Abstract
The IL-23 pathway is genetically linked to autoimmune disease in humans and is required for pathogenic Th17 cell functions in mice. However, as IL-23R-expressing mature Th17 cells are rare and poorly defined in mice at steady-state, little is known about IL-23 signaling. Here we show that the endogenous CCR6+ memory T cell compartment present in peripheral lymphoid organs of unmanipulated mice expresses Il23r ex vivo, displays marked pro-inflammatory responses to IL-23 stimulation in vitro, and is capable of transferring experimental autoimmune encephalomyelitis (EAE). The prolyl-tRNA synthetase inhibitor, halofuginone (HF), blocks IL-23-induced Stat3 phosphorylation and IL-23-dependent pro-inflammatory cytokine expression in endogenous CCR6+ Th17 cells via activation of the amino acid starvation response (AAR) pathway. In vivo, HF shows therapeutic efficacy in EAE, reducing both established disease progression and local Th17 cell effector function within the CNS. Mechanistically, AAR activation impairs Stat3 responses downstream of multiple cytokine receptors via selective, post-transcriptional suppression of Stat3 protein levels. Thus, our study reveals latent pathogenic functions of endogenous Th17 cells that are regulated by both IL-23 and AAR pathways, and identifies a novel regulatory pathway targeting Stat3 that may underlie selective immune regulation by the AAR.
Keywords: Halofuginone, amino acid starvation, AAR, T cell, Memory T cells, CCR6, Th17, Stat3, IL-17A, IL-17F, IL-22, IL-23, inflammation, autoimmunity, EAE
Introduction
T helper 17 (Th17) cells accumulate at mucosal surfaces where they regulate inflammation and immunity via the production of IL-17A, IL-17F and IL-22 (reviewed in (1)). They are also widely involved in the pathogenesis of many common autoimmune disorders, including rheumatoid arthritis, multiple sclerosis, psoriasis and inflammatory bowel disease. Thus, understanding points of Th17 control is important for the rational design of new and more selective autoimmune therapies.
Th17 cells differentiate from naïve precursors following T cell antigen receptor (TCR) stimulation in the presence of IL-6, together with TGFβ or IL-1β (2–4). These signals converge to induce expression of the retinoic acid-related orphan nuclear receptor, Rorc (RORγt), in a Stat3-dependent manner (5,6). RORγt and Stat3, in turn, synergize to activate Il17a gene expression and locus remodeling (7). Accordingly, T cells lacking Rorc or Stat3 display impaired Th17 differentiation in vitro, and these cells fail to induce Th17-associated pathologies when adoptively transferred in vivo (3,5). Numerous other transcription factors and signaling pathways have been implicated in the control of Th17 differentiation in recent years (reviewed in (8,9)).
Much less is known about the regulation of pre-established memory Th17 cells and their effector functions, though this is undoubtedly an important aspect of clinical autoimmunity. Local cytokine production by mature Th17 cells within autoimmune lesions is both a biomarker for and a proximal cause of tissue damage (1,10). IL-23 is a Stat3-activating cytokine that acts on memory Th17 cells to enforce expression of inflammatory cytokines – including IL-17A, IL-17F and IL-22 – and it is necessary for the pathogenic functions of murine Th17 cells in vivo (4,11,12). Moreover, genome-wide association studies (GWAS) have linked polymorphisms in the IL23R gene to several human autoimmune disorders, including Crohn’s disease, psoriasis, psoriatic arthritis, ankylosing spondylitis, spondyloarthritis, and Behcet’s disease (13–16).
Despite its established roles in autoimmune pathogenesis, the pathways that regulate IL-23 signaling in Th17 cells are unknown. In large part, this is because IL-23R is only expressed on pro-inflammatory subsets of mature Th17 cells, which are thought to be extremely rare in mice at steady-state (17). The few endogenous Th17 cells that are present in unmanipulated mice reside in portions of the gut and develop in response to colonization by specific microbiota (3,18,19), though it is unclear if these or other endogenous Th17 cell populations express IL-23R and have pathogenic potential (18,20). Because of these logistical issues, Th17 cell responses to IL-23 have only been studied in T cell populations generated in vitro or through analysis of Il23r−/− mice, where direct molecular analysis of IL-23-mediated signal transduction is not possible (12,21,22).
Clues from the human immune system provide insights into the potential identity of IL-23-responsive Th17 cells. Human peripheral blood contains a larger reservoir of endogenous Th17 cells than the peripheral lymphoid organs of wild-type mice. Human Th17 cells are not only characterized by expression of IL-17A, but also by expression of the Th17-associated chemokine receptor CCR6 (23–25). More recent evidence suggests that human CCR6+ memory T cells can express Th17-lineage markers, including Rorc and Il23r, independent of IL-17A expression (25). Thus, endogenous IL-23-responsive Th17 cells in mice may be underestimated by analyses of IL-17A expression alone.
Here we show that steady-state expression of CCR6 in unmanipulated healthy mice discriminates an endogenous Th17 cell compartment that includes IL-23-responsive memory Th17 cells. These cells, like their human counterparts, express Rorc and Il23r ex vivo, and upregulate IL-17A, IL-17F and IL-22 expression upon in vitro IL-23 stimulation. As predicted by their responsiveness to IL-23, we show that endogenous mouse CCR6+ memory Th17 cells have latent pathogenic functions and induce experimental autoimmune encephalomyelitis (EAE) following transfer into lymphopenic recipients. In leveraging this platform to investigate IL-23 signaling in Th17 cells, we describe a novel link between the IL-23 and amino acid starvation response (AAR) pathways.
The AAR pathway is an evolutionarily conserved, cytoprotective stress response that is activated by un-aminoacylated (uncharged) tRNA molecules (reviewed in (26)). AAR activation is controlled by binding of uncharged tRNAs to the protein kinase Gcn2, which in turn phosphorylates eIF2α to promote translation of the functional open reading frame of the stress-activated transcription factor Atf4. Whereas eIF2α phosphorylation limits amino acid demand by reducing protein synthesis, Atf4 increases amino acid supply by inducing transcription of gene products involved in amino acid transport and biogenesis (27,28). The AAR pathway is activated physiologically when amino acid concentrations are limiting, but it can also be induced pharmacologically via treatment of cells with tRNA synthetase inhibitors, such as the plant natural product derivative and prolyl-tRNA synthetase inhibitor, halofuginone (HF) (29,30).
We have previously shown that HF blocks Th17 differentiation in vitro and protects mice from developing IL-17A-associated autoimmune pathology in vivo (31). We have further demonstrated that HF activates the AAR by directly binding to and inhibiting the enzymatic function of the mammalian prolyl-tRNA synthetase, EPRS (29,31). Here we show that HF-induced AAR activation: (1) selectively blocks IL-23-mediated Stat3 signaling and downstream induction of inflammatory cytokines in endogenous CCR6+ Th17 cells; (2) acts therapeutically in vivo, limiting both established EAE disease severity and local IL-17A production by Th17 cells in the CNS; and (3) broadly impairs Stat3-dependent T cell responses downstream of multiple cytokine receptors by suppressing Stat3 expression at a post-transcriptional level. Collectively, these findings provide important insights into IL-23R/Stat3 signaling in Th17 cells and establish a novel platform for assessing IL-23-dependent Th17 cell function in vitro and in vivo.
Materials and Methods
Mice
Mice were housed in specific pathogen-free barrier facilities at Tempero Pharmaceuticals or Harvard Medical School and were used in accordance with protocols approved by respective animal care and use committees. C57BL/6J (B6), BALB/C, 2D2 TCR transgenic, and Rag1−/− mice were purchased from The Jackson Laboratory (Bar Harbor, ME). IL17A-IRES-eGFP transgenic reporter mice were purchased from Biocytogen, Inc. (Worcester, MA), and were bred to 2D2 TCR transgenic mice at The Jackson Laboratory. IL-23R-GFP reporter mice were licensed from Prof. Vijay Kuchroo (Brigham and Women’s Hospital) and bred at the Jackson laboratory.
EAE
Mice were immunized by subcutaneous injection of 200μg MOG33–55 peptide emulsified in IFA (DIFCO) supplemented with 5mg/ml heat killed M. tuburculosis strain H37Ra (DIFCO) in dorsal flanks. Mice were injected i.p. with 300ng B. pertussis toxin on day 0 and day 1. HF (0.3 mg/kg) or vehicle (DMSO) diluted in PBS were injected daily i.p. (6 μg/mouse; ~ 0.3 mg/kg) beginning at the onset of clinical symptoms. Disease was scored daily as follows: 0, asymptomatic; 1, limp tail; 2, hind-limb weakness; 3, partial hind-limb paralysis; 4, complete paralysis of one or more limbs; and 5, moribund state. Cell analyses were performed after 1 week of treatment by isolating lymph nodes (axillary, inguinal, brachial and cervical), spleen and CNS. For EAE transfer experiments, 105 day 5-expanded naïve, CCR6− memory, or CCR6+ memory T cells were injected i.p. into Rag1−/− mice.
Cell isolation
T cells were magnetically isolated from pooled lymph node and spleen cells using CD4+ negative isolation kits from either EasySep or Miltenyi according to the manufacturer’s instructions. Naïve (CD25−CD62LhiCD44lo) or memory (CD25−CD62LloCD44hi) T cells were further sorted for CCR6 expression by FACS sorting (FACS Aria II; BD). Cells were ≥ 98% pure following isolation as determined by FACS analyses. CNS-infiltrating mononuclear cells were isolated from brain and spinal cord after intracardiac perfusion with PBS. Tissue was minced and digested with 0.33mg/ml Liberase TL and 20U/ml DNase (Roche) for 30 min at 37 °C. Digested tissue was forced through 70μm nylon filters (BD). Mononuclear cells were isolated from the interface following 30%/70% Percoll (Sigma) gradient centrifugation.
Cell culture
Cells were cultured in DMEM or IMDM medium (Gibco) supplemented as described (31) at 37°C/10% CO2. For ex vivo activation of FACS-sorted naïve and memory T cell subsets, 2–3 × 104 T cells were added to round-bottom tissue culture treated plates (Corning) together with anti-CD3/anti-CD28-coated beads (Invitrogen; 3 beads:1 cell), and recombinant human IL-2 (BD Bioscience; 10 Units/mL). Recombinant human IL-23 (R&D Systems; 20 ng/mL) was added as indicated. For intracellular cytokine staining, cultured cells were washed once in complete medium and resuspended in medium containing PMA (10 nM), Ionomycin (1 μM) and Brefeldin A (10 μg/ml) for 3–4 hr.
Inhibitors and amino acids
Halofuginone (HF) was obtained from the chemical repository at GlaxoSmithKline, was reconstituted in DMSO, stored at −20°C, and diluted in culture medium for in vitro experiments. 1M L-proline (Sigma-Aldrich) was dissolved in sterile water to make 100x working stock solution and stored at 4°C. L-proline was added to cells following 1:20 dilution (50 mM) in culture medium. An equal volume of sterile water was added to cells as the control. MG132 was purchased from Tocris Bioscience, and was stored at −80°C at a stock concentration of 1 mg/mL in DMSO.
FACS analysis
Cell surface FACS staining was performed in FACS buffer (PBS + 2% FBS; HyClone) by incubating cells with fluorochrome-conjugated antibodies for 20 minutes at room temperature. Intracellular cytokine, and phospho-Stat3 (pY705) stains were performed as previously described (31). After staining, cells were washed with PBS and stored in PBS plus 1% PFA prior to acquisition on an LSRII-Fortessa Flow Cytometer (BD Bioscience). Data was analyzed using FlowJo software (Tree Star, Inc.). Antibodies against IL17A, IL17F, IL22, IFNγ, GM-CSF, TCRβ, CD4, CD25, CD44, CD45, CD62L, CD19, Gr-1, CD11b, NK1.1, and CCR6 were from Biolegend. Anti-phospho Stat3 (Y705), Stat1 (Y701) and Stat5 (Y694) were from BD Biosciences.
qPCR
Total RNA was isolated from flash-frozen T cell pellets using RNeasy mini kit (Qiagen). 1–10 ng of RNA quantified by a NanoDrop spectrophotometer (Thermo Scientific) was used for cDNA synthesis using a High Capacity RNA-to-cDNA kit (Applied Biosystems). Taqman qPCR was performed on a 7900HT Fast Real Time PCR System (Applied Biosystems). All taqman primer/probe sets were from Applied Biosystems and assay IDs were: Mm00437762_m1, Mm01261022_m1, Mm00519943_m1, Mm00439619_m1, Mm00521423_m1, Mm00444241_m1, Mm01268754_m1, Mm01168134_m1, Mm01290062_m1 and Mm01219775_m1.
Immunoblotting
Cultured T cells were harvested at the indicated time points, washed once in PBS, and lysed on ice at 5–10 × 107 cells/ mL in Laemmli sample buffer (Bio Rad) supplemented with protease inhibitor tablets (Roche) and phosphatase inhibitors as described in (31). Whole cell lysates cleared by centrifugation at 12,000 × g for 10 min at 4°C were quantified by Bradford Assay and stored at −80°C. 10–20 mg of total protein was resolved via SDS-PAGE. Protein was transferred to nitrocellulose membranes, blocked, and blotted with primary and secondary antibodies per manufacturer’s instructions. Antibodies against phospho-Stat3 (9138), total Stat3 (9139) and IκBα (4812) were from Cell Signaling. Antibodies against Stat1 (sc-346), Stat4 (sc-486), Stat5 (sc-835), Stat6 (sc-1689) and βactin (sc-1616) were from Santa Cruz Biotechnology. Anti-mouse, anti-goat, and anti-rabbit secondary antibodies were from GE Life Sciences. Western blots were visualized using the ECL Western Blotting System (GE Life Sciences) on a FluorChem Q (Cell Biosciences).
Results
Characterization of endogenous IL-23-responsive Th17 cells in mice
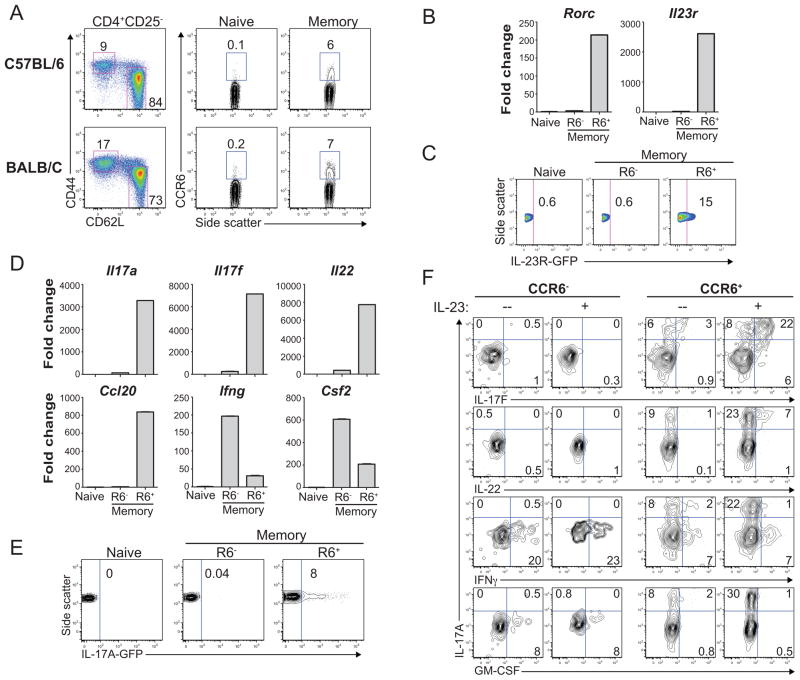
Endogenous Th17 cells in humans and mice are defined by ex vivo expression of IL-17A, and these cells uniformly express the inflammatory chemokine receptor CCR6 (17,23,24). However, human CCR6+ memory T cells can express other Th17-lineage markers independent of ex vivo IL-17A expression (25). We reasoned that CCR6 expression might similarly discriminate IL-23-responsive Th17 memory T cells in mice independent of IL-17A. We confirmed that unmanipulated wild-type mice, on either C57BL/6 or BALB/c backgrounds, possessed a small but readily detectable population of CD4+CD25−CCR6+ memory T cells in spleen and lymph nodes (Figure 1A); CCR6 expression was restricted to the CD62LloCD44hi effector/memory T cell compartment, and was observed, on average, on 5–10% of these T cells.
Figure 1. The endogenous CCR6+ memory T cell population in mice contains IL-23-responsive Th17 cells.
(A) Ex vivo CCR6 expression determined by FACS analysis in CD4+ naïve or memory T cell subsets from spleen and peripheral lymph nodes of wild-type C57BL/6 or BALB/c mice. Naïve and memory T cell subsets were distinguished within CD4+CD25− T cells based on expression of CD44 and CD62L (Left). Data represent more than 5 independent staining experiments on each mouse strain. (B) Expression of Rorc (RORγt) and Il23r mRNAs in FACS-sorted naïve, CCR6− memory, or CCR6+ memory T cells (see Figure S1 for FACS sorting/gating strategy). Rorc and Il23r gene expression was normalized to b2M, and is presented as mean differential (fold-change) expression ± SD from triplicate samples. Data represent 3 qPCR experiments. (C) GFP expression in naïve, CCR6− memory, or CCR6+ memory T cells, as determined by FACS analysis of GFP levels in CD4+ T cells isolated from Il23r+/GFP reporter mice. IL-23R-GFP expression is shown in cells gated as in (A). Data represent 5 experiments. (D) Cytokine and chemokine gene expression determined by qPCR in FACS-sorted naïve, CCR6− memory, or CCR6+ memory T cells (as in [B]). RNA was isolated from cells stimulated ex vivo with PMA and Ionomycin. Data are normalized and presented as fold change in triplicate samples as in (B). Data represent 3 experiments. (E) GFP expression in naïve, CCR6− memory, or CCR6+ memory T cells determined by FACS analysis in CD4+ T cells isolated from Il17a-eGFP transgenic reporter mice. IL-17A-GFP expression is shown in cells gated as in (A). Data represent 5 experiments. (F) FACS-sorted CCR6− or CCR6+ memory T cells were activated with anti-CD3/CD28-coated beads +/− IL-23. Cells were restimulated with PMA and ionomycin on day 2 for intracellular cytokine staining and FACS analysis.
Sorted splenic and lymph node CCR6+ memory T cells, but not naïve or CCR6− memory cells, expressed Rorc and Il23r, as determined by qPCR (Figure 1B). Selective expression of Il23r was also evident by GFP expression in a portion of endogenous CCR6+ memory T cells from Il23r+/eGFP reporter mice (32) (Figure 1C). In contrast to naïve or CCR6− memory T cells, ex vivo-stimulated CCR6+ memory T cells expressed characteristic Th17 cytokines (Il17a, Il17f, Il22), as well as the Th17-associated chemokine Ccl20 (Figure 1D). CCR6− memory T cells preferentially expressed Ifng and Csf2 (the gene encoding GM-CSF) (Figure 1D), whereas Il2 and Tnf were similarly expressed in CCR6+ and CCR6− memory subsets (data not shown). We confirmed that IL-17A was selectively expressed by a portion of ex vivo-stimulated CCR6+ memory T cells via intracellular cytokine staining (data not shown) and by analysis of GFP expression in endogenous naïve or memory T cell subsets from IL-17A-GFP transgenic reporter mice (Figure 1E).
To test the impact of IL-23 on endogenous memory T cell cytokine production, CCR6+ and CCR6− memory cells were FACS-sorted from wild-type mice (Figure S1), and these cells were CD3/CD28-stimulated for two days +/− IL-23. Upon restimulation and intracellular cytokine staining, only CCR6+ memory cells produced IL-17A, consistent with our ex vivo analyses, whereas both CCR6+ and CCR6− memory subsets produced IFNγ (Figure 1F). CCR6+ memory T cells activated in the absence of IL-23 produced IL-17A, but they expressed little to no IL-17F or IL-22 (Figure 1F). Addition of IL-23 to CCR6+ memory T cells induced marked increases in IL-17A levels, and more cells now also expressed IL-17F and IL-22 (Figure 1F). Consistent with their lack of Il23r expression, IL-23 had no effect on cytokine expression in CCR6− memory T cells; IL-23 also failed to influence GM-CSF or IFNγ production by CCR6+ memory T cells (Figure 1F). Thus, as in humans, CCR6 expression broadly discriminates endogenous mouse memory T cells with Th17 characteristics and can be used to isolate IL-23-responsive Th17 cells from mice at steady-state.
In vivo pathogenic functions of endogenous CCR6+ Th17 cells
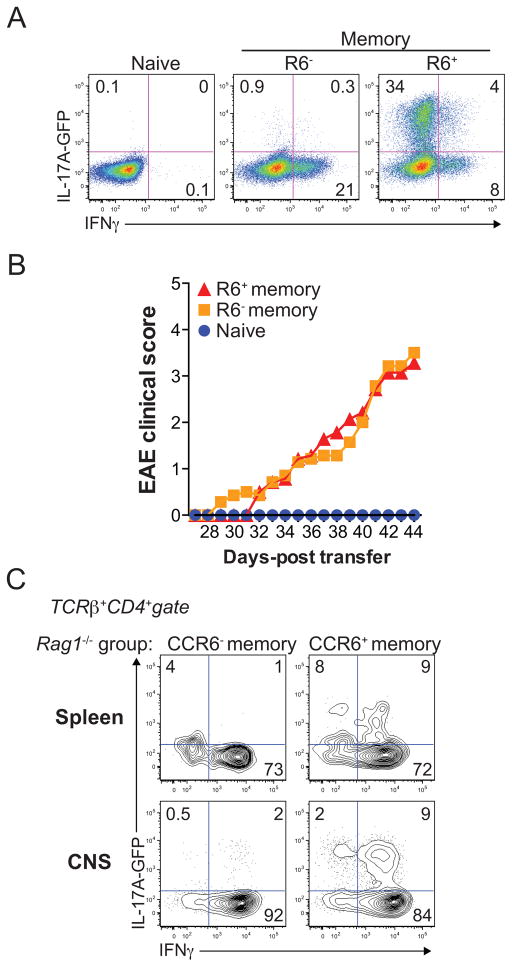
IL23R polymorphisms are linked to human autoimmune disease (13–15). Further, Il23r expression is both indicative of, and required for, pathogenic Th17 cell function in mouse models of autoimmunity (2,4,11,12). Thus, our findings that endogenous mouse CCR6+ memory T cells express Il23r ex vivo and display functional responses to IL-23 stimulation in vitro suggested that at least a subset of these cells have pathogenic potential. To test the pro-inflammatory functions of endogenous CCR6+ Th17 cells in vivo, we isolated these cells, as well as naïve and CCR6− memory cells, from the spleen and lymph notes of 2D2 TCR transgenic/IL-17A-GFP reporter mice, where 2D2 transgenic T cells are specific for the myelin antigen, MOG33–55, and are capable of inducing experimental autoimmune encephalomyelitis (EAE) following transfer into lymphopenic hosts (4,33). Because endogenous memory T cell numbers were limiting, we expanded 2D2+ naïve, CCR6− memory, or CCR6+ memory T cells in vitro for 5 days prior to transfer, using CD3/CD28-stimulation plus IL-2. As a control, a portion of these in vitro-expanded cells were restimulated and analyzed by intracellular cytokine staining to confirm that expanded CCR6+ memory T cells stably and selectively expressed IL-17A relative to naïve or CCR6− memory T cells (Figure 2A). These cells were then transferred into Rag1−/− mice, and recipients were followed for the development of EAE. Whereas Rag1−/− mice receiving activated naïve 2D2+ T cells failed to develop EAE, those injected with either CCR6+ or CCR6− memory T cells developed a chronic and progressive EAE disease, with symptoms developing around 4 weeks post transfer (Figure 2B). Transferred CCR6− memory T cells recovered from either the periphery (spleen) or CNS of sick Rag1−/− mice produced IFNγ, but not IL-17A, following ex vivo stimulation (Figure 2C), whereas transferred CCR6+ memory cells recovered from Rag1−/− recipient organs produced IFNγ and IL-17 (Figure 2C). Collectively, these data indicate that the endogenous CCR6+ memory T cell compartment present in healthy mice contains stable, IL-23-responsive Th17 cells with pathogenic in vivo functions.
Figure 2. The endogenous CCR6+ memory T cell compartment contains pathogenic Th17 cells.
(A) Cytokine expression in in vitro-expanded naïve, CCR6− memory or CCR6+ memory T cells, as determined by FACS analysis in FACS-sorted (as in Figure S1) cells from 2D2 TCR/l17a-eGFP transgenic reporter mice. Cells were expanded with anti-CD3/CD28-coated beads plus IL-2 and restimulated on day 5 with PMA and ionomycin (P+I). (B) Mean clinical EAE scores of Rag1−/− mice injected with naïve (n=6), CCR6− memory (n=7), or CCR6+ memory (n=7) T cells from 2D2 TCR/IL-17A-GFP transgenic mice expanded as in (A). Days-post transfer is relative to the day of cell injection (day 0). (C) Cytokine expression in total TCRβ+CD4+ T cells recovered from the spleen or CNS of Rag1−/− mice at day 50 following injection with CCR6− or CCR6+ memory T cells. All data represent independent 2 experiments.
Halofuginone regulates IL-23-dependent cytokine expression and Stat3 phosphorylation
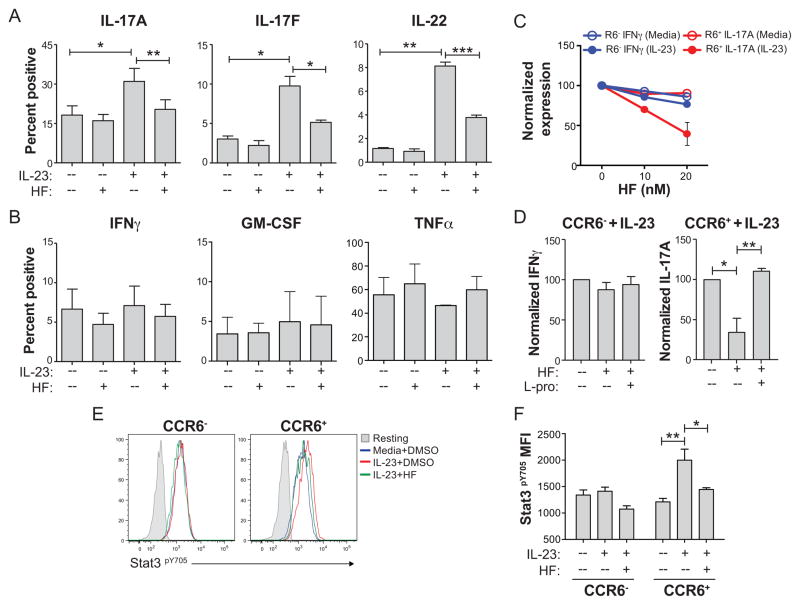
The prolyl-tRNA synthetase inhibitor halofuginone (HF) selectively blocks Th17 cell development via activation of the amino acid starvation response (AAR) pathway (29,31). We therefore tested if HF-induced AAR activation also regulates IL-23 responses in CCR6+ memory Th17 cells. FACS-sorted CCR6+ or CCR6− memory T cells were activated with anti-CD3/anti-CD28 for two days +/− IL-23, and these cells were further treated with either HF or vehicle (DMSO). HF treatment of CCR6+ memory T cells selectively impaired IL-23-dependent induction of IL-17A, IL-17F, and IL-22, but had no effect on the baseline expression of these cytokines in the absence of IL-23 (Figure 3A, Figure S2). Further, HF did not affect expression of IFNγ, GM-CSF or TNFα which were expressed in both CCR6+ and CCR6− memory T cells in an IL-23-independent manner (Figure 3B, Figure S2A). The effect of HF treatment on IL-23-induced IL-17A expression in CCR6+ memory T cells was dose-dependent at concentrations (5–20 nM) that had little to no effect on basal expression of IL-17A in CCR6+ memory cells or IFNγ production by CCR6+ or CCR6− memory T cells (Figure 3C, data not shown). Moreover, HF-mediated inhibition of IL-23 inducible IL-17A expression was abolished following addition of excess L-proline to culture medium (Figure 3D), confirming that HF modulates IL-23 signaling through inhibition of its cognate receptor, the prolyl-tRNA synthetase EPRS, and subsequent activation of the AAR pathway (29,30). These data establish that HF-induced AAR activation selectively represses pro-inflammatory IL-23 responses in mature Th17 memory cells.
Figure 3. HF regulates IL-23 signaling and downstream cytokine expression in endogenous Th17 cells.
Day 2 cytokine production determined by intracellular staining and FACS analysis in ex vivo-sorted and CD3/CD28-stimulated naïve, CCR6− memory, or CCR6+ memory T cells cultured +/− IL-23 and treated with vehicle (DMSO(or HF (20 nM). Cells were washed to remove HF and restimulated with PMA and ionomycin (P+I) on day 2 for analyses. Mean percentage of IL-17A, IL-17F, and IL-22 (A), or IFNγ, GM-CSF, and TNFα (B) positive cells ± SD from 3–4 biological replicates is shown. * P < .05; ** P < .01; *** P < .001 by paired student’s t test. (C) Dose-response of HF on IL-17A or IFNγ expression by CCR6+ or CCR6− memory T cells, respectively. Mean normalized percentages of cytokine positive cells ± SD from biological duplicates relative to vehicle-treated cultures are shown. IL-23-induced IL-17A expression by CCR6+ cells is shown after subtracting from baseline IL-17A expression in cells stimulated without IL-23. (D) Mean IFNγ expression in CCR6− (left) or IL-17A expression in CCR6+ (right) memory T cells ± SD from 3 independent experiments in cells stimulated with anti-CD3/CD28 plus IL-23 for 2 days +/− 20 nM HF with or without 50 mM L-proline. Controls for HF or L-proline were equal volumes of DMSO or sterile water, respectively. Cytokine expression was determined by intracellular cytokine staining on day 2 following P+I restimulation. * P < .05; ** P < .01 by paired student’s t test. (E) Stat3 (Y705) phosphorylation determined by phospho-specific intracellular staining and FACS analysis in CD3/CD28-stimulated CCR6− (left) or CCR6+ (right) memory T cells cultured for 2 days +/− IL-23 with or without 20 nM HF. Unstimulated cells (no anti-CD3/anti-CD28; Resting – shaded areas) were used to determine background. Data represent 3 experiments. (F) Phospho-Stat3 MFI from 3 experiments ± SD, where background staining in unstimulated cells is subtracted. * P < .05; ** P < .01 by paired student’s t test.
HF treatment inhibits Stat3 tyrosine (Y705) phosphorylation during Th17 cell differentiation (31). Because IL-23 also activates Stat3, and Stat3 is critical for the pro-inflammatory effects of IL-23 on Th17 cells (34,35), we next determined the effects of IL-23 and HF-induced AAR activation on Stat3 phosphorylation in endogenous memory T cell subsets. CCR6+ or CCR6− memory T cells were activated as above +/− IL-23, treated with or without HF, and Stat3 phosphorylation was determined by phospho-specific intracellular staining and FACS analysis. Consistent with their selective expression of Il23r and their functional responses to IL-23, only CCR6+ memory T cells displayed increased Stat3 phosphorylation in the presence of IL-23 (Figure 3E, 3F). HF treatment reduced IL-23-dependent Stat3 activation in CCR6+ memory T cells to near baseline levels observed in cells cultured without IL-23 (Figure 3E, 3F). As with its effects on Th17 cytokine production, the impact of HF treatment on Stat3 phosphorylation was abolished by the addition of excess L-proline (data not shown). HF treatment had no effect on the phosphorylation of Stat5 (Y694) or Stat1 (Y701) in stimulated CCR6+ or CCR6− memory cells cultured with or without IL-23 (Figure S2B, S2C). Thus, HF-directed suppression of both Th17 cell development and IL-23-driven Th17 cell effector function are associated with reduced Stat3 phosphorylation (31).
Halofuginone restricts established inflammation and modulates local Th17 cell function in EAE
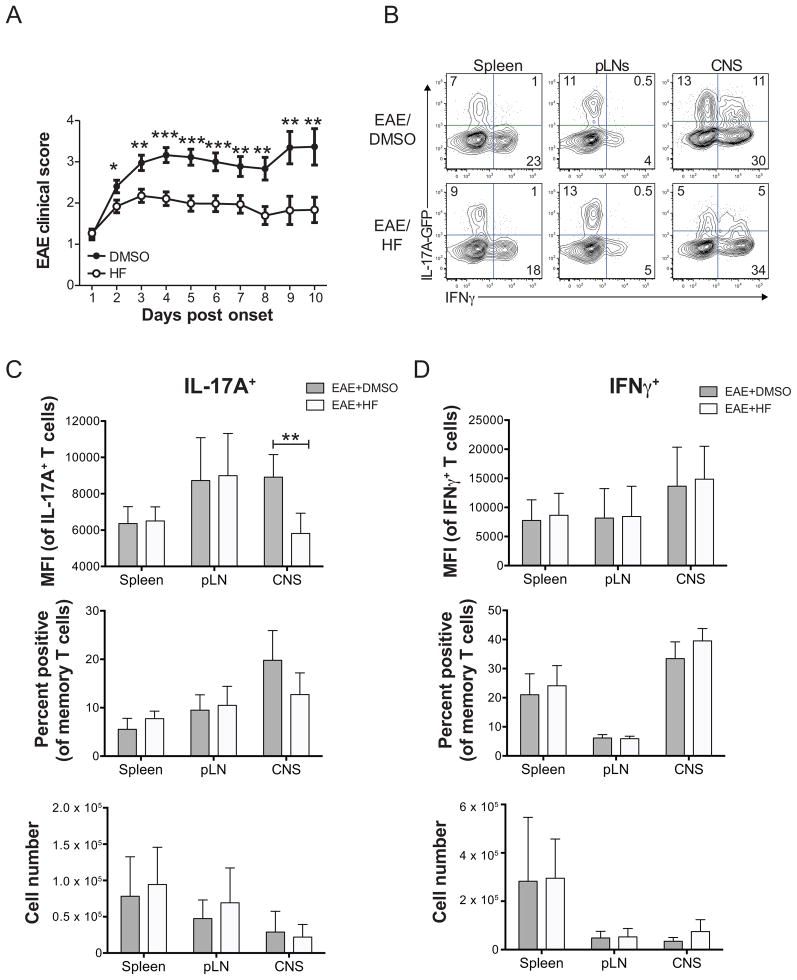
The central role of IL-23 in autoimmune pathogenesis (36), along with our studies showing that HF regulates pro-inflammatory IL-23 responses in mature endogenous Th17 cells, suggested that HF may have therapeutic effects on established autoimmune inflammation. To test this, wild-type or IL-17A-GFP mice were immunized with MOG33–55/CFA to induce EAE; animals were injected daily with HF or vehicle (DMSO) beginning at disease onset, and disease progression was monitored over a 10-day period. Notably, administration of HF rapidly and significantly attenuated the progression of pre-established EAE symptoms (Figure 4A). To assess if the therapeutic efficacy of HF was associated with perturbed T cell effector function, we determined the cytokine expression profiles of memory T cells isolated from MOG33–55/CFA-immunized IL-17A-GFP reporter mice following treatment with DMSO or HF. HF had no effect on cytokine expression (IL-17A, IFNγ) in CD4+CD44hi effector/memory T cells in non-target organs, such as the spleen or draining lymph nodes (Figure 4B–D). In contrast, HF suppressed the level of IL-17A expression in IL-17A+ cells in the CNS (as seen by reduced MFI) and modestly, but consistently, reduced the proportion of CNS-infiltrating memory T cells that expressed IL-17A (Figure 4B, 4C). Consistent with the fact that the strongest effect of HF treatment were on the level of IL-17A expression per cell, and not the frequency of cells expressing IL-17A, absolute numbers of Th17 cells within the CNS (and peripheral lymphoid organs) of HF-treated mice were not significantly different from vehicle-treated animals (Figure 4C). In contrast, HF had no effect on the expression levels, percentages, or absolute numbers of IFNγ-expressing effector/memory T cells (Figure 4B, 4D). Therapeutic HF treatment did not influence the infiltration of total or effector/memory CD4+ T cells into the CNS (Figure S3, S4), or recruitment/localization of other immune cell types, including B cells, NK cells, or neutrophils (Figure S3, S4A). Thus, therapeutic efficacy of HF in MOG/CFA-induced EAE is specifically associated with reduced IL-17A expression by Th17 cells in the CNS. Interestingly, in contrast to MOG/CFA-induced EAE, therapeutic HF dosing in Rag1−/− mice injected with endogenous 2D2 TCR transgenic CCR6+ effector/memory T cells did not prevent the progression of clinical EAE symptoms (Figure S4B, S4C).
Figure 4. HF treatment reduces established EAE disease and inhibits Th17 effector function in the CNS. (A).
Mean clinical EAE score ± SD in wild-type C57BL/6 mice immunized with MOG33–55 and treated daily with HF (0.3 mg/kg; n = 43) or vehicle (DMSO; n = 40) beginning at onset of disease symptoms. * P < .05; ** P < .01; *** P < .001 by paired student’s t test. (B) Cytokine expression in ex vivo-isolated and PMA+ionomycin-stimulated mononuclear cells from the spleen, peripheral lymph nodes (pLNs), or brain and spinal cord (CNS) of vehicle- (EAE score = 3) or HF- (EAE score = 2) treated mice with active EAE on day 6 post disease onset/HF treatment. Data shown are on CD45+TCRβ+CD4+CD44hi-gated memory T cells. Data represent 2 independent experiments, with each including analysis of 3 mice per group. Analysis of IL-17A (C) or IFNγ (D) expression by CD4+CD44hi effector/memory T cells from spleen, pLNs, or brain and spinal cord (CNS) of 6 DMSO- or HF-treated IL-17A-GFP reporter mice with active EAE on day 6 post disease onset/HF treatment. Top – mean fluorescence intensity of IL-17A (GFP) or IFNγ (intracellular cytokine staining) expression in effector/memory cells gated as positive for cytokine expression. Middle – percentage of effector/memory cells from each organ displaying detectable IL-17A or IFNγ expression. Bottom – absolute numbers of IL-17A+ or IFNγ+ effector/memory T cells, calculated as follows: total number of mononuclear cells recovered x % of live lymphocytes x % of CD4+CD44hi effector/memory T cells x % of cytokine positive cells. All data are mean values ± SD from 6 mice per group across two independent experiments. ** P < .01 by paired student’s t test.
Cytokine-independent regulation of Stat3 by the AAR
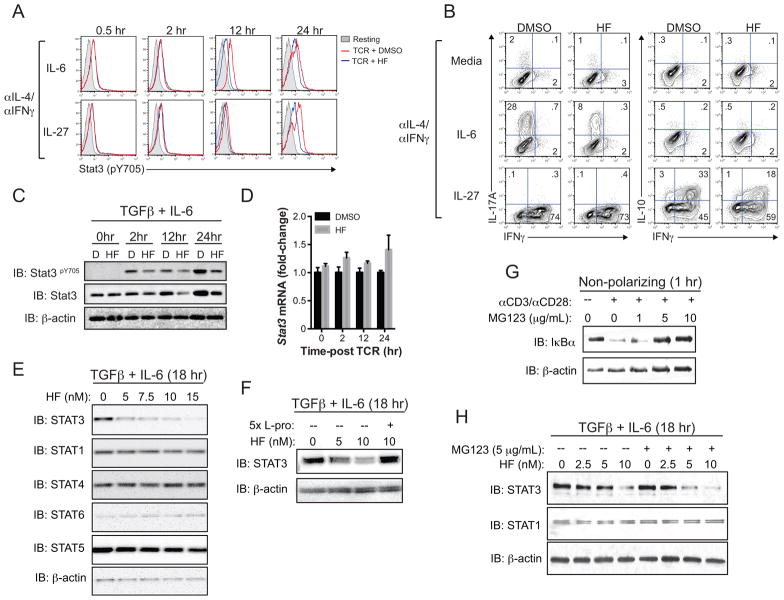
Stat3 is necessary for Th17 cell development and is important for IL-23-mediated inflammatory function of mature Th17 cells (5–7,12,34). As both our previous (31) and current findings indicate that HF-dependent inhibition of Th17 responses is associated with decreased Stat3 activation, we sought to determine the mechanism by which AAR activation regulates Stat3 in T cells. We used naïve CD4+ T cells for these experiments because of the limiting numbers of endogenous effector/memory T cells in unmanipulated mice; we first asked whether HF treatment regulates Stat3 responses in a cytokine-specific manner. This was not the case, as HF treatment reduced both IL-6- and IL-27-mediated Stat3 (Y705) phosphorylation in naïve T cells (Figure 5A). Specifically, and consistent with the effects of HF of Stat3 activation in naïve CD4 T cells treated with TGFβ plus IL-6 (31), HF treatment reduced the levels of sustained phosho-Stat3 in cells treated with IL-6 or IL-27 alone (e.g., 12–24 hours after adding ligand), without affecting Stat3 phosphorylation within the first 2 hours of adding ligand (Figure 5A). As expected, sustained Stat3 phosphorylation in IL-6- or IL-27-stimulated naïve T cells treated with HF was restored by adding excess proline (data not shown), confirming that, as with IL-23 stimulation, HF regulates IL-6 and IL-27-mediated Stat3 activation via inhibition of EPRS and activation of the AAR pathway.
Figure 5. Selective, post-transcriptional regulation of Stat3 expression by the AAR.
(A) Stat3 (Y705) phosphorylation, detected by phospho-specific intracellular staining and FACS analysis, in CD4+CD25− T cells cultured for the indicated times with anti-IL-4 and anti-IFNγ antibodies (αIL-4/αIFNγ) plus either IL-6 or IL-27 alone. Grey shaded peaks – resting T cells (no CD3/CD28 stimulation); red and blue peaks – CD3/CD28-stimulated T cells cultured with vehicle (DMSO) or 10 nM HF, respectively. Data represent 2 independent experiments, each with 2–3 replicates per condition. (B) Intracellular cytokine expression (IL-17A vs. IFNγ – left; IL-10 vs. IFNγ – right) by CD3/CD28-stimulated CD4+CD25− T cells cultured for 4 days with αIL-4/αIFNγ in the absence (Media) or presence of IL-6 or IL-27. Cells were restimulated with PMA and ionomycin in the presence of brefeldin A prior to intracellular staining. Data represent 2 independent experiments, each containing 2–3 replicates per condition. (C) Total Stat3, phospho (Y705)-Stat3, and β-actin protein levels in CD4+CD25− T cells activated for up to 24 hours in Th17-polarizing conditions (TGFβ + IL-6) plus DMSO “(D)” or 10 nM HF, as determined by western blotting. Data represent 3 independent experiments. (D) Mean relative (fold-change) Stat3 mRNA expression ± SD from duplicate samples determined by qPCR in CD4+CD25− T cells cultured for the indicated timepoints (as in (C)). Stat3 expression was normalized to B2M; data represent two experiments. (E) Effects of the indicated doses of HF on Stat protein levels in CD4+CD25− T cells cultured for 18 hours in Th17-polarizing cytokine conditions, as determined by western blotting. Data represent 2 independent experiments. (F) Stat3 and β-actin protein levels, determined by western blotting, in Th17-polarized CD4+CD25− T cells treated with titrating concentrations of HF as indicated for 18 hr. Some cultures were supplemented with 5x (50 mM L-proline (L-Pro)). Data represent 3 independent experiments. (G) IκBα and β-actin protein levels in resting or CD3/CD28-stimulated CD4+CD25− T cells cultured for 2 hours with or without titrating concentrations of MG132. Data represent 2 experiments. (H) Stat3, Stat1, and β-actin protein levels in CD4+CD25− T cells activated in Th17-polarizing conditions for 18 hours in the presence of titrating concentrations of HF +/− 5 μg/mL MG132 as indicated. MG132 was added 12 hours post-activation. Data represent 3 experiments.
Consistent with previous reports, we found that IL-6, when combined with anti-IL-4 and anti-IFNγ antibodies, was sufficient to induce Th17 differentiation and IL-17A expression, whereas IL-27 promoted expression of both IL-10 and IFNγ, but not IL-17A (7,37,38) (Figure 5B). IL-27-induced Stat3 activation directs IL-10 expression, whereas IL-27-induced IFNγ requires Stat1 (39,40). HF treatment attenuated both IL-6-induced IL-17A expression and IL-27-induced IL-10 expression, but did not affect induction of IFNγ by IL-27 (Figure 5B), suggesting that HF selectively targets Stat3 activation. Together with our previous studies (31), these findings suggest that HF treatment selectively inhibits Stat3 activation and downstream transcriptional responses in T cells, but does so irrespective of the activating cytokine/cytokine receptor.
Inhibition of Stat3 phosphorylation by HF was also evident by western blot analyses of naïve T cells cultured in Th17-polarizing cytokines (i.e., TGFβ + IL-6) (Figure 5C). However, these experiments also revealed that reduced phospho-Stat3 in HF-treated cells coincided with less total Stat3 protein (Figure 5C). In line with the effects of HF on Stat3 phosphorylation, HF treatment blocked increases in Stat3 protein observed at 12 and 24 hours post-activation, but did not affect steady-state Stat3 protein levels between 0–2 hours post-T cell activation (Figure 5C). In contrast to Stat3 protein, HF did not influence the levels of Stat3 mRNA; thus, the AAR regulates Stat3 at a post-transcriptional level (Figure 5D). Further, HF treatment reduced Stat3 protein levels in differentiating Th17 cells in a dose-dependent manner, and at concentrations that also regulate both Th17 differentiation (31) and IL-23-induced cytokine expression in endogenous CCR6+ memory cells (see above) (Figure 5E). Here again, effects of HF on total Stat3 protein levels were abolished by addition of excess proline (Figure 5F). Importantly, the inhibition of Stat3 protein expression by low-dose HF treatment was selective, as other Stat proteins were not similarly affected (Figure 5F).
The reduced abundance of Stat3 protein in HF-treated T cells could be due to decreased protein synthesis or to increased turnover. To address if the AAR regulates Stat3 protein abundance via a degradation pathway, control or HF-treated Th17 cells were cultured with or without the proteasome inhibitor MG132. Whereas MG132 blocked CD3/CD28 stimulation-induced IκBα degradation in naïve T cells (Figure 5G), it did not restore Stat3 protein levels in HF-treated Th17 cells (Figure 5H), indicating that HF-induced AAR activation regulates Stat3 protein abundance independent of the proteasome.
Discussion
We have shown that the CCR6+ memory T cell compartment present in mice at steady-state contains IL-23-responsive Th17 cells with latent pathogenic functions. Like their human counterparts (23–25), a portion of murine CCR6+ memory T cells express Rorc and Il23r ex vivo, and produce IL-17A, IL-17F and IL-22 following acute TCR stimulation. Further, CCR6+, but not CCR6−, memory cells display marked pro-inflammatory functional responses to IL-23 stimulation in vitro. Although CCR6+ memory T cells are likely heterogeneous and not all of these cells are “pathogenic Th17 cells” – as indicated by IL-17A- or IL-23R-GFP reporter gene expression in only a fraction of CCR6+ memory T cells – the frequency of CCR6+ memory cells that produce Th17 cytokines following in vitro culture, particularly in the presence of IL-23, is far greater than that predicted based on ex vivo IL-17A expression. Thus, as with human CCR6+ memory T cells, it appears the expression of Th17 cytokines in mature CCR6+ Th17 cells can be activated as readily as they are repressed (25). Presumably, this dynamic control of Th17 cytokine expression, generally referred to as “plasticity”, depends on cues present in local microenvironments or tissues (9,20,41).
The ability of endogenous CCR6+ Th17 cells to respond functionally to IL-23 is particularly significant given the well-established links of IL-23 to autoimmunity (36). Indeed, and despite their presence in healthy mice, we confirmed that endogenous CCR6+ Th17 cells have pathogenic potential in vivo. The fact that CCR6+ Th17 cells, like CCR6− memory T cells, can induce autoimmune inflammation following transfer into lymphopenic hosts, likely speaks to the importance of endogenous mechanisms of T cell tolerance, such as those provided by regulatory T cells, as well as of antigen specificity, in controlling pathologic immune responses (42,43). Our results suggest that Th17 cells, even those expressing IL-23R and displaying overt pro-inflammatory functions, only promote autoimmune tissue damage when other tolerance checkpoints are breached.
In assessing pathways that modulate IL-23 signaling in CCR6+ Th17 cells, we found that the small molecule halofuginone (HF) potently blocks both IL-23-mediated Stat3 activation and downstream Th17 cytokine expression. Akin to its regulation of Th17 cell development, the effects of HF on IL-23 signaling in mature Th17 cells was abolished by addition of excess L-proline; thus, HF-dependent regulation of mature Th17 cell effector function requires inhibition of the prolyl-tRNA synthetase, EPRS, and activation of the AAR (29–31). Notably, regulation of IL-23 signaling by HF was highly selective, as HF had no effect on: (1) basal expression of IL-17A in CCR6+ memory T cells; (2) other cytokines expressed by CCR6+ or CCR6− memory T cells independent of IL-23; and (3) activation of other Stat proteins, namely Stat5 and Stat1. These data support the notion that the AAR pathway regulates IL-17A expression in a context-dependent manner. In line with this, our in vivo experiments revealed that HF inhibits T cell expression of IL-17A locally in the CNS, but not systemically in peripheral organs. Collectively, these findings support the hypothesis that although HF activates the AAR systemically, this pathway acts preferentially on T cells at sites of inflammation, where pro-inflammatory cytokines such as IL-6 and IL-23 accumulate. Indeed, previous studies have shown that the expression levels of IL-23 subunits are elevated within active human autoimmune lesions, compared to healthy tissue (44). Given that HF-dependent AAR activation regulates both the development and the IL-23-directed effector function of Th17 cells, further insight into the selective anti-inflammatory mechanism of AAR action in vivo is important.
It is unclear whether perturbation of effector/memory Th17 cell function by HF is sufficient to explain its therapeutic effects in EAE. On one hand, HF treatment did not markedly reduce Th17 cell numbers in the CNS of mice with EAE, though the effect of HF on the level of IL-17A expression per memory cell (gated as IL-17A+) indicates an approximate 2-fold decrease in the amount of IL-17A in the CNS of sick mice. Potentially more importantly however is that HF did not show therapeutic efficacy in EAE where disease was induced in lymphopenic Rag1−/− animals by injection of endogenous CCR6+ effector/memory T cells from 2D2 TCR transgenic mice. These results may suggest that the regulatory activity of HF in EAE manifest in MOG33–55/CFA-immunized wild-type mice requires regulation of other cells, for example de novo Th17 cell differentiation from naïve cells following disease onset. However, at least two alternative explanations maintain that the therapeutic efficacy of HF in EAE observed in, in tact mice is linked to regulation of the endogenous CCR6+ Th17 effector/memory cell compartment. First, HF may influence disease activity in MOG33–55/CFA-immunized mice via regulation of “bystander” Th17 cells present in inflamed CNS tissue that are not specific for myelin antigens. Alternatively, lymphopenic expansion may enable pathogenic functions of CCR6+ Th17 cells that are not present in wild-type mice following MOG33–55/CFA-immunization and are not sensitive to HF and the AAR. Clearly, further insight into the settings where HF possesses or lacks immunoregulatory activity will be important to inform it’s potential therapeutic utility.
In beginning to explore the mechanism by which the AAR selectively regulates Th17 responses, we show that HF-induced AAR activation restricts Stat3-dependent T cell function downstream of multiple cytokines. In addition to repressing IL-23-mediated Stat3 activation and downstream cytokine expression in endogenous CCR6+ memory T cells, we show that HF treatment inhibits Stat3 activation in naïve T cells following IL-6 or IL-27 stimulation. In the context of IL-6 stimulation, the result of AAR activation is reduced IL-17A expression; when IL-27 is the principal activator of Stat3, the AAR represses IL-10 but not IFNγ expression. These results are consistent with Stat3-selective regulation by the AAR; both IL-6-dependent IL-17A expression and IL-27-induced IL-10 production require Stat3, whereas IL-27 activates IFNγ expression via Stat1 (39,40). Further, the AAR regulates Stat3 phosphorylation indirectly, by decreasing Stat3 protein abundance. HF treatment reduced Stat3 protein levels in a dose-dependent manner, despite normal, high-level Stat3 mRNA expression. Further, AAR-dependent regulation of Stat3 was selective and did not similarly reduce the expression of other Stat proteins, consistent with the fact that HF treatment preferentially affects Stat3-dependent cytokine expression. Finally, we show that the AAR does not influence Stat3 protein levels via activation of a proteasome-mediated degradation pathway, leading us to suggest that AAR activation likely targets Stat3 protein synthesis.
Examples of transcript-selective translational control have emerged in recent years. For example, Sabatini and colleagues have shown that the key nutrient sensing kinase, mTOR, preferentially regulates translation of mRNAs that encode ribosomal proteins and which contain a 5′ terminal oligopyrimidine (TOP) motif (45). In addition, VEGFA protein synthesis is selectively regulated in monocytes by both inflammation (in the form of IFNγ) and hypoxia. Here, distinct structural elements in the 3′ UTR of VEGFA mRNA dictate translation, where the gamma interferon-activated inhibitor of translation (GAIT) complex binds to a 3′ GAIT element in VEGFA mRNA and blocks translation initiation, while the hypoxia-inducible hnRNP L–DRBP76–hnRNP A2/B1 (HILDA) complex blocks GAIT complex binding and facilitates VEGFA mRNA translation (46). The GAIT complex may be particularly relevant to the effects of HF treatment on Stat3 protein levels in T cells, as the RNA-binding subunit of this complex is EPRS, the cellular receptor of HF (47).
In summary, our study suggests that both the size and the pro-inflammatory potential of the endogenous Th17 cell compartment in mice are greater than previously recognized. This new knowledge, together with the fact that endogenous Th17 cells can be readily discriminated and isolated based on steady-state expression of CCR6, will facilitate further study of mature Th17 cell inflammatory function. As established by IL-23, Stat3, and the AAR, these pathways are likely to play important roles in persistent inflammatory responses, including those that are manifest in patients with autoimmune disease.
Supplementary Material
Acknowledgments
Tempero pharmaceuticals, Inc. is majority-owned and funded by GlaxoSmithKline. Partial financial support by NIH grants AI40127 and CA42471 (to A.R). Additional support (to M.S.S.) was provided to Scripps Florida from The State of Florida
We thank Drs. Derya Unutmaz, Victor Torres, John Cleveland and members of Tempero pharmaceuticals, Inc. for helpful discussion and critical review of the manuscript.
Abbreviations
- AAR
Amino acid starvation response
- Atf4
activating transcription factor-4
- CNS
central nervous system
- EAE
experimental autoimmune encephalomyelitis
- eIF2α
eukaryotic translation initiation factor 2A
- EPRS
glutamyl-prolyl-tRNA synthetase
- GWAS
genome-wide association studies
- HF
halofuginone
- RORγt
retinoic acid-related orphan nuclear receptor
- Stat3
signal transducer activator of transcription 3
- TCR
T cell receptor
- Th17
T helper 17
Footnotes
Disclosures
The authors declare no competing financial interests.
References
- 1.Miossec P, Korn T, Kuchroo VK. Interleukin-17 and type 17 helper T cells. N Engl J Med. 2009;361:888–898. doi: 10.1056/NEJMra0707449. [DOI] [PubMed] [Google Scholar]
- 2.Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, Ramos HL, Wei L, Davidson TS, Bouladoux N, Grainger JR, Chen Q, Kanno Y, Watford WT, Sun HW, Eberl G, Shevach EM, Belkaid Y, Cua DJ, Chen W, O’Shea JJ. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 2010;467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, Sobel RA, Regev A, Kuchroo VK. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13:991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, Takahashi H, Sun HW, Kanno Y, Powrie F, O’Shea JJ. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–615. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, Dong C. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 2007;282:9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 7.Zhou L, I, Ivanov I, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 8.Hirahara K, Ghoreschi K, Laurence A, Yang XP, Kanno Y, O’Shea JJ. Signal transduction pathways and transcriptional regulation in Th17 cell differentiation. Cytokine Growth Factor Rev. 2010;21:425–434. doi: 10.1016/j.cytogfr.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters A, Lee Y, Kuchroo VK. The many faces of Th17 cells. Curr Opin Immunol. 2011;23:702–706. doi: 10.1016/j.coi.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pene J, Chevalier S, Preisser L, Venereau E, Guilleux MH, Ghannam S, Moles JP, Danger Y, Ravon E, Lesaux S, Yssel H, Gascan H. Chronically inflamed human tissues are infiltrated by highly differentiated Th17 lymphocytes. J Immunol. 2008;180:7423–7430. doi: 10.4049/jimmunol.180.11.7423. [DOI] [PubMed] [Google Scholar]
- 11.Ahern PP, Schiering C, Buonocore S, McGeachy MJ, Cua DJ, Maloy KJ, Powrie F. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity. 2010;33:279–288. doi: 10.1016/j.immuni.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, Blumenschein WM, McClanahan TK, O’Shea JJ, Cua DJ. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009;10:314–324. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellinghaus D, Ellinghaus E, Nair RP, Stuart PE, Esko T, Metspalu A, Debrus S, Raelson JV, Tejasvi T, Belouchi M, West SL, Barker JN, Koks S, Kingo K, Balschun T, Palmieri O, Annese V, Gieger C, Wichmann HE, Kabesch M, Trembath RC, Mathew CG, Abecasis GR, Weidinger S, Nikolaus S, Schreiber S, Elder JT, Weichenthal M, Nothnagel M, Franke A. Combined analysis of genome-wide association studies for Crohn disease and psoriasis identifies seven shared susceptibility loci. Am J Hum Genet. 2012;90:636–647. doi: 10.1016/j.ajhg.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Remmers EF, Cosan F, Kirino Y, Ombrello MJ, Abaci N, Satorius C, Le JM, Yang B, Korman BD, Cakiris A, Aglar O, Emrence Z, Azakli H, Ustek D, Tugal-Tutkun I, Akman-Demir G, Chen W, Amos CI, Dizon MB, Kose AA, Azizlerli G, Erer B, Brand OJ, Kaklamani VG, Kaklamanis P, Ben-Chetrit E, Stanford M, Fortune F, Ghabra M, Ollier WE, Cho YH, Bang D, O’Shea JJ, Wallace GR, Gadina M, Kastner DL, Gul A. Genome-wide association study identifies variants in the MHC class I, IL10, and IL23R-IL12RB2 regions associated with Behcet’s disease. Nat Genet. 2010;42:698–702. doi: 10.1038/ng.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherlock JP, Joyce-Shaikh B, Turner SP, Chao CC, Sathe M, Grein J, Gorman DM, Bowman EP, McClanahan TK, Yearley JH, Eberl G, Buckley CD, Kastelein RA, Pierce RH, Laface DM, Cua DJ. IL-23 induces spondyloarthropathy by acting on ROR-gammat+ CD3+CD4-CD8- entheseal resident T cells. Nat Med. 2012;18:1069–1076. doi: 10.1038/nm.2817. [DOI] [PubMed] [Google Scholar]
- 16.Coffre M, Roumier M, Rybczynska M, Sechet E, Law HK, Gossec L, Dougados M, Bianchi E, Rogge L. Combinatorial control of Th17 and Th1 cell functions by genetic variations in genes associated with the interleukin-23 signaling pathway in spondyloarthritis. Arthritis Rheum. 2013;65:1510–1521. doi: 10.1002/art.37936. [DOI] [PubMed] [Google Scholar]
- 17.Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, Cua DJ, Ahlfors H, Wilhelm C, Tolaini M, Menzel U, Garefalaki A, Potocnik AJ, Stockinger B. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol. 2011;12:255–263. doi: 10.1038/ni.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esplugues E, Huber S, Gagliani N, Hauser AE, Town T, Wan YY, O’Connor W, Jr, Rongvaux A, Van Rooijen N, Haberman AM, Iwakura Y, Kuchroo VK, Kolls JK, Bluestone JA, Herold KC, Flavell RA. Control of TH17 cells occurs in the small intestine. Nature. 2011;475:514–518. doi: 10.1038/nature10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanov II, de Frutos RL, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirota K, Turner JE, Villa M, Duarte JH, Demengeot J, Steinmetz OM, Stockinger B. Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat Immunol. 2013;14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Codarri L, Gyulveszi G, Tosevski V, Hesske L, Fontana A, Magnenat L, Suter T, Becher B. RORgammat drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol. 2011;12:560–567. doi: 10.1038/ni.2027. [DOI] [PubMed] [Google Scholar]
- 22.Wu C, Yosef N, Thalhamer T, Zhu C, Xiao S, Kishi Y, Regev A, Kuchroo VK. Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature. 2013;496:513–7. doi: 10.1038/nature11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, Sallusto F, Napolitani G. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. 2007;8:639–646. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- 24.Sallusto F, Zielinski CE, Lanzavecchia A. Human Th17 subsets. Eur J Immunol. 2012;42:2215–2220. doi: 10.1002/eji.201242741. [DOI] [PubMed] [Google Scholar]
- 25.Wan Q, Kozhaya L, ElHed A, Ramesh R, Carlson TJ, Djuretic IM, Sundrud MS, Unutmaz D. Cytokine signals through PI-3 kinase pathway modulate Th17 cytokine production by CCR6+ human memory T cells. J Exp Med. 2011;208:1875–1887. doi: 10.1084/jem.20102516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 27.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 28.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 29.Keller TL, Zocco D, Sundrud MS, Hendrick M, Edenius M, Yum J, Kim YJ, Lee HK, Cortese JF, Wirth DF, Dignam JD, Rao A, Yeo CY, Mazitschek R, Whitman M. Halofuginone and other febrifugine derivatives inhibit prolyl-tRNA synthetase. Nat Chem Biol. 2012;8:311–317. doi: 10.1038/nchembio.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou H, Sun L, Yang XL, Schimmel P. ATP-directed capture of bioactive herbal-based medicine on human tRNA synthetase. Nature. 2013;494:121–124. doi: 10.1038/nature11774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sundrud MS, Koralov SB, Feuerer M, Calado DP, Kozhaya AE, Rhule-Smith A, Lefebvre RE, Unutmaz D, Mazitschek R, Waldner H, Whitman M, Keller T, Rao A. Halofuginone inhibits TH17 cell differentiation by activating the amino acid starvation response. Science. 2009;324:1334–1338. doi: 10.1126/science.1172638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Awasthi A, Riol-Blanco L, Jager A, Korn T, Pot C, Galileos G, Bettelli E, Kuchroo VK, Oukka M. Cutting edge: IL-23 receptor gfp reporter mice reveal distinct populations of IL-17-producing cells. J Immunol. 2009;182:5904–5908. doi: 10.4049/jimmunol.0900732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Domingues HS, Mues M, Lassmann H, Wekerle H, Krishnamoorthy G. Functional and pathogenic differences of Th1 and Th17 cells in experimental autoimmune encephalomyelitis. PLoS One. 2010;5(11):e15531. doi: 10.1371/journal.pone.0015531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho ML, Kang JW, Moon YM, Nam HJ, Jhun JY, Heo SB, Jin HT, Min SY, Ju JH, Park KS, Cho YG, Yoon CH, Park SH, Sung YC, Kim HY. STAT3 and NF-kappaB signal pathway is required for IL-23-mediated IL-17 production in spontaneous arthritis animal model IL-1 receptor antagonist-deficient mice. J Immunol. 2006;176:5652–5661. doi: 10.4049/jimmunol.176.9.5652. [DOI] [PubMed] [Google Scholar]
- 35.Parham C, Chirica M, Timans J, Vaisberg E, Travis M, Cheung J, Pflanz S, Zhang R, Singh KP, Vega F, To W, Wagner J, O’Farrell AM, McClanahan T, Zurawski S, Hannum C, Gorman D, Rennick DM, Kastelein RA, de Waal Malefyt R, Moore KW. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol. 2002;168:5699–5708. doi: 10.4049/jimmunol.168.11.5699. [DOI] [PubMed] [Google Scholar]
- 36.Croxford AL, Mair F, Becher B. IL-23: one cytokine in control of autoimmunity. Eur J Immunol. 2012;42:2263–2273. doi: 10.1002/eji.201242598. [DOI] [PubMed] [Google Scholar]
- 37.Qin H, Wang L, Feng T, Elson CO, Niyongere SA, Lee SJ, Reynolds SL, Weaver CT, Roarty K, Serra R, Benveniste EN, Cong Y. TGF-beta promotes Th17 cell development through inhibition of SOCS3. J Immunol. 2009;183:97–105. doi: 10.4049/jimmunol.0801986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Awasthi A, Carrier Y, Peron JP, Bettelli E, Kamanaka M, Flavell RA, Kuchroo VK, Oukka M, Weiner HL. A dominant function for interleukin 27 in generating interleukin 10-producing anti-inflammatory T cells. Nat Immunol. 2007;8:1380–1389. doi: 10.1038/ni1541. [DOI] [PubMed] [Google Scholar]
- 39.Takeda A, Hamano S, Yamanaka A, Hanada T, Ishibashi T, Mak TW, Yoshimura A, Yoshida H. Cutting edge: role of IL-27/WSX-1 signaling for induction of T-bet through activation of STAT1 during initial Th1 commitment. J Immunol. 2003;170:4886–4890. doi: 10.4049/jimmunol.170.10.4886. [DOI] [PubMed] [Google Scholar]
- 40.Stumhofer JS, Silver JS, Laurence A, Porrett PM, Harris TH, Turka LA, Ernst M, Saris CJ, O’Shea JJ, Hunter CA. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat Immunol. 2007;8:1363–1371. doi: 10.1038/ni1537. [DOI] [PubMed] [Google Scholar]
- 41.Mukasa R, Balasubramani A, Lee YK, Whitley SK, Weaver BT, Shibata Y, Crawford GE, Hatton RD, Weaver CT. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity. 2010;32:616–627. doi: 10.1016/j.immuni.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miles JJ, Douek DC, Price DA. Bias in the alphabeta T-cell repertoire: implications for disease pathogenesis and vaccination. Immunol Cell Biol. 2011;89:375–87. doi: 10.1038/icb.2010.139. [DOI] [PubMed] [Google Scholar]
- 44.Duvallet E, Semerano L, Assier E, Falgarone G, Boissier MC. Interleukin-23: a key cytokine in inflammatory diseases. Ann Med. 2011;43:503–511. doi: 10.3109/07853890.2011.577093. [DOI] [PubMed] [Google Scholar]
- 45.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–13. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao P, Potdar AA, Ray PS, Eswarappa SM, Flagg AC, Willard B, Fox PL. The HILDA complex coordinates a conditional switch in the 3′-untranslated region of the VEGFA mRNA. PLoS Biol. 2013;11:e1001635. doi: 10.1371/journal.pbio.1001635. Epub 2013 Aug 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mukhopadhyay R, Jia J, Arif A, Ray PS, Fox PL. The GAIT system: a gatekeeper of inflammatory gene expression. Trends Biochem Sci. 2009;34:324–331. doi: 10.1016/j.tibs.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.