Abstract
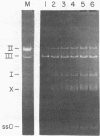
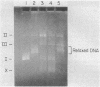
In the presence of adenosine 5'-[gamma-thio]triphosphate, a nonhydrolyzable analog of ATP, Escherichia coli recA protein extensively unwinds duplex DNA in a reaction that is strongly stimulated by either homologous or heterologous single-stranded DNA [Cunningham, R.P., Shibata, T., DasGupta, C. & Radding, C.M. (1979) Nature (London) 281, 191-195]. In the presence of ATP and homologous circular single-stranded DNA, recA protein also unwinds circular duplex DNA that is nicked at a heterologous site. When DNA ligase seals this nick, the product is a highly negatively superhelical molecule that can be relaxed by E. coli topoisomerase I. This unwinding requires a high degree of homology since phi X174 single-stranded DNA does not serve as a cofactor in the unwinding of G4 DNA, even though these molecules are 70% homologous. Like synapsis itself, and unlike strand exchange which follows synapsis, unwinding is sensitive to inhibition by ADP. Because recA protein unwinds duplex DNA when neither the single-stranded DNA nor the duplex DNA has a free end in the region of homology, unwinding can be initiated or mediated by a synaptic structure that differs from that of a simple D loop. The paired circular single strand in the synaptic structure behaves like one strand of an under-wound helix because E. coli topoisomerase I can interwind it with its complement.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cox M. M., Lehman I. R. Directionality and polarity in recA protein-promoted branch migration. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6018–6022. doi: 10.1073/pnas.78.10.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M. M., Lehman I. R. recA protein of Escherichia coli promotes branch migration, a kinetically distinct phase of DNA strand exchange. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3433–3437. doi: 10.1073/pnas.78.6.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M. M., Lehman I. R. recA protein-promoted DNA strand exchange. Stable complexes of recA protein and single-stranded DNA formed in the presence of ATP and single-stranded DNA binding protein. J Biol Chem. 1982 Jul 25;257(14):8523–8532. [PubMed] [Google Scholar]
- Cunningham R. P., DasGupta C., Shibata T., Radding C. M. Homologous pairing in genetic recombination: recA protein makes joint molecules of gapped circular DNA and closed circular DNA. Cell. 1980 May;20(1):223–235. doi: 10.1016/0092-8674(80)90250-0. [DOI] [PubMed] [Google Scholar]
- Cunningham R. P., Shibata T., DasGupta C., Radding C. M. Single strands induce recA protein to unwind duplex DNA for homologous pairing. Nature. 1979 Sep 20;281(5728):191–195. doi: 10.1038/281191a0. [DOI] [PubMed] [Google Scholar]
- Cunningham R. P., Wu A. M., Shibata T., DasGupta C., Radding C. M. Homologous pairing and topological linkage of DNA molecules by combined action of E. coli RecA protein and topoisomerase I. Cell. 1981 Apr;24(1):213–223. doi: 10.1016/0092-8674(81)90517-1. [DOI] [PubMed] [Google Scholar]
- DasGupta C., Radding C. M. Lower fidelity of RecA protein catalysed homologous pairing with a superhelical substrate. Nature. 1982 Jan 7;295(5844):71–73. doi: 10.1038/295071a0. [DOI] [PubMed] [Google Scholar]
- DasGupta C., Shibata T., Cunningham R. P., Radding C. M. The topology of homologous pairing promoted by RecA protein. Cell. 1980 Nov;22(2 Pt 2):437–446. doi: 10.1016/0092-8674(80)90354-2. [DOI] [PubMed] [Google Scholar]
- DasGupta C., Wu A. M., Kahn R., Cunningham R. P., Radding C. M. Concerted strand exchange and formation of Holliday structures by E. coli RecA protein. Cell. 1981 Aug;25(2):507–516. doi: 10.1016/0092-8674(81)90069-6. [DOI] [PubMed] [Google Scholar]
- Di Capua E., Engel A., Stasiak A., Koller T. Characterization of complexes between recA protein and duplex DNA by electron microscopy. J Mol Biol. 1982 May 5;157(1):87–103. doi: 10.1016/0022-2836(82)90514-9. [DOI] [PubMed] [Google Scholar]
- Dressler D., Potter H. Molecular mechanisms in genetic recombination. Annu Rev Biochem. 1982;51:727–761. doi: 10.1146/annurev.bi.51.070182.003455. [DOI] [PubMed] [Google Scholar]
- Dunn K., Chrysogelos S., Griffith J. Electron microscopic visualization of recA-DNA filaments: evidence for a cyclic extension of duplex DNA. Cell. 1982 Apr;28(4):757–765. doi: 10.1016/0092-8674(82)90055-1. [DOI] [PubMed] [Google Scholar]
- Flory J., Radding C. M. Visualization of recA protein and its association with DNA: a priming effect of single-strand-binding protein. Cell. 1982 Apr;28(4):747–756. doi: 10.1016/0092-8674(82)90054-x. [DOI] [PubMed] [Google Scholar]
- Kaguni J., Ray D. S. Cloning of a functional replication origin of phage G4 into the genome of phage M13. J Mol Biol. 1979 Dec 25;135(4):863–878. doi: 10.1016/0022-2836(79)90516-3. [DOI] [PubMed] [Google Scholar]
- Kahn R., Cunningham R. P., DasGupta C., Radding C. M. Polarity of heteroduplex formation promoted by Escherichia coli recA protein. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4786–4790. doi: 10.1073/pnas.78.8.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani T., Shibata T., Iwabuchi M., Watabe H., Iino T., Ando T. ATP-dependent unwinding of double helix in closed circular DNA by recA protein of E. coli. Nature. 1982 Sep 2;299(5878):86–89. doi: 10.1038/299086a0. [DOI] [PubMed] [Google Scholar]
- Radding C. M. Homologous pairing and strand exchange in genetic recombination. Annu Rev Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- Radding C. M. Recombination activities of E. coli recA protein. Cell. 1981 Jul;25(1):3–4. doi: 10.1016/0092-8674(81)90224-5. [DOI] [PubMed] [Google Scholar]
- Shibata T., Cunningham R. P., DasGupta C., Radding C. M. Homologous pairing in genetic recombination: complexes of recA protein and DNA. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5100–5104. doi: 10.1073/pnas.76.10.5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T., Cunningham R. P., Radding C. M. Homologous pairing in genetic recombination. Purification and characterization of Escherichia coli recA protein. J Biol Chem. 1981 Jul 25;256(14):7557–7564. [PubMed] [Google Scholar]
- Shibata T., Ohtani T., Chang P. K., Ando T. Role of superhelicity in homologous pairing of DNA molecules promoted by Escherichia coli recA protein. J Biol Chem. 1982 Jan 10;257(1):370–376. [PubMed] [Google Scholar]
- Shibata T., Ohtani T., Iwabuchi M., Ando T. D-loop cycle. A circular reaction sequence which comprises formation and dissociation of D-loops and inactivation and reactivation of superhelical closed circular DNA promoted by recA protein of Escherichia coli. J Biol Chem. 1982 Dec 10;257(23):13981–13986. [PubMed] [Google Scholar]
- Stasiak A., Di Capua E. The helicity of DNA in complexes with recA protein. Nature. 1982 Sep 9;299(5879):185–186. doi: 10.1038/299185a0. [DOI] [PubMed] [Google Scholar]
- Stettler U. H., Weber H., Koller T., Weissmann C. Preparation and characterization of form V DNA, the duplex DNA resulting from association of complementary, circular single-stranded DNA. J Mol Biol. 1979 Jun 15;131(1):21–40. doi: 10.1016/0022-2836(79)90299-7. [DOI] [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. The structure of DNA. Cold Spring Harb Symp Quant Biol. 1953;18:123–131. doi: 10.1101/sqb.1953.018.01.020. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Interaction between DNA and an Escherichia coli protein omega. J Mol Biol. 1971 Feb 14;55(3):523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]
- West S. C., Cassuto E., Howard-Flanders P. Heteroduplex formation by recA protein: polarity of strand exchanges. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6149–6153. doi: 10.1073/pnas.78.10.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A. M., Kahn R., DasGupta C., Radding C. M. Formation of nascent heteroduplex structures by RecA protein and DNA. Cell. 1982 Aug;30(1):37–44. doi: 10.1016/0092-8674(82)90009-5. [DOI] [PubMed] [Google Scholar]