Abstract
The bacterial Type 6 Secretion System (T6SS) is an organelle that is structurally and mechanistically analogous to an intracellular membrane-attached contractile phage tail. Recent studies determined that a rapid conformational change in the structure of a sheath protein complex propels T6SS spike and tube components along with anti-bacterial and anti-eukaryotic effectors out of predatory T6SS+ cells and into prey cells. The contracted organelle is then recycled in an ATP-dependent process. T6SS is regulated at transcriptional and post-translational levels, the latter involving detection of membrane perturbation in some species. In addition to directly targeting eukaryotic cells, the T6SS can also target other bacteria co-infecting a mammalian host, highlighting the importance of the T6SS not only for bacterial survival in environmental ecosystems but also in the context of infection and disease. This review highlights these and other advances in our understanding of the structure, mechanical function, assembly, and regulation of the T6SS.
Introduction
Several different types of protein secretion systems exist in Gram-negative bacteria that function to translocate proteins outside of their cells, into the extracellular milieu, and sometimes into adjacent prokaryotic or eukaryotic cells. The type 6 secretion system (T6SS) represents one of the most recently recognized examples of these organelles. It was defined functionally in 2006 in Vibrio cholerae through genetic identification of several of its critical components and canonical substrates (Pukatzki et al., 2006). However, genes now known to be integrally associated with the T6SS had been identified as playing roles in virulence nearly a decade ago for Salmonella (Folkesson et al., 2002), Rhizobium (Bladergroen et al., 2003), Fracisella (Nano et al., 2004) and Edwardsiella (Rao et al., 2004), while several bioinformatics studies had identified their high conservation and broad distribution in nearly 25% of all Gram-negative bacteria (Das and Chaudhuri, 2003; Pallen et al., 2002; Schlieker et al., 2005). An explosion of interest in T6SS has led to its rapid study in Pseudomonas (Mougous et al., 2006), Escherichia (Dudley et al., 2006), Burkholderia (Schell et al., 2007), Agrobacterium (Wu et al., 2008), Aeromonas (Suarez et al., 2008), Helicobacter (Bartonickova et al., 2012)and Campylobacter (Lertpiriyapong et al., 2012) as well as other organisms. Although these initial studies were understandably focused on the role of T6SS in virulence (Ma et al., 2009a) or host immunomodulation (Chow and Mazmanian, 2010), more recently, T6SSs have been implicated in inter-bacterial interactions ranging from bactericidal activity (Hood et al., 2010; MacIntyre et al., 2010) and competitive growth in mixed-culture biofilms (Schwarz et al., 2010) to self versus non-self discrimination (Alteri et al., 2013; Wenren et al., 2013). Like the type 4 secretion system (T4SS) of Gram-negative bacteria, T6SS can translocate proteins into both prokaryotic and eukaryotic cells, underlining the versatility of the T6SS nanomachine. This review focuses on advances in understanding the structure, mechanical function, assembly, and regulation of this remarkable secretion organelle.
T6SS components, structure, and energetics
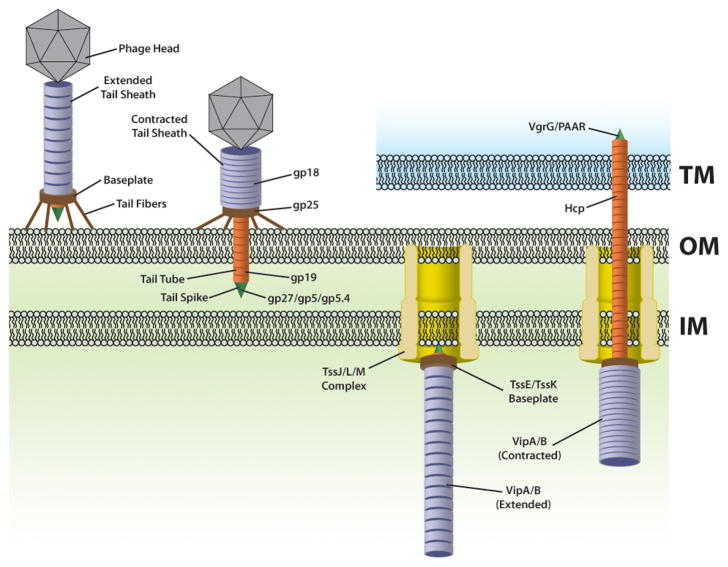
Among the first identified canonical substrates of the T6SS were those belonging to protein superfamilies commonly called Hcp (Haemolysin co-regulated protein) and VgrG (Valine-glycine repeat G) (Pukatzki et al., 2006). These proteins are unusual in that they are both secreted and required for T6SS apparatus functionality (Mougous et al., 2006; Pukatzki et al., 2006). Structure prediction algorithms indicated that VgrG proteins show significant structural homology to a complex called (gp27)3-(gp5)3, which corresponds to the tail spike or “needle” of the T4 phage. Like many other bacteriophages, the T4 phage tail structurally consists of sheath that is joined to tail fibers via a baseplate (Figure 1). When the tail fibers make contact with target bacteria cells, contraction of the tail sheath delivers a tube and spike that are thought to penetrate target bacterial cell membranes, facilitating the delivery of phage genetic material (Leiman and Shneider, 2012). Similar to the T4 tail spike, early evidence suggested that different VgrG proteins could form complexes (Pukatzki et al., 2007) and eventual evidence for homotrimeric complexes was obtained through crystallographic (Leiman et al., 2009) and biochemical analyses (Hachani et al., 2011). Crystallization of the Hcp1 T6SS protein of P. aeruginosa, and its atomic structure suggested that this protein could form tubes composed of stacked rings of Hcp hexamers (Mougous et al., 2006). Additional evidence for Hcp tube formation was obtained through cross-linking studies (Ballister et al., 2008) and further structural analysis of the Hcp protein fold showed its similarity to a duplicated ‘β-tube’ domain within the N-terminal gp27-like domain of VgrG (Leiman et al., 2009). These analyses predicted that if Hcp formed a tube in the context of the functional apparatus, then that tube would likely dock with the ‘pseudo hexameric’β-tube domain of a VgrG trimer (Leiman et al., 2009). Consistent with this idea, the NMR structure of the lambda phage tube protein p5 revealed it to be a structural homolog of Hcp (Pell et al., 2009), and indeed bioinformatic analysis of genes encoding Hcp orthologs suggests that they are as closely related to predicted phage tube proteins as other T6SS-encoded Hcp orthologs (Leiman et al., 2009). Such phage tube proteins include gp19 of T4 phage, which lacks a crystal structure but forms tubes similar in appearance to Hcp tubes (Leiman et al., 2009).
Figure 1. Contractile phage tails and the contractile T6SS organelle.
T6SS and contractile phage share a number of core structural components. Homologous components are depicted in the same color. While phage attach to the outer membrane (OM) via tail fibers connected to its baseplate, T6SS baseplate attaches to an inner membrane (IM) complex that spans the periplasm and associates with the outer membrane. Contraction of phage sheath delivers the phage spike into a target cell, while contraction of T6SS sheath forces the T6SS spike out of the cell and potentially across a target membrane (TM). Proteins conserved in T4 phage and the T6SS are labeled.
A third component of the T6SS apparatus, now known as TssE (Type six subunit E), showed significant homology to the T4 phage base plate component gp25 (Leiman et al., 2009), which is known to reside near the interface between the phage spike and the tube complex (Kanamaru et al., 2002). These similarities between T6SS components and phage pointed to a model predicting that the T6SS represented at least in part a phage tail-like structure in an orientation that is opposite that of phage infection (Leiman et al., 2009) (Figure 1) and supported earlier suggestions (Mougous et al., 2006) that Hcp in its tubular conformation might act as a conduit for protein transport by the T6SS machine once extruded from the cell.
Support for the phage tail model for the T6SS organelle came through a series of observations that revolved around three additional essential T6SS components. In a seminal study, Mogk and colleagues determined that in V. cholerae, the Clp family AAA+ ATPase ClpV recognized two other T6SS components, VipA and VipB (named for ClpV-interacting protein), which formed tubular structures in both Escherichia coli and V. cholerae (Bonemann et al., 2009). When viewed down the long axis under electron microscopy, VipA/B tubules formed 12-tooth cogwheel-like shapes that were completely disintegrated by a process dependent on ClpV-mediated ATP hydrolysis (Bonemann et al., 2009). It was first noted by Leiman et al. (2009) that the VipA/B tubule structures described by Bonemann et al. (2009) were highly similar to ‘contracted’ T4 phage tail sheaths, further suggesting that a VipA/B sheath contraction mechanism might provide the energy for T6SS protein transport. With this information, several models appeared envisioning how the apparatus might be organized and function (Bonemann et al., 2010; Filloux, 2009; Records, 2011). However, further insights into the functional mechanism of protein translocation by the T6SS organelle would require cell biological analysis and visualization of the dynamic action of intact organelles in living cells as well as super-high resolution visualization of flash-frozen cells.
Basler et al. (2012) directly visualized the T6SS organelle dynamics in V. cholerae using a combination of time-lapse fluorescence light microscopy and electron cryotomography. Utilizing functional, fluorescent VipA-GFP fusion proteins, these investigators showed that a large VipA-containing sheath structure exists inside cells and undergoes cycles of extension, contraction, disassembly, and re-assembly. The T6SS sheath polymerizes from a membrane-bound complex in an extended conformation, and like phage, the extended sheath structure then undergoes a rapid contraction event, estimated to occur in less than 5 milliseconds (Basler et al., 2012) (Figure 1). Disassembly of the contracted sheath structure is driven by ClpV, which recognizes only the contracted form of the T6SS sheath in both V. cholerae and P. aeruginosa (Basler and Mekalanos, 2012; Basler et al., 2012). The contraction event is correlated with protein secretion and delivery of effectors and attack signals into target cells (Basler et al., 2013; Basler and Mekalanos, 2012; Basler et al., 2012; Ho et al., 2013). Both the extended and contracted sheath structures are observed attached to the membrane and can be differentiated from each other by their dimensions, surface topology and internal density, the last of which has been hypothesized to correspond minimally to the Hcp tube (Basler et al., 2012) but may also reflect the density of effectors that are occupying the Hcp tube luminal channel based on more recent insights (Silverman et al., 2013).
As with phage, the T6SS apparatus needs a way to attach the tube and sheath complex to a membrane in order to translocate molecules across it. In phage, this is achieved via a large baseplate complex that serves both as a platform for assembly of the tube and sheath, as well as an attachments site for tail fibers, which directly contact target cells (Kostyuchenko et al., 2003) (Figure 1). For T6SS, gp25-like TssE is the only homolog to phage baseplate components (Leiman et al., 2009; Lossi et al., 2011). Instead of a fully phage-like baseplate complex, the T6SS tube and sheath are anchored to the membrane via a complex related to the Legionella Dot/Icm T4SS that injects effector proteins into host cells (Figure 1). This complex consists of two inner membrane components, TssL (DotU-related) and TssM (IcmF-related) (Durand et al., 2012; Ma et al., 2009b; VanRheenen et al., 2004), and an outer-membrane lipoprotein (TssJ) (Felisberto-Rodrigues et al., 2011) (Figure 2). In some organisms this complex is thought to be anchored to the peptidoglycan through an extension domain of TssL or through an additional accessory protein (Aschtgen et al., 2010). Although it remains unclear how the T6SS tube and sheath attach to the baseplate, two-hybrid interaction studies indicate that the essential cytoplasmic protein TssK interacts with both the membrane-bound TssJ-TssL-TssM complex as well as the Hcp tube and VipA/B sheath and may serve as an adaptor between the two (Zoued et al., 2013). There are 3 other essential proteins that are virtually universally conserved, TssF, TssG, and TssA (Boyer et al., 2009; Zheng et al., 2011; Zheng and Leung, 2007), but it is unclear what roles they play in T6SS assembly, structure, and/or function.
Figure 2. Model for T6SS assembly, effector translocation, and component recycling.
(A) Baseplate complex forms consisting of TssE, TssJ, TssK, TssL and TssM. Other components not pictured include TssA, TssF, and TssG. In some T6SSs, Fha is an essential part of this complex. TssJ, TssK, TssL and TssM are placed in this drawing based on protein localization and interaction studies (Felisberto-Rodrigues et al., 2011; Zoued et al., 2013), while TssE position is inferred from phage homolog (Kostyuchenko et al., 2003) (B) VgrG, PAAR, and effector proteins are recruited to this complex and assemble into the structure. VgrG interaction with PAAR or effectors likely contributes to the overall stability of the apparatus assembly. Although these components are pictured here as being cytosolic, it is unclear whether there is an opening into the periplasm. (C) Hcp tube polymerizes from VgrG while the VipA/VipB sheath polymerizes around it. (D) Analogous to phage, a conformation change in the sheath structure results in a contraction event that launches the Hcp tube out of the cell and across a target membrane. This contraction event delivers the loaded VgrG-effector “warhead” through the layers of the cell envelope, however it is not known how often penetration into the cytosol occurs, if at all. It is also unknown how much Hcp is lost outside the cell and how much is retained within the cytosol. (E) ClpV uses ATP to remodel the contracted sheath, restoring the pool of available sheath subunits. The now unsheathed Hcp tube disassembles; parts of the tube that not expelled from the cell are recycled within the cytosol. (F) The naked baseplate complex is then ready to be recycled or disassembled, depending on the T6SS and its activation state.
Other genes are clearly accessory in regard to their contribution to T6SS function. For example, the protein Fha (Forkhead-associated) (Pallen et al., 2002) is absent in more than half of the bioinformatically identified T6SSs (Boyer et al., 2009) (Figure 5A), several of which have been confirmed to be functional and active without it, such as in A. baylyi (Basler et al., 2013) and E. tarda (Zheng and Leung, 2007). However, in at least some of the T6SS that do have Fha, such as P. aeruginosa (Mougous et al., 2007) and V. cholerae (Zheng et al., 2011), the Fha protein has been shown to be essential. In a subset of these organisms (Figure 5A), Fha has evolved to play a critical role in mediating post-translational regulation of T6SS activity (Basler et al., 2013; Fritsch et al., 2013; Ho et al., 2013; Mougous et al., 2007).
Figure 5.
T6SS counterattack sensing pathway. (A) The program Hmmsearch (Finn et al., 2011) was used to identify 761 VipA (pfam05591)/VipB (pfam05943)-containing gene loci identified in sequenced bacterial genomes (ftp://ftp.ncbi.nih.gov/genomes/bacteria). 128 of these loci contained nearby (within 40kb) Fha (pfam00498), PpkA (PF00069 or PF13519), and PppA (PF13672) genes. Subsets of these loci carrying different combinations of TagQ (pfam13488), TagR (pfam03781), TagS (pfam02687), or TagT (pfam00005) homologs were also identified. All loci with TagS also had TagT (*). (B and C) Various signals involving membrane perturbation including exogenous T6SS attack, T4SS mating pair formation and certain membrane disrupting antibiotics like polymyxin B can trigger Fha phosphorylation by PpkA. TagR directly activates PpkA, while TagQ positions TagR in the periplasm and associates it with the outer membrane. TagS and TagT comprise an inner membrane ABC transporter. It is possible that TagS and TagT directly sense the membrane perturbation signal (B) or are responsible for localization of the actual signal sensor (C). It should be noted that TagS and TagT are not required for delivery of TagQ and TagR to the periplasmic space (Casabona et al., 2012).
Based on early data showing subcellular co-localization of Fha and ClpV (Mougous et al., 2007), most models that followed hypothesized that Fha orthologs trigger formation of a membrane-associated complex that is critical to formation of a functional T6SS organelle. As such, one way to conceptualize the T6SS organelle is as a complex composed of two distinct assemblies–one analogous to a contractile phage tail and baseplate and the other a trans-membrane complex (Cascales and Cambillau, 2012; Silverman et al., 2012). However, other than using structural or sequence homology as a guide to differentiate the proteins belonging to these two complexes, there are no data supporting the concept that these two assemblies can form independently. For this reason, we prefer to refer to the entire membrane-associated complex excluding the spike, tube, sheath and ClpV components as the “baseplate complex.”
Clearly, energy is required for the function of the T6SS organelle as a protein translocation machine. In this regard, there has been significant controversy in the T6SS field as to what role ClpV plays in the process. ClpV was initially identified as a member of the Clp family of AAA+ ATPases found almost exclusively in pathogenic bacteria (Schlieker et al., 2005) before being implicated in T6SS function (Mougous et al., 2006) and recognized as being widely conserved in nearly all T6SSs (Boyer et al., 2009). Early localization studies of fluorescent ClpV-GFP fusions suggested that ClpV was recruited to specific complexes on the bacterial membrane containing Fha (Mougous et al., 2007). Members of the Clp family form hexameric ring structures that bind and unfold protein substrates by threading them through their central channel (Mogk et al., 2008). The elegant biochemical studies of Bonemann et al. (2009) confirmed these activities in the context of T6SS, but some models for T6SS function have perhaps overreached on these observations by suggesting that ClpV also drives effector recognition and translocation. For example, one model postulates that after contraction of the VipA/B sheath and subsequent launching of the Hcp tube into a target cell to form an intercellular bridge, ClpV is recruited to power the translocation of effector proteins by threading them down the Hcp tube (Silverman et al., 2012). However, we now know this model is likely incorrect. First, ClpV is not essential for T6SS-dependent bacterial killing, but rather it merely increases T6SS efficiency (Basler et al., 2012; Zheng et al., 2011). Second, ClpV specifically recognizes and remodels VipB (and by association, VipA) in contracted sheaths and not other secreted substrates (Bonemann et al., 2009; Pietrosiuk et al., 2011). Third, when observed intime -lapse, ClpV-GFP visibly coats the entire length of the contracted sheath (Basler and Mekalanos, 2012), an observation further confirmed through high-resolution thin-section electron microscopy (Kapitein et al., 2013). Binding along the length of a contracted sheath would be unnecessary if ClpV were merely translocating effectors down the channel of the Hcp tube. Rather than playing a direct role in substrate translocation, these observations suggest that the role of ClpV may actually be to recycle T6SS sheath components, allowing for efficient turnover of at least the contracted sheath if not the entire organelle (Basler and Mekalanos, 2012; Basler et al., 2012) (Figure 2). All of the energy required for protein secretion and translocation is contained intrinsically within the assembly of the extended sheath structure. All available evidence indicates ClpV simply allows the cell to re-access this energy potential by ATP-driven remodeling of VipA/VipB that are trapped in contracted, low energy sheaths (Basler et al., 2012; Bonemann et al., 2009; Pietrosiuk et al., 2011).
The presence of ClpV in the T6SS represents an interesting evolutionary adaptation from its phage relatives. Contractile phages utilize a “one-and-done” mechanism, where a single contraction event is sufficient to penetrate a host membrane and deliver components needed to initiate a successful infection. Unlike phage, bacterial cells benefit greatly from being able to reuse intracellular components of the T6SS. Indeed, fluorescence microscopy has revealed that sheath components can be limiting for V. cholerae T6SS. Complementation of a vipA mutant with low levels of VipA expression from an inducible plasmid reduced the number of T6SS structures per cell significantly (Basler et al., 2012). Furthermore, nonfunctional ClpV mutants or ClpV mutants unable to recognize VipB show accumulation of contracted sheath structures as well as reduced formation of new T6SS structures (Basler and Mekalanos, 2012). This observation also explains why residual T6SS activity is still observed in ClpV knockouts (Basler et al., 2012; Zheng et al., 2011). In such situations, T6SS structures still form, and assembled sheaths still contract, however after this process occurs once, the T6SS structure is locked in a contracted form and cannot be recycled. Indeed several organisms including Campylobacter jejuni lack genes in their T6SS cluster that encodes a ClpV ortholog yet encode perfectly functional organelles (Bleumink-Pluym et al., 2013). Interestingly, the anti-cellular function of the C. jejuni organelle is limited by the presence of its capsule suggesting that extracellular matrixes can preclude strikes by the T6SS organelle even when prey and predator cells are in relatively close contact (Bleumink-Pluym et al., 2013).
Another key difference between phage and T6SS is the inherent difference in penetration depth provided by the contraction event. T4 phage tails, for example, are only about 100 nm in length (Duda et al., 1985), while T6SS sheaths can be more than 10 times that length, extending across the entire width and sometimes length of a cell (Basler et al., 2012) (Figure 3A). This difference in assembly length results in a contraction-driven tube extrusion for phages, such as T4, that is only about 50 nanometers. In contrast, T6SS can attack an area up to half the width of the cell and therefore possibly penetrate target cells in close contact as deep as 500 nanometers (Figure 3B). Like phage, which must ultimately deliver genetic information into the cytosol of the target bacteria, some T6SS nuclease effectors have clear cytosolic targets (Koskiniemi et al., 2013). Although direct delivery into the cytosol by a single T6SS contraction event has never been shown, the calculated penetration depth of the T6SS spike/tube complex suggests that it could commonly occur. If so, one wonders how the tube/spike complex is able to cross the rigid peptidoglycan. Many phage (e.g., T4) carry lysozyme domains fused to their tail spike complexes, which help break down peptidoglycan and thus facilitate phage contact with the bacterial cell wall, (Arisaka et al., 2003). However, T6SS VgrG proteins lack equivalent domains (Pukatzki et al., 2007) and those that have lysozyme-like effector domains do not need them for target cell intoxication (Zheng et al., 2011). It is possible that phages need to digest their way through the peptidoglycan barrier, while the T6SS can use brute force to puncture this layer and gain access to the cytosol in one contraction event. Ultimately, high-resolution imaging and more precise biophysical measurements of the forces involved will be needed to fully resolve these possibilities.
Figure 3.
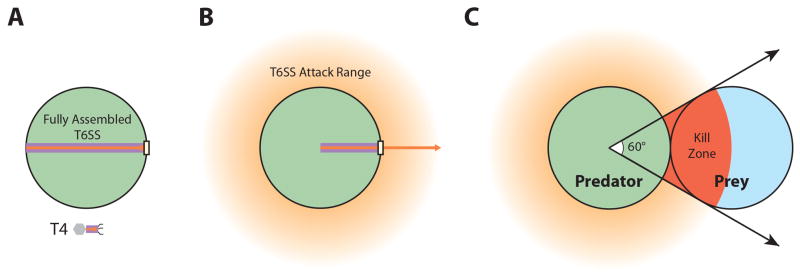
Spatial geometry of anti-bacterial T6SS attacks. (A) Fully assembled T6SS tube (orange arrow) and sheath (purple rectangle) can extends across the diameter of the cell. By comparison, T4 phage tails are typically only ~100 nm (~1/10 cell width). Phage tail length is drawn approximately to scale. (B) After contraction, sheath length is reduced by close to 50%. Assuming the inner Hcp completely fills the sheath, the full range of the T6SS (orange halo) is a zone approximately 500 nm wide (~1/2 cell width) surrounding the cell. (C) Assuming perfectly cylindrical cellular geometry and tight spatial packing, at most 1/6th of the T6SS+ attack range will contain a given prey cell. This number is even smaller the larger the distance between the T6SS+ predator and the prey cell, highlighting the need for proper aiming of T6SS attacks. If a prey cell cannot be sensed, the attacker must fire repeatedly in all directions, wasting a majority of the attack potential.
Effector identification, T6SS secretion recognition signals, and mechanism of translocation
Shortly after the discovery of T6SS, various effectors with enzymatic activity were identified, notably the VgrG-1 protein of V. cholerae (Pukatzki et al., 2007). This VgrG has become the prototypical example of “evolved VgrGs” that carry additional enzymatic domains fused to the C-terminus of their gp5-like β-helix domain (Leiman et al., 2009) (Figure 4). Other examples include V. cholerae VgrG-3, which carries a lysozyme-like domain (Brooks et al., 2013; Dong et al., 2013), A. hydrophila VgrG-1, which carries an ADP-ribosylating toxin domain (Suarez et al., 2010), and bioinformatically identified S-type pyocin nuclease VgrG in Salmonella (Blondel et al., 2009) and VgrG-lipase in Burkholderia (Pukatzki et al., 2009). These domain extensions can be further modified through interaction with additional effector proteins (Figure 4). For example, in V. cholerae, T6SS effector TseL (Type six effector Lipase) interacts with VgrG-3 and requires it for its secretion (Dong et al., 2013). The modular nature of these C-terminal extension domains and interacting proteins suggests that VgrG may be employed as a delivery vehicle for translocation of other heterologous functional domains. Indeed, in V. choleraea β-lactamase domain replacing the actin-crosslinking domain of VgrG-1 can be delivered into host cells by the T6SS (Ma et al., 2009a).
Figure 4. Adapted Multiple Effector Translocation VgrG (MERV) model for effector loading and delivery.
T6SS effectors are loaded onto the VgrG spike complex or within the distal end of the Hcp tube (left panel). Sheath contraction leads to the simultaneous delivery of the VgrG spike and all associated effectors (middle panel), abrogating the need for the Hcp tube to be stably maintained (right panel). Prototypical examples of effector classes 1–3 have been characterized: V. cholerae VgrG-1 and VgrG-3 (Class 1), V. cholerae TseL (Class 2), D. dadantii RhsA (Class 3). Effector classes 4 and 5 represent hypothetical mechanisms that are likely to exist. Class 4 effectors associate with PAAR protein extensions such as transthyretin-like domains (Shneider et al., 2013) or Rhs-repeat domains (Koskiniemi et al., 2013). Class 5 effectors bind to the lumenal side of the Hcp tube; although it has not yet been experimentally confirmed, it is likely that P. aeruginosa effector Tse2 falls into this category.
In both V. cholerae (Pukatzki et al., 2007) and P. aeruginosa (Hachani et al., 2011), there is evidence that VgrG forms a trimeric complex with other non-identical VgrG proteins (Hachani et al., 2011). Indeed, both hetero- and homo-trimeric VgrG complexes can be observed in P. aeruginosa (Hachani et al., 2011), while in V. cholerae, T6SS activity requires that at least two distinct VgrG proteins be present for T6SS activity to occur. V. cholerae VgrG-2 is required along with at least one of VgrG-1 and VgrG-3 (Zheng et al., 2011), suggesting that formation of hetero-trimeric complexes of VgrG-2 and VgrG-1 or VgrG-2 and VgrG-3 is required. Differences in spike complex assembly in different organisms may reflect differences in how the complex integrates itself within the T6SS apparatus along with other associated effector proteins. Indeed, in V. cholerae deletion of VgrG-3 along with secreted effector VasX abolishes T6SS activity including Hcp secretion (Dong et al., 2013), suggesting that the associations with VgrG complexes may play a direct role in T6SS assembly. These effector-VgrG interactions may partially explain why some organisms only have a single copy of VgrG (Rhizobium leguminosarum), while others have 3 (V. cholerae), 10 (P. aeruginosa), or even 32 (Sorangium cellulosum) distinct VgrG genes, as incorporation and delivery of different effectors may require a specifically adapted VgrG protein. Collectively, these observations suggest that heterotrimeric VgrG complexes have, in theory, the ability to deliver more than one effector domain in the context of a single spike trimer.
Mougous and colleagues identified cytotoxic non-VgrG effectors (Hood et al., 2010) that included enzymes that attacked glycan and peptide bonds in bacterial peptidoglycan (Russell et al., 2011). Collectively, VgrG and non-VgrG T6SS effectors exhibit a diverse range of functions targeting both eukaryotic hosts and bacterial competitors. Known effector activities include actin modification (Pukatzki et al., 2007; Suarez et al., 2010), muramidases and peptidases that attack the bacterial cell wall (Chou et al., 2012; Dong et al., 2013; English et al., 2012; Hood et al., 2010; Russell et al., 2011; Russell et al., 2012; Whitney et al., 2013), nucleases (Koskiniemi et al., 2013), lipases (Dong et al., 2013; Russell et al., 2013), and proteins facilitating eukaryotic membrane fusion (French et al., 2011). In order to prevent self or sister cell intoxication, T6SS+ organisms also encode immunity proteins to neutralize their cognate antibacterial effectors (Brooks et al., 2013; Dong et al., 2013; English et al., 2012; Hood et al., 2010).
Effectors have been identified using mass spectrometry-based approaches in a number of bacteria, including P. aeruginosa (Hood et al., 2010), Serratia marcescens (Fritsch et al., 2013), and V. cholerae (Miyata et al., 2011). Interestingly, P. aeruginosa mutants lacking all three effectors that were identified using this approach (Hood et al., 2010) were still able to kill V. cholerae and E. coli (Basler et al., 2013; Ho et al., 2013). Cell envelope puncturing can lead to cell death as in the case of the P. aeruginosa R-type pyocins that kill bacteria by depolarizing membranes in conjunction with pore formation (Michel-Briand and Baysse, 2002). However, Gram-negative bacteria have been reported to survive multiple cell envelope puncturing events where an open channel is not maintained (Suo et al., 2009), suggesting that in the case of P. aeruginosa (Basler et al., 2013; Ho et al., 2013), Tse1-, Tse2-, and Tse3-independent killing may either involve VgrG/Hcp channel formation or alternatively, other effectors that have escaped detection by the previously employed proteomic approaches due to either low expression levels or their intrinsic properties. Tn-seq (transposon mutagenesis coupled with massively parallel sequencing) has recently been employed as an alternative to proteomics to detect three pairs of effectors and their cognate immunity proteins in V. cholerae (Dong et al., 2013). This method takes advantage of the incidental sister cell-sister cell T6SS attacks, as cells carrying transposon disruptions of effector immunity genes drop out of mixed populations of T6SS+ but not T6SS− cells in a manner detectable by deep sequencing.
Bioinformatic analyses using the physical properties (i.e., molecular weight, isoelectric point and operon structure) of known P. aeruginosa effectors have predicted a large group of potential T6SS effectors (Russell et al., 2012). However, these analyses did not identify all of the effectors in V. cholerae and S. marcescens, indicating again the diversity of T6SS effectors. Notably, T6SS effector genes have been often found immediately downstream of orphan hcp-vgrG gene pairs not encoded within the main T6SS gene clusters in several organisms, including V. cholerae (Dong et al., 2013; Miyata et al., 2011; Zheng et al., 2011), Dickeya dadantii (Koskiniemi et al., 2013) and Proteus mirabilis (Alteri et al., 2013; Wenren et al., 2013). By extension, genes downstream of these hcp-vgrG gene pairs may encode T6SS effectors in other organisms as well. Indeed, many of these downstream genes belong to a diverse family of lipase effectors (Russell et al., 2013).
Recently, another gene product frequently found adjacent to T6SS genes has been implicated as both a structural component of the T6SS organelle and as a secreted substrate. Specifically, PAAR (Proline Alanine-Alanine-aRginine)-motif-containing proteins have been identified as metal-binding, cone shaped proteins that effectively sharpen the β-helical tip of the VgrG trimer (Shneider et al., 2013) (Figure 4). Like VgrG, PAAR proteins can also have additional extension domains with various predicted effector activities, suggesting that the hundreds of PAAR proteins identifiable in genome databases may be T6SS effectors as well (Shneider et al., 2013). Because the interaction between PAAR proteins and the C-terminal end of the VgrG trimer is driven by hydrogen bonding between the backbones of the respective proteins, it has been proposed that various PAAR proteins might be able to bind to any given VgrG trimer (Shneider et al., 2013). Indeed, an effector nuclease RhsA delivered to target cells by the T6SS D. dadantii (Koskiniemi et al., 2013) has been identified to be a PAAR protein (Shneider et al., 2013). This nuclease effector requires either of two VgrG genes to be delivered to sensitive target cells (Koskiniemi et al., 2013), suggesting that this PAAR protein is capable of recognizing different VgrG trimers. Furthermore, as with Hcp and VgrG, there are frequently multiple genes encoding PAAR proteins in bacterial species that encode T6SS (Shneider et al., 2013), suggesting that like VgrG effectors, genes encoding PAAR effector proteins have been subject to extensive horizontal transmission among organisms.
The T6SS is also capable of translocating proteins lacking identifiable signals for VgrG association such as PAAR motifs. Although it has been suggested that the Hcp tube may function as a passive conduit through which these effectors might be translocated (Mougous et al., 2006; Silverman et al., 2012), the opening in the Hcp tube is only 40Å wide (Mougous et al., 2006) and is too small for all but the smallest effectors to pass through in a folded state (Benz et al., 2012). Early on, the triple AAA+ ATPase ClpV was suggested to power unfolded proteins down an intercellular Hcp tube (Mougous et al., 2006; Silverman et al., 2012). In such models, proteins such as ClpV (Bonemann et al., 2009) or IcmF (Ma et al., 2012) that have ATP hydrolytic activity are envisioned as the source of energy to transport effectors down a conduit formed by the Hcp tube. However, such a mechanism would require unsheathed extracellular Hcp tubes to remain assembled during the transport process, and such structures have never been observed. Recent elegant biochemical and electron-microscopy evidence (Silverman et al., 2013) suggests that some effectors can bind to residues displayed on the inside of Hcp1 hexamers. Given that this interaction stabilizes some effectors such as Tse2, Hcp has been designated to have chaperone function (Silverman et al., 2013). Additionally, alteration of residues on the lumenal side of the Hcp tube prevents secretion of Tse2 and Tse3 and reduces secretion of Tse1. Given that Tse3 (44.4 kDa) is too large to fit into the Hcp tube in a folded conformation, Silverman et al. (2013) suggested that Tse3 may be interacting with lumenal Hcp residues while in an unfolded state. These Hcp-effector interactions are not without precedent as early biochemical analysis of the T6SS in E. tarda showed that secreted T6SS substrate EvpP also interacted with Hcp (Zheng and Leung, 2007). Thus, Hcp “effector chaperone” activity may be the first step in a process that recruits effectors into fully assembled Hcp tubes. These observations suggest that some effectors may be loaded into the Hcp tube during assembly of the T6SS structure and may be injected into target cells along with the tube. The effectors that reside in the Hcp tube in an unfolded conformation may also act analogously to phage tail “tape measure” proteins (Rodriguez-Rubio et al., 2012) and thus control the length of the extended (i.e., uncontracted) T6SS tail/tube complex.
Another group of potential T6SS effectors whose members are also too large to reside within the Hcp tube in a folded confirmation include members of the extended Rhs-repeat-containing protein family (Koskiniemi et al., 2013). Recently, structural analysis has determined that the Rhs-repeat domain of one bacterial toxin forms a shell-like structure that completely encloses and protects the toxin’s folded enzymatically active effector domain (Busby et al., 2013). Given that many PAAR proteins display Rhs-repeat domains as well as putative effector domains (Shneider et al., 2013), it seems likely that these caged effector molecules are completely folded structures that decorate the VgrG tip. Together these data suggest a refinement of a model termed the Multiple Effector Translocation VgrG (MERV) model (Shneider et al., 2013). According to this model (Figure 4), effectors can be secreted and delivered to target cells by the T6SS organelle using as many as five functionally distinct mechanisms. In accordance with this model, we propose that effectors can be delivered to target cells as a complex of cargo proteins that decorate the VgrG/PAAR spike or reside within the proximal part of the Hcp tube lumen (Figure 4). A key aspect of this model is that the entire cytotoxic load can be delivered in a single T6SS firing event, thereby abrogating the need for a stable intercellular channel and prolonged cell-cell association. Rather than providing more time for additional effectors to be transferred through a tube, longer cell-cell associations allow for repeated T6SS firing events to hit a given prey cell. It is also worth noting that certain large toxins also utilize a strategy that involves delivery of multiple effector domains in a presumably a single membrane translocation event (Satchell, 2011).
Regulation of T6SS expression and assembly
Because the assembly, contraction, disassembly and re-assembly cycle of the T6SS organelle is likely to be energetically costly to the bacterial cell, it is not surprising that both expression and assembly of this organelle is tightly regulated. In most cases, bacteria have some sort of transcriptional control over T6SS tied to quorum sensing (Kitaoka et al., 2011; Salomon et al., 2013; Zheng et al., 2010), biofilm formation (Aubert et al., 2008; Moscoso et al., 2011), iron-depletion (Brunet et al., 2011; Chakraborty et al., 2011), thermoregulation (Pieper et al., 2009; Salomon et al., 2013), salinity (Salomon et al., 2013) or other stress responses (Brooks et al., 2013; Gueguen et al., 2013). In V. cholerae, the major T6SS cluster encodes an essential regulator of T6SS, VasH, which acts as an activator of the transcription initiator σ54 (RpoN). Indeed, both RpoN and VasH are required for T6SS function (Pukatzki et al., 2006; Zheng et al., 2011), but interestingly RpoN and VasH only control the transcription of the two hcp operons and not the major cluster (Dong and Mekalanos, 2012). It is worth noting that VasH and RpoN were originally reported to directly bind to the main T6SS gene cluster in addition to the hcp2 promoter (Bernard et al., 2011). However, the authors of this study reached this conclusion using in vitro gel-shift assays and heterologous expression of transcriptional reporter fusions in E. coli rather than assaying these regulators under native conditions. That RpoN and VasH only control the hcp operons and not the main cluster suggests a two-tiered regulatory cascade. Environmental signals first need to trigger the transcription of the major cluster so that vasH is expressed, which subsequently activates the transcription of the hcp operons by RpoN. Because the hcpoperons carry many of the secreted T6SS components, namely Hcp, VgrG and the ir downstream effector proteins, while the main cluster contains mostly structural components, this two-tiered regulation may be important for maintaining different levels of expression for components that can be internally recycled versus those that are secreted and thus consumed by the functioning system. Despite this tight control, transcriptional regulation still has its limitations. Given the number of independent components of the T6SS apparatus and the multiplicity of subunits required, transcriptional regulation does not allow for rapid response to new stimuli. Furthermore, because of cellular geometry, T6SS firing events are inherently inefficient at hitting target cells. Even assuming perfectly cylindrical geometry and tight spatial packing of adjacent cells, a T6SS+ attacker would require on average six randomly positioned T6SS firing events in order to actually hit a given neighboring cell (Figure 3C). The consequence is that cells are forced into one of two states, a docile one with an inactive or ineffective T6SS or an aggressive one with an excessively active T6SS that indiscriminately attacks all neighbors.
P. aeruginosa addresses both T6SS efficiency and target selection through a post-translational regulatory system. The T6SS in P. aeruginosa requires the phosphorylation of Fha by threonine kinase PpkA (Mougous et al., 2007) (Figure 5B, C). PpkA activity requires outer membrane lipoprotein TagQ (Type six secretion-associated gene Q) and outer membrane-associated protein TagR (Silverman et al., 2011). TagR was originally identified as a protein bioinformatically predicted to interact with the periplasmic domain of PpkA (Hsu et al., 2009), while TagQ was shown to be required for proper localization of TagR (Casabona et al., 2012). Two additional proteins, TagS and TagT, comprise a membrane-bound complex related to bacterial ABC transporters and are both required for full activation of PpkA (Casabona et al., 2012). Deactivation of the T6SS is achieved through dephosphorylation of Fha by protein phosphatase PppA (Mougous et al., 2007). Deletion mutants of PppA exhibit elevated levels of T6SS activity when measured by either the Hcp secretion level (Mougous et al., 2007) or by ClpV dynamics (Basler et al., 2013).
It was initially hypothesized that surface growth triggered this post-translational regulation of the T6SS (Silverman et al., 2011), however time-lapse visualization of T6SS dynamics through fluorescence microscopy has revealed P. aeruginosa to have a more fascinating regulatory system (Basler and Mekalanos, 2012). When grown on a solid surface, a P. aeruginosa cell that spontaneously fires its T6SS apparatus will frequently strike a neighboring sister cell. This attacked sister cell then builds its own T6SS organelle and fires a retaliatory T6SS counterattack. The initial attackers then sense this T6SS counterattack and retaliate in turn. Because P. aeruginosa encodes immunity proteins to its own T6SS effectors, none of these T6SS attacks between sister cells are lethal, allowing for this reciprocal interaction to be stably maintained for several minutes. This phenomenon has been named ‘T6SS dueling’ (Basler and Mekalanos, 2012). Further genetic analysis of this dueling activity confirmed the involvement of the threonine phosphorylation pathway (Basler et al., 2013). As expected, mutations in PpkA, TagQ, or TagR caused complete loss of all T6SS activation. However, deletion of TagT maintained basal levels of T6SS activity but completely suppressed dueling activity. In contrast, although a PppA deletion mutant exhibited high levels of T6SS activity in virtually all cells, almost none of this activity was associated with T6SS dueling, suggesting that dephosphorylation of Fha is required to reposition the T6SS apparatus in response to newly sensed attacks. Because dueling activity can only occur if neighboring cells remain adjacent to each other, it is not surprising that this phenomenon is manifest only on solid surfaces and not in liquid culture. As such, it is highly likely that the observed correlation between surface growth and increased PpkA activity (Casabona et al., 2012) is due to accumulation of dueling sister cells within a given surface-growing population.
Although sister cell dueling might play some role in cell signaling involving non-T6SS targets of PpkA regulation (Goldova et al., 2011), responding to sister cell attacks is probably not the intended use of the T6SS response pathway. Indeed, P. aeruginosa T6SS is capable of responding to T6SS attack from heterologous organisms lacking cognate immunity proteins to P. aeruginosa T6SS effectors. When grown in competition with P. aeruginosa, T6SS+ V. cholerae and A. baylyi but not T6SS− mutants of these organisms were both killed in a TagT-dependent manner (Basler et al., 2013). A similar differentiation between T6SS+ and T6SS− Burkholderia thailandensis has been reported, although this observation was attributed by the authors to different degrees of susceptibility to P. aeruginosa T6SS (Leroux et al., 2012). Perhaps the most critical aspect of this T6SS response mechanism is its ability to precisely aim the retaliatory T6SS response directly at the source of the initial attack. When mixtures of both T6SS+ and T6SS− V. cholerae were mixed with P. aeruginosa, only the T6SS+V. cholerae cells were killed, while the T6SS− cells were largely untouched (Basler et al., 2013). The lack of collateral damage associated with activation of P. aeruginosa T6SS is perhaps indicative of the prevalence of P. aeruginosa in multispecies communities (Ha et al., 2012; Tashiro et al., 2013), where it might stand to gain from coexistence with non-hostile cohabitants.
An interesting and still unanswered question that arises from these results is how P. aeruginosa is able to avoid being killed by the T6SS effectors of these heterologous species. The “T6SS effector armor” might be attributable to its notorious outer-membrane impermeability (Nikaido, 1994), but it is hard to imagine how any membrane structure could repel T6SS attacks. Perhaps modifications of peptidoglycan structure or robust repair mechanisms underlie this observed resistance. Thus, immunity to heterologous effectors coupled with T6SS counterattacks may be fueling an arms race in antagonistic bacterial cell-cell interactions.
Based on bioinformatic analysis of the genes needed for dueling, this post-translation T6SS regulation is likely limited to a subset of Fha-containing T6SSs (Figure 5A). Specifically, this phenomenon may be restricted to Pseudomonas species, as this genus is the only one so far identified to contain all of the requisite regulatory components. Indeed, other organisms such as S. marcescens that still require phosphorylation of Fha by PpkA for T6SS activation but lack the TagQRST system do not require cell-cell contact to fully activate their T6SS (Fritsch et al., 2013). This observation suggests that the rapid response associated with post-translational control is not necessarily limited to responding to attacking bacterial neighbors as has been observed in P. aeruginosa. Rather, T6SS in each of these bacterial species might be adapted to specifically respond to the threats each species is likely to encounter in nature.
Recently, the T6SS of E. coli EAEC strain 17-2 has been observed to exhibit what appeared to be T6SS dueling behavior (Brunet et al., 2013). However, given that none of the TagQRST system genes required for T6SS dueling in P. aerugionsa are found in sequenced E. coli strains (Figure 5A), it is unlikely that these bacteria are exhibiting the same process observed in P. aeruginosa. Indeed, the dueling cells presented in this study do not appear to exhibit the precise geometric orientation observed in P. aeruginosa (Basler et al., 2013; Basler and Mekalanos, 2012) and thus may be more a consequence of T6SS organelles coincidentally being aimed at each other in adjacent cells. Another important clarification between the putative dueling observed in the studies by Brunet et al. (2013) described above and the dueling exhibited by P. aeruginosa is that since there is little to no collateral damage from these retaliatory T6SS attacks, T6SS activity does not tend to spread through a population of P. aeruginosa cells (Basler et al., 2013; Basler and Mekalanos, 2012). The T6SS activity observed in EAEC was reported to propagate within a population of growing cells (Brunet et al., 2013), suggesting that this organism may be responding to other signals associated with growth on a solid surface as a community (e.g., quorum sensing) or some diffusible signal released by cells undergoing T6SS attack. These observations stand as a testament to both the diversity and specialization of T6SS regulation in diverse bacterial species.
Although a precise definition of the signal recognized by P. aeruginosa that triggers T6SS assembly remains elusive, we now have some significant new clues. Recently, it was discovered that broad host range conjugation systems can also induce a P. aeruginosa T6SS in the form of a “donor-directed counterattack” (Ho et al., 2013). In this report, E. coli T4SS associated with the plasmid RP4 was found to render these bacteria 30-times more sensitive to killing by the P. aeruginosa T6SS. Because this RP4-dependent, T6SS-dependent killing involved precise target selection and required both TagT and PppA, the authors concluded that RP4 was triggering aimed assembly of the P. aeruginosa T6SS organelle in a manner perfectly analogous to the T6SS dueling response. Extensive genetic analysis of the RP4 conjugation system revealed that DNA transfer was not required for this response, but genes involved in sex pilus and mating pair formation were (Ho et al., 2013). Furthermore, the antibiotic polymyxin B, a cationic cyclic peptide that binds the outer membrane component lipid A to cause disruption of membrane integrity, also induced rapid TagT-dependent assembly and firing of the P. aeruginosa T6SS (Ho et al., 2013). Altogether, these observations suggested that the signal for T6SS assembly is a highly localized, physical or chemical signal associated with membrane perturbation. As such, the T6SS represents a generalized bacterial defense mechanism against foreign attack and acquisition of potentially infectious DNA elements. It remains unclear whether other membrane-penetrating processes such as phage infection, eukaryotic defensin or perforin might also induce similar T6SS counterattacks or if these hypothetical T6SS counterattacks can be used by bacteria to defend against phage or eukaryotic host cell responses.
As noted earlier, T6SS can also deliver effectors that are toxic to eukaryotic cells and indeed, evidence suggest that genes encoding this organelle are induced during infection and have been shown to induce host damage in experimental animals (Kapitein and Mogk, 2013; Ma and Mekalanos, 2010; Mandlik et al., 2011; Miyata et al., 2013; Zheng et al., 2010). The antibacterial activity associated with T6SS may also enhance colonization of the host by targeting heterologous or homologous bacterial cells. Recent evidence for the latter has been obtained in V. cholerae where a mutant defective in tsiV3, the cognate immunity protein to the bactericidal effector VgrG-3 (Dong et al., 2013), exhibited a defect in intestinal colonization (Yang et al., 2013). This in vivo colonization defect of the tsiV3 mutant depended on its co-colonization with T6SS+ strains carrying an intact VgrG3 gene. These data suggest that V. cholerae T6SS is functional during infection and that antagonistic sister cell-sister cell interactions occur during the infection process. Such a result predicts that heterologous species may also be subject to T6SS-dependent attacks and that these anti-microbiota interactions might thereby influence the virulence and in vivo fitness of T6SS+ species.
Conclusions
In summary, rapid progress has been made toward understanding the scope of biological activities associated with the bacterial T6SS, but many questions remain unanswered For example, what roles do TssF, TssG, and TssA play in structure, function, and assembly? What triggers sheath contraction? Is it regulated or spontaneous? How is tube/sheath length determined? Where are effectors initially delivered into target cells? Is the brute force generated by T6SS sheath contraction sufficient to penetrate the peptidoglycan? Can the T6SS tube/spike complex reach the cytosol and is its disassembly programmed to occur there? Do effectors with periplasmic targets (e.g., peptidoglycan) traffic from cytosol to periplasm after cytosolic delivery by the T6SS organelle? How are non-VgrG and non-PAAR effectors recognized by T6SS? How and where do effector complexes with VgrG and PAAR proteins form? How do effectors fit into the assembled baseplate? Do effectors that interact with Hcp rings fill the extended tube structure or reside only near the VgrG-Hcp interface? Can the genes for effectors be horizontally transferred among species and be quickly adapted to function with resident T6SS organelles in heterologous species? What mediates transmission of membrane perturbation signals to the TagQRST system? How are retaliatory T6SS attacks positioned in the cell spatially and temporally? Do host-derived signals (e.g., defensin, perforin, complement, etc.) also trigger post-translational assembly of the T6SS apparatus in bacteria harboring the TagQRST system? Do such signals result in efficient attack of eukaryotic target cells? What signals lead to induction of T6SS? Which host factors play a role in transcriptional activation? What role does anti-bacterial T6SS activity play within the microbiome of a eukaryotic host? Answering these questions will likely challenge T6SS researchers for years to come.
Besides acting in many species as virulence factors, T6SS is likely to be important for the ecological fitness of bacteria that live in consortia such as mixed biofilms (Miyata et al., 2013). Bacterial species that activate T6SS organelles and fire them in an unregulated manner (e.g., Vibrio and Acinetobacter species) are likely using them to kill all susceptible neighbors indiscriminately and thus may attack preformed biofilms efficiently. P. aeruginosa may have adapted to establishing beneficial complex biofilm communities (Kolenbrander et al., 2010; Peters et al., 2012), and therefore as a species P. aeruginosa would cooperate with other organisms so long as they do not pose a threat. On the other hand, if microbial neighbors threaten P. aeruginosa with either T6SS attack or with T4SS-mediated transfer of infectious conjugative elements, P. aeruginosa will use its T6SS as a primitive “innate immune system” (Ho et al., 2013) to neutralize these challenges. Understanding whether any of these ecological phenomenon lead to significant changes in the fitness of pathogens in vivo remains a priority for this rapidly moving field.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alteri CJ, Himpsl SD, Pickens SR, Lindner JR, Zora JS, Miller JE, Arno PD, Straight SW, Mobley HL. Multicellular Bacteria Deploy the Type VI Secretion System to Preemptively Strike Neighboring Cells. PLoS pathogens. 2013;9:e1003608. doi: 10.1371/journal.ppat.1003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisaka F, Kanamaru S, Leiman P, Rossmann MG. The tail lysozyme complex of bacteriophage T4. Int J Biochem Cell Biol. 2003;35:16–21. doi: 10.1016/s1357-2725(02)00098-5. [DOI] [PubMed] [Google Scholar]
- Aschtgen MS, Thomas MS, Cascales E. Anchoring the type VI secretion system to the peptidoglycan: TssL, TagL, TagP... what else? Virulence. 2010;1:535–540. doi: 10.4161/viru.1.6.13732. [DOI] [PubMed] [Google Scholar]
- Aubert DF, Flannagan RS, Valvano MA. A novel sensor kinase-response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia. Infection and immunity. 2008;76:1979–1991. doi: 10.1128/IAI.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballister ER, Lai AH, Zuckermann RN, Cheng Y, Mougous JD. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc Natl Acad Sci U S A. 2008;105:3733–3738. doi: 10.1073/pnas.0712247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartonickova L, Sterzenbach T, Nell S, Kops F, Schulze J, Venzke A, Brenneke B, Bader S, Gruber AD, Suerbaum S, et al. Hcp and VgrG1 are secreted components of the Helicobacter hepaticus type VI secretion system and VgrG1 increases the bacterial colitogenic potential. Cell Microbiol. 2013;15:992–1011. doi: 10.1111/cmi.12094. [DOI] [PubMed] [Google Scholar]
- Basler M, Ho BT, Mekalanos JJ. Tit-for-Tat: Type VI Secretion System Counterattack during Bacterial Cell-Cell Interactions. Cell. 2013;152:884–894. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Mekalanos JJ. Type 6 secretion dynamics within and between bacterial cells. Science. 2012;337:815. doi: 10.1126/science.1222901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz J, Sendlmeier C, Barends TR, Meinhart A. Structural Insights into the Effector - Immunity System Tse1/Tsi1 from Pseudomonas aeruginosa. PLoS One. 2012;7:e40453. doi: 10.1371/journal.pone.0040453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard CS, Brunet YR, Gavioli M, Lloubes R, Cascales E. Regulation of type VI secretion gene clusters by sigma54 and cognate enhancer binding proteins. J Bacteriol. 2011;193:2158–2167. doi: 10.1128/JB.00029-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bladergroen MR, Badelt K, Spaink HP. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol Plant Microbe Interact. 2003;16:53–64. doi: 10.1094/MPMI.2003.16.1.53. [DOI] [PubMed] [Google Scholar]
- Bleumink-Pluym NM, van Alphen LB, Bouwman LI, Wosten MM, van Putten JP. Identification of a Functional Type VI Secretion System in Campylobacter jejuni Conferring Capsule Polysaccharide Sensitive Cytotoxicity. PLoS pathogens. 2013;9:e1003393. doi: 10.1371/journal.ppat.1003393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel CJ, Jimenez JC, Contreras I, Santiviago CA. Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genomics. 2009;10:354. doi: 10.1186/1471-2164-10-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 2009;28:315–325. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonemann G, Pietrosiuk A, Mogk A. Tubules and donuts: a type VI secretion story. Mol Microbiol. 2010;76:815–821. doi: 10.1111/j.1365-2958.2010.07171.x. [DOI] [PubMed] [Google Scholar]
- Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics. 2009;10:104. doi: 10.1186/1471-2164-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks TM, Unterweger D, Bachmann V, Kostiuk B, Pukatzki S. Lytic Activity of the Vibrio cholerae Type VI Secretion Toxin VgrG-3 is Inhibited by the Antitoxin TsaB. J Biol Chem. 2013;288:7618–7625. doi: 10.1074/jbc.M112.436725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet YR, Bernard CS, Gavioli M, Lloubes R, Cascales E. An epigenetic switch involving overlapping fur and DNA methylation optimizes expression of a type VI secretion gene cluster. PLoS Genet. 2011;7:e1002205. doi: 10.1371/journal.pgen.1002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet YR, Espinosa L, Harchouni S, Mignot T, Cascales E. Imaging type VI secretion-mediated bacterial killing. Cell Reports. 2013;3:36–41. doi: 10.1016/j.celrep.2012.11.027. [DOI] [PubMed] [Google Scholar]
- Busby JN, Panjikar S, Landsberg MJ, Hurst MR, Lott JS. The BC component of ABC toxins is an RHS-repeat-containing protein encapsulation device. Nature. 2013;501:547–550. doi: 10.1038/nature12465. [DOI] [PubMed] [Google Scholar]
- Casabona MG, Silverman JM, Sall KM, Boyer F, Coute Y, Poirel J, Grunwald D, Mougous JD, Elsen S, Attree I. An ABC transporter and an outer membrane lipoprotein participate in posttranslational activation of type VI secretion in Pseudomonas aeruginosa. Environ Microbiol. 2012;15:471–486. doi: 10.1111/j.1462-2920.2012.02816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Cambillau C. Structural biology of type VI secretion systems. Philos Trans R Soc Lond B Biol Sci. 2012;367:1102–1111. doi: 10.1098/rstb.2011.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Sivaraman J, Leung KY, Mok YK. Two-component PhoB-PhoR regulatory system and ferric uptake regulator sense phosphate and iron to control virulence genes in type III and VI secretion systems of Edwardsiella tarda. J Biol Chem. 2011;286:39417–39430. doi: 10.1074/jbc.M111.295188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Bui NK, Russell AB, Lexa KW, Gardiner TE, Leroux M, Vollmer W, Mougous JD. Structure of a Peptidoglycan Amidase Effector Targeted to Gram-Negative Bacteria by the Type VI Secretion System. Cell Rep. 2012;1:656–664. doi: 10.1016/j.celrep.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J, Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe. 2010;7:265–276. doi: 10.1016/j.chom.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Chaudhuri K. Identification of a unique IAHP (IcmF associated homologous proteins) cluster in Vibrio cholerae and other proteobacteria through in silico analysis. In Silico Biol. 2003;3:287–300. [PubMed] [Google Scholar]
- Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc Natl Acad Sci U S A. 2013;110:2623–2628. doi: 10.1073/pnas.1222783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong TG, Mekalanos JJ. Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic acids research. 2012;40:7766–7775. doi: 10.1093/nar/gks567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda RL, Wall JS, Hainfeld JF, Sweet RM, Eiserling FA. Mass distribution of a probable tail-length-determining protein in bacteriophage T4. Proc Natl Acad Sci U S A. 1985;82:5550–5554. doi: 10.1073/pnas.82.16.5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol Microbiol. 2006;61:1267–1282. doi: 10.1111/j.1365-2958.2006.05281.x. [DOI] [PubMed] [Google Scholar]
- Durand E, Zoued A, Spinelli S, Watson PJ, Aschtgen MS, Journet L, Cambillau C, Cascales E. Structural characterization and oligomerization of the TssL protein, a component shared by bacterial type VI and type IVb secretion systems. J Biol Chem. 2012;287:14157–14168. doi: 10.1074/jbc.M111.338731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English G, Trunk K, Rao VA, Srikannathasan V, Hunter WN, Coulthurst SJ. New Secreted Toxins and Immunity Proteins Encoded within the Type VI Secretion System Gene Cluster of Serratia marcescens. Mol Microbiol. 2012;86:921–36. doi: 10.1111/mmi.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felisberto-Rodrigues C, Durand E, Aschtgen MS, Blangy S, Ortiz-Lombardia M, Douzi B, Cambillau C, Cascales E. Towards a structural comprehension of bacterial type VI secretion systems: characterization of the TssJ-TssM complex of an Escherichia coli pathovar. PLoS Pathog. 2011;7:e1002386. doi: 10.1371/journal.ppat.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filloux A. The type VI secretion system: a tubular story. The EMBO journal. 2009;28:309–310. doi: 10.1038/emboj.2008.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkesson A, Lofdahl S, Normark S. The Salmonella enterica subspecies I specific centisome 7 genomic island encodes novel protein families present in bacteria living in close contact with eukaryotic cells. Res Microbiol. 2002;153:537–545. doi: 10.1016/s0923-2508(02)01348-7. [DOI] [PubMed] [Google Scholar]
- French CT, Toesca IJ, Wu TH, Teslaa T, Beaty SM, Wong W, Liu M, Schroder I, Chiou PY, Teitell MA, et al. Dissection of the Burkholderia intracellular life cycle using a photothermal nanoblade. Proc Natl Acad Sci U S A. 2011;108:12095–12100. doi: 10.1073/pnas.1107183108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch MJ, Trunk K, Alcoforado Diniz J, Guo M, Trost M, Coulthurst SJ. Proteomic identification of novel secreted anti-bacterial toxins of the Serratia marcescens Type VI secretion system. Mol Cell Proteomics. 2013;12:2735–2749. doi: 10.1074/mcp.M113.030502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldova J, Ulrych A, Hercik K, Branny P. A eukaryotic-type signalling system of Pseudomonas aeruginosa contributes to oxidative stress resistance, intracellular survival and virulence. BMC Genomics. 2011;12:437. doi: 10.1186/1471-2164-12-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueguen E, Durand E, Zhang XY, d’Amalric Q, Journet L, Cascales E. Expression of a Type VI Secretion System Is Responsive to Envelope Stresses through the OmpR Transcriptional Activator. PLoS One. 2013;8:e66615. doi: 10.1371/journal.pone.0066615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha C, Park SJ, Im SJ, Lee JH. Interspecies signaling through QscR, a quorum receptor of Pseudomonas aeruginosa. Mol Cells. 2012;33:53–59. doi: 10.1007/s10059-012-2208-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachani A, Lossi NS, Hamilton A, Jones C, Bleves S, Albesa-Jove D, Filloux A. Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. J Biol Chem. 2011;286:12317–12327. doi: 10.1074/jbc.M110.193045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho BT, Basler M, Mekalanos JJ. Type 6 secretion system-mediated immunity to type 4 secretion system-mediated gene transfer. Science. 2013;342:250–253. doi: 10.1126/science.1243745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F, Schwarz S, Mougous JD. TagR promotes PpkA-catalysed type VI secretion activation in Pseudomonas aeruginosa. Mol Microbiol. 2009;72:1111–1125. doi: 10.1111/j.1365-2958.2009.06701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru S, Leiman PG, Kostyuchenko VA, Chipman PR, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415:553–557. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- Kapitein N, Bonemann G, Pietrosiuk A, Seyffer F, Hausser I, Locker JK, Mogk A. ClpV recycles VipA/VipB tubules and prevents non-productive tubule formation to ensure efficient type VI protein secretion. Mol Microbiol. 2013;87:1013–1022. doi: 10.1111/mmi.12147. [DOI] [PubMed] [Google Scholar]
- Kapitein N, Mogk A. Deadly syringes: type VI secretion system activities in pathogenicity and interbacterial competition. Curr Opin Microbiol. 2013;16:52–58. doi: 10.1016/j.mib.2012.11.009. [DOI] [PubMed] [Google Scholar]
- Kitaoka M, Miyata ST, Brooks TM, Unterweger D, Pukatzki S. VasH is a transcriptional regulator of the type VI secretion system functional in endemic and pandemic Vibrio cholerae. J Bacteriol. 2011;193:6471–6482. doi: 10.1128/JB.05414-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Palmer RJ, Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8:471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- Koskiniemi S, Lamoureux JG, Nikolakakis KC, T’Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A. 2013;110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyuchenko VA, Leiman PG, Chipman PR, Kanamaru S, van Raaij MJ, Arisaka F, Mesyanzhinov VV, Rossmann MG. Three-dimensional structure of bacteriophage T4 baseplate. Nat Struct Biol. 2003;10:688–693. doi: 10.1038/nsb970. [DOI] [PubMed] [Google Scholar]
- Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A. 2009;106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman PG, Shneider MM. Contractile tail machines of bacteriophages. Adv Exp Med Biol. 2012;726:93–114. doi: 10.1007/978-1-4614-0980-9_5. [DOI] [PubMed] [Google Scholar]
- Leroux M, De Leon JA, Kuwada NJ, Russell AB, Pinto-Santini D, Hood RD, Agnello DM, Robertson SM, Wiggins PA, Mougous JD. Quantitative single-cell characterization of bacterial interactions reveals type VI secretion is a double-edged sword. Proc Natl Acad Sci U S A. 2012;109:19804–19809. doi: 10.1073/pnas.1213963109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lertpiriyapong K, Gamazon ER, Feng Y, Park DS, Pang J, Botka G, Graffam ME, Ge Z, Fox JG. Campylobacter jejuni type VI secretion system: roles in adaptation to deoxycholic acid, host cell adherence, invasion, and in vivo colonization. PLoS One. 2012;7:e42842. doi: 10.1371/journal.pone.0042842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossi NS, Dajani R, Freemont P, Filloux A. Structure-function analysis of HsiF, a gp25-like component of the type VI secretion system in Pseudomonas aeruginosa. Microbiology. 2011;157:3292–3305. doi: 10.1099/mic.0.051987-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma AT, McAuley S, Pukatzki S, Mekalanos JJ. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe. 2009a;5:234–243. doi: 10.1016/j.chom.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma AT, Mekalanos JJ. In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc Natl Acad Sci U S A. 2010;107:4365–4370. doi: 10.1073/pnas.0915156107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma LS, Lin JS, Lai EM. An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its walker a motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J Bact. 2009b;191:4316–4329. doi: 10.1128/JB.00029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma LS, Narberhaus F, Lai EM. IcmF family protein TssM exhibits ATPase activity and energizes type VI secretion. J Biol Chem. 2012;287:15610–15621. doi: 10.1074/jbc.M111.301630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci U S A. 2010;107:19520–19524. doi: 10.1073/pnas.1012931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandlik A, Livny J, Robins WP, Ritchie JM, Mekalanos JJ, Waldor MK. RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe. 2011;10:165–174. doi: 10.1016/j.chom.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel-Briand Y, Baysse C. The pyocins of Pseudomonas aeruginosa. Biochimie. 2002;84:499–510. doi: 10.1016/s0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- Miyata ST, Bachmann V, Pukatzki S. Type VI secretion system regulation as a consequence of evolutionary pressure. J Medical Microbiol. 2013;79:2941–2949. doi: 10.1099/jmm.0.053983-0. [DOI] [PubMed] [Google Scholar]
- Miyata ST, Kitaoka M, Brooks TM, McAuley SB, Pukatzki S. Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect Immun. 2011;79:2941–2949. doi: 10.1128/IAI.01266-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Haslberger T, Tessarz P, Bukau B. Common and specific mechanisms of AAA+ proteins involved in protein quality control. Biochem Soc Trans. 2008;36:120–125. doi: 10.1042/BST0360120. [DOI] [PubMed] [Google Scholar]
- Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environmental microbiology. 2011;13:3128–3138. doi: 10.1111/j.1462-2920.2011.02595.x. [DOI] [PubMed] [Google Scholar]
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol. 2007;9:797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- Nano FE, Zhang N, Cowley SC, Klose KE, Cheung KK, Roberts MJ, Ludu JS, Letendre GW, Meierovics AI, Stephens G, et al. A Francisella tularensis pathogenicity island required for intramacrophage growth. J Bacteriol. 2004;186:6430–6436. doi: 10.1128/JB.186.19.6430-6436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- Pallen M, Chaudhuri R, Khan A. Bacterial FHA domains: neglected players in the phospho-threonine signalling game? Trends in Microbiol. 2002;10:556–563. doi: 10.1016/s0966-842x(02)02476-9. [DOI] [PubMed] [Google Scholar]
- Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc Natl Acad Sci U S A. 2009;106:4160–4165. doi: 10.1073/pnas.0900044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW, Shirtliff ME. Polymicrobial interactions: impact on pathogenesis and human disease. Clin Microbiol Rev. 2012;25:193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper R, Huang ST, Robinson JM, Clark DJ, Alami H, Parmar PP, Perry RD, Fleischmann RD, Peterson SN. Temperature and growth phase influence the outer-membrane proteome and the expression of a type VI secretion system in Yersinia pestis. Microbiology. 2009;155:498–512. doi: 10.1099/mic.0.022160-0. [DOI] [PubMed] [Google Scholar]
- Pietrosiuk A, Lenherr ED, Falk S, Bonemann G, Kopp J, Zentgraf H, Sinning I, Mogk A. Molecular Basis for the Unique Role of the AAA+ Chaperone ClpV in Type VI Protein Secretion. J Biol Chem. 2011;286:30010–30021. doi: 10.1074/jbc.M111.253377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A. 2007;104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, McAuley SB, Miyata ST. The type VI secretion system: translocation of effectors and effector-domains. Curr Opin Microbiol. 2009;12:11–17. doi: 10.1016/j.mib.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Rao PS, Yamada Y, Tan YP, Leung KY. Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol Microbiol. 2004;53:573–586. doi: 10.1111/j.1365-2958.2004.04123.x. [DOI] [PubMed] [Google Scholar]
- Records AR. The type VI secretion system: a multipurpose delivery system with a phage-like machinery. Mol Plant Microbe Interact. 2011;24:751–757. doi: 10.1094/MPMI-11-10-0262. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Rubio L, Gutierrez D, Martinez B, Rodriguez A, Gotz F, Garcia P. The tape measure protein of the Staphylococcus aureus bacteriophage vB_SauS-phiIPLA35 has an active muramidase domain. Appl Environ Microbiol. 2012;78:6369–6371. doi: 10.1128/AEM.01236-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Leroux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, Mougous JD. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature. 2013;11:538–549. doi: 10.1038/nature12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Singh P, Brittnacher M, Bui NK, Hood RD, Carl MA, Agnello DM, Schwarz S, Goodlett DR, Vollmer W, et al. A Widespread Bacterial Type VI Secretion Effector Superfamily Identified Using a Heuristic Approach. Cell Host Microbe. 2012;11:538–549. doi: 10.1016/j.chom.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon D, Gonzalez H, Updegraff BL, Orth K. Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS One. 2013;8:e61086. doi: 10.1371/journal.pone.0061086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchell KJ. Structure and function of MARTX toxins and other large repetitive RTX proteins. Annu Rev Microbiol. 2011;65:71–90. doi: 10.1146/annurev-micro-090110-102943. [DOI] [PubMed] [Google Scholar]
- Schell MA, Ulrich RL, Ribot WJ, Brueggemann EE, Hines HB, Chen D, Lipscomb L, Kim HS, Mrazek J, Nierman WC, et al. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol Microbiol. 2007;64:1466–1485. doi: 10.1111/j.1365-2958.2007.05734.x. [DOI] [PubMed] [Google Scholar]
- Schlieker C, Zentgraf H, Dersch P, Mogk A. ClpV, a unique Hsp100/Clp member of pathogenic proteobacteria. J Biol Chem. 2005;386:1115–1127. doi: 10.1515/BC.2005.128. [DOI] [PubMed] [Google Scholar]
- Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD. Burkholderia Type VI Secretion Systems Have Distinct Roles in Eukaryotic and Bacterial Cell Interactions. PLoS Pathog. 2010;6:e1001068. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JM, Agnello DM, Zheng H, Andrews BT, Li M, Catalano CE, Gonen T, Mougous JD. Haemolysin Coregulated Protein Is an Exported Receptor and Chaperone of Type VI Secretion Substrates. Mol Cell. 2013;51:584–593. doi: 10.1016/j.molcel.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JM, Austin LS, Hsu F, Hicks KG, Hood RD, Mougous JD. Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol Microbiol. 2011;82:1277–1290. doi: 10.1111/j.1365-2958.2011.07889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JM, Brunet YR, Cascales E, Mougous JD. Structure and Regulation of the Type VI Secretion System. Annu Rev Microbiol. 2012;66:453–472. doi: 10.1146/annurev-micro-121809-151619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez G, Sierra JC, Erova TE, Sha J, Horneman AJ, Chopra AK. A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J Bacteriol. 2010;192:155–168. doi: 10.1128/JB.01260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez G, Sierra JC, Sha J, Wang S, Erova TE, Fadl AA, Foltz SM, Horneman AJ, Chopra AK. Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb Pathog. 2008;44:344–361. doi: 10.1016/j.micpath.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo Z, Avci R, Deliorman M, Yang X, Pascual DW. Bacteria survive multiple puncturings of their cell walls. Langmuir. 2009;25:4588–4594. doi: 10.1021/la8033319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro Y, Yawata Y, Toyofuku M, Uchiyama H, Nomura N. Interspecies Interaction between Pseudomonas aeruginosa and Other Microorganisms. Microbes Environ. 2013;28:13–24. doi: 10.1264/jsme2.ME12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRheenen SM, Dumenil G, Isberg RR. IcmF and DotU are required for optimal effector translocation and trafficking of the Legionella pneumophila vacuole. Infect Immun. 2004;72:5972–5982. doi: 10.1128/IAI.72.10.5972-5982.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenren LM, Sullivan NL, Cardarelli L, Septer AN, Gibbs KA. Two Independent Pathways for Self-Recognition in Proteus mirabilis Are Linked by Type VI-Dependent Export. MBio. 2013;4 doi: 10.1128/mBio.00374-13. pii: e00374-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney JC, Chou S, Russell AB, Biboy J, Gardiner TE, Ferrin MA, Brittnacher M, Vollmer W, Mougous JD. Identification, structure and function of a novel type VI secretion peptidoglycan glycoside hydrolase effector-immunity pair. J Biol Chem. 2013;288:26616–26624. doi: 10.1074/jbc.M113.488320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HY, Chung PC, Shih HW, Wen SR, Lai EM. Secretome analysis uncovers an Hcp-family protein secreted via a type VI secretion system in Agrobacterium tumefaciens. Journal of bacteriology. 2008;190:2841–2850. doi: 10.1128/JB.01775-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Waldor MK, Mekalanos JJ. Activation of T6SS-mediated antibacterial activity in the host revealed through Tn-seq analysis of Vibrio cholerae intestinal colonization. Cell Host Microbe. 2013 doi: 10.1016/j.chom.2013.11.001. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Ho B, Mekalanos JJ. Genetic analysis of anti-amoebae and antibacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One. 2011;6:e23876. doi: 10.1371/journal.pone.0023876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Leung KY. Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol. 2007;66:1192–1206. doi: 10.1111/j.1365-2958.2007.05993.x. [DOI] [PubMed] [Google Scholar]
- Zheng J, Shin OS, Cameron DE, Mekalanos JJ. Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc Natl Acad Sci U S A. 2010;107:21128–21133. doi: 10.1073/pnas.1014998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoued A, Durand E, Bebeacua C, Brunet YR, Douzi B, Cambillau C, Cascales E, Journet L. TssK is a trimeric cytoplasmic protein interacting with components of both phage-like and membrane anchoring complexes of the Type VI secretion system. J Biol Chem. 2013;288:27031–27041. doi: 10.1074/jbc.M113.499772. [DOI] [PMC free article] [PubMed] [Google Scholar]