauma models is driven by the recognition of endogenous molecules by the pattern recognition receptors.
The 3 questions you asked are related, so I will answer all 3 first by saying that we hypothesize that damage to mitochondrial DNA and the circulating mitochondrial DNA DAMPs are the core signals of this pathway, which induces MODS. We have focused our research in this one area.
To specifically answer your first question, we did not measure other markers of cell death, such as lactate dehydrogenase, uric acid, or nuclear DNA because we are focused on the mitochondrial genome, which, as you note, is nearly identical to bacterial genome. One thing that is very important to understand is that the mitochondrial DNA is 50 times more sensitive to oxidative stress than nuclear DNA. So, it leads me to suggest that just as at one point in time, centuries ago, all roads led to Rome, now we believe that MODS in trauma can be initiated by damage to the mitochondrial DNA genome. After all, DNA is the fundamental basis for all life, and the notion that DNA of the mitochondrial genome functions as a sentinel molecule in severe injury has intrinsic appeal. We extended Hauser's previous observations identifying elevations in mitochondrial DAMPs after trauma in human subjects to show that mitochondrial DNA DAMP elevations in plasma are associated with outcome.
We believe that this mitochondrial genome directs the inflammatory response through 2 different pathways. One is release of mitochondrial DNA DAMPs into the systemic circulation, which induces the innate immune response that you alluded to. So, to answer your second question, although we did not measure circulating cytokines, it would not be surprising if plasma cytokine levels were related in time or amount to mitochondrial DNA levels.
The other pathway through which the mitochondrial genome could direct the inflammatory response is inside the cell through the NLRP 3 inflammasome. Here too, we did not address that mechanism in this study, partly because the methods to do so—to unambiguously define intracellular mitochondrial DNA trafficking—have not yet been developed. But the motives for developing these methods, as you have alluded, are compelling.
The third question asked about the mechanisms of mitochondrial DNA DAMP release. You make the assumption that cell death is the primary source for DAMP release, but while this is intuitively reasonable, it has not been proven. We just do not have any information on the specific mechanisms of mitochondrial DNA DAMP release and why patients with similar levels of trauma had such variable levels of plasma mitochondrial DNA DAMPs. Future efforts in our laboratory will focus on the time course and mechanisms of mitochondrial genomic damage and the release of mitochondrial DNA DAMPs.
DISCUSSANTS
C.J. Hauser (Boston, MA)
This work is particularly gratifying to me because it is a clinical demonstration of something we have been working on in the laboratory for a long time. I have some issues with the work that still need to be brought up.
First of all, mitochondrial DNA is a biomarker and a DAMP, and that is one of the things that you have looked at here. But in our experience, that of Karim Brohi's group in London, and now yours, this is a loose association. I am just not sure that your clinical significance is as strong as your statistical significance. There is a lot of overlap between your data sets. You have only looked at generally less injured versus less injured patients and they are obviously both different from uninjured controls.
Practically speaking, how valuable will this biomarker be, say, as a predictor of ICU use rather than as a target for intervention with specific therapies against it?
Next, release of mitochondrial DNA is associated with the release of many other mitochondrial products. And many of those may be important. Formylated peptides, for instance, are classic chemoattractants and will likely be critical activators of peripheral neutrophil function.
Third, mitochondrial DNA is probably released with molecules that will turn out to be necessary “co-DAMPs” like TFAM, which binds mitochondrial DNA and seems important for it getting into the cell. So, like Dr Billiar, I would like to know which of these other mitochondrial species you have looked at or are you planning to look at. Which of these will be better or worse associated with disease?
Last, we have also recently shown that mitochondrial DNA is released in high-grade primate sepsis and is associated with the persistence of SIRS after sepsis has been treated with antibiotics and the blood stream becomes culture-negative. Have you looked at that phenomenon in your ICU patients? Can you distinguish, or are you trying to distinguish sepsis from sterile SIRS by other methods, where circulating mitochondrial DNA may actually be characteristic of both states?
Response from W.O. Richards (Mobile, AL)
Let me answer the first question, how much overlap is there between mitochondrial DNA DAMP levels in the patients who develop MODS and those who do not? It looks like there is a lot of overlap on our graphs, but you have to remember that each Tc represents a doubling in quantity. In addition, our original power analysis suggested that we needed a minimum of 70 patients and this was the interim analysis. So, we were shocked that 14 patients gave us this level of significance with as much as a 40-fold difference in mitochondrial DNA DAMP levels. We are in the process of quantifying mitochondrial DNA DAMPs and increasing our sample size.
You asked how valuable is this as a biomarker, say a predictor of ICU use rather than specifically as a target for intervention? Mitochondrial DNA DAMP levels are elevated within 8 hours of admission in a severely traumatized patient who will subsequently develop MODS days later. Our data suggest that patients with mitochondrial DNA DAMP Tc greater than 30 at admission will not develop organ failure whereas all the patents with a Tc less than 30 developed MODS. We believe that this may become a very valuable biomarker but much more study must be done to identify the best predictive biomarker.
You noted in your discussion that mitochondrial DNA DAMPs may need co-DAMPs, things like TFAM, to bind the mitochondrial DNA and get it into the cell. You asked which of these other things are we looking at? We agree that there are a multitude of mitochondrial DNA DAMPs that could be vitally important in the pathogenesis of MODs; however, we did not measure TFAM or any other co-DAMPs because we are focused on demonstrating a relationship between mitochondrial DNA DAMPs and clinical outcome.
Finally, you asked if we have measured mitochondrial DNA DAMPs in sepsis and can we differentiate sterile sepsis from bacterial sepsis? Our studies of bacterial and oxidant challenges in isolated perfused lungs, cells, and animals suggest there are very similar changes in mitochondrial DNA DAMPs in sterile and bacterial sepsis but we have not yet looked at that in human subjects. However, we agree in principle with your recent paper that there are differences in mitochondrial DAMPs between sterile and bacterial sepsis that could be used to differentiate sterile from bacterial sepsis. We are anxious to further our studies in this field.
DISCUSSANTS
C.E. Lucas (Detroit, MI)
I would like you to tell us which cells, in your opinion, are the source for this DNA DAMPs release. And second, in your opinion, whether the release is due to the underlying shock or septic insult or due to some alteration in capillary permeability and the resuscitation regimen.
Response from W.O. Richards (Mobile, AL)
So, which cells are responsible for mitochondrial DNA DAMP formation? What I can tell you is that we do not really know. Drs Hauser and Billiar have done experiments in which cells taken from liver and muscle have been used to supply the mitochondrial DAMPs and they all seem to elicit the same pathway, so it is possible that these mitochondrial DNA DAMPs in the blood stream are derived from a multitude of cell types. But, neither we nor other investigators have any data in human subjects to identify the cells responsible for the elevation in mitochondrial DNA DAMPs found in this study.
Your second question asked whether the release is due to the underlying shock or due to alterations in capillary permeability and resuscitation. Certainly, we believe that it is not only direct trauma that causes release of the mitochondrial DAMPs, but we also believe that a large portion of the release of mitochondrial DAMPs is induced by shock and oxidative stress. I want to stress this point again that the mitochondrial genome is 50 times more sensitive to oxidative stress than is the nuclear genome. We believe that one mechanism to protect the nuclear genome from oxidant-mediated mutation involves the oxidant damage-triggered mitochondrial genome, wherein oxidative damage stimulates a spectrum of intra- and extracellular responses to danger. One of the big surprises about the data we presented was the marked elevation in mitochondrial DNA DAMPs at the onset, which did not change much with resuscitation in the 4 patients who developed MODS. This suggests that the levels of mitochondrial DNA DAMPs are not influenced heavily by our resuscitation but by unknown factors.
DISCUSSANTS
D.E. Fry (Chicago, IL)
I am coming up at this time, Dr Richard, to the 40th anniversary of my first mitochondria experiments. And I find it amazing that this subject continues to gain life.
I am really curious as to why mitochondrial DNA are not products of cellular necrosis. The broad spectrum of alarmins that have been described as a subset of danger-associated pathogens include elevated HMGB1 and heat shock proteins, uric acid, as Tim Billiar referred to, and the nuclear DNA elements, representing the content of cells that have been disrupted due to ischemia and injury. C-reactive protein is probably up as well.
I think it really is going to be important for you to choose some of these other markers to see whether they go up in parallel with mitochondrial DNA, because I think you are going to find that they will.
And, I guess I would issue one word of caution about looking for the silver bullet in human sepsis. You correctly identify that the current scoreboard is the pharmaceutical industry, zero; shock, multiple organ failure, and sepsis, 40. Clinical interventions that have been focused on a single target have been failed treatments. So, I am concerned that there is just an avalanche of cellular alarmins, and that mitochondrial DNA may be one of them.
I would caution you to really try to expand your studies in looking at these other alarmin signals that may be rising and falling in concert with the DNA, so you do not waste all of your time and effort on fusion proteins targeting DNA from the mitochondria that may lead to the dead end that I and many others have experienced for 40 years.
Response from W.O. Richards (Mobile, AL)
I can understand your skepticism about finding the magic bullet after everyone involved in the field has failed to identify a single successful drug during the past 40 years of study. I also agree that it is not just one DAMP that is responsible for human pathology and there are numerous examples of alarmins that have been targeted. However, we believe mitochondrial DNA DAMPs are more important than the other alarmins for a variety of reasons including the fact that it is DNA that is at the fundamental basis of life. It stands to reason that the basic coding for life should initiate the danger signals during oxidant damage. The mitochondria are known as the organelle that is responsible for carrying out orders for cell death, and thus it stands to reason that mitochondrial DNA DAMPs may be more important than other nonmitochondria alarmins in the initiation of apoptosis and cell death.
Finally, our cell, isolated perfused organ, and whole animal studies show that mitochondrial DNA repair proteins can prevent and reverse oxidant damage; thus we believe, on the basis of this work, that mitochondrial DNA DAMPs are the major initiators of the inflammatory cascade and worthy of investigation as a pharmacologic target. The study I presented today demonstrates that there is a strong correlation between elevations of mitochondrial DNA DAMPs in severely injured human subjects who develop MODs compared with those patients just as severely injured who do not develop MODS.
If MODS is initially triggered by disruption of the mitochondrial DNA genome and increases in circulating mitochondrial DNA DAMPs, then we have a therapeutic target. We should know in a few years if we are on the right track.
Acknowledgments
This research was supported by grants from the National Institutes of Health (RO1 HL058234, R01 HL113614).
Footnotes
Disclosure: M.N. Gillespie has licensed a patent on fusion proteins targeting DNA repair enzymes to mitochondria. The mitochondrial-targeted DNA repair enzymes are currently under development for use in lung transplant, and for treatment of trauma/sepsis-related multiple organ dysfunction. The authors declare no conflicts of interest.
REFERENCES
- 1.Zhou Z, Daugherty WP, Sun D, et al. Protection of mitochondrial function and improvement in cognitive recovery in rats treated with hyperbaric oxygen following lateral fluid-percussion injury. J Neurosurg. 2007;106:687–694. doi: 10.3171/jns.2007.106.4.687. [DOI] [PubMed] [Google Scholar]
- 2.Weinbroum AA, Hochhauser E, Rudick V, et al. Multiple organ dysfunction after remote circulatory arrest: common pathway of radical oxygen species? J Trauma. 1999;47:691–698. doi: 10.1097/00005373-199910000-00013. [DOI] [PubMed] [Google Scholar]
- 3.Fredriksson K, Flaring U, Guillet C, et al. Muscle mitochondrial activity increases rapidly after an endotoxin challenge in human volunteers. Acta Anaesthesiol Scand. 2009;53:299–304. doi: 10.1111/j.1399-6576.2008.01851.x. [DOI] [PubMed] [Google Scholar]
- 4.Fredriksson K, Tjader I, Keller P, et al. Dysregulation of mitochondrial dynamics and the muscle transcriptome in ICU patients suffering from sepsis induced multiple organ failure. PLoS One. 2008;3:e3686. doi: 10.1371/journal.pone.0003686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fredriksson K, Rooyackers O. Mitochondrial function in sepsis: respiratory versus leg muscle. Crit Care Med. 2007;35:S449–S53. doi: 10.1097/01.CCM.0000278048.00896.4B. [DOI] [PubMed] [Google Scholar]
- 6.Cheng G, Kong RH, Zhang LM, et al. Mitochondria in traumatic brain injury and mitochondrial-targeted multipotential therapeutic strategies. Br J Pharmacol. 2012;167:699–719. doi: 10.1111/j.1476-5381.2012.02025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Q, Raoof M, Chen Y, et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krysko DV, Agostinis P, Krysko O, et al. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends Immunol. 2011;32:157–164. doi: 10.1016/j.it.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Sun S, Sursal T, Adibnia Y, et al. Mitochondrial DAMPs increase endothelial permeability through neutrophil dependent and independent pathways. PLoS One. 2013;8:e59989. doi: 10.1371/journal.pone.0059989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sursal T, Stearns-Kurosawa DJ, Itagaki K, et al. Plasma bacterial and mitochondrial DNA distinguish bacterial sepsis from sterile systemic inflammatory response syndrome and quantify inflammatory tissue injury in nonhuman primates. Shock. 2013;39:55–62. doi: 10.1097/SHK.0b013e318276f4ca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q, Itagaki K, Hauser CJ. Mitochondrial DNA is released by shock and activates neutrophils via p38 map kinase. Shock. 2010;34:55–59. doi: 10.1097/SHK.0b013e3181cd8c08. [DOI] [PubMed] [Google Scholar]
- 12.Al-Mehdi AB, Pastukh VM, Swiger BM, et al. Perinuclear mitochondrial clustering creates an oxidant-rich nuclear domain required for hypoxia-induced transcription. Sci Signal. 2012:5:ra47. doi: 10.1126/scisignal.2002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pastukh VM, Zhang L, Ruchko MV, et al. Oxidative DNA damage in lung tissue from patients with COPD is clustered in functionally significant sequences. Int J Chron Obstruct Pulmon Dis. 2011;6:209–217. doi: 10.2147/COPD.S15922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sauaia A, Moore EE, Johnson JL, et al. Validation of postinjury multiple organ failure scores. Shock. 2009;31:438–447. doi: 10.1097/SHK.0b013e31818ba4c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference: definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Crit Care Med. 1992;20:864–874. [PubMed] [Google Scholar]
- 16.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grishko V, Solomon M, Wilson GL, et al. Oxygen radical-induced mitochondrial DNA damage and repair in pulmonary vascular endothelial cell pheno-types. Am J Physiol Lung Cell Mol Physiol. 2001;280:L1300–L1308. doi: 10.1152/ajplung.2001.280.6.L1300. [DOI] [PubMed] [Google Scholar]
- 18.Ruchko MV, Gorodnya OM, Zuleta A, et al. The DNA glycosylase Ogg1 defends against oxidant-induced mtDNA damage and apoptosis in pulmonary artery endothelial cells. Free Radic Biol Med. 2011;50:1107–1113. doi: 10.1016/j.freeradbiomed.2010.10.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shokolenko I, Venediktova N, Bochkareva A, et al. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009;37:2539–2548. doi: 10.1093/nar/gkp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chouteau J, Gorodnya O, Ruchko M, et al. Novel fusion protein constructs targeting DNA repair enzymes to mitochondria protect against pseudomonas aeruginosa-induced acute lung injury in intact rats [Abstracts, 2011 ATS International Conference]. Am J Resp Crit Care Med. 2011;183:A3763. [Google Scholar]
- 21.Hill JK, Obiako B, Gorodnya O, et al. A positive feedback cycle involving oxidative mitochondrial (mt) DNA damage and mtDNA damage associated molecular patterns (DAMPs) contributes to bacteria-induced endothelial barrier dysfunction in isolated rat lungs. Am J Resp Crit Care Med. 2013;187:A4978. [Google Scholar]
- 22.Gill R, Ruan X, Menzel CL, et al. Systemic inflammation and liver injury following hemorrhagic shock and peripheral tissue trauma involve functional TLR9 signaling on bone marrow-derived cells and parenchymal cells. Shock. 2011;35:164–170. doi: 10.1097/SHK.0b013e3181eddcab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edmonds RD, Vodovotz Y, Lagoa C, et al. Transcriptomic response of murine liver to severe injury and hemorrhagic shock: a dual-platform microarray analysis. Physiol Genomics. 2011;43(20):1170–1183. doi: 10.1152/physiolgenomics.00020.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeifer R, Kobbe P, Darwiche SS, et al. Role of hemorrhage in the induction of systemic inflammation and remote organ damage: analysis of combined pseudo-fracture and hemorrhagic shock. J Orthop Res. 2011;29:270–274. doi: 10.1002/jor.21214. [DOI] [PubMed] [Google Scholar]
- 25.Seok J, Warren HS, Cuenca AG, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao W, Mindrinos MN, Seok J, et al. A genomic storm in critically injured humans. J Exp Med. 2011;208:2581–2590. doi: 10.1084/jem.20111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koczor CA, Snyder JW, Shokolenko IN, et al. Targeting repair proteins to the mitochondria of mammalian cells through stable transfection, transient transfection, viral transduction, and TAT-mediated protein transduction. Methods Mol Biol. 2009;554:233–249. doi: 10.1007/978-1-59745-521-3_15. [DOI] [PubMed] [Google Scholar]
- 28.Dobson AW, Grishko V, LeDoux SP, et al. Enhanced mtDNA repair capacity protects pulmonary artery endothelial cells from oxidant-mediated death. Am J Physiol Lung Cell Mol Physiol. 2002;283:L205–L210. doi: 10.1152/ajplung.00443.2001. [DOI] [PubMed] [Google Scholar]
- 29.Rachek LI, Thornley NP, Grishko VI, et al. Protection of INS-1 cells from free fatty acid-induced apoptosis by targeting hOGG1 to mitochondria. Diabetes. 2006;55:1022–1028. doi: 10.2337/diabetes.55.04.06.db05-0865. [DOI] [PubMed] [Google Scholar]
- 30.Koczor CA, Shokolenko IN, Boyd AK, et al. Mitochondrial DNA damage initiates a cell cycle arrest by a Chk2-associated mechanism in mammalian cells. J Biol Chem. 2009;284:36191–36201. doi: 10.1074/jbc.M109.036020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Druzhyna NM, Hollensworth SB, Kelley MR, et al. Targeting human 8-oxoguanine glycosylase to mitochondria of oligodendrocytes protects against menadione-induced oxidative stress. Glia. 2003;42:370–378. doi: 10.1002/glia.10230. [DOI] [PubMed] [Google Scholar]
- 32.Rachek LI, Grishko VI, Ledoux SP, et al. Role of nitric oxide-induced mtDNA damage in mitochondrial dysfunction and apoptosis. Free Radic Biol Med. 2006;40:754–762. doi: 10.1016/j.freeradbiomed.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 33.Chouteau JM, Obiako B, Gorodnya OM, et al. Mitochondrial DNA integrity may be a determinant of endothelial barrier properties in oxidant-challenged rat lungs. Am J Physiol Lung Cell Mol Physiol. 2011;301:L892–L898. doi: 10.1152/ajplung.00210.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hashizume M, Mouner M, Chouteau JM, et al. Mitochondrial-targeted DNA repair enzyme 8-oxoguanine DNA glycosylase 1 protects against ventilator-induced lung injury in intact mice. Am J Physiol Lung Cell Mol Physiol. 2013;304:L287–L297. doi: 10.1152/ajplung.00071.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gebb SA, Decoux A, Waggoner A, et al. Mitochondrial DNA damage mediates hyperoxic dysmorphogenesis in rat fetal lung explants. Neonatology. 2012;103:91–97. doi: 10.1159/000342632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia N, Chavez E. Mitochondrial DNA fragments released through the permeability transition pore correspond to specific gene size. Life Sci. 2007;81:1160–1166. doi: 10.1016/j.lfs.2007.08.019. [DOI] [PubMed] [Google Scholar]