Abstract
Tumor metastasis is the leading cause of death among breast cancer patients. PELP1 is a nuclear receptor coregulator that is upregulated during breast cancer progression to metastasis and is an independent prognostic predictor of shorter survival of breast cancer patients. Here, we show that PELP1 modulates expression of metastasis-influencing microRNAs (miRs) to promote cancer metastasis. Whole genome miR array analysis using PELP1 over expressing and under expressing model cells revealed that miR-200a and miR-141 levels inversely correlated with PELP1 expression. Consistent with this, PELP1 knockdown resulted in lower expression of miR-200a target genes ZEB1 and ZEB2. PELP1 knockdown significantly reduced tumor growth and metastasis compared with parental cells in an orthotopic xenograft tumor model. Furthermore, re-introduction of miR-200a and miR-141 mimetics into PELP1 overexpressing cells reversed PELP1 target gene expression, decreased PELP1 driven migration/invasion in vitro, and significantly reduced in vivo metastatic potential in a preclinical model of experimental metastasis. Our results demonstrated that PELP1 binds to miR-200a and miR-141 promoters and regulates their expression by recruiting chromatin modifier HDAC2 as revealed by ChIP, siRNA and HDAC inhibitor assays. Taken together, our results suggest that PELP1 regulates tumor metastasis by controlling the expression and functions of the tumor metastasis suppressors miR-200a and miR-141.
Keywords: ER-coactivators, PELP1, Breast cancer, Metastasis, EMT, MiR
Introduction
Breast cancer is the most frequently diagnosed cancer in women. Metastases spawned by malignant tumors are responsible for the majority of breast cancer-related morbidity and mortality (1). Tumor metastasis comprises a series of discrete biological processes that move tumor cells from the primary neoplasm to a distant location (2) and involves a multi-step cascade of coordinated cell adhesion and contractility as well as a proteolytic remodeling of the extra-cellular matrix (3). Even though currently used breast cancer targeted therapies directed against nuclear receptors such as the ER and growth factor receptors such as HER2 are effective in curbing the localized disease, these therapies are less effective in treating metastases. Further, a substantial number of patients treated with targeted therapies acquire resistance over a period of time and the tumor recur as metastases. Even though significant information is available on the process of metastasis; a critical need still exists to identify novel targets that can be used to curb the progression of breast cancer metastasis.
MiR-mediated regulation of tumorigenesis is emerging as a new paradigm in the field of cancer biology. Misexpression of miR occurs in many cancers, including breast cancer. MiRs play critical roles in diverse biological processes, aberrantly expressed in breast tumors and function as regulators of tumor behavior and progression (4;5). MiRs are also involved in multiple steps in the metastatic cascade by influencing cancer cell adherence, migration, invasion, motility, and angiogenesis (6). The class of miRs associated with metastatic process was recently termed metastamir (7). The metastasis-promoting metastamir (such as miR-21, miR -373, and miR -155) enhance breast cancer metastasis (8), while metastasis-suppressing metastamirs (such as miR -200, miR -145, and miR -661) inhibit metastasis with minimum effects on orthotropic tumor growth (8). The molecular mechanism of miR deregulation and how such deregulation contributes to breast cancer metastasis remains elusive and is an area of significant importance.
Nuclear receptors (NR) play an important role in breast cancer progression and their signaling is complex, involving coregulators (9). Emerging evidence suggests that metastatic tumors express increased levels of coregulators, that NR coregulators have the potential to activate an appropriate set of genes to produce a desired goal such as cell growth via their interactions with multiple NRs (10–12) and that their deregulation provides the cancer cells an advantage in growth and metastasis. Accordingly, several NR-coregulators including steroid receptor coactivator-3 (SRC-3)/AIB1, metastasis tumor antigen 1 (MTA1), nuclear receptor corepressor (NCoR) complex and its paralog, silencing mediator of retinoid and thyroid hormone receptor (SMRT), and histone deacetylase (HDAC) are shown to play important roles during key steps in the invasion-metastasis cascade (13;14). How the coregulator signaling influences metastasis and whether the coregulator signaling involves miRs remain poorly understood.
Proline glutamic acid leucine rich protein (PELP1) is an NR coregulator that interacts with multiple hormonal receptors and exhibits aberrant expression in many hormone-related cancers (15;16). PELP1 functions as proto-oncogene (17) and its expression is upregulated in metastatic tumors. It is a prognostic indicator of shorter breast cancer-specific survival and disease-free interval. Recent studies from our lab have uncovered a pivotal role for PELP1 signaling in breast cancer cell migration and metastasis (18) and demonstrated PELP1 has the potential to modulate expression of genes involved in metastasis. PELP1 overexpression is equally observed in both in ER-positive and ER-negative metastatic tumors, suggesting that PELP1 may have functions independent of the ER in metastatic cells(17;18). Collectively, these published studies strongly suggest a role of PELP1 in metastasis; however, the mechanism by which PELP1 modulates metastasis genes and whether PELP1 signaling involves regulation of miR remain unknown.
In this study, we investigated the molecular mechanism and significance of PELP1-miR signaling axis in the regulation of breast cancer metastasis. Our results suggest that PELP1 plays a role in breast cancer metastasis by modulating expression of tumor suppressor miR-200a and miR-141. These novel findings suggest that PELP1-miR axis may be a crucial stimulus for promoting breast cancer metastasis.
Results
PELP1 modulates expression of miR-200a and miR-141
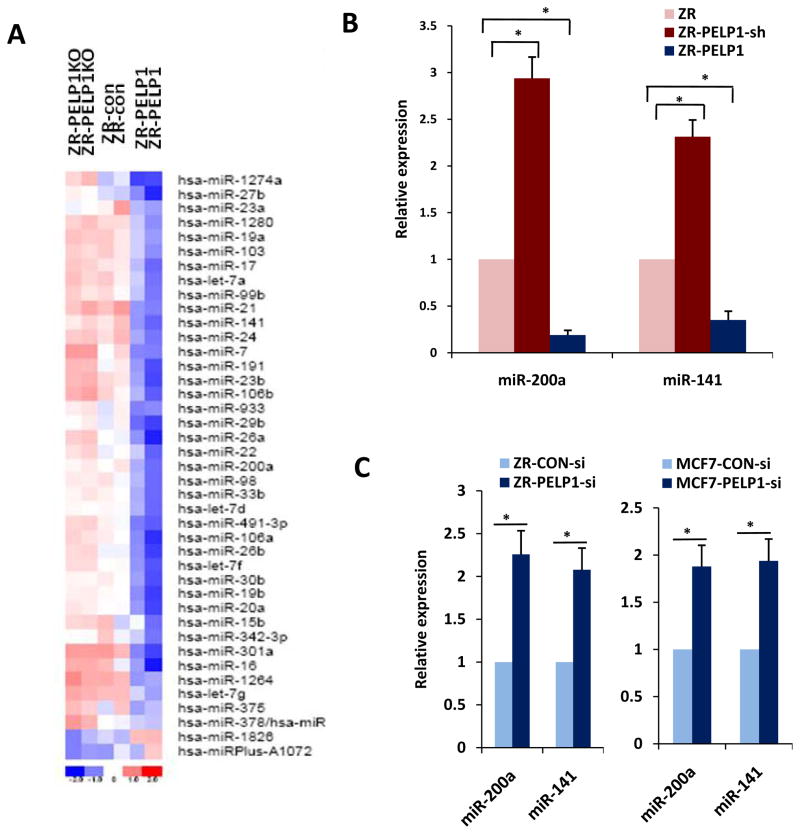
PELP1 is a proto-oncogene that functions as coregulator of several nuclear receptors with deregulated expression in metastastic tumors. We tested the hypothesis that PELP1 modulation of miRs contributes to its metastasis potential. For these assays, we used ZR75 models that stably express the PELP1, PELP1-shRNA or control-shRNA vector. Compared to the ZR75 vector cells, the ZR75PELP1-shRNA cells express an 80% reduction in endogenous PELP1 expression, while PELP1-overexpressing cells had an 8-fold increase in PELP1 transcript over endogenous PELP1 (Supplementary Figure S1a, b). We recently demonstrated that ZR-PELP1 model cells exhibit oncogenic properties and metastatic potential (17) while PELP1-shRNA cells exhibit decreased metastasis (18). To investigate the general effect of PELP1 deregulation on miR expression, whole-genome miR array analysis was performed by using the human miRCURY™ LNA microarrays. The PELP1 status significantly affected the miR expression profiles, as several miRs were found significantly downregulated in PELP1-overexpressing cells and PELP1 knockdown significantly increased the expression of these miRs (Figure 1a). We used real-time qPCR to validate the expression of the top 10 miRs with the highest fold change from array results (Supplementary Figure S1c). Results of confirmation for two representative miRs are shown in Figure 1b. RTqPCR results confirmed that PELP1 deregulation suppressed selective miRs while PELP1 knockdown enhanced their expression. Among the PELP1-regulated miRs, we focused on miR200a and miR141 as these two miRs are implicated in Epithelial to Mesenchymal transition (EMT) and the metastasis processes of the cells (19). To confirm the results obtained from PELP1 stable model cells, we further validated the results by using transient knockdown with PELP1 siRNA that target a different site on PELP1 than the PELP1-shRNA targeted site. In these assays, transient knockdown of PELP1 expression also substantially enhanced the expression miR-200a and miR-141 in two ER-positive model cell lines MCF7 and ZR75(Figure 1c). Interestingly, PELP1 also has the potential to regulate expression of miR200a and miR141 in ER-negative breast cancer model cells MDA-MB-231 and MDA-MB-468 (Supplementary Figure S1 d, e) suggesting PELP1 regulation of miR200a and miR141 is independent of ER-alpha. Collectively, these results suggest that the proto-oncogene PELP1 has the potential to regulate the expression of miR-200a and miR-141.
Figure 1.
PELP1 status influences miR expression. A, RNA isolated from ZR-control, ZR-PELP1-shRNA, and ZR-PELP1 cells were subjected to human miRCURY™ LNA Array miR profiling. All three samples were used in duplicate for the array analysis. The heat map shown depicts expression levels of the top most discriminatory up- and down-regulated miRs among three different samples. The blue color indicates down-regulation and the red color indicates up-regulation of miR expression. B, real-time qPCR validation of two different miRs in ZR-control vector, ZR-PELP1-shRNA, and ZR-PELP1 cells. C, ER-positive (ZR-75, MCF7) cells were transiently transfected with control siRNA and PELP1 siRNA, and after 72h, expression levels of miR-200a and miR-141 were measured by real-time qPCR analysis. The relative expression of each miR was quantified by measuring Ct values and normalized against RNU19.*, P ≤ 0.05, t test.
MiR-200a and 141 mimetics inhibit the PELP1-mediated migratory and invasion potential
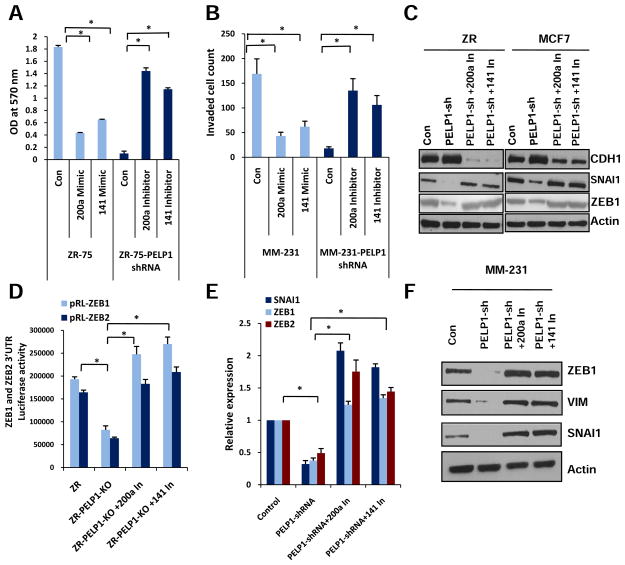
Because miR-200 family members are implicated in metastasis and because PELP1 has the potential to regulate expression of miR200a/141, we tested the significance of miR-200a and miR-141 on PELP1-mediated migratory and invasion functions by using mimetics and antagomirs of miR-200a and miR-141. For these assays we have used established ZR75 (20) and MDA-MB-231 (18) model cells that stably express PELP1 shRNA. Stable expression of PELP1 shRNA reduced PELP1 expression to 70–80% of endogenous PELP1 in these models. As seen in previous published studies(17;21), PELP1 knockdown alter cell morphology, reduces motile F-actin containing structures in MDA-MB-231 cells. Similarly, PELP1 overexpression in less motile ZR-75 cells altered cell morphology and promoted F-actin containing motile structures (Supplementary Figure S2a, b). In migration assays using the Boyden chamber, PELP1 knockdown significantly reduced migration of ZR-75 cells compared to the migration of the control vector-transfected cells (Figure 2a). Similarly, mimetics of miR200a and miR141 also inhibited the migration of ZR75 cells. Conversely, treatment of ZR75-PELP1-shRNA cells with antagomirs of miR-200a and miR-141 rescued the defective migration seen in ZR75-PELP1-shRNA cells. To test the PELP1 effect on invasion, we used highly invasive MDA-MB-231 cells. PELP1 knockdown cells (MDA-MB-231-PELP1shRNA) had significantly reduced invasion compared to the invasion of the control cells (MDA-MB-231-conshRNA) (Figure 2b), while treatment of control MDA-MB-231 shRNA vector cells with the mimetics of miR200a/141 resulted in less invasion and treatment with antagomirs of miR-200a and miR-141 restored the lost invasion potential of the MDA-MB-231-PELP1shRNA cells. The results observed in ZR75 and MDA-MB-231-PELP1shRNA stable clones were also validated using transient knock down of PELP1 by siRNA (data not shown). Published studies implicate a role of miR-200 family members in the regulation of EMT genes (19). Since our earlier studies showed that PELP1 has the potential to regulate genes involved in EMT (18), we tested the hypothesis that PELP1 regulation of EMT genes may involve miR-200 family members. Western analysis revealed that miR-200a and miR-141 antagomirs reverses PELP1-shRNA mediated alterations in EMT genes in ER-positive cells (Figure 2c, Supplementary Figure S2c). Accordingly, in reporter gene-based assays PELP1-shRNA cells exhibited less ZEB1 and ZEB2 3′UTR luciferase reporter activity than the activity in control-shRNA cells and the PELP1 knockdown-mediated repression of reporter activity was relieved by the miR200a and miR141 antagomirs (Figure 2d). Similarly, miR-200a and miR-141 antagomirs also reversed PELP1-shRNA-mediated changes in EMT genes in ER-negative model cells (Figure 2e and f). Collectively, these results suggest that PELP1-mediated cell migratory/invasion functions involve the miR-200a and miR-141 family members.
Figure 2.
MiRs influence the PELP1-mediated migratory and invasion potential. A, Boyden chamber analysis of the cell migration potential of the ZR cells transfected with mimetics and ZR-PELP1-shRNA cells transfected with inhibitors/antagomirs of miRs 200a and 141. B, MDA-MB-231 and MDA-MB-231-PELP1-shRNA cells were transfected with miRs 200a and 141 mimetics and inhibitors/antagomirs respectively and their effects on invasion were analyzed by using Matrigel invasion chamber assays. Mean and SDs are from three independent experiments. C, Western blot analysis of EMT genes expression in control, PELP1-shRNA cells and PELP1-shRNA clones treated with miR inhibitors/antagomirs. D, ZR-control-shRNA and ZR-PELP1-shRNA cells were transfected with ZEB1 and ZEB2 3′UTR reporter genes and treated with miR-200a and 141 inhibitors/antagomirs. The reporter activity was measured after 48 h. Mean and SDs are from 3 independent experiments *, P ≤ 0.05, t test. E, F, MDA-MB-231 cells stably expressing control-shRNA or PELP1-shRNA were used in these assays. Real-time qPCR (E) and Western blot analysis (F) of EMT gene expression in control shRNA, PELP1 shRNA model cells and PELP1 shRNA cells treated with miR inhibitors/antagomirs in MDA-MB-231 cells. *, P≤ 0.05; t test.
PELP1 modulates expression of miR-200 family members through epigenetic mechanisms
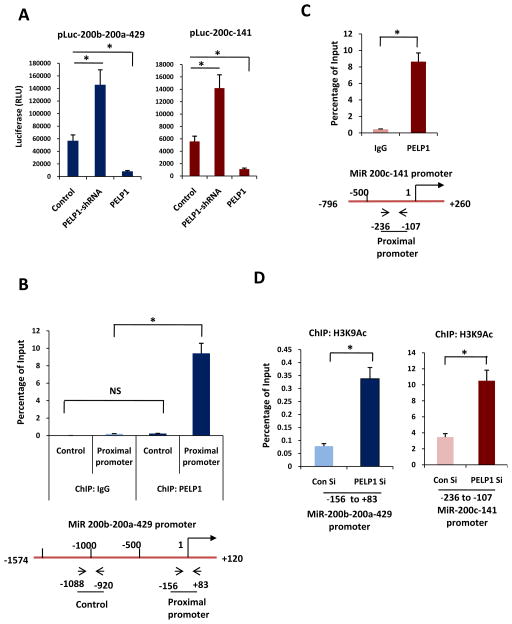
To elucidate the mechanism by which PELP1 regulates miR-200a and miR-141 expression, we performed miR-200-promoter-Luc assays. Over expression of PELP1 significantly reduced the activities of both 200b-200a-429 and 200c-141 promoter luciferase reporters (Figure 3a). Published studies suggested that PELP1 modulate expression of genes either by modulating epigenetic changes via its interaction with chromatin modifying enzymes or by promoting non-genomic actions via Src, PI3K and MAPK pathways. Since PELP1 regulated expression of miR family members in both ER+ve and ER-ve cells, we initially examined whether PELP1 mediated non-genomic actions may play a role in PELP1 regulation of miRs. To test this we have performed miR-200-promoter-Luc assays in the presence or absence of specific inhibitor of Src-, PI3K- and MAPK-pathways. All three inhibitors failed to block PELP1 mediated suppression of miRs (Supplementary Figure S3a, b). We then tested whether PELP1 mediated genomic actions may play a role in modulation of miR expression and tested if PELP1 is recruited to the promoter region of miR200. Previous studies have shown that the promoter region containing −153 to +83 relative to premiRNA200b-200a-429 as proximal promoter that contains regulatory elements for transcription of miR200b-200a-429 (22;23). ChIP results showed that PELP1 is recruited to the proximal promoter region −153 to +83 of 200b-200a-429. We have also used distal promoter region (−1088 to −920) as a control and no recruitment of PELP1 was observed in that region (Figure 3b). We next tested PELP1 recruitment to miR 200c-141 promoter using primers that encompass −236 to −107 promoter region of miR 200c-141. We focused our ChIP studies to this region based on earlier published studies that suggested −236 to −107 have regulatory elements which control miR200c-141 promoter and this region is subjected to epigenetic regulation (22;24). ChIP results confirmed PELP1 ability to recruit to miR200c-141 promoter (Figure 3c). These results suggest that PELP1 has the potential to recruit to miR-200 promoters and that PELP1-mediated genomic actions may play a role in the regulation of these miRs. PELP1 does not possess any known enzymatic activity. However, published studies demonstrated that PELP1 interacts with several chromatin-modifying enzymes including histone acetyl transferases and KDM1 (lysine-specific demethylase 1) and has the potential to alter epigenetic marks at the target gene promoters (25;26). We therefore, examined the status of the histone epigenetic modifications at the proximal promoters of miR-200a and miR-141. ChIP analysis revealed that PELP1 knockdown enhanced the active histone mark H3K9Ac at the promoters of both miR-200a and miR-141 (Figure 3d). ChIP analysis with H3K9Me2 also revealed that PELP1 knockdown reduces this inactive histone mark at the promoters of both miR-200a and miR-141 (Supplementary Figure S3c). These results suggest that PELP1-mediated alteration in epigenetic modifications may play a role in the modulation of the expression and function of miR-200 family genes by PELP1.
Figure 3.
PELP1 recruits and promotes epigenetic changes at the promoters of miR-200. A, ZR, ZR-PELP1-shRNA and ZR-PELP1 cells were transfected with the promoter reporters of the miR200c-141 and miR200b-200a-429 clusters, and the luciferase activity was measured after 72 h.B, ChIP assay was done by using the DNA isolated from the ZR cells with antibodies specific for PELP1 or the isotype rabbit IgG control. DNA recovered from ChIP or input controls was subjected to real-time qPCR with primers specific to −156 to +83 and −1088 to −920 that represent in the proximal and nonspecific promoter regions of miR200b-200a-429 promoter. Schematic diagram showing the location of primers with respect to transcription site of 200b-200a-429 is depicted at the bottom. C, ChIP DNA was prepared as described in B, and real-time qPCR was done using primers specific to −236 to −107 region of miR200c-141 promoter. Schematic diagram showing the location of primers with respect to transcription site of miR200c-141 is depicted at the bottom. D, ZR cells were transfected with control or PELP1-siRNA, and the ChIP assay was done with antibodies specific for H3K9Ac. DNA recovered from the ChIP or input controls was subjected to real-time qPCR using primers that detect −156 to +83 and −236 to −107 promoter region of miR200b-200a-429 and miR200c-141 respectively. The promoter occupancy was calculated based on the ratio of ChIP/input control. *, P≤ 0.05; t test.
HDAC2 and PELP1 interaction play a critical role in regulation of miR-200 family members
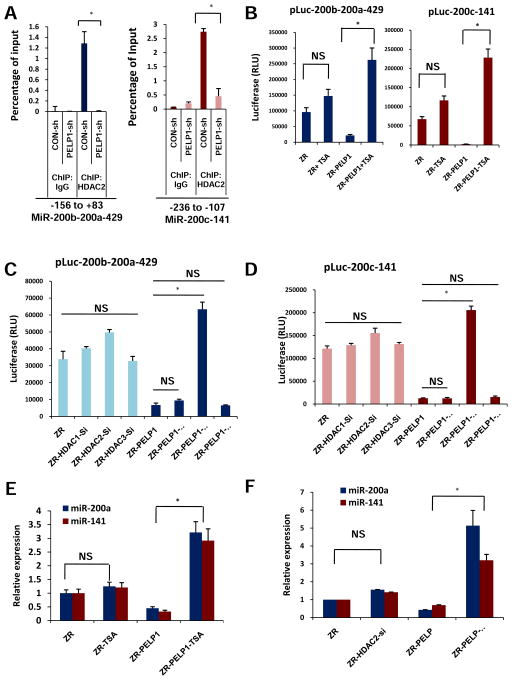
Previous published findings established that PELP1 interacts with the histone deacetylase HDAC2 (25). Upregulation of H3K9Ac at the miR promoter region in PELP1 knockdown cells suggest a possibility that the PELP1 interaction with the HDAC2 enzyme may contribute to the alteration in acetylation levels at the miR-200 promoters. The ChIP assay with HDAC2 demonstrated the recruitment of HDAC2 at the regulatory regions of both miR promoters (Figure 4a). The role of PELP1 in HDAC2 recruitment at the miR200 promoters was further examined by performing a ChIP assay with HDAC2 in ZR-PELP1-shRNA cells (Figure 4a). Diminished recruitment of HDAC2 to the miR promoter regions was found in the absence of PELP1. Accordingly, the HDAC inhibitor Trichostatin A (TSA) treatment reversed PELP1-driven repressive effects on both miR200c-141 and miR200b-200a-429 promoter activities (Figure 4b). To further confirm the role and specificity of HDAC2 in PELP1 mediated regulation of miR-200a and miR-141, we have used HDAC2-siRNA. Non-targeting-, HDAC1- and HDAC3-siRNAs were used as additional controls and siRNA specificity was validated by Western blotting (Supplementary Figure 3d). The results showed that only HDAC2 siRNA but not HDAC1 or HDAC3 siRNA was able to reverse PELP1 mediated repression of miR-200a and miR-141 in miR promoter reporter assays (Figure 4c, d). Further, treatment with either TSA or HDAC2 siRNA was able to reverse PELP1 mediated repression of miR-200a and miR-141 (Figure 4e, f). These results suggest that PELP1 down regulates the expression of miR-200a and miR-141 by binding to and recruiting HDAC2 to their promoters.
Figure 4.
PELP1-mediated epigenetic changes at miR-200 promoter involve HDAC2 and PELP1 interactions. A, ChIP assay was done using the DNA isolated from ZR and ZR-PELP1-shRNA cells with antibodies specific for HDAC2 or isotype rabbit IgG control. DNA recovered from the ChIP or input controls was subjected to real-time qPCR using primers that detect −156 to +83 and −236 to −107 promoter region of miR200b-200a-429 and miR200c-141 respectively. HDAC2 recruitment to promoter region of 200b-200a-429 and 200c-141 was determined. The promoter occupancy was calculated on the basis of the ratio of ChIP/input control. B, ZR and ZR-PELP1 cells were transfected with Luc-promoter reporters of miR200c-141 and miR200b-200a-429. After 48h the cells were treated with Trichostatin (TSA) for 24 h. Reporter activity was measured after 72 h. C, D, ZR and ZR-PELP1 cells were transfected with Luc-promoter reporters of miR200b-200a-429 (C) and miR200c-141 (D). After 24h the cells were transiently transfected with HDAC1, HDAC2, HDAC3 siRNAs. Reporter activity was measured after 72 h. E, F, Real-time qPCR analysis of miRs expression in ZR and ZR-PELP1 cells treated with Trichostatin (TSA) for 24 h (E) or HDAC2 siRNA for 72 h (F). The relative expression of each miR was quantified by measuring Ct values and normalizing against RNU19. *, P≤ 0.05; Student’s t test.
PELP1 knockdown significantly reduces breast tumor growth and metastasis
To test whether depletion of PELP1 expression reduce metastasis in vivo, we have used an orthotopic xenograft tumor model. MDA-MB-231-conshRNA and MDA-MB-231-PELP1shRNA cells were implanted into the mammary fat pad and tumor growth and metastases were monitored. Results from this experiment showed that PELP1 knock down significantly reduced tumor growth compared to control cells (66 %, P<0.001, Supplementary Figure S4a). Compared with control MDA-MB-231-shRNA cells injected mice, nude mice injected with PELP1-shRNA cells had a significant reduction in tumor metastatic signal (80%, P < 0.001, Supplementary Figure S4b, c). The presence of metastases in the lung was assessed by counting fluorescent foci using fluorescence microscopy. Results showed a significant reduction (75%, P < 0.001) in metastatic tumor nodule formation in the PELP1-shRNA treatment groups compared with the control group (Supplementary Figure S4d). IHC analyses showed decreased proliferation, increased apoptosis and decreased expression of EMT marker vimentin in MDA-MB-231-PELP1-shRNA tumors compared with control group (Supplementary Figure S4e, f, g). RTqPCR analysis showed decreased ZEB1, ZEB2 and increased expression of miR200a and miR141 in MDAMD-231-PELP1shRNA tumors compared to control group (Supplementary Figure S4h). These results suggest that PELP1 mediated signaling play a role in metastases of tumor cells from primary tumor site.
Therapeutic efficacy of PELP1-miR200a/141 axis on breast cancer cell colonization and outgrowth
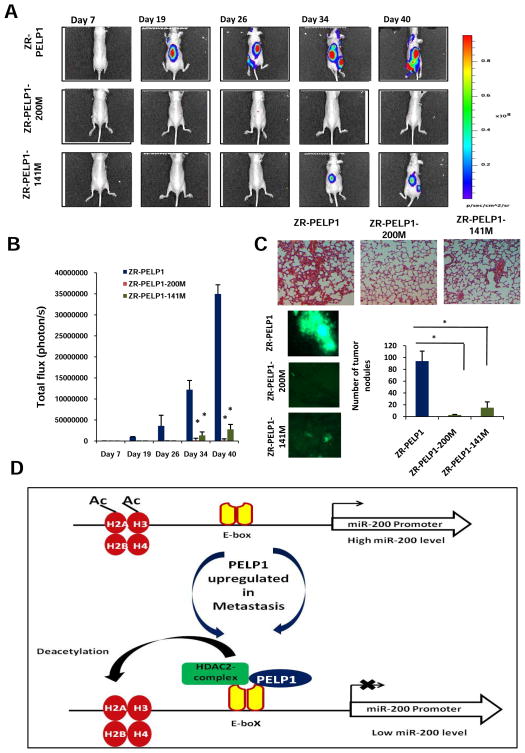
We next explored PELP1 regulated miRs (200 family members that target EMT) for their potential use as novel therapeutics to control PELP1-driven metastasis potential. To facilitate testing of their therapeutic potential in vivo, we generated ZR-PELP1 and MCF7-PELP1 cell lines that stably express miRIDIAN shMIMIC of miR-200a and miR-141 in PELP1 model cells (ZR-PELP1-200a-mimetic and ZR-PELP1-141-mimetic) via lentiviral transfection (Supplementary Figure S5a). Expression of mimetics of 200a and 141 reversed PELP1-mediated changes in EMT gene expression (Supplementary Figure S5b) and decreased PELP1-mediated migratory functions (Supplementary Figure S5c). To examine, whether ectopic expression of miR200a and 141 in PELP1-overexpressing cells reduces PELP1-driven metastasis potential in vivo, we performed studies using the ZR-PELP1-GFP-Luc, ZR-PELP1-miR200a-mimetic-GFP-Luc, and ZR-PELP1-miR141-mimetic-GFP-Luc cells in a preclinical model of experimental metastasis. For this assay, model cells (1 × 105 in 0.1 ml PBS/mouse) were injected into the left cardiac ventricle of nude mice (10 animals each group, total 30 mice). The Xenogen non-invasive optical imaging system was used for whole animal imaging. The mice injected with ZR-PELP1 cells had a greater propensity for the metastasis signal both in the dorsal and ventral views (Figure 5a, b and Supplementary Figure S6) than the mice injected with model cells that expressed mimetics of miR-200a and miR141. Mean body weights were not significantly different between the control and mimetics treatment groups (data not shown). To further examine the role of PELP1 in dissemination of tumor cells in vivo, the lungs were collected after euthanasia and the presence of metastases in the lung was assessed by counting fluorescent foci using fluorescence microscopy. Results showed significant reductions (97% and 84%; p < 0.0001) in tumor nodule formation in both treatment groups (Figure 5c). H&E staining of lungs also elucidated that more tumor burden was found in the ZR-PELP1 control mice than in the treatment groups (Figure 5c). Collectively, these proof-of-principle studies demonstrated the therapeutic efficacy of the mimetics of metastasis suppressors miR-200a and miR-141 on PELP1-driven in vivo breast cancer cell colonization and outgrowth.
Figure 5.
Significance of the PELP1-miR axis on the on breast cancer cell colonization and outgrowth. A, whole animal imaging analysis of mice (n = 10 per group) injected with ZR-PELP1, ZR-PELP1- shMIMIC of miR-200a or ZR-PELP1- shMIMIC of miR-141 stable cells. Dorsal views at indicated time points are shown for all three groups. B, luciferase signals were quantitated by using the software Living Image 3.2. Data are plotted as mean +/− s.e.m., n=10. C, Representative photomicrographs of metastases in the lung visualized by florescent foci and H&E staining (20X magnification). The number of tumor nodules in the lung were quantitated by counting fluorescent foci using fluorescence microscopy in the control and shMIMIC-treated mice groups. n=6 lungs per group; *, P < 0.05, t test. D, A putative model illustrating the interactions of PELP1/HDAC2 in regulating the EMT by modulating miR-200 transcription.
Discussion
Metastatic breast cancer is the leading cause of death in patients diagnosed with advanced-stage breast cancer. A critical need still exists to identify novel therapeutic targets to treat this metastasis. Metastasis is complex process, requiring coordinated activation of multiple genes and pathways (27). During the past decade, research has provided evidence to suggest that alterations in the levels of coregulator concentrations or the genetic dysfunction of NR-coregulators can contribute to a pathologic outcome by modulating genes and pathways that drive cancer cell proliferation and metastasis (28). Whether the NR-coregulator signaling is also involved in the activation of miRs that contribute to metastasis remains unknown. In this study, we found that (a) the NR-coregulator PELP1 modulates the expression of several metastasis suppressing miRs (b) PELP1 recruits promoters of the miR200 family members and promotes epigenetic changes, and (c) miR regulation plays a critical role in PELP1-mediated in vivo metastasis. Mechanistic studies suggested that under conditions of PELP1 deregulation, miR-200 is silenced by the PELP1-HDAC2 complex, which deacetylates histones. These findings were further validated in PELP1 knockdown experiments that showed the disruption of the interaction of the PELP1-HDAC2 complex, leading to re-expression of miR200a/141 by increasing acetylated histone marks at the miR-200 promoter (Figure 5d). Collectively, these novel findings demonstrate unknown new role for PELP1 in epigenetically controlling the functions of the tumor metastasis suppressors miR-200a and miR-141 and thus contributes to metastasis.
MiR-200 family comprises five members and clusters in two genomic loci (200b-200a-429 and 200c-141). The expression of miR-200 family is lost in the regions of metaplastic breast cancer specimens and metastases (19). The miR-200 family regulates EMT by targeting 3′UTR of ZEB1 and SIP1 (19). Some evidence suggests that ZEB1 suppresses the transcription of the miR-200 family members (22;29), indicating reciprocal repression between ZEB1 and members of the miR-200 family in EMT and invasion of cancer cells. Although the role of the miR-200 family members in EMT is well established, the mechanism by which oncogenes regulate miR-200 family members is elusive. Our data suggested that the proto-oncogene PELP1 has the potential to modulate expression of miR-200 family members. PELP1 interacts with several chromatin-modifying enzymes, including histone-modifying acetylases, deacetylases, methyltransferases, and demethylases (29, 30). Our mechanistic studies revealed that PELP1 is recruited to the promoters of miR-200 family members and modulates their expression by promoting repressing epigenetic marks. Further, these studies revealed that the PELP1 interactions with HDAC2 play a critical role in this process and HDAC inhibitor can reverse PELP1-mediated suppression of miR-200 family members. Collectively, these studies indicate PELP1 deregulation as having the potential to suppress expression of tumor suppressor miRs by facilitating repressive epigenetic changes.
Loss of the epithelial adhesion molecule E-cadherin is implicated as having a critical role in metastasis by disrupting intercellular contacts, an early step in metastatic dissemination (30). Functional or transcriptional loss is commonly associated with an invasive and poorly differentiated phenotype (31). Deregulation of NR-coregulator signaling can lead to aberrant expression of Snail, resulting in the loss of expression of E-cadherin and invasive growth. For example, MTA1, a commonly deregulated coregulator in breast cancer, promotes transcriptional repression of ER, leading to metastatic progression (32). The NR coregulator AIB1 amplified in breast cancer promotes breast cancer metastasis by activation of PEA3-mediated matrix metalloproteinase 2 (MMP2) and MMP9 expression (33). SRC-1, another NR coregulator, has also been shown to promote breast cancer invasiveness and metastasis by coactivating PEA3-mediated Twist expression (34). Recent studies have also found deregulation of the NR coregulator PELP1 in invasive and metastatic breast tumors (17), and its elevated expression is positively associated with markers of poor outcome (35). The results from our studies for the first time linked NR coregulator PELP1 with epigenetic silencing of breast cancer metastasis suppressing miRs.
PELP1 interacts with several proteins involved in migratory functions, including PI3K(21), four-and-a-half LIM protein 2 (36), and ILK1 (37), and metastasis associated antigen 1 (MTA1), which is a protein implicated in metastasis. Our recent studies demonstrated that PELP1 contributes to the metastatic potential of both ER+positive and ER-negative breast cancer cells. Such studies indicate that PELP1 regulation of metastasis may involve a mechanism that is independent of its ER-coactivaton functions. Our study results provided strong evidence that PELP1 directly recruits and suppress expression of several miRs that regulate metastasis including miR-200a and miR-141. Accordingly, miR-200a and miR-141 mimetics blocked PELP1-mediate migration and invasion in vitro and metastasis in vivo. PELP1 regulated expression of miR similarly in ER-positive and ER-negative cells, further confirming that this regulation is independent of the ER. Since PELP1 expression is commonly deregulated in metastatic tumors, PELP1 regulation of miRs may have implications in tumor progression to metastasis. In spite of significant progress in the understanding of the etiological role of PELP1 in breast cancer progression and its mechanism of action, no drugs are currently available to target PELP1. On the basis of our results, we speculate that drugs targeting PELP1-miR200 axis can be used to target PELP1-driven tumor progression to metastasis.
In summary, our study demonstrates for the first time that PELP1 modulates the expression of miRs and PELP1-regulated miRs play a critical role in breast cancer metastasis. Furthermore, we provided evidence that PELP1-mediated epigenetic changes have an important role in the modulation of miRs involved in metastasis. PELP1-modulated miR signatures may serve as useful biomarkers and novel targets for the therapeutic intervention of PELP1-driven tumors; however, further studies are needed to understand the prognostic significance of the PELP1-miR axis.
Materials and Methods
Cell cultures and reagents
ZR-75, MCF7, MDA-MB-231 and MDA-MB-468 cells were purchased from the American Type Culture Collection (Manassas, VA) and maintained in RPMI-1640 supplemented with 10% FBS. All model cells were passaged in the user’s laboratory for less than 6 months after receipt or resuscitation. PELP1, Snail, E-cadherin, Vimentin and ZEB1 antibodies were purchased from Bethyl Laboratories (Montgomery, TX) and Cell Signaling (Beverly, MA). Matrigel and n-cadherin antibodies were purchased from BD transduction (San Jose, CA). Occludin and HDAC 1,2,3 antibodies were purchased from Life technologies (Grand Island, NY) and Active Motif (Carlsbad, CA). TUNEL kit for apoptosis detection was purchased from Roche (Mannheim, Germany) and Ki-67 anti human Clone MiB-1 antibody was purchased from Dako (Carpinteria, CA). miRCURY LNA miR Inhibitor and miRIDIAN miR Mimics for hsa-miR-200a and hsa-miR-141 were purchased from Exiqon (Denmark) and Dharmacon (Lafayette, CO) respectively. Control, HDAC1, HDAC2, HDAC3, siRNA were purchased from Qiagen (Germantown, MD). The plasmids encoding the promoter reporter of 200c-141 containing −796 to +290 (38) and 200b-200a-429 clusters containing region −1574 to +120 (22) as well as the ZEB1 and ZEB2 3′UTR luciferase reporter (19) were described previously.
Generation of model cells
Non-metastastic, ER-positive (ZR75, MCF7) and metastatic, ER-negative (MDA-MB-231,MDA-MB-468) models that either stably overexpress PELP1 or PELP1-shRNA were developed as described (18). Transient knockdown of PELP1 was achieved using On-TargetPlusSMARTpool siRNA from Dharmacon. Breast cancer cells stably expressing PELP1 were transfected with human miRIDIAN shMIMIC lentiviral miR (Open Biosystems, Thermo-Fisher Scientifics; Huntsville, AL) for long-term expression of miR 200a and 141. Stable clones were selected with puromycin selection (1 μg/mL) and pooled clones were used for all studies. Human shMIMIC lentiviral miR Non -Silencing control was used to generate control cells.
Microarray studies
Total RNA that was isolated from the ZR control and ZR cells stably overexpressing PELP1cDNA or PELP1-shRNA were used for microarray analysis. MiR microarray profiling was done by Exiqon using the Human miRCURY™ LNA Array. Target miR whose expression was differentially regulated (at least 2-fold difference) by PELP1 expression was selected and validated by using real-time PCR analysis. Briefly, total RNA was extracted by using the miRNEasy Mini kit (Qiagen; Valencia, CA), 2 μg of total RNA was reverse transcribed by using the miScript RT kit (Qiagen) and real-time PCR was performed with a forward primer specific to miR by using the miScript SYBR Green PCR kit (Qiagen). Triplicate reactions were run for each cDNA sample. The relative expression of each miR was quantified by measuring Ct values and normalized by using RNU19. All real-time PCR primers that were used for validation of PELP1-regulated genes were purchased from RealTimePrimers (Elkins Park, PA).
Cell migration, invasion and luciferase reporter gene assays
The cell migration and invasion assays were carried out by using the calorimetric cell migration assay kit (Chemicon) and the BD Biocat growth factor reduced Matrigel invasion chamber kit (BD Biosciences), respectively, as described in the manufacturer protocol (21). The pRL-ZEB1 and ZEB2 3′UTR luciferase reporter and the promoter reporter for two clusters in the two different genomic loci miR200b-200a-429 and miR200c-141 were used for the reporter gene assays by using FuGENE6 (Roche, Indianapolis, IN) transient transfection method (18). Cells were lysed in passive-lysis buffer 36–48 h after transfection, and the luciferase assays were performed using a luciferase assay kit (Promega, Madison, WI). Each transfection was carried out in 6-well plates in triplicate and normalized with either β-gal activity or the total protein concentration.
Chromatin immunoprecipitation assays
ChIP analysis was performed as described previously (39). Briefly, cells were cross-linked using 1% formaldehyde, and the chromatin was subjected to immunoprecipitation using the indicated antibodies. Isotype-specific IgG was used as a control. DNA was re-suspended in 50 μl of TE buffer and used for real-time PCR amplification using the gene specific primers. 200b-200a-429 primer sequences: 156 forward: ctgcgtcaccgtcactgg; +83 reverse: aacactcgcccgtctctg; −1088 forward: accagtttccagcgagaaga; −920 reverse: cggaagagcccataatgaaa. 200c-141 primer sequences: −236 forward: aggggtgagactaggcaggt; −107 reverse: ccactgccttaaccccttc.
Nude mice studies
All animal experiments were performed after obtaining UTHSCSA IACUC approval. The animals were housed in accordance with UTHSCSA institution’s protocol for animal experiments. For orthotopic xenograft tumor model studies, model cells (2 × 106) were injected into mammary fatpad of 6-week-old female nude mice (n = 5 per group, 10 tumors) as described (26). Athymic nude mice (nu/nu) were injected with control MDAMB231-shRNA and MDAMB-231-PELP1 shRNA cells by mixing them with equal volume of MatrigelTM Matrix (BD Biosciences) to both left and right side of mammary fat pad. For experimental metastasis model, cells (1 × 105) in serum-free medium were injected into left cardiac ventricle of 5-week-old female athymic nude mice (n = 10 per group) as described (18). ZR-PELP1 cells and ZR-PELP1 model cells that stably express miR 200a and 141 were transfected with green fluorescent protein (GFP)-Luc plasmid to monitor metastasis with whole animal imaging (40–42). The mice were monitored daily for adverse effects and the body weight was recorded every week. The Xenogen Small-Animal Imaging System was used for subcellular imaging in live mice once a week. On day 40, mice were euthanized and the number of micrometastastic tumor nodules in the lungs were counted with an inverted microscope. Lung and tumor tissues were fixed in 10% neutral buffered formalin (Fisher Scientific, Pittsburgh, PA) for 48 h at room temperature and embedded in paraffin.
Immunofluorescence and immunohistochemical (IHC) analysis
Immunofluorescence was performed as described previously (37). The F-actin status was analyzed by phalloidin staining. IHC analysis was performed as described (26). Sections were stained with hematoxylin, and eosin using UTHSCSA Pathology core protocol. Antibody dilutions are vimentin (1:50), and Ki-67 (1:150). Proliferative index was calculated as percentage of Ki-67-positive cells in 10 randomly selected microscopic fields at 20X per slide. TUNEL analysis was done using the in situ Cell Death Detection Kit (Roche, Indianapolis, IN) as per the manufacturer’s protocol and 10 randomly selected microscopic fields in each group were used to calculate the relative ratio of TUNEL-positive cells.
Statistical analysis
Statistical differences among groups were analyzed with either a student’s t test or ANOVA as appropriate using SPSS software. P values less than 0.05 were considered significant. Error bars on the graphs show s.e.m.
Supplementary Material
Acknowledgments
This study was supported by the NIH/NCI grant CA095681 (R.K.V.) T32 training grant (SSR)), and the Cancer Therapy and Research Center at the University of Texas Health Science Center at San Antonio through the NCI Cancer Center Support Grant P30CA054174-17
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
Reference List
- 1.Eckhardt BL, Francis PA, Parker BS, Anderson RL. Strategies for the discovery and development of therapies for metastatic breast cancer. Nat Rev Drug Discov. 2012 Jun;11(6):479–97. doi: 10.1038/nrd2372. [DOI] [PubMed] [Google Scholar]
- 2.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006 Aug;12(8):895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 3.Stetler-Stevenson WG, Liotta LA, Kleiner DE., Jr Extracellular matrix 6: role of matrix metalloproteinases in tumor invasion and metastasis. FASEB J. 1993 Dec;7(15):1434–41. doi: 10.1096/fasebj.7.15.8262328. [DOI] [PubMed] [Google Scholar]
- 4.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005 Aug 15;65(16):7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 5.Khoshnaw SM, Green AR, Powe DG, Ellis IO. MicroRNA involvement in the pathogenesis and management of breast cancer. J Clin Pathol. 2009 May;62(5):422–8. doi: 10.1136/jcp.2008.060681. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, Wang J. MicroRNA-mediated breast cancer metastasis: from primary site to distant organs. Oncogene. 2012 May 17;31(20):2499–511. doi: 10.1038/onc.2011.444. [DOI] [PubMed] [Google Scholar]
- 7.Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009 Oct 1;69(19):7495–8. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi M, Liu D, Duan H, Shen B, Guo N. Metastasis-related miRNAs, active players in breast cancer invasion, and metastasis. Cancer Metastasis Rev. 2010 Dec;29(4):785–99. doi: 10.1007/s10555-010-9265-9. [DOI] [PubMed] [Google Scholar]
- 9.Saha RS, Vadlamudi RK. Role of estrogen receptor signaling in breast cancer metastasis. Int J Breast Cancer. 2012;2012:654698. doi: 10.1155/2012/654698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Malley BW. Molecular biology. Little molecules with big goals. Science. 2006 Sep 22;313(5794):1749–50. doi: 10.1126/science.1132509. [DOI] [PubMed] [Google Scholar]
- 11.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006 Jun 1;20(11):1405–28. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 12.Schiff R, Massarweh SA, Shou J, Bharwani L, Arpino G, Rimawi M, et al. Advanced concepts in estrogen receptor biology and breast cancer endocrine resistance: implicated role of growth factor signaling and estrogen receptor coregulators. Cancer Chemother Pharmacol. 2005 Nov;56(Suppl 1):10–20. 10–20. doi: 10.1007/s00280-005-0108-2. [DOI] [PubMed] [Google Scholar]
- 13.Mann M, Krishnan S, Vadlamudi RK. Emerging significance of estrogen cancer coregulator signaling in breast cancer. Minerva Ginecol. 2012 Feb;64(1):75–88. [PubMed] [Google Scholar]
- 14.O’Malley BW. Coregulators: from whence came these “master genes”. Mol Endocrinol. 2007 May;21(5):1009–13. doi: 10.1210/me.2007-0012. [DOI] [PubMed] [Google Scholar]
- 15.Vadlamudi RK, Kumar R. Functional and biological properties of the nuclear receptor coregulator PELP1/MNAR. Nucl Recept Signal. 2007 May 17;5:e004. doi: 10.1621/nrs.05004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakravarty D, Tekmal RR, Vadlamudi RK. PELP1: A novel therapeutic target for hormonal cancers. IUBMB Life. 2010 Mar;62(3):162–9. doi: 10.1002/iub.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajhans R, Nair S, Holden AH, Kumar R, Tekmal RR, Vadlamudi RK. Oncogenic potential of the nuclear receptor coregulator proline-, glutamic acid-, leucine-rich protein 1/modulator of the nongenomic actions of the estrogen receptor. Cancer Res. 2007 Jun 1;67(11):5505–12. doi: 10.1158/0008-5472.CAN-06-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy S, Chakravarty D, Cortez V, De MK, Bandyopadhyay A, Ahn JM, et al. Significance of PELP1 in ER-negative breast cancer metastasis. Mol Cancer Res. 2012 Jan;10(1):25–33. doi: 10.1158/1541-7786.MCR-11-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008 May;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 20.Nair BC, Nair SS, Chakravarty D, Challa R, Manavathi B, Yew PR, et al. Cyclin-dependent kinase-mediated phosphorylation plays a critical role in the oncogenic functions of PELP1. Cancer Res. 2010 Sep 15;70(18):7166–75. doi: 10.1158/0008-5472.CAN-10-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakravarty D, Roy SS, Babu CR, Dandamudi R, Curiel TJ, Vivas-Mejia P, et al. Therapeutic targeting of PELP1 prevents ovarian cancer growth and metastasis. Clin Cancer Res. 2011 Apr 15;17(8):2250–9. doi: 10.1158/1078-0432.CCR-10-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008 Oct 1;68(19):7846–54. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Ahn YH, Gibbons DL, Zang Y, Lin W, Thilaganathan N, et al. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J Clin Invest. 2011 Apr;121(4):1373–85. doi: 10.1172/JCI42579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiklund ED, Bramsen JB, Hulf T, Dyrskjot L, Ramanathan R, Hansen TB, et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int J Cancer. 2011 Mar 15;128(6):1327–34. doi: 10.1002/ijc.25461. [DOI] [PubMed] [Google Scholar]
- 25.Choi YB, Ko JK, Shin J. The transcriptional corepressor, PELP1, recruits HDAC2 and masks histones using two separate domains. J Biol Chem. 2004 Dec 3;279(49):50930–41. doi: 10.1074/jbc.M406831200. [DOI] [PubMed] [Google Scholar]
- 26.Cortez V, Mann M, Tekmal S, Suzuki T, Miyata N, Rodriguez-Aguayo C, et al. Targeting the PELP1-KDM1 axis as a potential therapeutic strategy for breast cancer. Breast Cancer Res. 2012 Jul 19;14(4):R108. doi: 10.1186/bcr3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009 Jun;119(6):1438–49. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Malley BW, Kumar R. Nuclear receptor coregulators in cancer biology. Cancer Res. 2009 Nov 1;69(21):8217–22. doi: 10.1158/0008-5472.CAN-09-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008 Jun;9(6):582–9. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008 May 15;68(10):3645–54. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 31.Beavon IR. The E-cadherin-catenin complex in tumour metastasis: structure, function and regulation. Eur J Cancer. 2000 Aug;36(13 Spec):1607–20. doi: 10.1016/s0959-8049(00)00158-1. [DOI] [PubMed] [Google Scholar]
- 32.Mazumdar A, Wang RA, Mishra SK, Adam L, Bagheri-Yarmand R, Mandal M, et al. Transcriptional repression of oestrogen receptor by metastasis-associated protein 1 corepressor. Nat Cell Biol. 2001 Jan;3(1):30–7. doi: 10.1038/35050532. [DOI] [PubMed] [Google Scholar]
- 33.Qin L, Liao L, Redmond A, Young L, Yuan Y, Chen H, et al. The AIB1 oncogene promotes breast cancer metastasis by activation of PEA3-mediated matrix metalloproteinase 2 (MMP2) and MMP9 expression. Mol Cell Biol. 2008 Oct;28(19):5937–50. doi: 10.1128/MCB.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin L, Liu Z, Chen H, Xu J. The steroid receptor coactivator-1 regulates twist expression and promotes breast cancer metastasis. Cancer Res. 2009 May 1;69(9):3819–27. doi: 10.1158/0008-5472.CAN-08-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Habashy HO, Powe DG, Rakha EA, Ball G, Macmillan RD, Green AR, et al. The prognostic significance of PELP1 expression in invasive breast cancer with emphasis on the ER-positive luminal-like subtype. Breast Cancer Res Treat. 2009 Jun 3;120:603–12. doi: 10.1007/s10549-009-0419-9. [DOI] [PubMed] [Google Scholar]
- 36.Nair SS, Guo Z, Mueller JM, Koochekpour S, Qiu Y, Tekmal RR, et al. Proline-, glutamic acid-, and leucine-rich protein-1/modulator of nongenomic activity of estrogen receptor enhances androgen receptor functions through LIM-only coactivator, four-and-a-half LIM-only protein 2. Mol Endocrinol. 2007 Mar;21(3):613–24. doi: 10.1210/me.2006-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chakravarty D, Nair SS, Santhamma B, Nair BC, Wang L, Bandyopadhyay A, et al. Extranuclear Functions of ER Impact Invasive Migration and Metastasis by Breast Cancer Cells. Cancer Res. 2010 May 11;70(10):4092–101. doi: 10.1158/0008-5472.CAN-09-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol. 2011 Mar;13(3):317–23. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nair SS, Nair BC, Cortez V, Chakravarty D, Metzger E, Schule R, et al. PELP1 is a reader of histone H3 methylation that facilitates oestrogen receptor-alpha target gene activation by regulating lysine demethylase 1 specificity. EMBO Rep. 2010 Jun;11(6):438–44. doi: 10.1038/embor.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bandyopadhyay A, Wang L, Agyin J, Tang Y, Lin S, Yeh IT, et al. Doxorubicin in combination with a small TGFbeta inhibitor: a potential novel therapy for metastatic breast cancer in mouse models. PLoS One. 2010;5(4):e10365. doi: 10.1371/journal.pone.0010365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldshaid L, Rubinstein E, Brandis A, Segal D, Leshem N, Brenner O, et al. Novel design principles enable specific targeting of imaging and therapeutic agents to necrotic domains in breast tumors. Breast Cancer Res. 2010;12(3):R29. doi: 10.1186/bcr2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jenkins DE, Hornig YS, Oei Y, Dusich J, Purchio T. Bioluminescent human breast cancer cell lines that permit rapid and sensitive in vivo detection of mammary tumors and multiple metastases in immune deficient mice. Breast Cancer Res. 2005;7(4):R444–R454. doi: 10.1186/bcr1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.