Abstract
Milk oligosaccharides (OS)—free complex carbohydrates—confer unique health benefits to the nursing neonate. Though human digestive enzymes cannot degrade these sugars, they provide nourishment to specific commensal microbes and act as decoys to prevent the adhesion of pathogenic micro-organisms to gastrointestinal cells. At present, the limited quantities of human milk oligosaccharides (HMO) impede research on these molecules and their potential applications in functional food formulations. Considerable progress has been made in the study of OS structures; however, the synthetic pathways leading to their synthesis in the mammary gland are poorly understood. Recent studies show that complex OS with fucose and N-acetyl neuraminic acid (key structural elements of HMO bioactivity) exist in goat milk. Polymorphisms in the CSN1S1 locus, which is responsible for synthesis of αs1-casein, affect lipid and casein micelle structure in goat milk. The present study sought to determine whether CSN1S1 polymorphisms also influence goat milk oligosaccharide (GMO) production and secretion. The GMO compositions of thirty-two goat milk samples, half of which were from genotype A/A (αs1-casein producers) and half from genotype O/O (αs1-casein non-producers), were determined with nanoflow liquid chromatography high-accuracy mass spectrometry. This study represents the most exhaustive characterization of GMO to date. A systematic and comprehensive GMO library was created, consolidating information available in the literature with the new findings. Nearly 30 GMO, 11 of which were novel, were confirmed via tandem mass spectrometric analyses. Six fucosylated OS were identified; 4 of these matched HMO compositions and three were identified for the first time in goat milk. Importantly, multivariate statistical analysis demonstrated that the OS profiles of the A/A and O/O genotype milks could be discriminated by the fucosylated OS. Quantitative analysis revealed that the goat milk samples contained 1.17 g/L of OS; however, their concentration in milks from A/A and O/O genotypes was not different. This study provides evidence of a genetic influence on specific OS biosynthesis but not total OS production. The presence of fucosylated GMO suggests that goat milk represents a potential source of bioactive milk OS suitable as a functional food ingredient.
Keywords: CSN1S1, Fucose, Genetic polymorphisms, Goat milk, Mass spectrometry, Oligosaccharides
1. Introduction
Although milk evolved as the sole nourishment of the mammalian neonate, all known milks contain free oligosaccharides (OS) that are indigestible by infants (Engfer et al., 2000; Gnoth et al., 2000). Rather than functioning as a direct nutrient for the infant, human milk OS enhance the growth of specific bacteria and deflect the adhesion of pathogens in the infant gut (LoCascio et al., 2007; Morrow et al., 2005). Therefore, an HMO-like product would be valuable as a supplement for infant or even adult nutrition.
Goat milk has smaller casein micelles and fat globules, higher concentration of some whey proteins and oligosaccharides than bovine milk (Silanikove et al., 2010). Several studies report the effects of goat CSN1S1 gene polymorphisms on milk production and composition, milk technological properties and on milk fatty acid content (Silanikove et al., 2010). Little is known, however, about the influence of these polymorphisms on milk oligosaccharides.
We were particularly interested in goat milk oligosaccharides (GMO) because of two studies, which suggested that GMO have anti-inflammatory effects and reduce intestinal inflammation in mice with induced colitis (Daddaoua et al., 2006; Lara-Villoslada et al., 2006). In addition, goat milk is known to contain fucosylated and sialylated OS, making them similar in composition to HMO (Chaturvedi and Sharma, 1990a; Urashima et al., 1994). Therefore, goat milk may represent an ideal source of OS for supplementary and therapeutic applications.
Recent advances in mass spectrometry have revealed the detailed structures of OS in mammalian milks. We employed high-throughput, high-resolution mass spectrometry to measure and identify OS from goat milk.
In addition to profiling the OS present in goat milk, we compared differences in OS production between goats of two different genotypes. αs1-Casein is major protein component of milks (including goat milk) and makes up part of the well-known milk macrostructure, the casein micelle. The CSN1S1 gene, which encodes for αs1-casein, is highly polymorphic in goats, with 15 alleles: A, B1, B2, B3, B4, C, E, F, G, H, I, L, M, O1 and O2 characterized so far (Bevilacqua et al., 2002). These polymorphisms drive the large variability in the amount of αs1-casein observed in individual goat milks (Moioli et al., 1998). The A/A genotype produces the most αs1-casein (approximately ~7 g/L). A large deletion in the CSN1S1 locus (called the O allele) precludes the expression of αs1-casein, which is therefore absent in milk of goats of the O/O genotype. Inability to produce αs1-casein, which is essential for casein micelle formation, results in the accumulation of large amounts of β- and κ-casein in the mammary epithelial cells’ ER, and the micelles are not transported to the Golgi bodies properly (Chanat et al., 1999).
These major changes in casein micelle production, unsurprisingly, have a variety of effects on milk composition and quality. These effects have been intensively studied and include milk composition, renneting properties, cheese yield, and milk fat content (Grosclaude and Martin, 1997; Martin and Leroux, 2000). For example, A/A goats produce milk with higher protein and lipid content than O/O goats (Pierre et al., 1998).
The effects of these CSN1S1 polymorphisms on OS synthesis remain uninvestigated. As OS synthesis occurs in the ER of the mammary epithelial cell (like αs1-casein production), we hypothesized that the build-up of caseins in the O/O genotype would negatively impact the production and secretion of GMO. Therefore, MS was employed to profile OS in milks from both the A/A and O/O goat genotypes at the CSN1S1 locus to see if the absence of αs1-casein has any effect on OS synthesis. This study is the first to examine the effects of these polymorphisms on GMO synthesis.
2. Materials and methods
2.1. Sample collection and standards
Morning and evening milk samples were collected from 16 Alpine goats (8 from genotype A/A and 8 from genotype O/O, for a total of 32 milk samples) for identification and statistical comparison of neutral GMO. An additional set of 8 goat milk samples (4 of each genotype) were collected for analysis of GMO concentration and identification of acidic GMO. Three commercial goat milks were purchased at a local supermarket (Davis, CA, USA). Three samples of bovine milk were obtained from the University of California, Davis dairy barn. Three samples of human milk were obtained from a UC Davis lactation study directed by Dr. Jennifer Smilowitz. Monosaccharide standards—fucose (Fuc), N-acetylglucosamine (GlcNAc), galactose (Gal), glucose (Glc), N-acetylneuraminic acid (NeuAc), N-glycolylneuraminic acid (NeuGc), and d-allose—were purchased from Sigma–Aldrich (St. Louis, MO, USA). Maltopentaose, maltohexaose, and maltoheptaose were purchased from Sigma–Aldrich. The OS 3′-sialyl-lactose, 6′-sialyl-lactose, 3′-sialyl-N-acetyllactosamine, 6′-sialyl-N-acetyllactosamine, 3′-fucosyllactose, 2′-fucosyllactose, and lacto-N-neohexaose were purchased from Dextra (Reading, UK).
2.2. Oligosaccharide isolation and purification
OS were isolated from milk samples and purified according to a previously described method (Ninonuevo et al., 2006). Briefly, milk samples (0.5 mL) were diluted with an equal volume of nanopure water (18.2 MΩ ionic purity) and centrifuged at 4 °C for 30 min at 4000 ×g to remove lipids. The fat-free fraction (infranate) was treated with 4 volumes of 2:1 (v/v) chloroform:methanol, and the emulsion was centrifuged at 4000 ×g for 30 min at 4 °C. The lower chloroform and protein pellet were discarded. The upper layer containing OS was collected. Two volumes of pure ethanol were added to the OS fraction and the remaining protein was precipitated at 4 °C overnight. The samples were then centrifuged at 4000 ×g for 30 min at 4 °C and the protein-free supernatant was collected and dried in a vacuum centrifuge at 37 °C. OS were rehydrated in 1 mL nanopure water. Residual peptides were removed using C8 columns (DSC-C8 Discovery, 3 mL tube capacity, 500 mg bed weight, Supelco, Bellefonte, PA, USA). The cartridges were conditioned with three column volumes (cv) of pure HPLC-grade acetonitrile (ACN) followed by three cv of nanopure water. The carbohydrate-rich solution was loaded onto the cartridge, and the peptide-free eluate was collected. The OS were further purified by nonporous graphitized carbon solid-phase extraction (GCC-SPE, 150 mg carbon, 4 mL tube capacity, Alltech, Deerfield, IL, USA). Prior to use, the GCC-SPE cartridge was activated with 3 cv of 80% ACN, 0.05% trifluoroacetic acid (TFA, v/v), and equilibrated with 3 cv nanopure water. The carbohydrate-rich solution was loaded onto the cartridge, and salts were removed by washing with three cv of nanopure water at a flow rate of 1 mL/min. For neutral OS profiling and statistical analysis, 32 milk samples were eluted from the cartridge using 2 cv of 20% ACN in water. For acidic OS profiling and total quantification, 8 milk samples were each eluted with 2 cv of 20% ACN in water followed by 2 cv of 40% ACN/0.1% TFA in water to collect the neutral and the acidic OS, respectively. Eluted fractions were dried prior to analysis by MS by vacuum centrifugation at 37 °C.
2.3. Nano-LC-Chip–Q-TOF MS
Prior to MS analysis, dried OS samples were reconstituted in 100 μL of nanopure water. MS analysis was performed with an Agilent 6520 accurate-mass Quadrupole-Time-of-Flight (Q-TOF) LC/MS with a microfluidic nano-electrospray chip (Agilent Technologies, Santa Clara, CA, USA) as previously described (Wu et al., 2011). The chip employed contained enrichment and analytical columns, both packed with graphitized carbon. Chromatographic elution was performed with a binary gradient of 3% ACN/0.1% formic acid in water (solvent A), and 90% ACN/0.1% formic acid in water (solvent B). The column was initially equilibrated and eluted with a flow rate of 0.3 μL/min for the nanopump and 4 μL/min for the capillary pump. The 65-min gradient was programmed as follows: 2.5–20 min, 0–16% B; 20–30 min, 16–44% B; 30–35 min, 44–100% B; 35–45 min, 100% B; and 45–65 min, 0% B (to equilibrate the chip column before the next sample injection). Data were acquired in the positive ionization mode with a 450–2500 mass/charge (m/z) range. The electrospray capillary voltage was 1600–1700 V. The acquisition rate was 0.63 spectra/s for both MS and MS/MS modes. Automated precursor selection was employed based on abundance, with up to 6 MS/MS per MS. The precursor isolation window was narrow (1.3 m/z). Fragmentation energy was set at 1.8 V/100 Da with an offset of −2.4 V.
2.4. OS identification and statistical analyses
The Molecular Feature Extraction function of Mass Hunter Qualitative Analysis Version B.04.00 (Agilent Technologies) was used to generate a list of deconvoluted masses selected to be in a range of 450–1500 m/z with a ≥1000 height count and a typical isotopic distribution of small biological molecules. Charge states allowed were +1 and +2. OS compositions were then determined from the deconvoluted mass list with an in-house program, Glycan Finder (Ninonuevo et al., 2006), with a mass error tolerance of ≤5 ppm Distinct OS structures were identified based on accurate mass and retention times (RT) compared with previously identified structures. Monosaccharide compositions were further confirmed by tandem MS (MS/MS) analysis. Statistical analyses were performed on the deconvoluted masses corresponding to known OS compositions within Agilent Mass Profiler Professional v2.2 software. The remaining compounds were matched and aligned with respect to RT. RT correction was performed without a standard. The retention time window allowed for compound-matching was ±0.25 min with the addition of ±0.25% of the RT at each timepoint. The retention time window was increased as retention time increased because the chromatogram was less reproducible toward the end of each run. Potential contaminants were removed with a filter that retained only compounds that were detected in at least two of the samples and present in at least 25% of the samples within a genotype group. For Principle Component Analysis (PCA), compound intensities were normalized on the median intensity of each compound. This normalization is intended to provide equal weight to both low and high abundance peaks for the PCA. An asymptotic ANOVA (paired conditions) test with the Benjamini–Hochberg false discovery rate (FDR) correction was performed to identify compounds that had significantly different intensity between the groups. A P-value cut-off of 0.05 was used. PCA was performed using intensity and volume of compounds (compounds were defined as a mass and a RT) to discriminate between the two genotype groups.
2.5. OS quantification via gas chromatography (GC)
2.5.1. Sample preparation for GC analysis
Methanolysis and trimethylsilylation were performed according to a published method (Bordiga et al., 2012). Briefly, 1 M anhydrous methanolic hydrochloric acid (MeOH:HCl) was prepared by adding 140 μL acetyl chloride to 1 mL anhydrous methanol. Dried OS samples were resuspended in 100 μL nanopure water. A volume of this mixture containing approximately 250 μg of OS (based on literature references for goat, human, and bovine milks) was mixed with 100 μg of internal standard (d-allose), dried, resuspended in 0.5 mL of 1 M MeOH:HCl, and heated at 80 °C for 24 h. The mixture was dried under a stream of nitrogen gas at room temperature. Twice, 250 μL of pure MeOH were added and dried under nitrogen. To re-N-acetylate, 200 μL of 10:1 MeOH:acetic anhydride (v/v) were added to each sample and incubated at 85 °C for 24 h. After the re-N-acetylation reaction was completed, samples were dried under nitrogen, washed twice with 250 μL of pure MeOH, and dried again. Derivatization was carried out according to a published method (Sweeley and Walker, 1964). Briefly, an excess (0.3 mL) of silylating reagent (10:2:1, v/v/v, pyridine:hexamethyldisilazane:chlorotrimethylsilane) was added. The solution was heated at 80 °C for 24 h. The reagent was removed by evaporation under nitrogen. The residue was extracted with hexane (1 mL), centrifuged, and transferred to a glass vial. The hexane solution containing silylated monosaccharides was concentrated to approximately 200 μL, and 3 μL were used for GC analysis. A Hewlett Packard HP-6890 gas chromatograph equipped with a capillary split/splitless inlet and a flame ionization detector was employed with a DB-1 fused-silica capillary column (30 m × 0.25 μm i.d., 0.25 μm film thickness (J&W Scientific, Folsom, CA)). Hydrogen was used as the carrier gas (at a flow rate of 2.5 mL/min, 17 psi). Samples were injected in the pulsed split mode with a split ratio of 5:1. The injector and the flame ionization detector were operated at 280 °C. The gas chromatograph was operated with temperature programming (120–200 °C at 1.5 °C/min, held at 200 °C for 5 min, and at 250 °C for a post run of 2 min). For each monosaccharide, a response factor of the specific peak was calculated using the d-allose standard peak as a reference (Merkle and Poppe, 1994). A final correction factor, which corresponded to the efficiency of the OS isolation and purification procedure used, was applied to the concentrations of OS calculated. To determine extraction efficiency of the method, a 0.5 mg mixture of 50 μg each of ten pooled OS standards (3′-sialyl-lactose, 6′-sialyl-lactose, 3′-sialyl-N-acetyllactosamine, 6′-sialyl-N-acetyllactosamine, 3′-fucosyllactose, 2′-fucosyllactose, lacto-N-neohexaose, maltopentaose, maltohexaose, and maltoheptaose) were applied to all the same extraction methods of the actual samples in triplicate and analyzed by GC. Based on these experiments, the extraction efficiency of this method was 53 ± 3%. This extraction efficiency was applied to the GC determined OS concentration for each sample.
3. Results
Mammalian milk contains both neutral and acidic OS. Galactose (Gal), glucose (Glc), fucose (Fuc), N-acetylglucosamine (GlcNAc) and N-acetylgalactosamine (GalNAc) grafted on a lactose core make up neutral OS; these same monomers, plus N-acetylneuraminic acid (NeuAc) and, more rarely, N-glycolylneuraminic acid (NeuGc), comprise acidic OS. Table 1 presents a full library of GMO found in previous papers and in this manuscript. The table includes the composition stated as number of each monosaccharide residue that makes up the OS (RN), accurate neutral mass, concentration (g/L), conventional name (abbreviation in parentheses) and chemical structure. The compositions (RN) reported in the library refer to the sequential monomeric composition; for example, the first OS in the table, RN 21000, is made of 2 hexoses + 1 fucose + 0 HexNAc + 0 NeuAc + 0 NeuGC.
Table 1.
Library of goat milk oligosaccharides described in the literature and validated by MS/MS in the present study. Compositions (Comp.) are indicated as total residue numbers (RN) of hexose, fucose, Nacetylhexosamine, N-acetylneuraminic acid, and N-glycolylneuraminic acid.
| Comp. RN | Neutral (N) or acidic (A) OS? | Neutral mass (Da) | MS/MS | Conc. reported (g/L) | Name (abbreviation) | Structure | Ref.a |
|---|---|---|---|---|---|---|---|
| 21000 | N | 488.174 | ✓ | n.rb | α-2′-Fucosyl-lactose (2′-FL) | α-l-Fuc.p-(1-2)-β-d-Gal.p-(1-4)-d-Glc | 1 |
| 30000 | N | 504.169 | ✓ | 0.03–0.05 | α-3′-Galactosyl-lactose (3-GL) | α-d-Gal.p-(1-3)-β-d-Gal.p-(1-4)-d-Glc | 1-3 |
| 30000 | N | 504.169 | ✓ | n.rb | β-6′-Galactosyl-lactose (6-GL) | β-d-Gal.p-(1-6)-β-d-Gal.p-(1-4)-d-Glc | 1, 2 |
| 11100c | N | 529.201 | ✓ | Fucosyl-lactosamine | |||
| 20100 | N | 545.196 | ✓ | 0.02–0.04 | 6′-N-Acetyl-glucosaminyl-lactose (NAL) | β-d-GlcNAc-(1-6)-β-d-Gal.p-(1-4)-d-Glc | 2-4 |
| 20010 | A | 633.212 | ✓ | 0.05–0.07 | 6′-Sialyl-lactose (6-SL) | Neu5Ac-(α2-6)-Gal-(β1-4)-Glc | 2, 3, 5 |
| 20010 | A | 633.212 | ✓ | 0.03–0.05 | 3′-Sialyl-lactose (3-SL) | Neu5Ac-(α2-3)-Gal-(β1-4)-Glc | 2, 3, 5 |
| 20001 | A | 649.206 | ✓ | 0.04–0.06 | 6′-Glycolyl-neuraminyl-lactose (NGL) | Neu5Gc-(α2-6)-Gal-(β1-4)-Glc | 2, 3, 5 |
| 40000c | N | 666.222 | ✓ | ||||
| 10110 | A | 674.238 | ✓ | n.rb | 6′-Sialyl-lactosamine | Neu5Ac-(α2-6)-Gal-(β1-4)-GlcNAc | 3, 5 |
| 10101c | A | 690.233 | ✓ | Glycolyl-neuraminyl-lactosamine | |||
| 30100 | N | 707.248 | ✓ | Trace | N-Acetyl-glucosaminyl-hexosyl-lactose (NAHL) | Gal-(β1-4)-GIcNAc-(β1-6)-Gal-(1-4)-Glc | 2, 4 |
| 20200 | N | 748.275 | ✓ | Trace | Di-N-acetyl-glucosaminyl-lactose (DNAL) | 2 | |
| 30010 | A | 795.264 | ✓ | Trace | 3′-Sialyl-6′galactosyl-lactose (3-SHL) |
|
2, 6 |
| 30010 | A | 795.264 | ✓ | Trace | 6′-Sialyl-3′galactosyl-lactose (6-SHL) |
|
2, 5, 6 |
| 30001 | A | 811.259 | ✓ | Trace | N-Glycolyl-neuraminyl-hexosyl-lactose (SNGHL) | 2 | |
| 50000c | N | 828.275 | ✓ | ||||
| 20110c | A | 836.291 | ✓ | Sialyl-N-acetylglucosaminyl-lactose (SNAL) | |||
| 31100 | N | 853.306 | ✓ | n.rb | Lacto-N-fuco-pentaose III (LNFP III) |
|
7 |
| 31100 | N | 853.306 | ✓ | n.rb | Lacto-N-fuco-pentaose V (LNFP III) |
|
7 |
| 40100 | N | 869.301 | ✓ | Trace | N-Acetyl-glucosaminyl-dihexosyl-lactose (NADHL) |
|
2, 4 |
| 30200c | N | 910.328 | ✓ | ||||
| 20020 | A | 924.307 | ✓ | 0.001–0.005 | Disialyl-lactose (DSL) | Neu5Ac-(α2-8)-Neu5Ac-(α2-3)-Gal-(β1-4)-Glc | 2, 3 |
| 20011 | A | 940.302 | Trace | Sialyl-N-glycolyl-neuraminyl-lactose (SNGL) | 2 | ||
| 20002 | A | 956.297 | Trace | Di-N-glycolyl-neuraminyl-lactose (DNGL) | 2 | ||
| 40010 | A | 957.317 | Trace | Sialyl-di-hexosyl-lactose (SDHL) | 2 | ||
| 6000c | N | 990.327 | ✓ | ||||
| 40200 | N | 1072.381 | ✓ | 0.001–0.005 | Lacto-N-hexaose (LNH) |
|
2 |
| 30020 | A | 1086.360 | Trace | Disialyl-hexosyl-lactose | 2 | ||
| 30011 | A | 1102.355 | Trace | Sialyl-N-glycolyl-neuraminyl-hexosyl-lactose (SNGHL) | 2 | ||
| 30300c | N | 1113.407 | ✓ | ||||
| 30002 | A | 1118.350 | Trace | Di-N-glycolyl-neuraminyl-hexosyl-lactose (DNGHL) | 2 | ||
| 20400c | N | 1154.434 | ✓ | ||||
| 41200c | N | 1218.438 | ✓ | ||||
| 31300c | N | 1259.465 | ✓ | ||||
| 40300 | N | 1275.460 | Trace | N-Acetyl-glucosaminyl-lacto-N-hexaose (NALNH) | 2 | ||
| 40210 | A | 1363.476 | Trace | Sialyl-lacto-N-hexaose (SNLH) | 2 |
References numbers corresponds to the following literature citations: (1) Urashima et al. (1994); (2) Martinez-Ferez et al. (2006a,b); (3) Mehra and Kelly (2006); (4) Chaturvedi and Sharma (1988); (5) Urashima et al. (1997); (6) Viverge et al. (1997); and (7) Chaturvedi and Sharma (1990a,b).
Not reported.
Newly identified OS.
The OS compositions of 29 GMO were validated by MS/MS analysis (Table 1). Fig. 1 depicts a typical example of GMO compositional validation by tandem MS analysis.
Fig. 1.
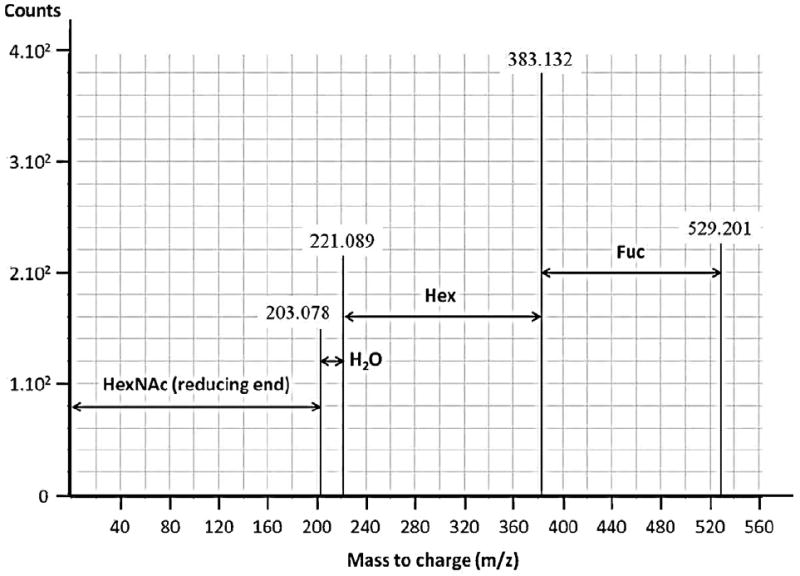
Nano LC-Chip–Q-TOF MS/MS deconvoluted spectra of the unreduced oligosaccharide 1Hex, 1Fuc, 1HexNAc (RN 11100, neutral mass was 529.2007). A 162.053 difference of mass corresponded to loss of one hexose (Hex), and a 146.058 difference of mass corresponded to loss of one fucose (Fuc).
The porous graphitized carbon nano-liquid-chromatography column separated both compositionally different glycans and structural isomers. The chip’s highly reproducible retention times allowed assignment of several chemical structures (e.g., 3′- and 6′-sialyl-lactose) based on identical retention times or accurate mass; these assignments were based on commercial OS standards and data from earlier publications on milk OS (Table 1) (Strum et al., 2012; Wu et al., 2011, 2010). In addition to the structures reported in the literature, 29 GMO were identified by MS/MS, 11 of which were novel as indicated in the footnote at Table 1. This study confirmed the presence of 18 of the 26 previously characterized OS in goat milk (Chaturvedi and Sharma, 1988, 1990b; Martinez-Ferez et al., 2006b; Mehra and Kelly, 2006; Urashima et al., 1994, 1997; Viverge et al., 1997). Combining the known and the new information, a total of 37 GMO are reported in the library. The 8 GMO in the library not found in our study were all found previously in only one study and at trace levels (Martinez-Ferez et al., 2006b).
3.1. Neutral GMO
Literature suggests that the three most abundant neutral GMO are 3-galactosyl-lactose (composition RN 30000 in Table 1), N-acetylglucosaminyl-lactose (RN 20100) and lacto-N-hexaose (RN 40200) (Martinez-Ferez et al., 2006b; Mehra and Kelly, 2006). The present study confirmed the presence of 3-galactosyl-lactose, N-acetylglucosaminyl-lactose, and lacto-N-hexaose in all goat milk samples analyzed. Additionally, we showed that N-acetyl-glucosaminyl-hexosyl-lactose (RN 30100) and the OS with composition RN 30300 were present in all the goat milk samples analyzed (Fig. 2). Fig. 2 displays five neutral OS that were present in all 32 goat milk samples and the commercial goat milk samples.
Fig. 2.
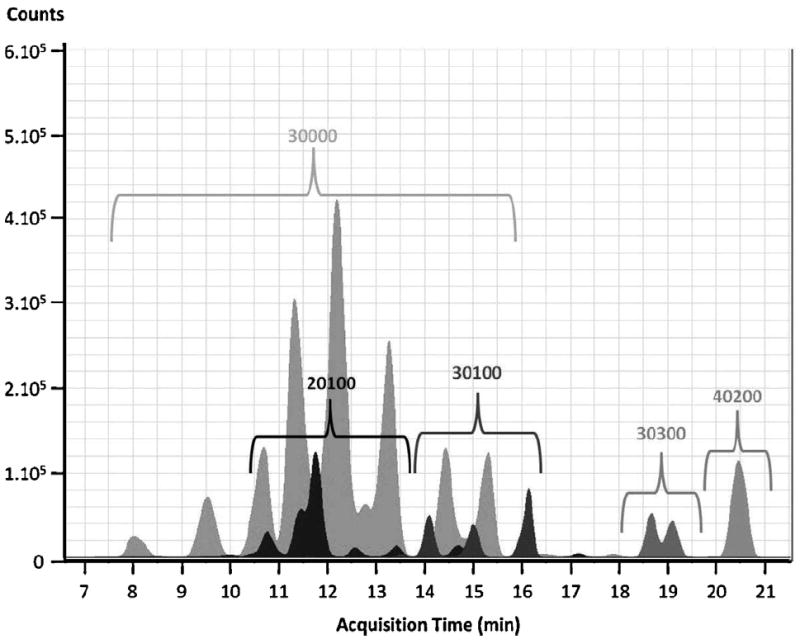
Typical extracted ion chromatograms of five neutral oligosaccharides in goat milk (neutral fraction, O/O genotype). 3Hex (RN 30000); 2Hex, 1HexNAc (RN 20100); 3Hex, 1HexNAc (RN 30100); 3Hex, 3HexNAc (RN 30300); and 4Hex, 2HexNAc (RN 40200). Multiple peaks for the same composition represent structural isomers.
3.2. Acidic GMO
In this study, 10 acidic GMO, including two previously unreported acidic species, were identified. As shown in Table 1, the two novel acidic GMO compositions motifs identified in this study were 1Hex, 1HexNAc, 1NeuGc (RN 10101), and 2Hex, 1HexNAc, 1NeuAc (RN 20110). Combining the newly identified acidic GMO with the 15 previously identified acidic GMO (Martinez-Ferez et al., 2006b; Mehra and Kelly, 2006; Viverge et al., 1997), a total of 17 acidic GMO are now known to exist in goat milk. Of the 17 acidic GMO, 5 contain only NeuGc, 10 contain only NeuAc, and 2 have both NeuGc and NeuAc. Only 8 of the 15 previously known acidic GMO were found in this study. The absence of the 7 acidic GMO previously identified is unsurprising since Martinez-Ferez et al. (2006a,b) detected those GMO only in trace amounts. Both 3′-sialyl-lactose (SL) and 6′-SL were present in all eight samples after elution with 40% ACN/0.1% TFA, in agreement with a previous report (Martinez-Ferez et al., 2006b).
3.3. Fucosylated GMO
Six of the 29 GMO identified in this work contained fucose (Fuc). The OS called α-2′-fucosyl-lactose (RN 21000) and two isomers of lacto-N-fucopentaose (RN 31100) were previously identified in goat milk using NMR (Chaturvedi and Sharma, 1990b). Three of the fucosylated GMO identified in this study have novel GMO compositions: 1Hex, 1HexNAc, 1Fuc; 4Hex, 1Fuc, 2HexNAc and 3Hex, 1Fuc, 3HexNAc.
3.4. Influence of Genotype on oligosaccharide composition
Interestingly, the GMO with composition 1Hex, 1Fuc, 1HexNAc (RN 11100) was present exclusively in the O/O genotype milk samples. Two peaks were consistently observed for RN11100 at RT 9.7 and 11.2 min Another fucosylated GMO, with composition 3Hex, 1Fuc, 3HexNAc (RN 31300) was present in milk from the O/O genotype goats, whereas it was found only in trace amounts in A/A genotype goats (approximately twenty-fold less abundant than in milk from O/O genotype goats). These two GMO compositions could serve as markers to discriminate between O/O and A/A genotype goats. Interestingly, both RN 11100 and RN 31300 were also detected in the commercial goat milk samples, but genotype information for these milks was not available. For all other OS found, abundances were not statistically significant between the O/O and A/A genotype goat’s milks. Base peak chromatograms of representative samples from one O/O and one A/A goat are shown in Fig. 3.
Fig. 3.
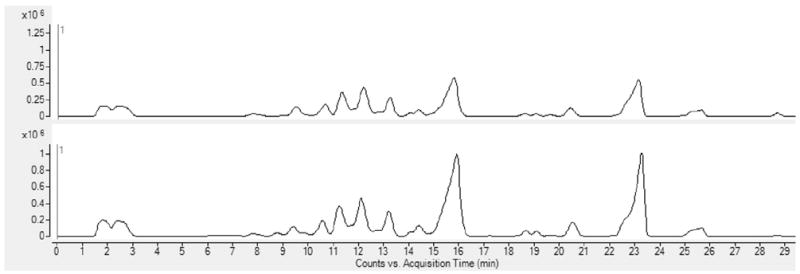
Representative base peak chromatogram from one A/A goat sample (top) and one O/O goat sample (bottom).
PCA of the entire dataset revealed a clear separation of the GMO according to CSN1S1 genotype (Fig. 4). The PCA model successfully clustered the milk samples according to the genotype using the first three principal components that explained, respectively, 17.8%, 9.1%, and 8.7% of the variance. Morning and evening milk GMO were also compared by PCA, but there was no time-dependent clustering of the data. There were no significantly different peak volumes between morning and evening milks.
Fig. 4.
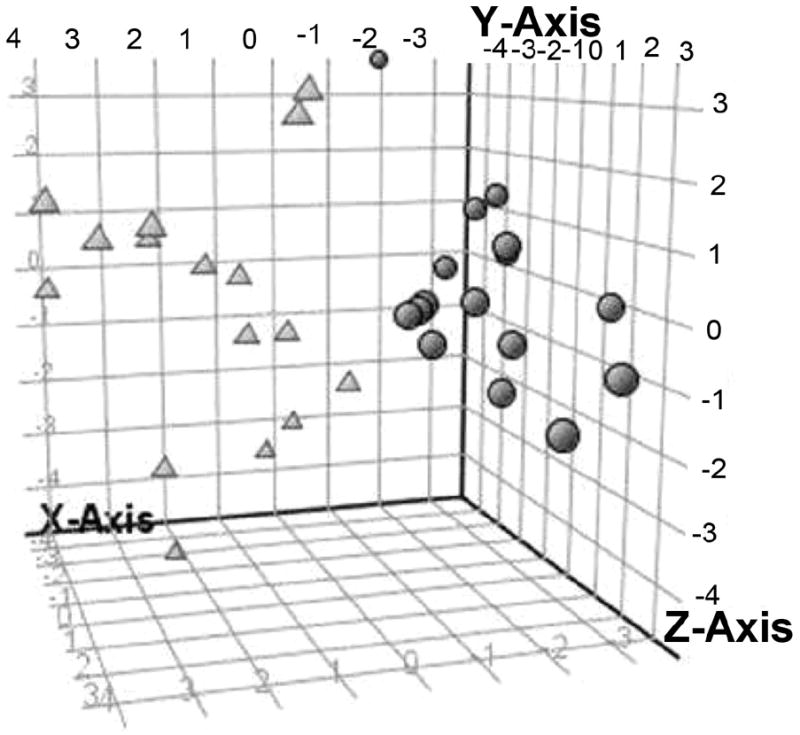
Tri-dimensional scatter plot showing grouping of principle component analysis. Scores for the goat milk oligosaccharides from two genotypes. Circles correspond to the A/A genotype and triangles to O/O genotype.
Standard unpaired t-tests were performed on the 32 milk samples. The abundance of the fucosylated OS RN 11100 was significantly different between the two genotype groups. Both peaks of this composition discriminated between the two genotypes: the peaks at 9.7 min and 11.2 min had P-values of 2.7 × 10−7 and 1.2 × 10−7, respectively. The t-tests revealed that the abundance of both isomers of fucosylated OS, RN 31300 (at 13.7 and 18.0 min), also significantly differed between the two genotypes.
3.5. OS quantification
GMO concentrations were determined by GC for eight of the goat milk samples (four from each genotype group) and from the commercial goat milk samples as previously described (Bordiga et al., 2012); the data are presented in Table 2. As an example, Fig. 5 presents the GC chromatogram for one commercial goat milk sample.
Table 2.
Relative abundance of the monosaccharides composing goat milk oligosaccharides analyzed by gas chromatography. Data are the averages of eleven goat milk samples (three commercial milks, four A/A genotype, and four O/O genotype).
| Commercial goat milk | O/O goat milk | A/A goat milk | |
|---|---|---|---|
| Number of samples | 3 | 8 | 8 |
| Fucose | Trace | Trace | Trace |
| N-Acetylglucosamine | 4.7 ±2.7% | 9.3 ±6.2% | 8.4 ±5.5% |
| Galactose | 44.4 ±1.4% | 44.2 ±4.9% | 43.3 ±4.6% |
| Glucose | 46.9 ±4.1% | 44.8 ±4.3% | 46.2 ±3.7% |
| N-Acetylneuraminic acid/N-glycolylneuraminic acid | 2.8 ±2.1% | 1.2 ±0.6% | 1.5 ±0.7% |
| Total concentration (g/L) | 1.35 ±0.19 | 1.25 ±0.32 | 1.11 ±0.29 |
Fig. 5.
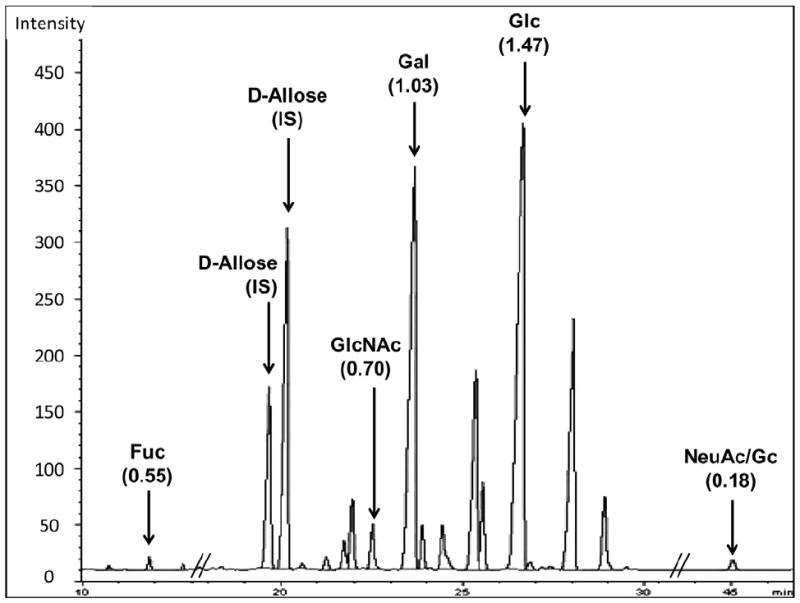
Typical gas chromatography profile of the trimethylsilyl methyl glycoside derivatives generated after methanolic HCl treatment of a mixture of purified goat milk oligosaccharides. The monosaccharides analyzed were fucose (Fuc), N-acetylglucosamine (GlcNAc), galactose (Gal), glucose (Glc), N-acetylneuraminic acid (NeuAc), and N-glycolylneuraminic acid (NeuGc). Response factors for each monosaccharide were calculated based on d-allose, the internal standard (IS) and indicated in parentheses.
Milks from A/A and O/O genotype goats had GMO concentrations of 1.11 ±0.29 g/L (range, .71–1.47 g/L) and 1.25 ±0.32 g/L (range, .90–2.00 g/L), respectively, which were not significantly different (t-test, P = 0.34); these concentrations are on the same order as that found in the commercial goat milk sample (1.35 ±0.19 g/L; range, 1.17–1.54 g/L). For comparison, OS concentrations in bovine and human milk samples were 1.09 ±0.13 g/L and 10.19 ±0.43 g/L, respectively.
4. Discussion
Goat milk has many health-benefits. One of the most significant benefits of goat milk is that it is hypoallergenic in comparison with bovine milk (Park, 1994). Other nutritional and therapeutic features of goat milk include its higher buffering capacity; short-chain fatty acid content; and zinc, iron, and magnesium content than bovine milk (Slacanac et al., 2010). Goat milk also has stronger immunological and antibacterial effects than bovine milk (Slacanac et al., 2010).
The discovery of a large number of potentially functional OS in goat milk opens the door to translational opportunities to improve human health. In particular, the presence of fucosylated and sialylated OS is of special interest because these residues provide health benefits such as prebiotic and pathogen binding activities. A limited number of studies have demonstrated direct benefits of GMO in vivo. Two studies suggested that GMO have anti-inflammatory effects and reduce intestinal inflammation in mice with induced colitis (Daddaoua et al., 2006; Lara-Villoslada et al., 2006). In order to perform functional testing, larger amounts of GMO need to become available.
To detect a large number of GMO, this study employed high-sensitivity, high-accuracy MS. LC–MS is evolving as the preferred method for analyses of OS because of its high sensitivity and capacity for compositional verification via MS/MS analysis. Moreover, the micro-chip packed with graphitized carbon solid phase employed provides reproducible retention times and separates structural isomers, enabling matching with libraries of known OS structures. MS/MS analysis can provide useful information about the arrangement of the monosaccharides within the OS, but it cannot provide linkage-specific information. However, MS can be used to solve OS structure in conjunction with systematic, sequential enzyme digestion steps.
4.1. GMO quantification
Using high-pH anion-exchange chromatography with pulsed amperometric detection, Martinez-Ferez et al. (2006a) found GMO concentrations of 0.25–0.30 g/L—5–8 times more than the OS concentration they found in bovine milk (0.03–0.06 g/L). The concentration of OS in goat milk is far below that in human milk, in which 8–13 g/L are reported for mature milk (Coppa et al., 1993b; Kunz et al., 2000) and 20–23 g/L in colostrum (Coppa et al., 1999). Our gas chromatography method revealed a GMO concentration of 1.11–1.35 g/L—roughly 4–5 times higher than previously reported (Martinez-Ferez et al., 2006b)—and a BMO concentration of 1.09 g/L—up to 18 times higher than previously reported (Fong et al., 2011; Martin-Sosa et al., 2003). However, this measurement is similar to the 0.7–1.2 g/L of OS measured in bovine colostrum (Veh et al., 1981). These measurement differences likely resulted from the different sample preparation and analysis methods employed and the different lactation stages of the samples. However, this technique found an HMO concentration (10.19 ±0.43 g/L) in agreement with previously reported concentrations (Coppa et al., 1993a; Erney et al., 2000; Kunz et al., 2000; Martinez-Ferez et al., 2006b; Nakhla et al., 1999; Viverge et al., 1985)).
The GC OS quantification method used in the present study was chosen because it does not require the use of commercial intact glycan standards. In the quantification by other methods, such as HPLC and MS, each intact OS molecule is individually quantified using the few available internal standards, whereas in this GC method, the OS are hydrolyzed prior to the analysis and only the monosaccharide standards (all commercially available) are required; therefore, the information about the relative abundance of individual species is lost, but the total concentration is determined.
4.2. CSN1S1 locus
O/O genotype goats are known to produce no αs1-casein. This lack of αs1-casein production is known to influence other aspects of goat milk composition and properties (Grosclaude and Martin, 1997; Martin and Leroux, 2000). As OS synthesis also occurs in the ER, we hypothesized that the build-up of caseins would negatively impact the production and secretion of GMO. However, in this study, the concentration of GMO secreted in milk was unaffected by the CSN1S1 genotype. Differences between the GMO profiles of the different genotypes on the individual glycan species level, however, were present. One GMO (RN 11100) was present only in the O/O milk and absent in A/A milk and another (RN 31300) was present in abundance in the O/O milk, but found only in trace amounts in the A/A milks. These differences point to a relationship between the CSN1S1 genotype and GMO synthesis.
4.3. Goat milk as a source of HMO-like compounds
HMO have a wide variety of beneficial functions. HMO exhibit an array of protective activities in the gastro-intestinal tract, and possibly promote infant brain growth (Wang et al., 2003). Fucosylated OS from human milk, in particular α-1,2-linked fucose, inhibit adhesion of pathogens that cause gastrointestinal disorders (e.g. Campylobacter jejuni, Escherichia coli) (Morrow et al., 2005; Newburg et al., 2005). Isolated acidic OS block adhesion of Helicobacter pylori which causes peptic ulcers and other gastric diseases (Simon et al., 1997). As adhesion to intestinal cells is the first step of invasion and infection, fucosylated and acidic OS protect the infant against infections, including diarrhea, which is one of the most common causes of infant mortality (Black et al., 2003; Morrow et al., 2005; Newburg et al., 2004).
At present, human milk remains the only known source of complex fucosylated and sialylated OS; this limits further functional and translational research. Currently, HMO structural complexity precludes chemical synthesis of all but the simplest HMO. A source of OS similar to HMO would be of great value as a nutritional supplement for infant and even adult formulations.
Although the number (37) of GMO reported here is lower than the number of known HMO structures (75), these GMO have structural elements critical to HMO bioactivity, including fucosylation and sialylation. Six fucosylated GMO were found, three of which were identified for the first time in goat milk (RN 11100, RN 41200, and RN 31300). Four of the six fucosylated OS found also exist in human milk: RN 21000, two isomeric forms of RN 31100 and RN 41200 (Wu et al., 2010).
The GMO with composition RN 41200 is a good example of the resemblance between GMO and HMO. Six structural isomers of this fucosylated OS were identified in human milk using HPLC-Chip–Q-TOF MS (Wu et al., 2010). In the present study, two isomers of this composition were identified in goat milk. Future structural characterization is required to determine whether these isomers match the structures found in human milk.
This study reveals that goat milk represents a source of complex HMO-like OS, which can be used as a food ingredient. Though A/A and O/O CSN1S1 genotype did not alter total GMO concentration, glycan species differences were clear.
Acknowledgments
The authors thank APIS-GENE (a French simplified joint stock company gathering professional stakeholders) for partly funding this project. The authors acknowledge Agilent for the technical assistance with the Nano LC-Chip–Q-TOF MS/MS and Mass Profiler Professional Software Analyses. The authors warmly thank C.J. Dillard for editorial assistance.
References
- Bevilacqua C, et al. Interallelic recombination is probably responsible for the occurrence of a new αs1-casein variant found in the goat species. Eur J Biochem. 2002;269:1293–1303. doi: 10.1046/j.1432-1033.2002.02777.x. [DOI] [PubMed] [Google Scholar]
- Black RE, Morris SS, Bryce J. Where and why are 10 million children dying every year? Lancet. 2003;361:2226–2234. doi: 10.1016/S0140-6736(03)13779-8. [DOI] [PubMed] [Google Scholar]
- Bordiga M, et al. Identification and characterization of complex bioactive oligosaccharides in white and red wine by a combination of mass spectrometry and gas chromatography. J Agric Food Chem. 2012;60:3700–3707. doi: 10.1021/jf204885s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanat E, Martin P, Ollivier-Bousquet M. Alpha (S1)-casein is required for the efficient transport of beta-and kappa-casein from the endoplasmic reticulum to the Golgi apparatus of mammary epithelial cells. J Cell Sci. 1999;112:3399. doi: 10.1242/jcs.112.19.3399. [DOI] [PubMed] [Google Scholar]
- Chaturvedi P, Sharma C. Purification, by high-performance liquid chromatography, and characterization, by high-field 1H-nmr spectroscopy, of two fucose-containing pentasaccharides of goat’s milk. Carbohydr Res. 1990a;203:91–101. doi: 10.1016/0008-6215(90)80048-8. [DOI] [PubMed] [Google Scholar]
- Chaturvedi P, Sharma CB. Goat milk oligosaccharides: purification and characterization by HPLC and high-field 1H-NMR spectroscopy. Biochim Biophys Acta. 1988;967:115–121. doi: 10.1016/0304-4165(88)90196-1. [DOI] [PubMed] [Google Scholar]
- Chaturvedi P, Sharma CB. Purification, by high-performance liquid chromatography, and characterization, by high-field 1H-n.m.r. spectroscopy, of two fucose-containing pentasaccharides of goat’s milk. Carbohydr Res. 1990b;203:91–101. doi: 10.1016/0008-6215(90)80048-8. [DOI] [PubMed] [Google Scholar]
- Coppa G, Gabrielli O, Pierani P, Giorgi P. New Perspectives in Infant Nutrition. Georg Thieme Verlag; Stuttgart: 1993a. Oligosaccharides in human milk and their role in bacterial adhesion; pp. 43–49. [Google Scholar]
- Coppa G, et al. Oligosaccharides in human milk during different phases of lactation. Acta Paediatr. 1999;88:89–94. doi: 10.1111/j.1651-2227.1999.tb01307.x. [DOI] [PubMed] [Google Scholar]
- Coppa GV, et al. Changes in carbohydrate composition in human milk over 4 months of lactation. Pediatrics. 1993b;91:637–641. [PubMed] [Google Scholar]
- Daddaoua A, et al. Goat milk oligosaccharides are anti-inflammatory in rats with hapten-induced colitis. J Nutr. 2006;136:672–676. doi: 10.1093/jn/136.3.672. [DOI] [PubMed] [Google Scholar]
- Engfer MB, Stahl B, Finke B, Sawatzki G, Daniel H. Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am J Clin Nutr. 2000;71:1589–1596. doi: 10.1093/ajcn/71.6.1589. [DOI] [PubMed] [Google Scholar]
- Erney RM, et al. Variability of human milk neutral oligosaccharides in a diverse population. J Pediatr Gastroenterol Nutr. 2000;30:181. doi: 10.1097/00005176-200002000-00016. [DOI] [PubMed] [Google Scholar]
- Fong B, Ma K, McJarrow P. Quantification of bovine milk oligosaccharides using liquid chromatography-selected reaction monitoring-mass spectrometry. J Agric Food Chem. 2011;59:9788–9795. doi: 10.1021/jf202035m. [DOI] [PubMed] [Google Scholar]
- Gnoth MJ, Kunz C, Kinne-Saffran E, Rudloff S. Human milk oligosaccharides are minimally digested in vitro. J Nutr. 2000;130:3014–3020. doi: 10.1093/jn/130.12.3014. [DOI] [PubMed] [Google Scholar]
- Grosclaude F, Martin P. IV. 1-Casein polymorphisms in the goat. International Dairy Federation (special issue) 1997:241–253. [Google Scholar]
- Kunz C, Rudloff S, Baier W, Klein N, Strobel S. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu Rev Nutr. 2000;20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- Lara-Villoslada F, et al. Oligosaccharides isolated from goat milk reduce intestinal inflammation in a rat model of dextran sodium sulfate-induced colitis. Clin Nutr. 2006;25:477–488. doi: 10.1016/j.clnu.2005.11.004. [DOI] [PubMed] [Google Scholar]
- LoCascio RG, et al. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem. 2007;55:8914–8919. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- Martin-Sosa S, Martin MJ, Garcia-Pardo LA, Hueso P. Sialyloligosaccharides in human and bovine milk and in infant formulas: variations with the progression of lactation. J Dairy Sci. 2003;86:52–59. doi: 10.3168/jds.S0022-0302(03)73583-8. [DOI] [PubMed] [Google Scholar]
- Martin P, Leroux C. Le gène caprin spécifiant la caséine αs1: un suspect tout désigné aux effets aussi multiples qu’inattendus. INRA Prod Anim HS2000. 2000:125–132. [Google Scholar]
- Martinez-Ferez A, Guadix A, Guadix EM. Recovery of caprine milk oligosaccharides with ceramic membranes. J Membr Sci. 2006a;276:23–30. [Google Scholar]
- Martinez-Ferez A, et al. Goats’ milk as a natural source of lactose-derived oligosaccharides: isolation by membrane technology. Int Dairy J. 2006b;16:173–181. [Google Scholar]
- Mehra R, Kelly P. Milk oligosaccharides: structural and technological aspects. Int Dairy J. 2006;16:1334–1340. [Google Scholar]
- Merkle RK, Poppe I. Carbohydrate composition analysis of glycoconjugates by gas-liquid chromatography/mass spectrometry. Methods Enzymol. 1994;230:1–15. doi: 10.1016/0076-6879(94)30003-8. [DOI] [PubMed] [Google Scholar]
- Moioli B, Pila F, Tripaldi C. Detection of milk protein genetic polymorphisms in order to improve dairy traits in sheep and goats: a review. Small Rumin Res. 1998;27:185–195. [Google Scholar]
- Morrow AL, Ruiz-Palacios GM, Jiang X, Newburg DS. Human-milk glycans that inhibit pathogen binding protect breast-feeding infants against infectious diarrhea. J Nutr. 2005;135:1304–1307. doi: 10.1093/jn/135.5.1304. [DOI] [PubMed] [Google Scholar]
- Nakhla T, Fu D, Zopf D, Brodsky NL, Hurt H. Neutral oligosaccharide content of preterm human milk. Br J Nutr. 1999;82:361–367. doi: 10.1017/s0007114599001609. [DOI] [PubMed] [Google Scholar]
- Newburg DS, et al. Human milk alpha 1,2-linked fucosylated oligosaccharides decrease risk of diarrhea due to stable toxin of Ecoli in breastfed infants. Protect Infants through Hum Milk. 2004;554:457–461. doi: 10.1007/978-1-4757-4242-8_64. [DOI] [PubMed] [Google Scholar]
- Newburg DS, Ruiz-Palacios GM, Morrow AL. Human milk glycans protect infants against enteric pathogens. Annu Rev Nutr. 2005;25:37–58. doi: 10.1146/annurev.nutr.25.050304.092553. [DOI] [PubMed] [Google Scholar]
- Ninonuevo MR, et al. A strategy for annotating the human milk glycome. J Agric Food Chem. 2006;54:7471–7480. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- Park Y. Hypo-allergenic and therapeutic significance of goat milk. Small Rumin Res. 1994;14:151–159. [Google Scholar]
- Pierre A, Le Quéré J-L, Famelart M-H, Riaublanc A, Rousseau F. Composition, yield, texture and aroma compounds of goat cheeses as related to the A and O variants of αsl casein in milk. Lait. 1998;78:291–301. [Google Scholar]
- Silanikove N, Leitner G, Merin U, Prosser C. Recent advances in exploiting goat’s milk: quality, safety and production aspects. Small Rumin Res. 2010;89:110–124. [Google Scholar]
- Simon PM, Goode PL, Mobasseri A, Zopf D. Inhibition of Helicobacter pylori binding to gastrointestinal epithelial cells by sialic acid-containing oligosaccharides. Infect Immun. 1997;65:750–757. doi: 10.1128/iai.65.2.750-757.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slacanac V, et al. Nutritional and therapeutic value of fermented caprine milk. Int J Dairy Technol. 2010;63:171–189. [Google Scholar]
- Strum J, Aldredge D, Barile D, Lebrilla C. Coupling Flash LC with MS for enrichment and isolation of milk oligosaccharides for functional studies. Anal Biochem. 2012;424:87–96. doi: 10.1016/j.ab.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeley CC, Walker B. Determination of carbohydrates in glycolipids and gangliosides by gas chromatography. Anal Chem. 1964;36:1461–1466. [Google Scholar]
- Urashima T, Bubb WA, Messer M, Tsuji Y, Taneda Y. Studies of the neutral trisaccharides of goat (Capra hircus) colostrum and of the one- and two-dimensional 1H and 13C NMR spectra of 6′-N-acetylglucosaminyllactose. Carbohydr Res. 1994;262:173–184. doi: 10.1016/0008-6215(94)84177-2. [DOI] [PubMed] [Google Scholar]
- Urashima T, Murata S, Nakamura T. Structural determination of monosialyl trisaccharides obtained from caprine colostrum. Comp Biochem Physiol B: Biochem Mol Biol. 1997;116:431–435. doi: 10.1016/s0305-0491(96)00269-6. [DOI] [PubMed] [Google Scholar]
- Veh RW, et al. New chromatographic system for the rapid analysis and preparation of colostrum sialyloligosaccharides. J Chromatogr A. 1981;212:313–322. doi: 10.1016/s0021-9673(01)84044-9. [DOI] [PubMed] [Google Scholar]
- Viverge D, et al. Variations of lactose and oligosaccharides in milk from women of blood types secretor A or H, secretor Lewis, and secretor H/nonsecretor Lewis during the course of lactation. Ann Nutr Metab. 1985;29:1–11. doi: 10.1159/000176947. [DOI] [PubMed] [Google Scholar]
- Viverge D, Grimmonprez L, Solere M. Chemical characterization of sialyl oligosaccharides isolated from goat (Capra hircus) milk. Biochim Biophys Acta Gen Subjects. 1997;1336:157–164. doi: 10.1016/s0304-4165(97)00021-4. [DOI] [PubMed] [Google Scholar]
- Wang B, Staples A, Hunter A, Yu B, Brand Miller J. Effect of experimental oligosaccharide on brain and body weight. Asia Pac J Clin Nutr. 2003;12(Suppl):S60. [Google Scholar]
- Wu S, Grimm R, German JB, Lebrilla CB. Annotation and structural analysis of sialylated human milk oligosaccharides. J Proteome Res. 2011;10:856–868. doi: 10.1021/pr101006u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Tao N, German JB, Grimm R, Lebrilla CB. Development of an annotated library of neutral human milk oligosaccharides. J Proteome Res. 2010;9:4138–4151. doi: 10.1021/pr100362f. [DOI] [PMC free article] [PubMed] [Google Scholar]


