Abstract
Toll-like receptor 3 (TLR3) activation plays an important role in the innate immune responses to viral infections. We show here that the activation of TLR3 signaling pathway by poly I:C, a synthetic mimic of dsRNA, could induce high-level expression of interferon (IFN)-λ1 in a hepatoma cell line. The induced IFN-λ1 contributed to poly I:C-mediated inhibition of hepatitis C virus (HCV) Japanese fulminant hepatitis-1 (JFH-1) replication in Huh7 cells. This inhibitory effect of poly I:C on HCV replication, however, was compromised by HCV infection of Huh7 cells. Investigation of the mechanisms showed that HCV infection suppressed the expression of poly I:C-induced IFN-λ1 and IFN-stimulated genes [IFN-stimulated gene 56 (ISG-56), myxovirus resistance A (MxA) and 2′-5′-oligoadenylate synthetase 1 (OAS-1))], the key antiviral elements in IFN signaling pathway. Among the HCV nonstructural (NS) proteins tested, NS3/4A, NS5A and NS5B had the ability to inhibit poly I:C-induced IFN-λ1 expression in Huh7 cells. These observations provide the experimental evidence that HCV and its proteins impair TLR3 signaling and inhibit intracellular IFN-λ1/ISG expression in a hepatoma cell line, which may account for HCV persistence in the liver.
Keywords: Hepatitis C virus, interferon-λ, IFN-stimulated genes, poly I:C, Toll-like receptor 3
Introduction
Hepatitis C virus (HCV) infection is a major cause of chronic hepatitis, liver cirrhosis and hepatocellular carcinoma. 1 The clinical outcome of HCV infection and the degree of liver damage are the result of complicated interactions between the virus and host immune responses.2 Although cellular and humoral immune responses are present during acute and chronic HCV infection, these immune responses appear to be ineffective in eradicating the virus.3 The majority of HCV-infected patients develop chronic infection, suggesting that HCV has evolved strategies to overcome or evade host immune responses.4–8
Viral infections result in induction of interferons (IFNs) that are the key players in antiviral responses.9 Induction of the IFN-based antiviral innate immunity depends on several Toll-like receptors (TLRs), i.e. TLR3, 7 and 9.10 TLR3 is a major intracellular sensor that recognizes dsRNA and triggers the IFN signaling pathway. However, persistent HCV infection is associated with its ability to disrupt and evade TLR3-mediated immune responses. HCV nonstructural (NS)3/4A protease ablates TLR3 signaling by cleaving the TLR3 adaptor protein Toll-IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF) to evade type I IFN-mediated innate immunity.11 Recent studies have demonstrated that, in addition to IFN-α/β, IFN-λ also has the ability to inhibit HCV replication in both in vitro and in in vivo systems.12–17 In the present study we examined whether TLR3 signaling induces IFN-λ expression and whether HCV inhibits IFN-λ expression in hepatoma cell line.
Materials and methods
Cells, plasmids and virus
A hepatoma cell line (Huh7) provided by Dr Charles Rice (The Rockefeller University, New York, NY, USA) was maintained in DMEM with 10% FBS, penicillin (100 U/ml) and streptomycin (100 μg/ml). HCV NS3, NS3/4A, NS5A and NS5B plasmids, and control plasmid (pcDNA 3.1) were obtained from Dr Michael Gale (University of Washington, Seattle, WA, USA).5 The generation of infectious HCV JFH-1 and infection of Huh7 cells [multiplicity of infection (MOI) of 0.01] were carried out as described previously.18,19 HCV JFH-1 infection of Huh7 was analyzed by the immunostaining with the mouse anti-HCV core Ab or by real time RT-PCR for HCV RNA.
Reagents
LyoVec transfection reagent, poly I:C (high molecular mass) were purchased from Invivogen (San Diego, CA, USA). ELISA kit for IFN-λ1 was purchased from eBioscience Inc. (San Diego, CA, USA). Mouse Ab against HCV core antigen was purchased from ABR Affinity BioReagents, Thermo Scientific (Rockford, IL, USA). Rabbit Abs against IFN-stimulated gene 56 (ISG56), myxovirus resistance A (MxA), 2′-5′-oligoadenylate synthetase 1(OAS-1) and actin were purchased from Sigma-Aldrich (St Louis, MO, USA). Rabbit anti-IFN stimulated transcription factor 3 gamma (ISGF3G) Ab was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Hoechst 33342 was purchased from Molecular Probes (Carlsbad, CA, USA).
Poly I:C stimulation
Huh7 cells were seeded at a density of 105 each well in a 24 well-plate. After culturing for 24 h, the cells were stimulated with poly I:C (0.2–5 μg/ml) using LyoVec transfection reagent. Cells were collected for mRNA extraction after 24 h stimulation and supernatant (SN) was collected for ELISA assay after 24–48 h stimulation. For the anti-HCV experiment, the cells were stimulated with poly I:C for 8 h prior to HCV JFH-1 infection. Culture SN and cells were collected for mRNA extraction at JFH-1 72 h post-infection. For the poly I:C stimulation of HCV JFH-1-infected Huh7 cells experiment, JFH-1-infected Huh7 cells (72 h post-infection) were stimulated with poly I:C (0.2–5 μg/ml) using LyoVec transfection reagent. Culture SN and cells were collected for mRNA extraction after 48 h stimulation. As a negative control of the transfection experiment, cells were incubated with the LyoVec transfection reagent only. For the blocking experiments with Bafilomycin A1, a vacuolar H +- ATPase inhibitor that inhibits the acidification of endosome,20 Huh7 cells were treated with 100 nM of Bafilomycin A1 for 1 h prior to poly I:C stimulation.
Plasmid transfection
Transfection of the plasmid DNA was carried out with LyoVec transfection reagent, as recommended by the manufacturer. The Huh7 cells were transfected with the plasmids (HCV NS3, NS3/4A, NS5A and NS5B) using LyoVec transfection reagent at a ratio of LyoVec: plasmid 50:1 (μl:μg) at a final plasmids concentration of 0.5 μg/ml of each cell culture well. The empty vector (pcDNA 3.1) for HCV NS genes containing constructs was used as the control and included in parallel for each transfection experiment.
RNA extraction and real time RT-PCR
Total RNA from culture cells or SN was extracted with Tri-Reagent (Molecular Research Center, Cincinnati, OH, USA) as described previously.21 Total RNA (1 μg) was subjected to RT using the RT system (Promega, Madison, WI, USA) with random primers for 1 h at 42°C. The reaction was terminated by incubating the reaction mixture at 99°C for 5 min, and the mixture was kept at 4°C. The resulting cDNA was used as a template for real time PCR quantification. Real time PCR was performed with 1/10 of the cDNA with the iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA) as described previously. 22 The amplified products were visualized and analyzed using the software MyiQ provided with the thermocycler (iCycler iQ real time PCR detection system; Bio-Rad Laboratories). The oligonucleotide primers were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA, USA) and sequences will be available upon request. The cDNA was amplified by PCR and the products were measured using SYBR green I (Bio-Rad Laboratories). The data were normalized to GAPDH and presented as the change in induction relative to that of untreated control cells.
ELISA
IFN-λ1 gene expression analyzed by the real time RT-PCR was evaluated by ELISA for protein expression. SN collected from poly I:C-stimulated Huh7 cultures was tested directly for IFN-λ1 protein levels by ELISA, which was performed according to the manufacturer’s instructions.
Statistical analysis
Student’s t-test was used to evaluate the significance of difference between groups, and multiple comparisons were performed by regression analysis and ANOVA. P-Values< 0.05 were considered significant. All data are presented as mean ± SD. Statistical analyses were performed with SPSS 11.5 for Windows (SPSS, Chicago, IL, USA). Statistical significance was defined as P<0.05.
Results
TLR3 signaling induces IFN-λ1 expression
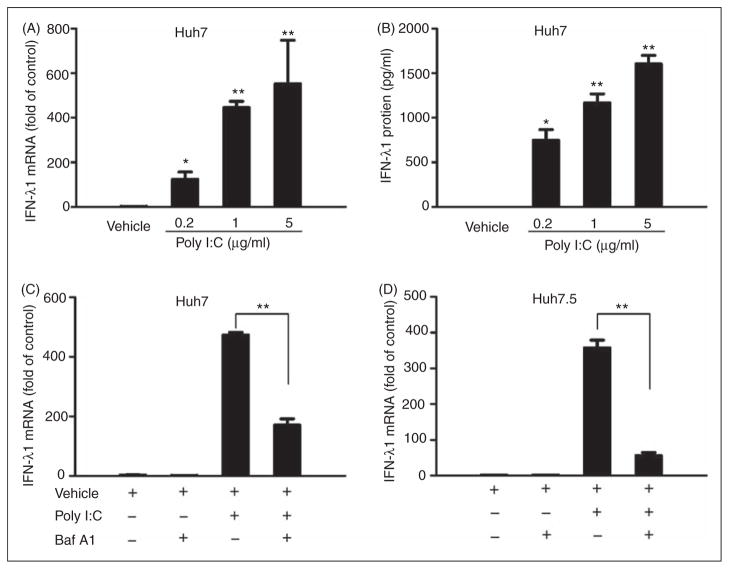
It is known that the activation of TLR3 can induce type I IFNs in human hepatocytes.23 However, it is unclear whether TLR3 signaling has an effect on IFN-λ expression. We showed that poly I:C stimulation of Huh7 cells significantly induced the expression of IFN-λ1 at both mRNA (Figure 1A) and protein (Figure 1B) levels, and this effect was dose dependent. To investigate whether TLR3 activation is responsible for poly I:C-induced IFN-λ1 expression, Bafilomycin A1, an inhibitor of the TLR3 signaling pathway, was used to block the poly I:C action. As shown in Figure 1C, Bafilomycin A1 treatment compromised the poly I:C-induced IFN-λ1 expression in Huh7 cells. To determine whether retinoic acid-inducible gene I (RIG-I) signaling is involved in poly I:C-mediated IFN-λ induction, we used Huh7.5 cells that have a defect in the RIG-I signaling pathway18,24 in the experiments. We showed that poly I:C could induce IFN-λ1 expression in Huh7.5 cells, which could be largely blocked by Bafilomycin A1 treatment (Figure 1D).
Figure 1.
TLR3 activation induces IFN-λ1 expression. (A) Effect of poly I:C on IFN-λ1 mRNA expression. Huh7 cells were stimulated with poly I:C at the indicated concentrations for 24 h. (B) Effect of poly I:C on IFN-λ1 protein production. Huh7 cells were stimulated with poly I:C at the indicated concentrations for 48 h. SN was collected from the cell cultures for ELISA to measure the protein levels of IFN-λ1. (C, D) Blocking TLR3 signaling compromises poly I:C-induced IFN-λ1 expression. Huh7 (C) or Huh7.5 (D) cells were pretreated with or without Bafilomycin A1 (100 nM) for 1 h prior to poly I:C stimulation (24 h). Total RNA extracted from cells was subjected to the real time RT-PCR for the mRNA levels of IFN-λ1 and GAPDH. The data are expressed IFN-λ1 mRNA levels relative (fold) to the control (vehicle only, which defined as 1). The results shown are mean ± SD of triplicate cultures, representative of three experiments (poly I:C stimulated Huh7 cell vs vehicle only, **P<0.01, *P<0.05).
TLR3 signaling induces IFN-stimulated gene expression
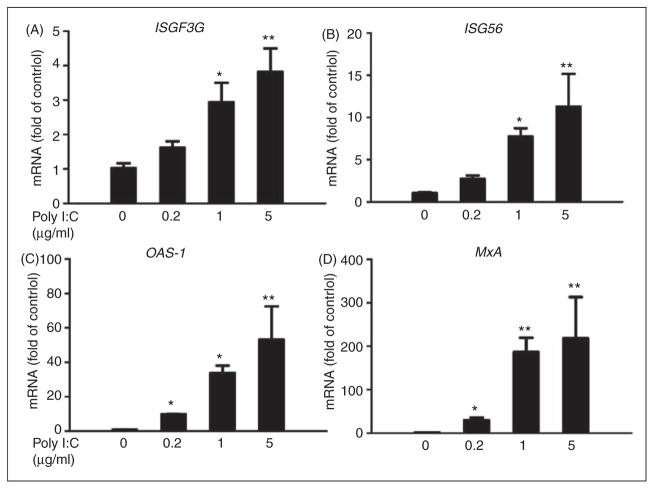
ISGF3G also known as IFN regulatory factor 9 (IRF-9), plays a critical role in ISGs induction.25 We observed that poly I:C induced ISGF3G expression in Huh7 cells at both mRNA (Figure 2A) and protein (Figure 3) levels. We next examined the effect of poly I:C on the ISGs that have antiviral effects. As shown in Figure 2B–D, poly I:C significantly induced ISG56, MxA and OAS-1 mRNA expression in a dose-dependent manner. We also examined the protein levels of these ISGs; as shown in Figure 3, poly I:C could increase the protein levels of ISG 56, OAS-1 and MxA, the key antiviral elements in the IFN signaling pathway.26–29
Figure 2.
Effect of poly I:C on the mRNA expression of ISGs. Huh7 cells were stimulated with poly I:C at the indicated concentrations for 24 h. Total RNA extracted from cells was subjected to the real time RT-PCR for the mRNA levels of ISGF3G (A), ISG56 (B), OAS-1 (C), MxA (D) and GAPDH. The data are expressed as mRNA levels relative (fold) to the control (vehicle only, which defined as 1). The results shown are mean ± SD of triplicate cultures, representative of three experiments (poly I:C stimulated Huh7 cell vs vehicle only, **P<0.01, *P<0.05).
Figure 3.
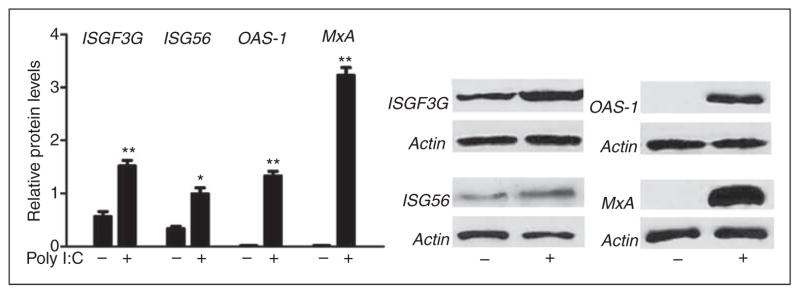
Poly I:C induces ISGs protein expression. Huh7 cells were stimulated with poly I:C (1μg/ml) for 48 h. Total proteins extracted from cell cultures were subjected to Western blot assay using the Abs against ISGF3G, ISG56, OAS-1, MxA and β-actin. Representative blots from three independent experiments were shown. Densitometry analysis of the blot was performed using ImageJ 1.44 software (National Institutes of Health, Bethesda, MD, USA) and plotted into graphs from data collected from triplicate experiments (poly I:C stimulated Huh7 cell vs vehicle only, **P<0.01, *P<0.05).
TLR3 signaling inhibits HCV replication
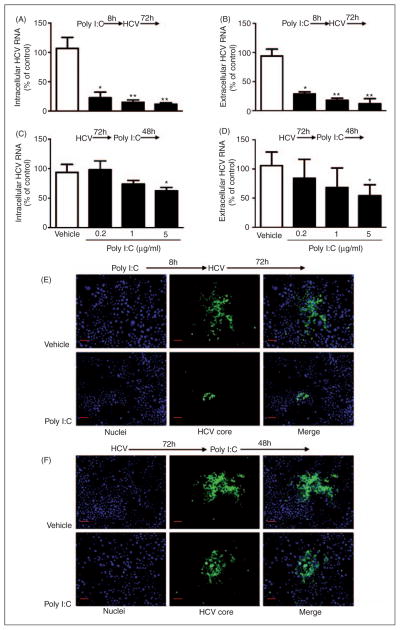
Knowing that TLR3 signaling could induce the expression of IFN-λ1 and the antiviral ISGs, we then examined whether TLR3 signaling of Huh7 cells can inhibit HCV replication. As shown in Figure 4A and B, poly I:C significantly inhibited HCV JFH-1 infection of Huh7 cells and the degree of the suppression was correlated positively with the doses of poly I:C. This inhibitory effect of poly I:C on HCV could be observed before or after HCV infection (Figure 4). However, TLR3 signaling by poly I:C prior to HCV infection (Figure 4A, C, E) was much more potent in HCV inhibition than that after HCV infection (Figure 4B, D, F).
Figure 4.
Effect of poly I:C on HCV replication. (A–D) Effect of Poly I:C on HCV RNA replication. Huh7 cells were stimulated with poly I:C at indicated concentration for 8 h prior to HCV JFH-1 infection (A, B) or stimulated with poly I:C at 72 h post-infection (C, D). Intracellular (A, C) and extracellular (B, D) RNA extracted from Huh7 cells or culture SN was subjected to the real time RT-PCR for HCV and GAPDH RNA quantification. HCV RNA level is expressed as HCV RNA levels relative (%) to the control (vehicle only, which are defined as 100%). The results shown are mean ± SD of triplicate cultures, representative of three experiments (poly I:C-stimulated Huh7 cell vs vehicle-treated cells only: **P<0.01, *P<0.05). (E, F) Effect of poly I:C on HCV core protein expression. Huh7 cells were stimulated with or without poly I:C (1 μg/ml) for 8 h prior to HCV JFH-1 infection (E) or 72 h post-infection for 48 h (F). HCV core protein expression was determined by immunofluorsence staining with Ab against HCV core protein (green). The nuclei were stained with Hoechst 33342 (blue). One representative experiment is shown (original magnification 200×, scale bar: 20 μM).
HCV suppresses IFN-λ1 and ISG expression
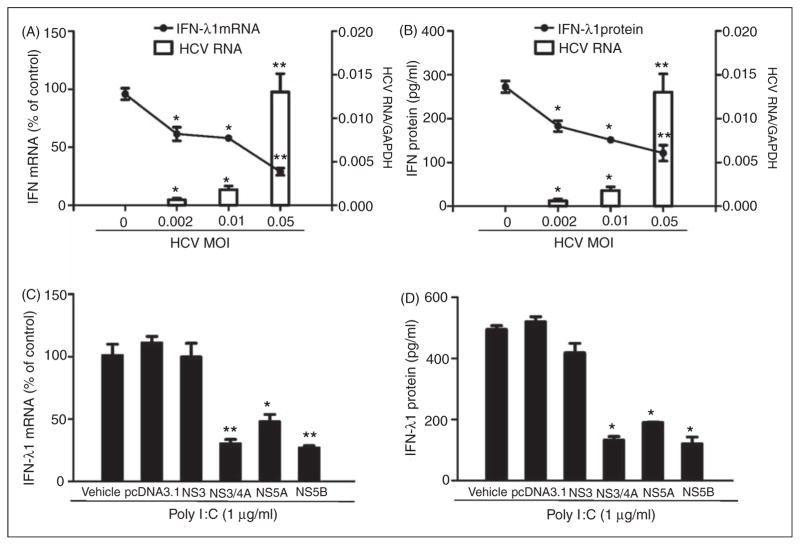
We next examined whether HCV has the ability to inhibit TLR3 activation-induced IFN-λ1 and ISG expression. We infected Huh7 cells with HCV JFH-1 at different doses (MOI 0.002, 0.01 and 0.05) for 72 h prior to poly I:C stimulation. We observed that the poly I:C-induced IFN-λ1 expression was decreased at both mRNA (Figure 5A) and protein (Figure 5B) levels in HCV-infected cells. Similarly, HCV infection inhibited the expression of the ISG induced by poly I:C (Figure 6A–C). To determine which HCV protein(s) play a major role in HCV-mediated down-regulation of intracellular IFN-λ1 expression induced by poly I:C in Huh7 cells, we transfected Huh7 cells with the plasmids containing HCV genes that encode NS proteins for 18 h prior to poly I:C stimulation. The expression of the HCV NS3 protein had little effect on poly I:C-induced IFN-λ1 expression. In contrast, the expression of HCV NS3/4A, NS5A and NS5B proteins in Huh7 cells resulted in the significant suppression of poly I:C-induced IFN-λ1 expression at both mRNA (Figure 5C) and protein (Figure 5D) levels.
Figure 5.
HCV inhibits poly I:C-induced IFN-λ1 expression. (A, B) Huh7 cells were infected with HCV JFH-1 at indicated MOI for 72 h, then stimulated with poly I:C (1 μg/ml) for 24 h. (C, D) Effect of HCV NS proteins on poly I:C induced IFN-λ1 expression. Huh7 cells were stimulated with the plasmids containing HCV NS genes for 18 h prior to poly I:C (1 μg/ml) stimulation for 24 h. Total RNA extracted from cells was subjected to real time RT-PCR for the mRNA levels of IFN-λ1 and GAPDH. The data are expressed as IFN-λ1 mRNA (A, B) levels relative (fold) to the control (without HCV infection or without plasmids transfection, which defined as 100%). SN collected from cell cultures was assayed by ELISA to measure the IFN-λ1 protein (B, D). The results shown are mean ± SD of triplicate cultures, representative of three independent experiments (poly I:C-stimulated Huh7 cell versus vehicle only: **P<0.01, *P<0.05).
Figure 6.
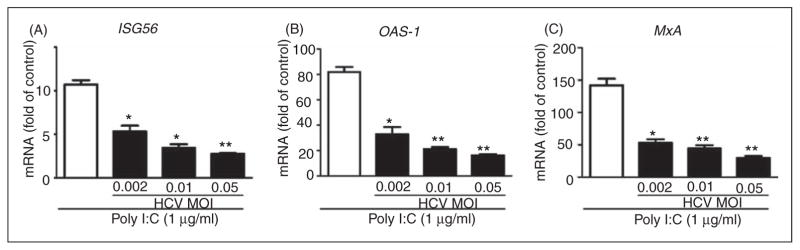
HCV suppresses poly I:C-induced ISGs expression. Huh7 cells were infected with HCV JFH-1 at the indicated MOI for 72 h, then stimulated with poly I:C for 24 h. Total RNA extracted from cells was subjected to real time RT-PCR for the mRNA levels of ISG56, OAS-1, MxA and GAPDH. The data are expressed as ISG56 (A), OAS-1 (B) and MxA (C) mRNA level relative (fold) to the control (vehicle only, which defined as 1). The results shown are mean ± SD of triplicate cultures, representative of three experiments (poly I:C-stimulated infected Huh7 cells vs poly I:C-stimulated uninfected Huh7 cells, **P<0.01, *P<0.05).
Discussion
The interaction of HCV and host immune responses is an important determinant of the outcome of HCV infection. Innate immunity is the first line of defense against viral infections. IFNs are the key players in host innate immunity, as they possess antiviral activity against a number of viruses,9 including HCV.30,31 In addition to type I and type II IFNs, which have been known for decades as the antiviral cytokines, type III IFNs (IFN-λ) are also shown to have a strong antiviral function.32 Although IFN-λ1 and IFN-λ2/3 exert biological activities similar to type I IFNs, they appear to have a more specialized role in innate antiviral defense.32 IFN-λ exhibits antiviral action on HCV replication in both replicon and cell culture infectious virus model systems.12–14 TLR3 plays a crucial role in the innate immunity against viral infection, as its activation induces type I IFN-based antiviral immunity.33–35 However, little is known about whether TLR3 signaling of hepatocytes can induce IFN-λ expression against HCV infection/replication. We demonstrated that poly I:C could induce intracellular IFN-λ1 expression in Huh7 cells at both mRNA and protein levels. This induction of IFN-λ was associated with the decrease of HCV RNA and core protein expression in Huh7 cells activated by poly I:C. This finding is supported by a recent report that HCV could induce the expression of IFN-λ1 and ISGs in primary liver cultures, which limited the growth and spread of the virus.36
Poly I:C, a synthetic analog of dsRNA to mimic viral infection, has been used widely in TLR3 signaling related studies for years. There are two commercially available poly I:Cs [high (HMM) or low molecular mass (LMM)]. Studies have shown that stimulation of cells with HMM or LMM poly I:C resulted in different cytokine profile.37,38 We demonstrated recently that the efficiency of TLR3 signaling of Huh7 cells by HMM poly I:C was significantly higher than that by LMM poly I:C.39 In addition to being recognized by TLR3, poly I:C could be recognized by RIG-I or melanoma differentiation-associated protein 5 (MDA-5). To determine whether the TLR3 signaling is critical in HMM poly I:C-mediated IFN induction, we used Bafilomycin A1, an inhibitor of the TLR3 signaling pathway to block the poly I:C effect. We found that Bafilomycin A1 could largely block poly I:C-induced IFN-λ1 expression in Huh7 cells. Furthermore, to determine the role of RIG-I in poly I:C-mediated IFN-λ1 induction, we performed the experiments with Huh7.5 cells that have a defect in the RIG-I signaling pathway.18,24 We observed that poly I:C could induce IFN-λ1 expression in Huh7.5 cells, suggesting that poly I:C induces IFN-λ1 via a RIG-I independent mechanism. With regard to the role of MDA-5 in poly I:C-mediated action on IFN-λ1, our previous study39 showed that the knock down of MDA-5 by siRNA had little effect on poly I:C-mediated IFN induction in Huh7.5 cells. Taken together, these results indicate clearly that TLR3 is the major sensor for the poly I:C action on IFN-λ1 induction in Huh7 cells.
In order to establish a persistent infection in the liver, HCV has evolved several strategies to evade host immune response, including inhibition of intracellular innate immunity.40 Our previous study22 demonstrated that HCV inhibits intracellular IFN-α and IRF-7 expression in human hepatic cell lines. Others have documented that HCV NS3/4A protease is able to impair TLR3 by cleaving the TRIF adaptor protein and block RIG-I signaling by cleaving the mitochondrial antiviral signaling protein off the mitochondria to inhibit IRF-3 activation and IFN-λ expression.11,41,42 In addition, HCV infection could impair IRF-7 translocation and IFN-α synthesis in immortalized human hepatocytes.43 In agreement with these findings, we showed that HCV replication impaired poly I:C-activated TLR3 signaling of Huh7 cells, which is evidenced by the observation that poly I:C-induced intracellular IFN-λ1 and ISG expression was inhibited by HCV in infected Huh7 cells.
HCV-encoded proteins, including structural proteins (E2, core) and NS proteins (NS3/4A, NS5A and NS5B), are immunosuppressive.22,40,44–46 Among these HCV immunosuppressive proteins, NS3/4A has the ability to impair TLR3 and RIG-I signaling pathways and inhibits IFN-β expression.5,11,41,42 In agreement with these findings, we showed that NS3/4A had an important role in HCV-mediated suppression of intracellular IFN-λ1 expression. In addition, HCV NS5A and NS5B were also involved in the HCV-mediated suppression of intracellular IFN-λ1 expression in hepatic cells. NS5B has been identified as a RNA-dependent RNA polymerase responsible for the synthesis of negative-strand HCV RNA.47 Inhibition of NS5B has long been considered an attractive target for therapeutic intervention in HCV-infected patients.47,48 Our finding that there was a negative effect of NS5B on IFN-λ-mediated host cell innate immunity in Huh7 cells further highlights the important role of NS5B in HCV-mediated evasion of host cell immunity.
Taken together, we have provided further evidence to demonstrate the ability of HCV to evade the antiviral mechanisms in the host cells. Our data indicate that HCV not only inhibits intracellular type I IFN (IFN-α) expression,22 but also suppresses TLR3 activation- induced type III IFN (IFN-λ1) expression in hepatoma cell line. Thus, HCV and its proteins use the complex mechanisms to target multiple steps of the TLR3/IFN signaling pathway in hepatocytes, compromising the intracellular innate immunity. Further studies are needed to understand the in vivo interplays between HCV and host cell innate immunity in the context of persistent infection in the liver.
Acknowledgments
Funding
This work was supported by the grants (DA12815, DA22177, and DA27550) from the National Institutes of Health.
We thank Dr Charles Rice (The Rockefeller University, New York, NY, USA) for providing Huh7. We also thank Dr Michael Gale (University of Washington, Seattle, WA, USA) for providing the HCV NS3, NS3/4A, NS5A and NS5B plasmids.
Footnotes
Conflict of interest
The authors do not have any potential conflicts of interest to declare.
References
- 1.Alter MJ. Prevention of spread of hepatitis C. Hepatology. 2002;36:S93–S98. doi: 10.1053/jhep.2002.36389. [DOI] [PubMed] [Google Scholar]
- 2.Chen SL, Morgan TR. The natural history of hepatitis C virus (HCV) infection. Int J Med Sci. 2006;3:47–52. doi: 10.7150/ijms.3.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battaglia AM, Hagmeyer KO. Combination therapy with interferon and ribavirin in the treatment of chronic hepatitis C infection. Ann Pharmacother. 2000;34:487–494. doi: 10.1345/aph.19183. [DOI] [PubMed] [Google Scholar]
- 4.Hu KQ, Vierling JM, Redeker AG. Viral, host and interferon-related factors modulating the effect of interferon therapy for hepatitis C virus infection. J Viral Hepat. 2001;8:1–18. doi: 10.1046/j.1365-2893.2001.00253.x. [DOI] [PubMed] [Google Scholar]
- 5.Foy E, Li K, Wang C, Sumpter R, Jr, Ikeda M, Lemon SM, et al. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- 6.He Y, Katze MG. To interfere and to anti-interfere: the interplay between hepatitis C virus and interferon. Viral Immunol. 2002;15:95–119. doi: 10.1089/088282402317340260. [DOI] [PubMed] [Google Scholar]
- 7.Racanelli V, Rehermann B. Hepatitis C virus infection: when silence is deception. Trends Immunol. 2003;24:456–464. doi: 10.1016/s1471-4906(03)00178-9. [DOI] [PubMed] [Google Scholar]
- 8.Tan SL, Katze MG. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology. 2001;284:1–12. doi: 10.1006/viro.2001.0885. [DOI] [PubMed] [Google Scholar]
- 9.Katze MG, He Y, Gale M., Jr Viruses and interferon: a fight for supremacy. Nat Rev Immunol. 2002;2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 10.Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13:816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 11.Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, et al. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pagliaccetti NE, Eduardo R, Kleinstein SH, Mu XJ, Bandi P, Robek MD. Interleukin-29 functions cooperatively with interferon to induce antiviral gene expression and inhibit hepatitis C virus replication. J Biol Chem. 2008;283:30079–30089. doi: 10.1074/jbc.M804296200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, et al. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 14.Doyle SE, Schreckhise H, Khuu-Duong K, Henderson K, Rosler R, Storey H, et al. Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology. 2006;44:896–906. doi: 10.1002/hep.21312. [DOI] [PubMed] [Google Scholar]
- 15.Zhu H, Butera M, Nelson DR, Liu C. Novel type I interferon IL-28A suppresses hepatitis C viral RNA replication. Virol J. 2005;2:80. doi: 10.1186/1743-422X-2-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005;79:3851–3854. doi: 10.1128/JVI.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muir AJ, Shiffman ML, Zaman A, Yoffe B, de la Torre A, Flamm S, et al. Phase 1b study of pegylated interferon lambda 1 with or without ribavirin in patients with chronic genotype 1 hepatitis C virus infection. Hepatology. 2010;52:822–832. doi: 10.1002/hep.23743. [DOI] [PubMed] [Google Scholar]
- 18.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye L, Wang X, Wang S, Wang Y, Song L, Hou W, et al. CD56+ T cells inhibit hepatitis C virus replication in human hepatocytes. Hepatology. 2009;49:753–762. doi: 10.1002/hep.22715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vijay-Kumar M, Gentsch JR, Kaiser WJ, Borregaard N, Offermann MK, Neish AS, et al. Protein kinase R mediates intestinal epithelial gene remodeling in response to double-stranded RNA and live rotavirus. J Immunol. 2005;174:6322–6331. doi: 10.4049/jimmunol.174.10.6322. [DOI] [PubMed] [Google Scholar]
- 21.Yang JH, Lai JP, Douglas SD, Metzger D, Zhu XH, Ho WZ. Real-time RT-PCR for quantitation of hepatitis C virus RNA. J Virol Methods. 2002;102:119–128. doi: 10.1016/s0166-0934(02)00007-1. [DOI] [PubMed] [Google Scholar]
- 22.Zhang T, Lin RT, Li Y, Douglas SD, Maxcey C, Ho C, et al. Hepatitis C virus inhibits intracellular interferon alpha expression in human hepatic cell lines. Hepatology. 2005;42:819–827. doi: 10.1002/hep.20854. [DOI] [PubMed] [Google Scholar]
- 23.Li K, Chen Z, Kato N, Gale M, Jr, Lemon SM. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon- beta production in hepatocytes. J Biol Chem. 2005;280:16739–16747. doi: 10.1074/jbc.M414139200. [DOI] [PubMed] [Google Scholar]
- 24.Bartenschlager R, Pietschmann T. Efficient hepatitis C virus cell culture system: what a difference the host cell makes. Proc Natl Acad Sci U S A. 2005;102:9739–9740. doi: 10.1073/pnas.0504296102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bluyssen AR, Durbin JE, Levy DE. ISGF3 gamma p48, a specificity switch for interferon activated transcription factors. Cytokine Growth Factor Rev. 1996;7:11–17. doi: 10.1016/1359-6101(96)00005-6. [DOI] [PubMed] [Google Scholar]
- 26.Justesen J, Hartmann R, Kjeldgaard NO. Gene structure and function of the 2′-5′-oligoadenylate synthetase family. Cell Mol Life Sci. 2000;57:1593–1612. doi: 10.1007/PL00000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sen GC, Sarkar SN. The interferon-stimulated genes: targets of direct signaling by interferons, double-stranded RNA, and viruses. Curr Top Microbiol Immunol. 2007;316:233–250. doi: 10.1007/978-3-540-71329-6_12. [DOI] [PubMed] [Google Scholar]
- 28.Garcia MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Carroll TD, Matzinger SR, Genesca M, Fritts L, Colon R, McChesney MB, et al. Interferon-induced expression of MxA in the respiratory tract of rhesus macaques is suppressed by influenza virus replication. J Immunol. 2008;180:2385–2395. doi: 10.4049/jimmunol.180.4.2385. [DOI] [PubMed] [Google Scholar]
- 30.Castet V, Fournier C, Soulier A, Brillet R, Coste J, Larrey D, et al. Alpha interferon inhibits hepatitis C virus replication in primary human hepatocytes infected in vitro. J Virol. 2002;76:8189–8199. doi: 10.1128/JVI.76.16.8189-8199.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dash S, Prabhu R, Hazari S, Bastian F, Garry R, Zou W, et al. Interferons alpha, beta, gamma each inhibit hepatitis C virus replication at the level of internal ribosome entry site-mediated translation. Liver Int. 2005;25:580–594. doi: 10.1111/j.1478-3231.2005.01082.x. [DOI] [PubMed] [Google Scholar]
- 32.Ank N, Paludan SR. Type III IFNs: new layers of complexity in innate antiviral immunity. Biofactors. 2009;35:82–87. doi: 10.1002/biof.19. [DOI] [PubMed] [Google Scholar]
- 33.West J, Damania B. Upregulation of the TLR3 pathway by Kaposi’s sarcoma-associated herpesvirus during primary infection. J Virol. 2008;82:5440–5449. doi: 10.1128/JVI.02590-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starace D, Galli R, Paone A, De Cesaris P, Filippini A, Ziparo E, et al. Toll-like receptor 3 activation induces antiviral immune responses in mouse sertoli cells. Biol Reprod. 2008;79:766–775. doi: 10.1095/biolreprod.108.068619. [DOI] [PubMed] [Google Scholar]
- 35.Kumar A, Zhang J, Yu FS. Toll-like receptor 3 agonist poly(I:C)-induced antiviral response in human corneal epithelial cells. Immunology. 2006;117:11–21. doi: 10.1111/j.1365-2567.2005.02258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marukian S, Andrus L, Sheahan TP, Jones CT, Charles ED, Ploss A, et al. Hepatitis C virus induces interferon-lambda and interferon-stimulated genes in primary liver cultures. Hepatology. 2011;54:1913–1923. doi: 10.1002/hep.24580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang M, Osterlund P, Sarin LP, Poranen MM, Bamford DH, Guo D, et al. Innate immune responses in human monocyte-derived dendritic cells are highly dependent on the size and the 5′ phosphorylation of RNA molecules. J Immunol. 2011;187:1713–1721. doi: 10.4049/jimmunol.1100361. [DOI] [PubMed] [Google Scholar]
- 38.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Y, Guo M, Wang X, Li J, Wang Y, Ye L, et al. TLR3 activation efficiency by high or low molecular mass poly I:C. Innate Immun. doi: 10.1177/1753425912459975. Epub ahead of print 3 October 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gale M, Jr, Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436:939–945. doi: 10.1038/nature04078. [DOI] [PubMed] [Google Scholar]
- 41.Foy E, Li K, Sumpter R, Jr, Loo YM, Johnson CL, Wang C, et al. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci USA. 2005;102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raychoudhuri A, Shrivastava S, Steele R, Dash S, Kanda T, Ray R, et al. Hepatitis C virus infection impairs IRF-7 translocation and Alpha interferon synthesis in immortalized human hepatocytes. J Virol. 2010;84:10991–10998. doi: 10.1128/JVI.00900-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor DR, Shi ST, Romano PR, Barber GN, Lai MM. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999;285:107–110. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- 45.Gale M, Jr, Blakely CM, Kwieciszewski B, Tan SL, Dossett M, Tang NM, et al. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18:5208–5218. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melen K, Fagerlund R, Nyqvist M, Keskinen P, Julkunen I. Expression of hepatitis C virus core protein inhibits interferon-induced nuclear import of STATs. J Med Virol. 2004;73:536–547. doi: 10.1002/jmv.20123. [DOI] [PubMed] [Google Scholar]
- 47.Behrens SE, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 48.Reed KE, Rice CM. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr Top Microbiol Immunol. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]