Abstract
Salmonella enterica serovar Typhimurium (S. Typhimurium) is an intracellular pathogen that has evolved to survive in the phagosome of macrophages. The periplasmic copper-binding protein CueP was initially known to confer copper resistance to S. Typhimurium. Crystal structure and biochemical studies on CueP revealed a putative copper binding site surrounded by the conserved cysteine and histidine residues. A recent study reported that CueP supplies copper ions to periplasmic Cu, Zn-superoxide dismutase (SodCII) at a low copper concentration and thus enables the sustained SodCII activity in the periplasm. In this study, we investigated the role of CueP in copper resistance at a high copper concentration. We observed that the survival of a cueP-deleted strain of Salmonella in macrophage phagosome was significantly reduced. Subsequent biochemical experiments revealed that CueP specifically mediates the reduction of copper ion using electrons released during the formation of the disulfide bond. We observed that the copper ion-mediated Fenton reaction in the presence of hydrogen peroxide was blocked by CueP. This study provides insight into how CueP confers copper resistance to S. Typhimurium in copper-rich environments such as the phagosome of macrophages.
Keywords: copper resistance, CueP, Fenton reaction, Salmonella
INTRODUCTION
Copper is ubiquitous in nature and is required for all living organisms (Crichton and Pierre, 2001). Copper ions are involved in catalyzing many biochemical reactions due to easy conversion between the cuprous (Cu+) and cupric (Cu2+) forms (Leary and Winge, 2007). However, they are also involved in generating free radicals in the presence of oxygen or hydrogen peroxide and excess copper ions (Halliwell and Gutteridge, 1984). To cope with the dual nature of copper ions, all living organisms tightly control cellular copper pools with elaborate copper homeostatic mechanisms (O’Halloran and Culotta, 2000), including copper-responsive transcriptional regulators, membrane transporters and copper chaperones (Franke et al., 2003; Osman and Cavet, 2008; Pontel and Soncini, 2009; Rensing et al., 2000).
Salmonella enterica serovar Typhimurium (S. Typhimurium) is an important human pathogen that causes worldwide food-associated Salmonella gastrointestinal disease (Coburn et al., 2007). The ability of the bacteria to survive in macrophage phagosomes is associated with its virulence (Fields et al., 1986). Since copper ions participate in killing the invading pathogens in the phagosomes of the activated macrophages and neutrophils in the Fenton reaction (Dupont et al., 2011; Hodgkinson and Petris, 2012), copper resistance of the bacteria has been implicated in the ability of the bacteria to survive in the phagosomes of macrophages (Achard et al., 2012; White et al., 2009b). Thus copper resistance of some intracellular pathogens is now considered an important virulence factor (Prohaska and Lukasewycz, 1981; White et al., 2009a; Wolschendorf et al., 2011).
In S. Typhimurium, elevated copper ions are exported via either one of two related P1B-type ATPases, CopA and GolT, which are essential in the survival of the bacteria in excessive copper environments (Osman et al., 2010). The expression of CopA and GolT in S. Typhimurium is triggered by the copper-sensing transcriptional regulator CueR leading to copper-responsive expression of the proteins (Espariz et al., 2007). CueR also mediated the expression of the multi-copper oxidase CueO (Rensing et al., 2000) and the periplasmic copper-binding protein CueP (Espariz et al., 2007; Osman et al., 2010; Pontel and Soncini, 2009).
CueP was initially known to confer copper resistance to S. Typhimurium as a functional replacement of the Cu+-specific exporter CusAFBC found in most Gram-negative bacteria (Pontel and Soncini, 2009). The CueP-deleted strain of S. Typhimurium was highly susceptible to copper, particularly in anaerobiosis (Pontel and Soncini, 2009). Crystal structure and biochemical studies of CueP revealed a putative copper binding site surrounded by the conserved cysteine and histidine residues (Yoon et al., 2013). A recent study reported that the CueP protein in complex with copper ion supplies copper ion to periplasmic Cu, Zn-superoxide dismutase (SodCII) at a low concentration of copper, and thus sustains the SodCII activity that is implicated in the bacterial virulence (Osman et al., 2013). However, the biochemical role of CueP remains to be elucidated in excessive copper environments. In this study, we investigate the role of CueP in scavenging formation of the hydroxyl radical from H2O2 in the presence of high concentrations of copper ion.
MATERIALS AND METHODS
Bacterial strains and plasmids
The DNA construction, protein expression, and protein purification of S. Typhimurium CueP (residues 22–179 in the numbering of the precursor CueP) has been previously described (Yun et al., 2011). Salmonella Typhimurium SL1344 deleted for cueP was obtained by the λ Red method (Datsenko and Wanner, 2000) using primers that have been previously used by Osman et al. (2010): 5′- GGAACCCCTATAGTAGGCAGGGAGATTGT TCACAAGGAATTGAAGTTATGGTGTAGGCTGGAGCTGCT T-3′ and 5′-GATAACCCATTATGTTATCGGGCATTTTTTTAA CGTAATGGTAATTCCGTCATATGAATATCCTCCTTA-3′. To construct plasmids for expression of cueP and its variants in E. coli, full-length S. Typhimurium CueP, including the promoter region, was amplified using PCR primers 5′-GCATGAATTCAC TTTACCCTGCGTCCCT-3′, and 5′-GCATCCATGGTAACCCA TTATGTTATCG-3′, digested with EcoRI and NcoI, and ligated into pACYC184 vector. The mutations H94A, C96S, H99A, C104S and C172S were introduced into the cloned wild-type cueP gene by two subsequent PCR reactions (Landt et al., 1990). To construct a plasmid for expression of cueP in S. Typhimurium, plasmid pACYC177-CueP was constructed by ligating PCR DNA digested with BamHI and HindIII into the same site of pACYC177. PCR DNA containing the coding region of CueP was synthesized using two primers, 177cuePF (5′-GATCAAGCTTACTTTACCCTGCGTCCCT-3′) and 177cue PR (5′-GATCGGATCCTAACCCATTATGTTATCG-3′).
Size exclusion chromatography
To determine the molecular size of CueP or to confirm its dimeric form in solution, size-exclusion chromatography was performed at a flow rate of 0.5 ml/min on Superdex S-200 HR 10/30 (GE Healthcare) equilibrated with 20 mM Tris buffer (pH 8.0) containing 150 mM NaCl and 2 mM 2-mercaptoethanol.
Preparation of the CueP protein sample
Since the wild-type and mutant CueP proteins contain 2 mM 2-mercaptoethanol to prevent oxidation, 2-mercaptoethanol was removed from the sample buffer by extensive dialysis against highly-degassed 20 mM Tris buffer (pH 8.0) containing 150 mM NaCl. After dialysis, the protein samples were concentrated and stored at −80°C until use.
Measurement of free sulfhydryl contents by using DTNB
Free cysteine residues of wild-type and mutant CueP proteins were determined by using the 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) assay (Ellman, 1959) with modification. The purified protein samples (0.2 mM) were incubated with 0.5 mM CuCl2 at room temperature for 1 min. Then, 10 μl of the incubated sample were taken in 75 μl of 30 mM Tris buffer (pH 8.2) containing 3 mM EDTA solution. Subsequently, 5 mM DTNB and 99% methanol (vol) were added and incubated for 15 min at room temperature. After centrifugation at 13,000 rpm for 5 min, 2-nitro-5-thiobenzoate produced from the disulfide bond cleavage of DTNB by thiol group of cysteine was quantified by measurement of the absorption at 412 nm [ɛ = 14,150/M cm: (Ellman, 1959)].
Electron paramagnetic resonance spectroscopy
Copper sulfate standard solution (0.2 mM) in 20 mM Tris buffer (pH 8.0) containing 150 mM NaCl was prepared and the EPR spectra were recorded at −196°C with 0.01 mW microwave power on a Beckman X-band CW-EPR (Beckman Coulter). Upon performing baseline correction, the spectra were double-integrated from 2500 to 3500 G. The copper peak at 3250 G was clearly identified. The protein sample (70 μM) was added to the CuCl2-containing buffer and incubated for 5 min at room temperature, and then the EPR spectra were collected. The concentration of Cu2+ was calculated by integrating the peak around 3250 G corresponding to Cu2+.
Hydroxyl radical scavenging assay
To prepare the CueP protein in the reduced or oxidized state, the purified protein was treated with 2 mM 2-mercatoethanol or 0.5 mM CuCl2, respectively, and then dialyzed against highly degassed 20 mM Tris buffer containing 150 mM NaCl at 4°C. The protein was used for this study within 1 h after dialysis. One μl of CuCl2 (5 μM as a final concentration) was added into 10 μl of each protein sample (5 μM) in 20 mM Hepes buffer (pH 7.0) and incubated for 10 min at room temperature. Then, 10 μl of supercoiled plasmid (1 μg; pPROEX-HTA, Invitrogen) containing 2 mM H2O2 was added and incubated for 5 min at 22°C and applied to 0.8% agarose gel electrophoresis to examine the levels of intact (supercoiled) and open circle plasmid bands.
Sample preparation for mass spectrometry
The mature form of CueP (residues 22–179) was incubated at a concentration of 1 mg/mL (56.7 μM monomer) in 1× PBS buffer (8.5 g NaCl, 0.2 g KCl, 1.78 Na2HPO4, 0.27 g KH2PO4 in 1 L H2O, pH 7.4) with or without addition of 0.1 mM CuCl2 for 15 min at room temperature. Each sample was treated with 6 M urea and 100 mM N-ethylmaleimide (NEM) for 1 h in the dark to derivatize free thiol groups of cysteine residues and then treated with 2 M trichloroacetic acid (TCA) to stop the NEM reaction and induce the acidolytic cleavage of the Cu-thiolate bond in CueP, which was then allowed to precipitate for 15 min on ice. After centrifugation for 10 min, the supernatant was removed and the pellet was washed twice with pre-chilled 80% acetone. The air-dried protein was dissolved in 50 mM Tris/HCl (pH 8.0) containing 6 M urea and 100 mM iodoacetamide (IAA) to derivatize the Cu-free thiol groups for 1 h in the dark. Then, the protein was precipitated with 2 M TCA and 80% acetone. For the analysis of disulfide bond in the CueP monomer excised from a non-denaturing SDS-PAGE gel, in-gel protein digestion was performed with trypsin and chemotrypsin, as described previously (Yoon et al., 2013), and peptides were cleaned up using ZipTip® C18 for mass spectrometry.
Mass spectrometry
Peptide samples were dissolved in 0.4% acetic acid and analyzed as previously described with an LTQ-Velos mass instrument (Thermo Fisher Scientific Inc.) equipped with EASY-nLC 1000 Liquid Chromatograph (Yoon et al., 2013). The acquired mass spectral data were analyzed by SEQUEST search algorithm (Eng et al., 1995) with the options of average mass (m/z); precursor mass tolerance, 0.8 Da; fragment mass tolerance, 0.8 Da; variable modifications for cysteine, including NEM (m/z difference, 125.125), carbamidomethylation with IAA (57.021), oxidation (15.995) and dioxidation (31.999), and histidine protonation (1.008). Disulfide-bonded peptides were analyzed by in-house Excel program with the option of 2 H loss (−2.016) from all combinations of 2 peptides, each containing cysteine. The extracted ion chromatograms (XIC) were generated by Thermo Xcalibur Qual Browser 2.1 with the precursor ion mass (m/z) and mass tolerance of 0.2 Da.
Sensitivity of E. coli harboring S. Typhimurium CueP to the Fenton reaction
To see the effect of S. Typhimurium CueP in E. coli strain BW25113 under cusB-deleted background, the E. coli strain BW25113 ΔcusB was obtained from KEIO collection (Yamamoto et al., 2009). Wild-type or mutant S. Typhimurium cueP gene was inserted into a pACYC184 vector, as described above. The vector was transformed into the E. coli strain, and the E. coli was grown in LB at 37°C until OD600 range of 0.7–0.9. The cells were immediately washed with pre-warmed 20 mM Hepes buffer (pH 7.5) two times and then incubated with 0.1 mM CuCl2 in 20 mM Hepes buffer for 15 min at room temperature. After washing the cells two times with 20 mM Hepes buffer (pH 7.5), 30 mM H2O2 was added to the cells for 5 min. To measure the survival of the bacteria, the cells were spread on LB agar plates.
Cell culture and macrophage infections
Macrophages were infected with S. Typhimurium strain SL1344 and its isogenic strain deleted for the cueP gene (ΔcueP) by the method described by Osman et al. (2010). To restore the cueP gene, pACYC177-CueP was transformed to ΔcueP or wild-type strain of S. Typhimurim. For infection, RAW264.7 macrophage cells obtained from the Korean Cell Line Bank (Korea) were cultivated under 5% CO2 at 37°C in RPMI 1640 (Welgene) supplemented with 10 % (v/v) fetal bovine serum (Welgene). Wild-type and ΔcueP S. Typhimurium strains were grown overnight in LB media at 37°C and 200 rpm. The bacteria were washed and resuspended in sterile D-PBS (8.5 mM sodium phosphate, 1.5 mM potassium phosphate, 137 mM NaCl, pH 7.4) to mix into the RAW264.7 cell cultures at a multiplicity of infection (MOI) of 10:1 (bacteria:macrophage). At 2-h post-infection, the cell culture media were replaced with serum-free media supplemented with 200 μg/ml gentamicin to kill extracellular bacteria for 2 h. The infected RAW264.7 cells were then washed with D-PBS and intracellular bacteria were released with 0.1% Triton X-100, followed by incubation at 37°C for 5 min. The number of intracellular bacteria was assessed by viable counts on non-selective and selective LB agar plates.
Western blot analysis
Protein concentrations of CueP and ribosomal protein S1 in whole-cell extracts were determined using a rabbit polyclonal anti-CueP and aniti-S1 antibody. Standard ECL reagents and films (GE Healthcare Life Sciences, USA) were used for Western blot detection with horseradish peroxidase (HRP)-conjugated anti-rabbit IgG antibody (Santa Cruz Biotechnology, Inc., USA). Specific proteins were imaged by using a VersaDoc 100 instrument (Bio-Rad, USA) and quantified by using Quantity One (Bio-Rad). The polyclonal anti-CueP antibody was raised in rabbits, which were immunized with the purified recombinant CueP protein (Ab Frontier, Korea). Polyclonal antibodies to S1 were obtained from Dr. Stanley N. Cohen (Stanford University).
RESULTS
The survival of cueP-deleted strain of S. Typhimurium in macrophage is significantly decreased
Salmonella can survive within macrophage phagosomes, and this survival is critical for bacterial virulence during systemic disease (Fields et al., 1986). To examine the role of CueP in the survival of the bacteria in the macrophage phagosomes, we deleted the cueP gene in virulent S. Typhimurium (strain SL1344) to compare its survivals with that of the wild-type strain when infecting the murine macrophage cell line RAW264.7. As shown in Fig. 1, the survival of the ΔcueP strain was markedly decreased in the mouse macrophage cell line (by about 60%). However, the survivals of the bacteria were lower than those of the wild-type strain when the cueP gene was overexpressed in the wild-type or cueP-deleted strain of S. Typhimurium. This observation suggests that the cueP gene is constitutively expressed in Salmonella, which has evolved to survive in the macrophage phagosome.
Fig. 1.
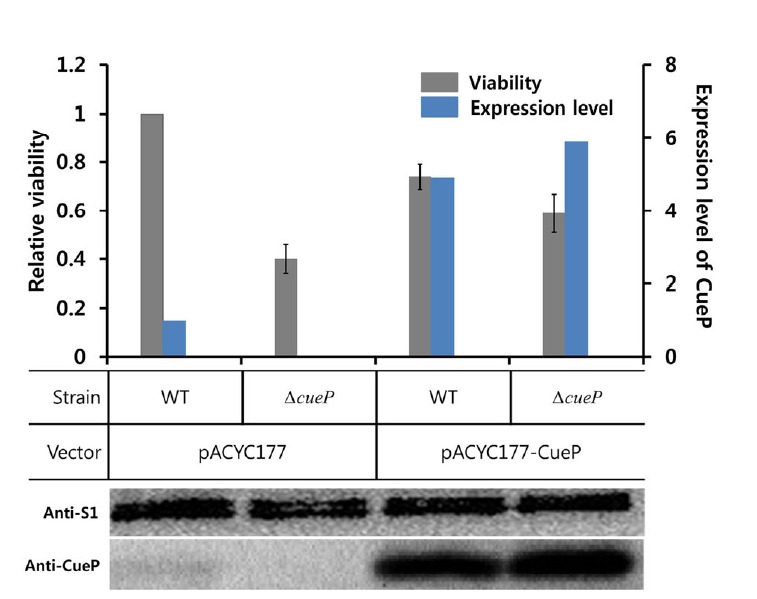
The role of CueP in the survival of Salmonella in macrophage phagosomes. Relative viability of Salmonella strains was measured in murine macrophage RAW264.7 cells. Infection assays were performed with wild-type S. Typhimurium (WT) and cueP-deleted (ΔcueP) strains, transformed with pACYC177 vector or pAcyC177-CueP that harbors the wild cueP gene. Relative viability index was defined as the colony-forming unit ratio of mutant and wild-type strains. In each case, bacterial strains were used at a final MOI of 10:1. Data points represent the mean (± S.E) for at least three independent experiments. Total proteins of wild-type and cueP-deleted (ΔcueP) strains of S. Typhimurium were separated by SDS-PAGE and analyzed in Western blot using anti-CueP and anti-S1 antibodies. The expression level of CueP, assessed by the Western blot analysis, is displayed in the right y axis.
A disulfide bond between Cys96 and Cys104 is formed by Cu2+
To examine how CueP biochemically contributes to the pathogenesis of S. Typhimurium, we investigated the involvement of the CueP protein with copper ion. CuCl2 specifically induces a disulfide bond of the protein according to SDS-PAGE under non-reducing conditions (Pontel and Soncini, 2009). In this study, mass spectrometric analysis was employed to determine which cysteine residues were engaged in the disulfide bond bridge induced by Cu2+. When exposed to CuCl2, the CueP protein formed the disulfide bond between Cys96 and Cys104 in the monomer subunits separated by non-reducing SDS-PAGE (Fig. 2A). In comparison with the untreated sample, the CuCl2-treated CueP protein had marked decreases in the free thiol groups of Cys96 and Cys104 derivatized with N-ethylmaleimide [denoted by the subscripts of NEM in Figs. 2A(d) and 2A(e)], while their metal-binding parts derivatized with iodoacetamide (IAA) remained constant. In contrast, the CuCl2 addition increased the metal-binding portion of Cys172 (cIAA) while decreasing the free thiol level (cNEM), as shown in Fig. 2A(f). Otherwise, a certain amount of Cys172 formed the disulfide bond with Cys96 in the absence of CuCl2 [Fig. 2A(a)]. The mass spectral data showed that the disulfide bond between the conserved residues Cys96 and Cys104 was newly made by addition of Cu2+ (Fig. 2B) and differed from the disulfide bond (Cys96 and Cys172) induced by molecular oxygen (Yoon et al., 2013). This implies that copper ion induces a conformational change of the CueP protein to generate the disulfide bond between Cys96 and Cys104 instead of Cys172, which participated in metal binding or spontaneous oxidation of the CueP protein. The cysteine oxidation by CuCl2 did not change the oligomeric state of the CueP protein, as assessed by size exclusion chromatography (Fig. 2C), indicating that the disulfide bond is formed in an intramolecular manner.
Fig. 2.
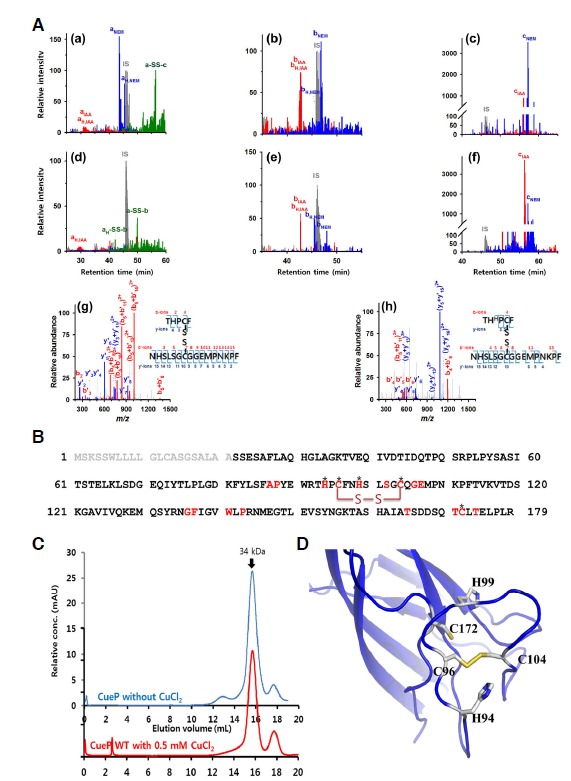
Disulfide bond between Cys96 and Cys104, induced by CuCl2. (A) Merged graphs of extracted ion chromatograms (XICs) of cysteine-containing peptides in the CueP monomers excised from a non-denaturing SDS PAGE gel. Prior to gel separation, the CueP proteins were incubated for 15 min in the absence (a–c) or presence (d–f) of 0.1 mM CuCl2 at room temperature. The peaks of precursor ions of 93THPCF97, 98NHSLSG CQGEMPNKPF113 and 162TASHA IATSDDSQTCLTELPLR183 are shown with the respective letters, a, b and c, followed by the subscripts: H, histidine protonation at His94 (aH) or His99 (bH); NEM or IAA, alkylation of cysteine with N-ethylmaleimide or iodoacetamide; and -SS-, disulfide bond between two peptides. The relative intensity of a unique peptide, LSDGEQIY (m/z = 462.99, z = +2 at RT 46.0 min), used as internal standard (IS), is expressed as 100 for each chromatogram. Disulfide-bonded peptides, a-SS-b and aH-SS-b, derived from the CuCl2-treated CueP protein, are identified by tandem mass spectra assigned to the b- and y-ions generated from collision-induced fragmentation of the precursor ions at m/z = 759.85 and 760.19 in the charge states of +3, and tandem mass spectral data for the other peptide peaks are shown in Supplementary Fig. S1. (B) Amino acid sequence of S. Typhimurium CueP. The key conserved residues are in red, of which the strictly conserved Cys and His residues are indicated by *. The disulfide bond bridge is shown in a red line with ‘S-S’, which is induced by CuCl2. The signal sequence is shown in gray. (C) The molecular size of the CueP protein when oxidized by Cu2+, measured by size exclusion chromatography. The reduced form of wild-type CueP protein (CueP without CuCl2) and the oxidized protein by addition of 0.5 mM CuCl2 (CueP with CuCl2) are subject to size exclusion chromatography (Superdex 200 10/30) in a highly-degassed buffer containing 20 mM Tris-HCl and 150 mM NaCl at a flow rate of 0.5 ml/min. To oxidize the protein with CuCl2, the wild-type CueP protein (1 mg/ml) was incubated with 0.5 mM CuCl2 for 30 min at room temperature (CueP with CuCl2). The elution profile of CueP with CuCl2 was offset for clarity. (D) A molecular model for the Cys-rich region of CueP in the oxidized state, induced by CuCl2. The side chains of His94, Cys96, His99, Cys104, and Cys172 are shown.
The crystal structure of CueP showed a putative copper binding site, consisting of the conserved residues Cys96, Cys104, Cys172, His94, and His99 (Yoon et al., 2013). Based on the crystal structure (Yoon et al., 2013), we modeled the putative copper binding site (referred to as cysteine-rich region in this study) in the oxidized state, which is induced by CuCl2 (Fig. 2D). The model seems reasonable because the crystal structure revealed that the distance between the thiol groups of Cys96 and Cys104 is relatively short, and the loop containing Cys104 is intrinsically flexible (Yoon et al., 2013).
CueP is rapidly oxidized by Cu2+ with a stoichiometry of 1:1
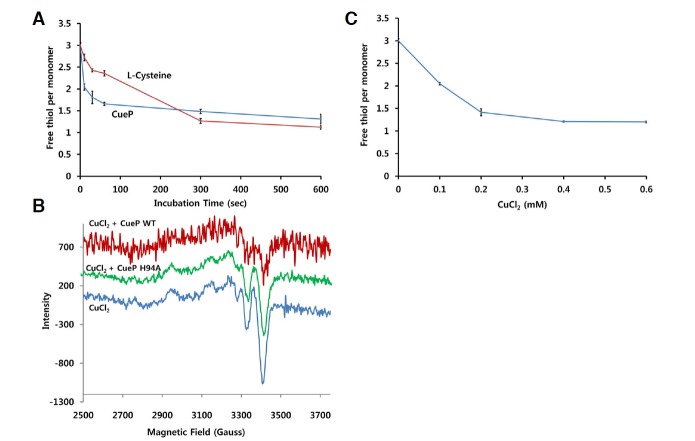
We next examined how fast the disulfide bond of CueP is formed in response to Cu2+, using the agent DTNB to selectively react with the free thiol groups with a short reaction time (Ellman, 1959). To compare the reactivity of the cysteine residues in CueP with the authentic cysteine in solution, three-times higher concentration of L-cysteine was used since one molecule of CueP has three cysteine residues. As shown in Fig. 3A, all the cysteine residues of the purified CueP protein are in the reduced state since the free thiol contents of CueP and L-cysteine were the same at the initial stage. When CuCl2 was added to the CueP protein in a reducing-agent-free buffer, two-thirds of the free thiol was rapidly consumed, which is consistent with the mass spectrometry result revealing the disulfide bond between Cys96 and Cys104 of CueP. In contrast, Cu2+ slowly decreased the free thiol content of L-cysteine (Fig. 3A).
Fig. 3.
The rapid reaction of CueP with Cu2+. (A) Comparison of the reactivity of CueP to Cu2+ with L-cysteine. Prior to the experiment, wild-type CueP protein was dialyzed against 20 mM Tris buffer (pH 8.0) containing 150 mM NaCl. The protein concentration of CueP was measured based on the molar extinction coefficient. The CueP protein (0.2 mM) or L-cysteine (0.6 mM), dissolved in Tris buffer, was incubated with 0.5 mM CuCl2 for a given time. The error bars indicate the standard deviation from three independent experiments. The free thiol group was quantified using the agent DTNB by measuring absorbance at 412 nm. (B) Representative EPR spectra of Cu2+ around 3400 G in the presence/absence of wild-type or variant CueP. The sample containing 0.5 mM CuCl2 and wild-type or H94A CueP protein (0.4 mM) was incubated for 5 min at room temperature and then applied to EPR at −197°C. More than three independent experiments were performed and produced similar results. Each spectrum was offset for clarity. (C) Titration with CuCl2 to the oxidation of CueP. Each amount of CuCl2 was incubated with 0.2 mM CueP protein for 15 min, and then the free thiol content was measured using the agent DTNB. The error bars indicate the standard deviation from three independent experiments.
Since Cu2+ brings about the oxidation of CueP, we undertook an electron paramagnetic resonance spectroscopy (EPR) investigation to verify the reduction of Cu2+ by CueP. The wild-type CueP protein decreased the concentration of Cu2+ by converting it to a more reduced form of Cu (Cu+ or Cu0) (Fig. 3B). However, a non-functional mutant (H94A; see below) CueP did not result in this reaction and was used as a control. Unfortunately, we were not able to distinguish whether the product of this reaction is Cu+ or Cu0 because both Cu+ and Cu0 are undetectable in EPR.
To further analyze the reaction product, we titrated the oxidation of CueP (200 μM) with CuCl2. When the molar ratio of CueP to Cu2+ was about 1, the oxidation of CueP was saturated (Fig. 3C). This result suggests that the stoichiometry of CueP to Cu2+ is ∼1, and one Cu2+ is required to form one disulfide bond in CueP. However, an NMR-based experiment indicated that one electron was transferred from L-cysteine to Cu2+, resulting in 1/2 cystine and Cu+ (Rigo et al., 2004). Therefore, our result suggests that the disulfide bond formation of CueP, which provides two electrons, is coupled to the transition of Cu2+ to Cu0.
His94, His99 and Cys104 play an important role in maintaining the reduced state of CueP in the absence of Cu2+
To investigate the role of the conserved cysteine and histidine residues at the cysteine-rich region, we mutated Cys96, Cys104, Cys172, His94, and His99 into serine or alanine and measured the free thiol concentrations of the proteins in reducing agent-free buffer. To see the changes in the oxidation states of the proteins, we also compared the free thiol concentrations of the proteins before and after exposure to 0.5 mM CuCl2 (Fig. 4A). Among the five mutants, H99A, C104S, and C172S exhibited significantly decreased free thiol concentrations, even prior to exposure to CuCl2, (Fig. 4A). H94A also displayed a decreased free thio concentration in the absence of CuCl2, albeit the decreasing amount was smaller than H994, C104S, and C172S variant protein. However, the C96S had significant fractions of free thiol before addition of CuCl2 and were partly oxidized by the addition of CuCl2 although the oxidation rates were less than those of the wild-type protein. The most protein of C172S mutant existed as a tetramer on a size exclusion chromatography, indicating that the intermolecular disulfide bonds were easily formed in this mutant protein (Supplementary Fig. S2).
Fig. 4.
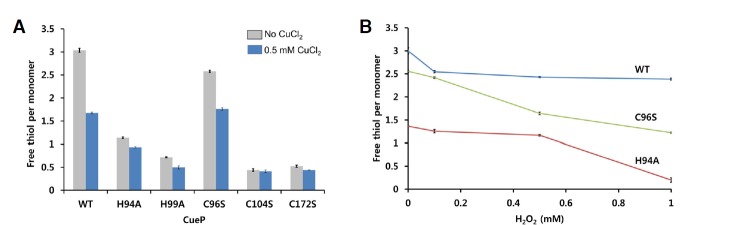
The role of the conserved residues in the cysteine-rich region. (A) The amount of free thiol group in the CueP proteins (wild-type and mutants), measured using the agent DTNB. Freshly prepared protein sample (0.2 mM) was incubated with 0 or 0.5 mM CuCl2 for 1 min in 20 mM Tris buffer (pH 8.0) containing 150 mM NaCl, and then subjected to the assays. (B) Reactivity of CueP to H2O2. Each CueP protein (0.2 mM) was incubated with the indicated concentration of H2O2 for 15 min, and then the free thiol contents were measured using the agent DTNB. Only wild-type (WT), H94A and C96S mutants are displayed. The mutant proteins H99A, C104S, and C172S are displayed in Supplementary Fig. S3.
We next measured the sensitivity of CueP to oxidative stress produced by H2O2. H2O2 did not efficiently decrease the amount of free thiol in wild-type CueP within 15 min (Fig. 4B). In this experiment, C96S and H94A made the mutant protein more sensitive to oxidation by H2O2 than wild-type CueP, although the mutant protein conferred some resistance to oxidation by molecular oxygen in reducing agent-free buffer. By contrast, the other mutant proteins (H99A, C104S, and C172S) had been already oxidized by oxygen during the dialysis against the reducing agent-free buffer, so the proteins were not further oxidized by H2O2 (Supplementary Fig. S3).
CueP efficiently scavenges Cu2+-mediated Fenton reaction
The Cu2+ titration indicated that CueP catalyzes the conversion of Cu2+ to Cu0. Since elemental copper Cu0 cannot catalyze the Fenton reactions, we expected that the CueP protein would inhibit the Fenton reactions by depleting copper ions. Thus we tested whether CueP can prevent the Cu2+/Cu+-mediated Fenton reaction in vitro. The CueP protein was incubated in a reaction mixture containing supercoiled plasmids and CuCl2, to which H2O2 was added in order to trigger the Fenton reaction. Supercoiled plasmids were used to probe the generation of hydroxyl radicals in this experiment, because hydroxyl radicals, but not H2O2, can make a single strand breakage in the plasmid, leading to the generation of an open-circle plasmid (Netto et al., 1996). As shown in Fig. 5A, the CueP protein in the reduced state protected more than 50% of the DNA from hydroxyl radicals, whereas the protein in the oxidized state resulted in the complete DNA cleavage. We next performed the same assay to observe the dose-dependent effect of the reduced CueP protein. Interestingly, CueP showed maximum hydroxyl radical scavenging activity at the molar ratio of 1:1 (CueP: Cu2+), and the activity disappeared when more CueP protein was added. At the molar ratio of 4:1, the scavenging activity was decreased by about 50%, compared to the maximum scavenging activity at the molar ratio of 1:1. These results indicate that the ratio of the protein to Cu2+ might affect the chemical reaction (Fig. 5B). These results show that the CueP protein could diminish the Fenton reaction by reducing the copper ion to elemental copper at the expense of a disulfide bond in a certain range of the protein concentration.
Fig. 5.
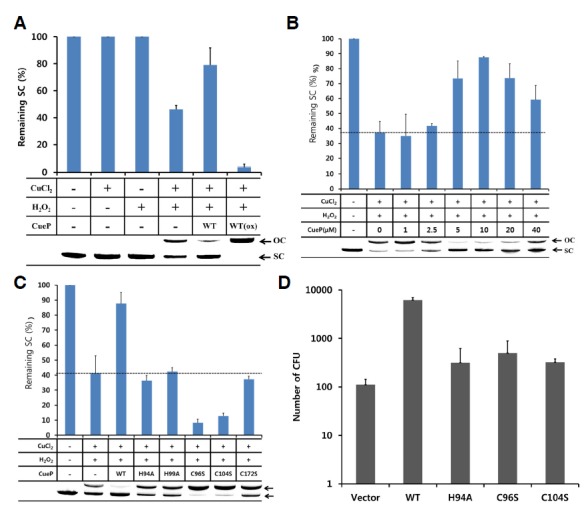
CueP prevents the copper ion-mediated Fenton reaction. (A) An assay for hydroxyl radicals generated from H2O2 in the presence of the copper ion using supercoiled plasmid. The hydroxyl radicals cleave the supercoiled plasmid (SC) into the open circle form (OC). After reaction of the reduced (CuePred) or oxidized (CuePOx) form of CueP protein with 10 μM CuCl2, the plasmid and 2 mM H2O2 was added and incubated for 5 min at room temperature and then immediately subjected to agarose gel electrophoresis (0.8%). The DNA was visualized by EtBr staining. Data points represent the mean for two independent experiments. A representative gel figure is shown in the bottom. (B) The hydroxyl radical formation assay in a dose-dependent manner of the CueP protein. The reduced form of CueP was added to the reaction mixture as indicated. The reaction conditions are the same as in Fig. 5A. Data points represent the mean (± S.E) for at least three independent experiments. The dashed-line indicates the level of the remaining supercoiled plasmid in the absence of CueP in our experimental condition for the Fenton reaction. A representative gel figure is shown in the bottom. (C) The role of the conserved cysteine or histidine residues in preventing the formation of hydroxyl radicals in the presence of Cu2+ and H2O2. The same amount of wild-type or mutant CueP proteins was added to the reaction mixture. The reaction conditions are the same as in Fig. 5A. Data points represent the mean (± S.E) for at least three independent experiments. The dashed-line indicates the level of the remaining supercoiled plasmid in the absence of CueP in our experimental condition for the Fenton reaction. A representative gel figure is shown in the bottom. (D) The effect of S. Typhimurium CueP in E. coli under cusB-deleted background. Wild-type or mutant S. Typhimurium cueP was inserted into pACYC177 vector, and the vector was transformed into the E. coli strain. Exponentially grown E. coli in LB was immediately washed with 20 mM Hepes buffer (pH 7.5) at room temperature. The cells were incubated with 0.1 mM CuCl2 in 20 mM Hepes buffer to allow the copper ions to diffuse into the bacterial cell and then washed with copper-free buffer. H2O2 (30 mM) was incubated with the cell for 5 min to induce the Fenton reaction within the cell, and then the cells were plated to measure the survival of the bacteria. Data points represent the mean (± S.E) for at least three independent experiments.
To examine the importance of the conserved cysteine and histidine residues, the hydroxyl radical scavenging assays were performed with mutant proteins. As shown in Fig. 5C, any single mutation at the conserved residues completely abolished the DNA protection activity of CueP. Some of the mutants (C96S and C104S) rather accelerated the reaction. Thus, these cysteine and histidine residues together play an important role in the inhibition of the Fenton reactions by the CueP action on the copper ion in the presence of H2O2.
CueP increases the survival of E. coli in Fenton reaction
Iron ions are sequestered to limit the growth of bacteria in macrophage phagosome, although iron ions mediate the Fenton reaction in the presence of H2O2 (Schaible and Kaufmann, 2004). Instead, copper ions are transported to the phagosome to enhance the toxicity of ROS or RNS (Leary and Winge, 2007; White et al., 2009b). To examine whether the purified CueP protein can protect the live bacteria from the Fenton reaction mediated by copper ions, we introduced the cueP gene, including its copper-responsive promoter region, into the cusB-deleted strain of E. coli. As experimental controls, the empty vector or the non-functional mutants (H94A, C96S and C104S) were also transformed to evaluate the role of wild-type CueP. Exponentially grown cells were washed with 20 mM Hepes buffer and incubated with 0.1 mM CuCl2 for diffusion into the bacterial cell. After washing the cells with copper-free buffer, the cells were treated with 30 mM H2O2 for 5 min to induce the Fenton reaction and then cultivated on LB agar plate to measure the survival of the bacteria.
Neither CuCl2 nor H2O2 treatment alone significantly killed the bacteria (Fig. 5D). In contrast, when treated with CuCl2 and H2O2 together, E. coli cells expressing wild-type CueP showed higher survivals by about 100 times than those harboring an empty vector or a mutant gene. This result indicated that wild-type CueP was able to protect E. coli cells against copper ion-mediated H2O2 decomposition, which generates hydroxyl radicals as the ultimate cell-killing agents. However, the mutant CueP proteins showed a marginal or similar effect to empty vector, which failed to protect against the cell-killing effects of CuCl2 and H2O2. Therefore, the conserved histidine and cysteine residues are crucial for CueP’s role.
We found an inconsistency in the experimental results using the mutants. In the DNA protection assay using the purified proteins, some of the mutant proteins showed the decreased DNA scavenge activities compared to that in the absence of the CueP protein used as a control (Fig. 5C). However, when the mutant cueP was expressed in E. coli, the mutant cueP partly contributed to the survival of the bacteria in the experimental condition (Fig. 5D). We speculate that this inconsistency might results from a beneficial role of the expressed CueP in E. coli. For example, the CueP mutants, expressed in the E. coli periplasm, could transfer a copper ion to the periplasmic SodCII, as observed in S. Typhimurium (Osman et al., 2013).
DISCUSSION
Many pathogenic bacteria can evade or resist the bacterial cell killing mechanism of macrophages. In the intracellular bacteria S. enterica, the periplasmic protein CueP was identified as a functional substitute for the Cus Cu+ efflux pump (Pontel and Soncini, 2009). Previously, CueP from S. Typhimurium was suggested to transfer a copper ion to SodCII in the periplasm at a low copper concentration, leading to activation of SodCII (Osman et al., 2013). In this study, we studied the biochemical role of CueP at a higher concentration of copper. Since the copper levels are elevated and accumulate in the phagosome of macrophages exposed to inflammatory cytokines via specific transporters (Achard et al., 2012; White et al., 2009a; Wolschendorf et al., 2011), our results suggest that CueP may be associated with the pathogenesis of the bacteria in the macrophage phagosomes.
Copper ions can catalyze the Fenton reaction by alternating between Cu+ and Cu2+, and consequently generate highly toxic hydroxyl radicals (Czech et al., 1974). Cu+ mediates the decomposition of H2O2 by transferring one electron, generating the hydroxyl radical, hydroxide ion, and Cu2+ (reaction 1); Cu2+ is then reduced back to Cu+ by a complex series of chain reactions from H2O2 (reaction 2). These reactions continue generating hydroxyl radicals until all H2O2 is consumed (Bae et al., 2011). Since the rate of reaction 1 is significantly greater than that of reaction 2, the generation rate of hydroxyl radical in solution is limited by the regeneration of Cu+. When L-cysteine is added, the conversion of Cu+ from Cu2+ is accelerated, resulting in formation of cysteine (Cys-Cys) (reaction 3).
| (reaction 1) |
| (reaction 2) |
| (reaction 3) |
| (reaction 4) |
Our results suggest that the reduced form of CueP (CueP2Cys) catalyzes the conversion of Cu2+ to Cu0, as described in reaction 4. Since Cu0 is not able to mediate the Fenton reaction in the presence of H2O2, the CueP reaction with Cu2+ attenuates the formation of hydroxyl radical. In a previous report, CueP partly restored the copper resistance of the cus-deleted strain of E. coli, particularly under anaerobic conditions (Pontel and Soncini, 2009). This observation is well explained by the Cu2+-reducing activity of CueP revealed in this study. Reduction of copper ions by CueP can protect the bacteria from the copper ion-mediated decomposition of H2O2 to hydroxyl radical, considered the ultimate cell-killing agent of the phagosome. Since the reaction of CueP with copper ion is free of an oxygen molecule and relatively insensitive to molecular oxygen and H2O2, it can act either under anaerobic or aerobic conditions. These properties confer CueP with a great advantage over the copper oxidase CueO (Grass and Rensing, 2001). In the presence of H2O2, the reaction product of CueO, Cu2+, contributes to Fenton chemistry (reaction 2), although it can reduce the rate of hydroxyl radical generation by Cu+ (reaction 1). However, CueP can block the total Fenton reactions by the conversion of copper ions to elemental copper atoms.
S. Typhimurium causes critical diseases for animals including humans, leading to substantial morbidity, mortality and a considerable disease burden globally (Coburn et al., 2007). Since the gene deletion or mutation of the highly conserved amino acids of CueP diminishes the ability of pathogens to live in the host immune cells, CueP may be a good target in the development of antibacterial agents. Furthermore, the results of this study will increase our understanding of the roles of copper tolerance at the host-pathogen interface.
Supplemental Figures
Acknowledgments
This research was supported by a 2-Year Research Grant of Pusan National University.
Note:
Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Achard M.E., Stafford S.L., Bokil N.J., Chartres J., Bernhardt P.V., Schembri M.A., Sweet M.J., McEwan A.G. Copper redistribution in murine macrophages in response to Salmonella infection. Biochem. J. 2012;444:51–57. doi: 10.1042/BJ20112180. [DOI] [PubMed] [Google Scholar]
- Bae Y.S., Oh H., Rhee S.G., Yoo Y.D. Regulation of reactive oxygen species generation in cell signaling. Mol Cells. 2011;32:491–509. doi: 10.1007/s10059-011-0276-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn B., Grassl G.A., Finlay B.B. Salmonella, the host and disease: a brief review. Immunol. Cell Biol. 2007;85:112–118. doi: 10.1038/sj.icb.7100007. [DOI] [PubMed] [Google Scholar]
- Crichton R.R., Pierre J.L. Old iron, young copper: from Mars to Venus. Biometals. 2001;14:99–112. doi: 10.1023/a:1016710810701. [DOI] [PubMed] [Google Scholar]
- Czech M.P., Lawrence J.C., Jr., Lynn W.S. Evidence for electron transfer reactions involved in the Cu2+-dependent thiol activation of fat cell glucose utilization. J. Biol. Chem. 1974;249:1001–1006. [PubMed] [Google Scholar]
- Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont C.L., Grass G., Rensing C. Copper toxicity and the origin of bacterial resistance--new insights and applications. Metallomics. 2011;3:1109–1118. doi: 10.1039/c1mt00107h. [DOI] [PubMed] [Google Scholar]
- Ellman G.L. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- Eng J.K., McCormack A.L., Yates J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc Mass Spectrom. 1995;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Espariz M., Checa S.K., Audero M.E., Pontel L.B., Soncini F.C. Dissecting the Salmonella response to copper. Microbiology. 2007;153:2989–2997. doi: 10.1099/mic.0.2007/006536-0. [DOI] [PubMed] [Google Scholar]
- Fields P.I., Swanson R.V., Haidaris C.G., Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke S., Grass G., Rensing C., Nies D.H. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 2003;185:3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass G., Rensing C. CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem. Biophys. Res. Commun. 2001;286:902–908. doi: 10.1006/bbrc.2001.5474. [DOI] [PubMed] [Google Scholar]
- Halliwell B., Gutteridge J.M. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson V., Petris M.J. Copper homeostasis at the host-pathogen interface. J. Biol. Chem. 2012;287:13549–13555. doi: 10.1074/jbc.R111.316406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt O., Grunert H.P., Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990;96:125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- Leary S.C., Winge D.R. The Janus face of copper: its expanding roles in biology and the pathophysiology of disease. Meeting on Copper and Related Metals in Biology. EMBO Rep. 2007;8:224–227. doi: 10.1038/sj.embor.7400915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netto L.E.S., Chae H.Z., Kang S.W., Rhee S.G., Stadtman E.R. Removal of hydrogen peroxide by thiol-specific antioxidant enzyme (TSA) is involved with its antioxidant properties. TSA possesses thiol peroxidase activity. J. Biol. Chem. 1996;271:15315–15321. doi: 10.1074/jbc.271.26.15315. [DOI] [PubMed] [Google Scholar]
- O’Halloran T.V., Culotta V.C. Metallochaperones, an intracellular shuttle service for metal ions. J. Biol. Chem. 2000;275:25057–25060. doi: 10.1074/jbc.R000006200. [DOI] [PubMed] [Google Scholar]
- Osman D., Cavet J.S. Copper homeostasis in bacteria. Adv. Appl. Microbiol. 2008;65:217–247. doi: 10.1016/S0065-2164(08)00608-4. [DOI] [PubMed] [Google Scholar]
- Osman D., Waldron K.J., Denton H., Taylor C.M., Grant A.J., Mastroeni P., Robinson N.J., Cavet J.S. Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. J. Biol. Chem. 2010;285:25259–25268. doi: 10.1074/jbc.M110.145953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman D., Patterson C.J., Bailey K., Fisher K., Robinson N.J., Rigby S.E., Cavet J.S. The copper supply pathway to a Salmonella Cu,Zn-superoxide dismutase (SodCII) involves P(1B)-type ATPase copper efflux and periplasmic CueP. Mol. Microbiol. 2013;87:466–477. doi: 10.1111/mmi.12107. [DOI] [PubMed] [Google Scholar]
- Pontel L.B., Soncini F.C. Alternative periplasmic copper-resistance mechanisms in Gram negative bacteria. Mol. Microbiol. 2009;73:212–225. doi: 10.1111/j.1365-2958.2009.06763.x. [DOI] [PubMed] [Google Scholar]
- Prohaska J.R., Lukasewycz O.A. Copper deficiency suppresses the immune response of mice. Science. 1981;213:559–561. doi: 10.1126/science.7244654. [DOI] [PubMed] [Google Scholar]
- Rensing C., Fan B., Sharma R, Mitra B., Rosen BP. CopA: An Escherichia coli Cu(I)-translocating P-type ATPase. Proc. Natl. Acad. Sci USA. 2000;97:652–656. doi: 10.1073/pnas.97.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigo A., Corazza A., di Paolo M.L., Rossetto M., Ugolini R., Scarpa M. Interaction of copper with cysteine: stability of cuprous complexes and catalytic role of cupric ions in anaerobic thiol oxidation. J. Inorg. Biochem. 2004;98:1495–1501. doi: 10.1016/j.jinorgbio.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Schaible U.E., Kaufmann S.H. Iron and microbial infection. Nat. Rev. Microbiol. 2004;2:946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- White C., Kambe T., Fulcher Y.G., Sachdev S.W., Bush A.I., Fritsche K., Lee J., Quinn T.P., Petris M.J. Copper transport into the secretory pathway is regulated by oxygen in macrophages. J. Cell Sci. 2009a;122:1315–1321. doi: 10.1242/jcs.043216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C., Lee J., Kambe T., Fritsche K., Petris M.J. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J. Biol. Chem. 2009b;284:33949–33956. doi: 10.1074/jbc.M109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolschendorf F., Ackart D., Shrestha T.B., Hascall-Dove L., Nolan S., Lamichhane G., Wang Y., Bossmann S.H., Basaraba R.J., Niederweis M. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci USA. 2011;108:1621–1626. doi: 10.1073/pnas.1009261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N., Nakahigashi K., Nakamichi T., Yoshino M., Takai Y., Touda Y., Furubayashi A., Kinjyo S., Dose H., Hasegawa M., et al. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 2009;5:335. doi: 10.1038/msb.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon B.Y., Kim Y.H., Kim N., Yun B.Y., Kim J.S., Lee J.H., Cho H.S., Lee K., Ha N.C. Structure of the periplasmic copper-binding protein CueP from Salmonella enterica serovar Typhimurium. Acta Crystallogr. D Biol. Crystallogr. 2013;69:1867–1875. doi: 10.1107/S090744491301531X. [DOI] [PubMed] [Google Scholar]
- Yun B.Y., Piao S., Kim Y.G., Moon H.R., Choi E.J., Kim Y.O., Nam B.H., Lee S.J., Ha N.C. Crystallization and preliminary X-ray crystallographic analysis of Salmonella Typhimurium CueP. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011;67:675–677. doi: 10.1107/S1744309111010645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.