Abstract
Cellular retinaldehyde-binding protein (CRALBP) chaperones 11-cis-retinal to convert opsin receptor molecules into photosensitive retinoid pigments of the eye. We report a thermal secondary isomerase activity of CRALBP when bound to 9-cis-retinal. UV/VIS and 1H-NMR spectroscopy were used to characterize the product as 9,13-dicis-retinal. The X-ray structure of the CRALBP mutant R234W:9-cis-retinal complex at 1.9 Å resolution revealed a niche in the binding-pocket for 9-cis-aldehyde different from that reported for 11-cis-retinal. Combined computational, kinetic, and structural data lead us to propose an isomerization mechanism catalyzed by a network of buried waters. Our findings highlight a specific role of water molecules in both CRALBP-assisted specificity towards 9-cis-retinal and its thermal isomerase activity yielding 9,13-dicis-retinal. Kinetic data from two point mutants of CRALBP support an essential role of Glu202 as the initial proton donor in this isomerization reaction.
Introduction
Retinal is the photon-absorbing partner of opsins in cone and rod cells of vertebrate eye retina. Absorption of a photon is coupled to cis-trans photoisomerization in the retinylidene moiety triggering conformational changes in the opsin core.1, 2 Regenerative trans-to-cis recycling occurs in the rod and cone cells and in the neighboring retinal pigment epithelium (RPE) and Müller cells, respectively.3, 4 The key step in the canonical visual cycle is the trans- to cis isomerisation catalyzed by RPE65 in the RPE.5 Recently, DES1, another isomerase has been implicated in the alternative cone visual cycle.6 Both these enzymes can produce substantial quantities of cis-retinoids other than 11-cis-retinal in vitro.6, 7 Their in vivo selectivity is probably controlled by mass action through interacting retinoid-binding proteins.8, 9
Cellular retinaldehyde-binding protein (CRALBP) chaperones 11-cis-retinoids for the conversion of opsin receptor molecules into photosensitive visual pigments. CRALBP is a water-soluble protein that specifically binds cis-retinoids with high affinity. Dissociation constants for 11-cis-retinal/ol (Kd ~ 21 nM/53 nM) and 9-cis-retinal (Kd ~ 51nM) have been reported.10, 11 By trapping these retinals, CRALBP protects the retina from side reactions of the chemically reactive aldehyde group.12 Simultaneously, it preserves the light-sensitive cis-chromophore from premature photoisomerization in the constantly illuminated eye. Because 11-cis-retinal is inherently unstable, this chromophore is essentially unsuitable for sustained replacement therapy.13 A solution to this dilemma is provided by the more conformationally stable 9-cis-retinal which allows larger quantities of iso-rhodopsin to exist in photoreceptors, thereby probably reducing the activity of the retinoid cycle.14 9-cis-Retinal has been successfully administered to animal models to restore visual function.15, 16 Despite the fact that the principal components of the classical visual cycle are known, a satisfactory explanation for cis-retinoid redundancy in retinoid-recycling has not been established at the molecular level.17 A full structural understanding of both the CRALBP:11-cis-retinal and CRALBP:9-cis-retinal complexes and their intermediates is therefore crucial for improving retinoid replacement therapies of inherited human blinding diseases. Numerous retinal pathologies are associated with mutations of the human gene encoding CRALBP (RLBP1) including autosomal recessive retinitis pigmentosa, night blindness, Bothnia dystrophy, retinitis punctata, Newfoundland rod-cone dystrophy, and the less severe phenotype of delayed “dark adaptation”.18, 19
We recently reported the crystal structures of both WT CRALBP and its Bothnia disease mutant R234W in complex with the highly abundant endogenous ligand 11-cis-retinal.20 Here, we investigated the molecular basis of CRALBP’s high specificity for the less abundant natural isomer 9-cis-retinal and report evidence for the first time that, when bound to CRALBP, 9-cis-retinal undergoes trans-cis isomerization to 9–13-dicis-retinal. The process is thermally driven, and does not require light absorption. To understand the mechanistic aspects of this phenomenon, we solved crystal structures of 9-cis-retinal in complex with both CRALBP and the Bothnia mutant R234W. Analysis of these X-ray structures revealed the presence of a binding mode for 9-cis-retinal distinct from that of the 11-cis conformer. In particular, binding of 9-cis-retinal was associated with presence of few ordered water molecules in the binding cavity. These formed a hydrogen-bond network connecting the aldehyde group of retinal to the side-chain of Glu202. Detection of a strong kinetic isotope effect in the process of isomerization leads us to propose a reaction mechanism involving catalytic proton shuttling. CRALBP’s secondary activity as a thermal isomerase of 9-cis-retinal yielding 9,13-dicis-retinal could imply a role in metabolic pathways not directly associated with the visual cycle.
Experimental section
Materials
11-cis-Retinal was obtained from the National Eye Institute, National Institutes of Health (NEI/NIH) and 9-cis-retinal was purchased from Sigma-Aldrich. Retinoids were dissolved in ethanol and kept at −20°C. D2O as well as CD3Cl were obtained from Cambridge Isotope Laboratories. Solvents for HPLC and spectroscopic experiments were of HPLC grade or better. Unless otherwise specified, procedures involving retinoids were performed under dim red illumination at 4°C.
Cloning, Expression and Purification of CRALBP
We used the pET28a(+) vector containing the RLBP1 gene as a template for introducing point mutations of CRALBP following the Quickchange protocol (Stratagene). For this PCR was carried out with primers 5’-CTTGCAGGCATtcTGCTTCATCCTGGAGAAGCTGCTGGAG-3’ and 5’-CCAGGATGAAGCAgaATGCCTGCAAGATCTCATCATCAAAGG-3’ or 5’-GGAAACTCAAATCAATGGCTTCTGTATCATTcAGAACTTCAAGG-3’ and 5’-GCTGCATGGTAAAGCCCTTGAAGTTCTgAATGATACAGAAGC-3’ containing the Y180F or the E202Q mutation, respectively. Mutagenized positions are denoted in lower-case. Resulting clones were confirmed by sequencing (Microsynth, Switzerland).
Expression and purification of recombinant CRALBP, R234W, E202Q and Y180F were carried out as previously described.20 Briefly, the corresponding pET28a vector constructs were transformed into E.coli Lemo21 (DE3) cells (New England Biolabs). Cells were cultured overnight with agitation at 37°C in 100 ml of LB medium (5 g/l NaCl, 5g/l yeast extract, 10 g/l tryptone) containing 50 µ g/ml kanamycin and 34 µg/ml chloramphenicol. For protein production, cultures were inoculated with 2.5 l of low salt LB medium (5 g/l yeast extract, 10 g/l tryptone) supplemented with a 50 fold dilution of 5052-stock solution (25% v/v glycerol, 2.5% v/v glucose, 10% v/v α-lactose), and a 20- fold dilution of a NPS-stock solution (0.5 M (NH4)2SO4, 1 M KH2PO4, 1 M Na2HPO4), 1 mM MgSO4, 50 µg/ml kanamycin, 34 µg/ml chloramphenicol, 0.5% v/v of trace metal solution (5 M HCl containing 36 mM FeSO4, 8 mM ZnSO4, 4 mM CuSO4, 2 mM MnSO4, 0.6 mM Na2B4O7, 14 mM CaCl2, 0.08 mM (NH4)6Mo7O24), and 2 µg/ml of both biotin and thiamine. Expression of both recombinant proteins was carried out in an INFORS HT Minifors bioreactor. Cells were continuously fed with a lactose solution (22% v/v α- lactose, 50% v/v glycerol, 0.46 M urea) once the medium reached an oxygen threshold of 12.5% and a pH of 6.83. After 24 h, cells were harvested by centrifugation at 5000×g for 25 min and 50 g batches of cells were resuspended in 400 ml of lysis buffer (20 mM Tris-HCl, pH 7.4, 100 mM NaCl, 20 mM imidazole). Cells were disrupted by homogenization and the lysate was centrifuged at 50,000×g for 30 min. (His)6-tagged proteins were purified from the supernatant by affinity chromatography on 20 ml of Ni-NTA SUPERFLOW (Qiagen). The lysate was loaded onto the column, washed with 20 column volumes of lysis buffer, and purified protein was eluted in 70 ml of elution buffer (20 mM Tris-HCl, pH 7.4, 100 mM NaCl, 200 mM imidazole). Protein concentrations in the eluate were 3–4 mg/ml as determined by absorbance at 280 nm using NanoDrop (ThermoScientific). For crystallisation experiments, the (His)6-tag was cleaved by adding 20 units of thrombin protease (GE Healthcare) to 20 ml of eluate at 4°C overnight and the digested eluate was reloaded on Ni-NTA to remove the tag. Ligand loading of affinity purified CRALBP and the R234W mutant was carried out by adding aliquots of a 20 mM 9-cis-retinal stock dissolved in ethanol to the protein solutions at a 1.5 molar excess with a final ethanol concentration of 2% v/v. Mixtures were concentrated by a Vivaspin 15R Hydrosart (Sartorius) to 30–50 mg/ml of protein with three washes in low salt buffer (20 mM Tris-HCl, pH 7.4). Desalted samples were centrifuged at 16,000×g for 10 min and loaded onto a self-packed 8 ml PorosHQ column (Applied Biosystems) previously equilibrated with low salt buffer. CRALBP proteins were eluted over a 40% v/v gradient of high salt buffer (20 mM Tris-HCl, pH 7.4, 1 M NaCl). Peak fractions representing monomeric ligand–protein complexes were concentrated and re-equilibrated in low salt buffer to a final concentration of 15 mg/ml of protein.
Protein Crystalization and Data Collection
Hexagonal crystals of wild-type CRALBP in complex with 9-cis-retinal were obtained at 18°C by hanging-drop vapour-diffusion. Drops were set up by mixing either 1, 2, or 3 µl of CRALBP:9-cis-retinal (15 mg/ml) with 1 µ l of reservoir solution (1.2 M K/Na tartrate, 0.2 M Li2SO4, 0.1 M Tris-HCl, pH 8.5) and were equilibrated against an 80 µ l reservoir solution. Crystals of 0.3×0.07×0.07 mm size were obtained after 3 days. These crystals belonged to space group P6522 with a = 71.9 Å, b = 71.9 Å, c = 303.2 Å; α = 90°, β = 90°, and γ = 120°. Before data collection, crystals were flash-cooled in a nitrogen stream at 110 K in the crystallization solution containing 15% v/v glycerol. Data were collected at the SLS synchrotron beamline X06DA PXIII (PSI Villigen) at 100 K, with a P2M pixel detector. The crystal- to-detector distance was 340 mm and the oscillation angle was 2° per frame.
Crystals of R234W CRALBP in complex with 9-cis-retinal were obtained by mixing 1 µL of protein- ligand solution (20 mg/ml) with 1 µL of reservoir solution (20% w/v PEG 3000, 0.1 M Hepes, pH 7.5, 0.2 M NaCl). After 2–3 weeks, monoclinic crystals were observed. These grew to an average size of 0.05 mm×0.05 mm×0.002 mm. These crystals belonged to space group C2 with a = 87.9 Å, b = 57.9 Å, c = 75.2 Å; α = 90°, β = 122.8°, and γ = 90°. Before data collection, crystals were flash-cooled in a nitrogen stream at 110 K in crystallization solution containing 15% v/v glycerol. Data were collected at the SLS synchrotron beamline X06DA PXIII (PSI Villigen) at 100 K with a P2M pixel detector. The crystal-to-detector distance was 156 mm, and the oscillation angle was 1.5° per frame.
Structure Solution and Refinement
The structure of R234W:9-cis-retinal was solved by molecular replacement with the structure of R234W:11-cis-retinal (PDB code 3HX3) used as a model. Following molecular replacement, ligand information was obtained from the PubChem Compound Database (http://pubchem.ncbi.nlm.nih.gov/) and 9-cis-retinal (entry CID 6436082) was added to the structure with the LigandFit module of the PHENIX program package.21 The structure then was refined to 1.9 Å by using the phenix.refine module of the PHENIX program package. The final model contained residues 57–306, 9-cis-retinal, and 315 water molecules. Procedures for solving the structure of WT CRALBP:9-cis-retinal were essentially the same as for R234W:9-cis-retinal with the exception that molecular replacement was carried out with the atomic model of CRALBP:11-cis-retinal (PDB code 3HY5). This structure was refined to a final resolution of 3.4 Å. Electron density for 9-cis-retinal was clearly visible in the R234W:9-cis-retinal complex whereas electron density for 9-cis-retinal in CRALBP was restricted to the β-ionone ring including the tail region up to atom C9. Therefore, the structure was finally refined with a truncated ligand model using phenix.refine and then evaluated with the model-building program COOT.22 Data collection and refinement statistics for both structures are given in SI Table 1.
Isomerase Activity Assay
To determine rate constants for the isomerisation reaction, CRALBP:9-cis-retinal stock solutions were prepared with 15 mg/ml of protein. Incubations were carried out at 37°C in low salt buffer at pH 7.4. Aliquots for HPLC analysis were removed at various time points during the incubation. To monitor a potential isotopic effect on the reaction rate, exchange of hydrogen to deuterium was carried out by dialysing CRALBP:9-cis-retinal against a D2O-based low salt protein buffer yielding an estimated final D2O concentration of ~95 %.
HPLC and Spectroscopic Analysis
For HPLC analysis samples were denatured by adding equal volumes of ethanol to each CRALBP:ligand complex. Subsequently, 80 µ l of hexane were added, vortexed for 30 s, and samples were centrifuged at 16,000×g for 10 min at 4°C. The hexane phase was carefully transferred into a fresh vial and used for injection. HPLC was carried out on a Jasco PU-980 equipped with a Jasco UV-975 UV detector. Samples were separated on a silica column (5µm, 250 mm×4.6 mm, Agilent Technology) with 98:2 (v/v) hexane/tert-butyl methyl ether as the mobile phase for isocratic elution.23 Peak fractions were collected and the solvent was evaporated under a nitrogen stream. For UV/VIS spectroscopy the residual oil film of dried peak fractions was dissolved in ethanol and analysed with an Evolution Array UV/VIS spectrophotometer (Thermo Scientific).
Hybrid QM/MM simulations
The X-ray structure of the R234W mutant bound to 9-cis-retinal was used as the starting configuration. Titratable groups were protonated at standard positions at pH 7.0 except for residue Glu202, which was modeled either as glutamic acid or glutamate, according to the chemical model for the pocket studied. The AMBER FF0324 and the GAFF25 force fields were used to parameterize the protein and ligands, respectively. The RESP charge fitting procedure26 was used to obtain atomic charges of the retinals from ab initio calculations at the HF/6-31G* level of theory. The system was solvated with ~23,000 TIP3P water molecules27 and eight/nine chloride anions were added to achieve neutrality. The system contained ~ 62,000 atoms in a box of 90.3×93.1×103.2 Å3.
First, 5 ns of classical MD was run to relax the structure. Nose-Hoover chain of thermostats28–30 and Langevin pistons31 were used to keep the system at constant temperature and pressure. The Particle-Mesh Ewald (PME) method32 was employed to treat long-range electrostatics. A time step of Δt = 1.5 fs was used in combination with the SHAKE algorithm33 on bonds involving hydrogens. Simulations were run with the NAMD package (http://www.ks.uiuc.edu/Research/namd). The system then was partitioned into two regions: the QM part, comprising the retinal and the three buried water molecules present in the binding pocket; and the MM part, consisting of the protein, counter- ions, and the other water molecules. The QM region was treated at the DFT level of theory. The exchange-correlation functional was described by the GGA/BLYP approximation.34, 35 Electronic wave functions were projected on a plane-wave basis of functions with a kinetic energy cutoff of 70 Ry. Core-electrons were integrated out by the use of norm-conserving Martins-Troullier pseudopotentials.36 Dispersion interactions were included by using dispersion-corrected atomic-centered potentials (DCACP).37 The classical part was described by the AMBER force–field.24 Interactions between the two regions were treated with the full-Hamiltonian scheme38, 39 implemented in the CPMD code (http://www.cpmd.org).
Three systems were investigated: i) the neutral, closed shell molecule; ii) the carbocation obtained by attachment of H+ at C12; and iii) the radical molecule obtained by attachment of an H atom at C12. In the last case, spin polarized DFT was used to solve the electronic problem. Self- interaction corrections were applied, following ref.40. Car-Parrinello MD simulations41 of each system using a fictitious electron mass of 600 a.u., and a time-step of 0.01 fs were performed. Systems were brought to 300 K by using a velocity-rescaling algorithm, and then cooled down by a simulated annealing procedure with a scaling factor of 0.999. Simulations were repeated multiple times to ensure that the annealed structures corresponded to the global minimum of the energy.
NMR analyses
For NMR analyses all HPLC purified retinoids were dissolved in CDCl3. All NMR data were acquired at a regulated temperature of 298 K with a Bruker AvanceII 500 MHz NMR spectrometer equipped with an inverse 1.7 mm triple channel (1H, 13C, 31P) microprobe head.
1D 1H NMR data were acquired with 512 or 1024 transients into 32k data points over a width of 14 ppm in a classical one-pulse experiment (acquisition time = 2.34 s). A relaxation delay of 6 s was applied between transients. Data sets were zero- filled to 64k points and multiplied by a 0.3 Hz line broadening factor before Fourier transformation.
2D 1H-1H NMR COSY data were acquired over a frequency width of 14 ppm in both F2 and F1 into 2k complex data points in F2 (acquisition time = 147 ms) by using 128 t1 increments. A relaxation delay of 1 s between transients was used for all experiments. The data were recorded using 64 or 128 transients. Spectra were zero filled to 2048×1024 points and processed with a sine-bell function in F2 and in F1. All data are displayed in the absolute value mode.
Results
Discovery of thermal isomerase activity in CRALBP:9-cis-retinal complex
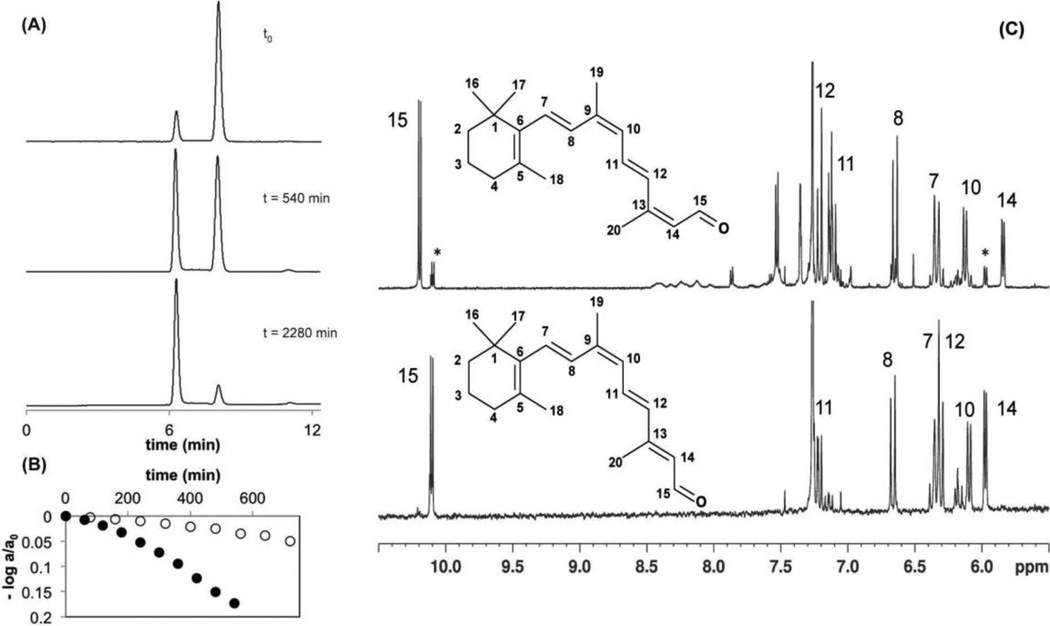
When extracts of freshly prepared complexes of CRALBP:9-cis-retinal were routinely checked for their isomeric purity by HPLC, a contaminating peak other than 9-cis-retinal consistently appeared. This peak eluted slightly earlier than 9-cis-retinal and had a very similar absorption spectrum indicative of a geometric retinal isomer (Figure 1A).
Figure 1. Characterization of a CRALBP:9-cis-retinal reaction product.
A) HPLC traces of retinoid extracts of CRALBP:9-cis-retinal freshly prepared (top), after 9 h incubation at 37°C (middle) and after 38 h incubation at 37°C (bottom). B) Kinetics of CRALBP:9-cis-retinal isomerization in H2O buffer (filled circles) and in D2O buffer (open circles) quantified by HPLC and plotted versus time. Rate constants (k) were determined from the fit of the data to the following equation: log (A380(t)/A380(t0)) = −kt/ln(10), where ΔA(t) = A380(t = 0) −A380(t). C) 500 MHz 1H-NMR spectra of 9,13-dicis-retinal (top) and 9-cis-retinal (bottom) produced by incubation of 9-cis-retinal in CRALBP. Both retinoids were extracted from the complex and dissolved in CDCl3. Allocations of the six vinyl protons 7, 8, 10, 11, 12, and 14, as well as of the aldehyde proton 15 are indicated in the spectra, according to the numbering given in the structures. A second set of resonances visible for protons 14 and 15 and marked with asterisks, indicates traces of residual 9-cis-retinal.
Accounting for 14–15% of total retinal, this peak appeared in the absence of light and oxygen thereby excluding any possible light-driven isomerization or oxidation of 9-cis-retinal. Incubation of freshly prepared CRALBP:9-cis-retinal at 37°C for 9 h revealed a virtual 1:1 ratio of the geometric isomer and 9-cis-retinal whereas prolongation of the incubation time to 38 h at 37°C yielded almost complete conversion of 9-cis-retinal to this isomer. When repeating the experiment using the corresponding complex of R234W with 9-cis-retinal, we observed a rapid equilibration to a 2:3 ratio for 9-cis/9,13-dicis-retinal, that remained virtually unchanged after 48h of incubation at physiological temperature (Figure S1, Supplementary information).
When the progress of the CRALBP:9-cis-retinal reaction was monitored by HPLC at various time points, the reaction was found to be (pseudo) first order with a rate constant of k = 1.1×10−5 s−1 and a half- life for CRALBP:9-cis-retinal of t½ = 17.5 h. When the reaction was carried out in D2O buffer under otherwise identical conditions, a kinetic rate constant of k = 0.23×10−5 s−1 was obtained. In both cases these measurements were repeated on two independent data sets, yielding a systematic correlation coefficient for the linear fitting of ≈ 0.95. The kinetic isotope effect of 4.8 suggested an isotopic substitution in a chemical bond of 9-cis-retinal within CRALBP that is directly involved in the rate-limiting step of this reaction (Figure 1B).
The identity of the geometric retinal isomer formed by the thermal process within the cavity of CRALBP was confirmed initially by UV/VIS spectroscopy of the complex and verified in organic solvent extracts (Figure S2). Freshly prepared CRALBP:9-cis-retinal complex had an absorption maximum at 415 nm. After incubation of the complex for 38 h at 37°C its absorption maximum revealed a blue-shift towards 400 nm. Extraction and HPLC purification of the corresponding free retinal isomers before and after incubation revealed absorption maxima of 373 nm and 368 nm, respectively. These values were identical to the ones previously reported for 9-cis-retinal and for 9,13-dicis-retinal.42 Control experiments were carried out to rule out the possibility of spontaneous isomerization of 9-cis-retinal in the absence of protein (Figure S3). Thus, when HPLC-purified 9-cis-retinal was dissolved in low salt buffer containing 2% ethanol at pH 7.4 and incubated for 24 h at 37°C in the dark without oxygen, no spontaneous formation of geometric isomers of 9-cis-retinal was detected by HPLC. Moreover, when CRALBP:11-cis-retinal was incubated under similar conditions but for 48h, no thermal isomers of 11-cis-retinal were observed.
Analyses of the 1H-NMR (Figure 1C and Figure S4) and 2D 1H-1H NMR COSY spectra (Figures S5-S8) recorded with HPLC purified samples of extracts from CRALBP containing 11-cis-retinal, 9-cis-retinal or the putative 9,13-dicis-retinal permitted an unambiguous attribution for the 1H-NMR resonances of the vinyl protons 7, 8, 10, 11, 12, 14, and of the aldehyde proton 15. In addition, differentiation between 9-cis-retinal, 11-cis-retinal and 9,13-dicis-retinal was enabled by use of simulated 1H-NMR spectra (Figures S9-S11). The structure of 9,13-dicis-retinal formed through thermal isomerization of 9-cis-retinal bound to CRALBP could thus be unambiguously confirmed. For example, according to the 1H-NMR simulations, proton 12 should be strongly low- field shifted in 9,13-dicis-retinal compared to 9-cis- and 11-cis-retinal. This strong low- field shift was experimentally confirmed: proton 12 resonated at 5.93 and 6.10 ppm in 9-cis-retinal and 11-cis-retinal, respectively, whereas it was low-field shifted by about 1 ppm in the 1H-NMR spectrum of 9,13-dicis-retinal where its resonance appeared at 7.21 ppm. Likewise, the 1H-NMR spectra, as well as the simulations, clearly revealed a trans conformation for the C12-C13 bond, thus ruling out the formation of 11-cis-retinal or 11,13-dicis-retinal.
1H NMR spectra of 9,13-dicis-retinal extracts from CRALBP samples incubated in H2O and in D2O buffer were recorded to gain further information about the reaction mechanism (Figure S12). Comparison of these spectra clearly indicated that substitution of a proton by a deuterium at position 14 could not be validated. Indeed, both spectra were identical, especially the resonances of both protons 14 and 15. If a deuterium substitution had occurred, the resonance of proton 14 would have vanished and that of proton 15 would have appeared as a singlet.
Structures of CRALBP and its Bothnia mutant R234W with 9-cis-retinal
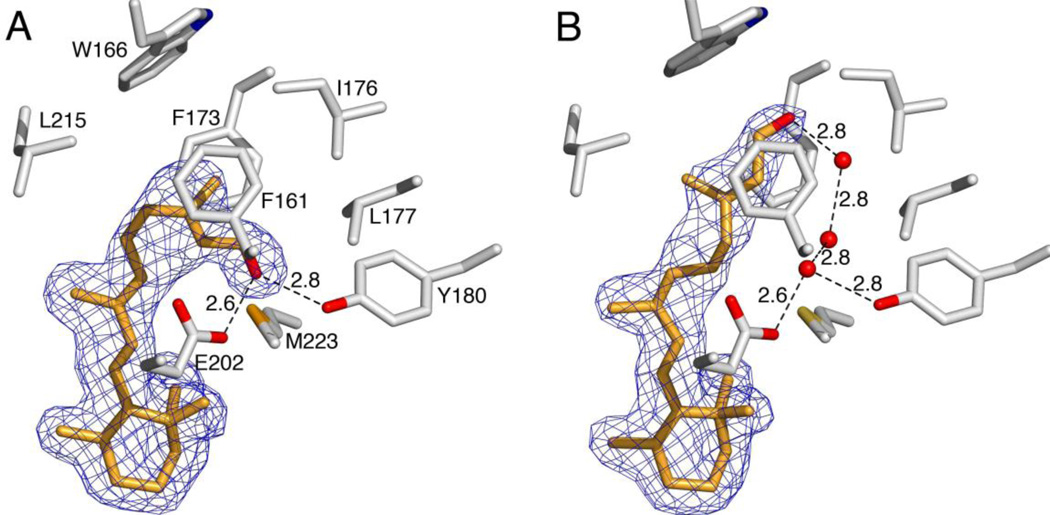
We solved the crystal structures of CRALBP and its Bothnia disease mutant R234W, both in complex with the less abundant natural isomer 9-cis-retinal. Details about data collection, refinement, and validation statistics are reported in Supporting Information (Table S1). Excellent resolution of the R234W:9-cis-retinal complex provided a detailed picture of the retinoid conformation and allowed a direct comparison with the known binding-site of R234W:11-cis-retinal. Electron densities of all ligand carbons from the β-ionone ring to the terminal aldehyde (C1–C15) were well defined in both complexes. In the R234W:11-cis-retinal complex the ligand was bound in a bent conformation (Figure 2A) with the terminal aldehyde occupying a hydrophilic pocket formed by Glu202 and Tyr180. In contrast, 9-cis-retinal adopted an elongated conformation within R234W (Figure 2B).
Figure 2. Close-up views of the R234W ligand-binding pockets.
A) R234W bound to 11-cis-retinal. B) R234W bound to 9-cis-retinal. Side chains (gray) that form van der Waals contacts with the cis-retinoid tails (orange) or participate in hydrogen bonding within the ligand-binding pockets are shown as sticks. The 2Fo–Fc electron densities for the ligands are shown as blue mesh at 1 σ. Dashed black lines indicate hydrogen bonds; distances are given in A.
The β-ionone ring conserved its position whereas the aldehyde-tail pointed towards an alternative niche within the binding-pocket formed by Trp166, F161, and F173. Interestingly, in this 9-cis-retinal complex the expected 11-trans bond was severely strained with a 90° twist suggesting at least partial loss of conjugation. But the most distinctive feature of the R234W:9-cis-retinal complex was a hydrogen-bonding network that included three ordered water molecules that connect Tyr180 and Glu202 to the aldehyde oxygen of 9-cis-retinal. In fact with 11-cis-retinal, the resulting binding pocket of R234W is completely dehydrated.20 Glu202 and Tyr180 trap a single water molecule through hydrogen bonding interactions precisely at the position occupied by the aldehyde oxygen in the R234W:11-cis-complex. The two other water molecules line up in a string along the retinoid-backbone stabilizing the aldehyde oxygen of 9-cis-retinal through coupled hydrogen-bonding interactions. This water network provides a means to compensate for ligand- induced rearrangements throughout the binding-pocket and provides the structural basis for the alternate 9-cis-/11-cis-retinal binding in the R234W mutant.
Recent studies have reported a high stereospecific preference for 11-cis-retinal and 9-cis-retinal by CRALBP and its mutant R234W as well as a lack of binding to either 13-cis-retinal or all-trans-retinal.43 Based on previously determined X-ray structures of CRALBP complexes, this geometric specificity for cis-retinoids was primarily assigned to shape complementarities between the binding pocket and ligand.20 However, complexes of CRALBP bound to either 11-cis-retinal or 9-cis-retinal have thus far yielded very poorly defined electron density maps of the retinal tail region (C11–C15) (Figure S13). These X-ray structural models of CRALBP implicated the presence of multiple conformations for the polyene tail region that did not allow its precise assignment within the cavity.
The above lack of information has been a major obstacle to a detailed molecular understanding of the alternate retinoid-binding modes manifested by CRALBP. However, computational studies based on classical MD simulations allowed us to confirm both the above described alternate binding mode of CRALBP to either 11-cis-retinal or 9-cis-retinal, and a not fully dehydrated cavity for the CRALBP:9-cis-retinal complex.44
Hybrid QM/MM simulations
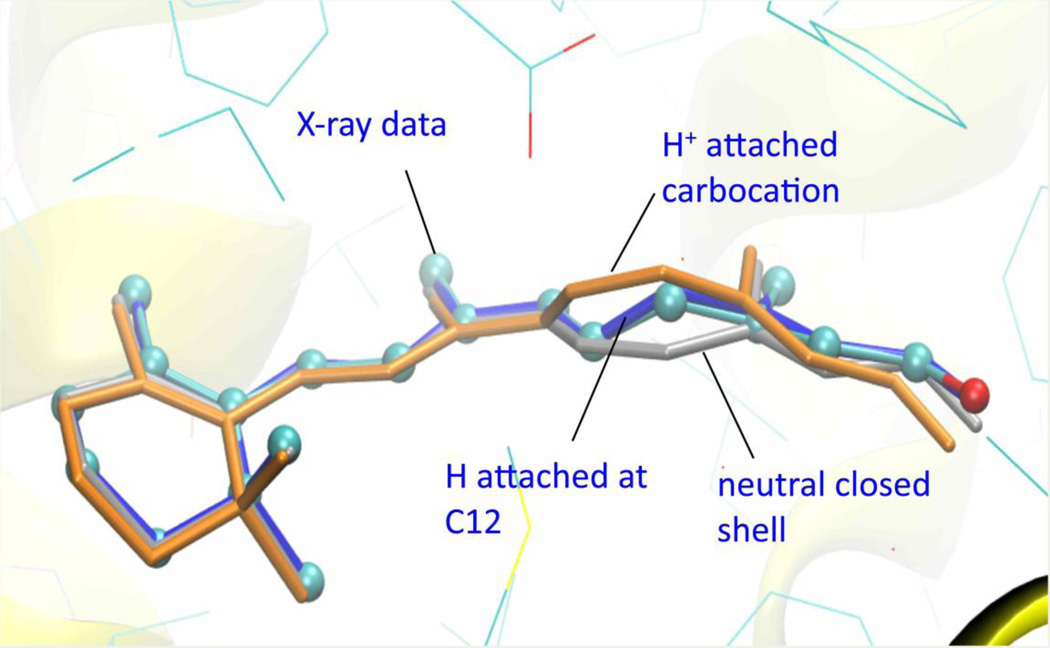
The X-ray structure of R234W:9-cis-retinal showed a strong geometrical distortion of the retinal at the C12 position suggesting a change in its hybridization. We tested three possible explanations for this finding by hybrid QM/MM simulations: i) a neutral, closed shell structure, ii) attachment of a H+ ion at C11/C12, or iii) trapping of an H• atom at the same position. Optimized geometries obtained from repeated simulated annealing cycles are shown in Figure 3, and compared with the atomic positions predicted by the experimental electron density.
Figure 3. Superposition of different computational models of 9-cis-retinal in R234W.
The retinal model fitting the experimental X-ray density is shown in cyan balls-and-sticks. The QM/MM optimized geometry obtained for the neutral closed-shell molecule is drawn in silver licorice; the carbocation with H+ attached at C12 in orange licorice; and the optimized structure of the radical obtained by H• capture at C12 in blue licorice.
Our calculations clearly show that the structure characterized by hydrogen attachment at C12 fits the experimental data significantly better than the two other hypotheses (Figure 3). Specifically, the RMSD between the retinal structure obtained by fitting the crystallographic electron density map and those of the different models was 0.66 Å for the H+ attached system, 0.33 Å for the neutral closed-shell molecule, but only 0.11 Å for the H• attached model. This last model fits the distorted C10-C11-C12-C13 dihedral angle especially well, featuring a calculated value of −76.9° compared to −71.8° in the X-ray structure. In contrast, the neutral closed-shell and cation species provided values of +4.8° and −10.0°, respectively for the same angle. A possible source of hydrogen radicals in the binding pocket is the hydroxyl group of Tyr180. However, Tyr180 sits relatively far from the retinal C12, thus making a direct hydrogen transfer improbable. Most likely the process is associated with X-ray- induced formation of solvated electrons in the protein crystal at 100K, which are captured by retinal. This possibility is supported by the pronounced color change from bright yellow to deep red observed in all our CRALBP:retinal crystals during X-ray exposure. The resulting radical anion is likely to be rapidly protonated to form the neutral radical species predicted by our QM/MM calculations. An analogous coupled electron-proton transfer mechanism was previously described for the pulse radiolysis-induced characteristics of free retinyl polyenes, which display pronounced red shifts and a major decay pathway through protonation in acidic solvents.45 In our case, migration of the proton is possibly mediated by the water network present in the binding pocket, and we suggest Glu202 as its primary acidic source.
Discussion
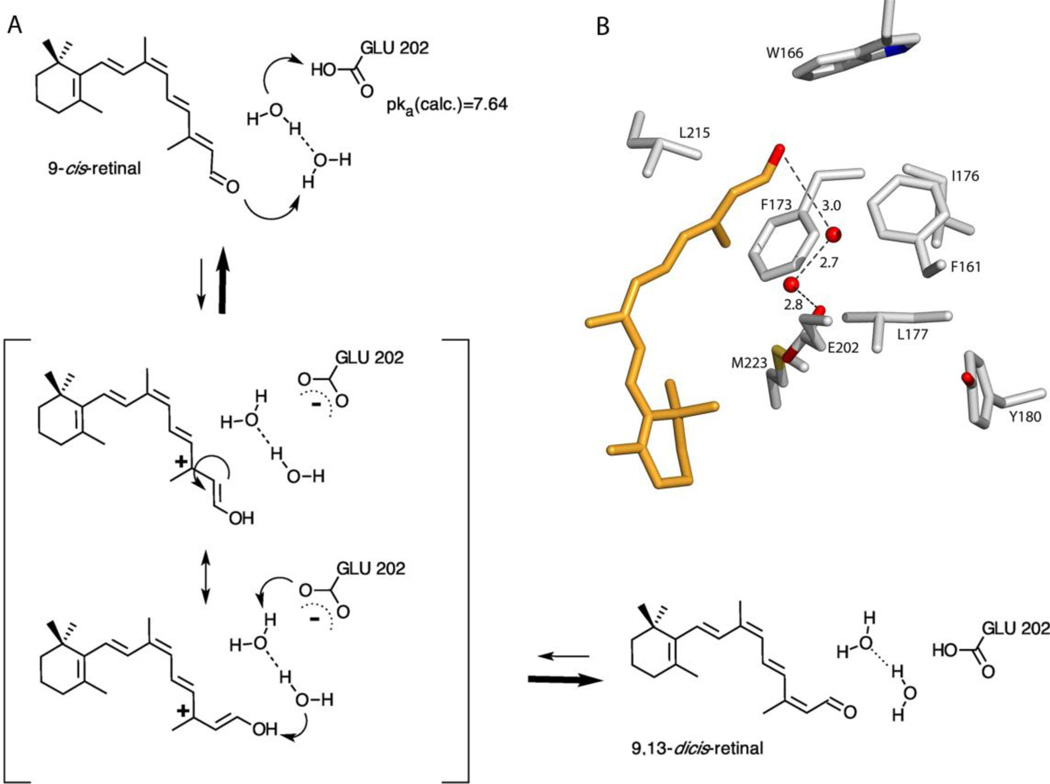
CRALBP is the major chaperone of freshly synthesized 11-cis-retinol/al in the RPE and Müller cells. It binds 11-cis-retinol/al with nanomolar affinity in the regenerative branch of the visual cycle and thus provides selectivity to the eye for the major visual chromophore.8–11 The geometric specificity of CRALBP is complemented by a steep concentration gradient towards the photoreceptors generated by the essentially irreversible binding of opsin to 11-cis-retinal. Surprisingly, CRALBP also binds with nanomolar affinity to the low abundance chromophore 9-cis-retinol/al.10 Here we provide compelling structural and functional evidence that CRALBP exhibits multiple functions including its catalytic isomerization activity. We have observed that 9-cis-retinal in complex with CRALBP upon incubation at 37°C slowly changes into a new chemical species identified by both UV/Vis and 1H-NMR spectroscopy as 9,13-dicis-retinal (Figure 1). This previously undetected isomerase function occurs in the dark, thus no photo-physics can be involved. The isomerase function was specific to 9-cis-retinal in that 11-cis-retinal was not affected. In addition, repetition of the experiment in deuterated water showed a kinetic isotope effect of 4.8. In the case of 11-cis-retinal, ordered water is completely excluded from the binding pocket of R234W.20 The X-ray structural model of the R234W:9-cis-retinal complex unexpectedly revealed a string of three buried waters in the binding pocket that hydrogen-bonded Glu202 to the aldehyde oxygen of the polyene. Intriguingly, the detected water structure strongly resembles the classic proton-wire observed in catalytically active proteins46–48 and also in nanotubes.49, 50 MD simulations based on the X-ray structures presented helped us determine the CRALBP:9-cis-retinal complex conformation (Figure 4B).44 In contrast to the string of three water molecules visible in the experimental binding-pocket of R234W:9-cis-retinal, the MD complex of CRALBP displayed two uncoordinated water molecules connecting Glu202 to the aldehyde. From this data, we propose a mechanism of reversible acid-catalyzed protonation of the aldehyde moiety by proton transfer via two water molecules leading to formation of a carbocation at C13 prior to isomerization (Figure 4A).
Figure 4. Mechanism of the isomerization reaction.
A) Proposed mechanism for the thermal isomerization of 9-cis-retinal to 9,13-dicis-retinal in CRALBP. The reaction proceeds via direct protonation of the polyene aldehyde, isomerization of the charge separated transition state and final deprotonation of the enol. B) Close-up view of the MD structural model of the dark state binding pocket of CRALBP:9-cis-retinal.44 Side chains and 9-cis-retinal are shown as gray and orange sticks, respectively. Dashed black lines indicate hydrogen bonds. Distances are given in A.
The strong isotope effect in the reaction kinetics confirms that the rate- limiting step must be a proton transfer event. The 1H-NMR isotope labeling experiment did not evidence accumulation of deuterated species in the product, strongly suggesting an enol cation as the intermediate protonated species. In this case, the same catalytic proton is transferred back after rotation of the C13-C14 bond.
Active protonation of the 9-cis-retinal polyene chain depends on the presence of a suitable proton donor in the protein system. Solving the linearized Poisson Boltzmann equation,51 we calculated a pKa of 7.64 for Glu202. This value would allow efficient proton shuttling onto the aldehyde oxygen via a water-mediated hydrogen bond pathway. Proton transfer could then occur either by a direct Grotthus- like mechanism, or by formation of a transient (H5O2)+ Zundel cation involving the two buried waters.52, 53 Protonation of the aldehyde moiety would complete the charge separation of the intermediate and stabilize the carbocation at the most substituted C13 atom. This would allow isomerization to proceed around the C13-C14 bond, followed by back-deprotonation of the retinal cation, thereby establishing the polyene configuration as 9,13-dicis-retinal.
The retinal-binding site of CRALBP appears to be a tight cavity that permits only limited retinoid motion primarily by restricting the longitudinal space for binding. This can be inferred by the geometric retinoid specificity of CRALBP that precludes binding of either all-trans- or 13-cis-retinal.54 Nevertheless, from the evidence of alternate binding modes reported here for 11-cis-retinal and for 9-cis-retinal or 9,13-dicis-retinal respectively, we infer that rotations of the polyene tail around the four terminal bonds must be sterically allowed. Data concerning I2-catalyzed reversible isomerization of free retinals show a 2:1 ratio for 9-cis/9,13-dicis-retinal,55 implying that free 9,13-dicis-retinal is higher in free energy than the 9-cis isomer. Nonetheless, we observed a constant accumulation of 9,13-dicis-retinal leading to an essentially quantitative conversion of 9-cis-retinal. Thus, CRALBP induced an irreversible transformation of this retinoid. In contrast, in the corresponding complex of R234W with 9-cis-retinal we found a rapid equilibration to a 2:3 ratio for 9-cis/9,13-dicis-retinal, that remained virtually unchanged after 48 h of incubation at physiological temperature (Figure S1). This means that the isomerization kinetics of the mutant protein is both faster and reversible. The fact that the ratio of the two isomers in R234W is qualitatively different from that of the free species could imply that the tighter cavity of the mutant protein favors the more compact dicis-retinal.
The appearance of ordered water molecules in the very hydrophobic cavity of CRALBP implies a general stochastic exchange mechanism from bulk solvent associated with ligand recognition and subsequent relaxation of the ligand-binding pocket. Both X-ray and MD models show subtle differences in the hydration pattern of various CRALBP:retinal complexes.44 This suggests that modulation of the hydrogen-bonding network in the binding cavity may have pronounced effects on the kinetics of isomerization, similar to what has been reported in several other systems of chemical and biochemical relevance.56–60
The absence of isomerization in 11-cis-retinal can be explained by an already optimal complementarity between the shape of the pocket and conformation of the ligand. The complex is also stabilized by the hydrogen-bond between Glu202 and the retinal carbonyl. This interaction would be disrupted by the isomerization of the 13-trans bond.
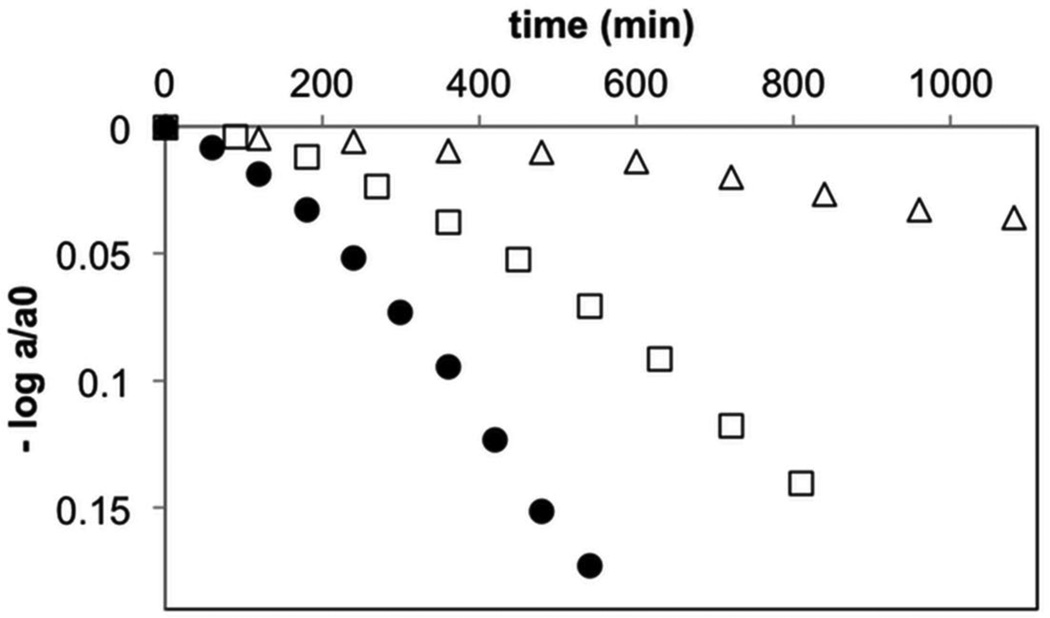
To assess both the mechanism and the relevance of disorder within the binding cavity on the reaction kinetics, we expressed two mutants of CRALBP, namely E202Q and Y180F. The first mutation should eliminate the primary source of catalytic protons from the binding cavity and thus block the isomerase activity. Previous MD simulations showed that Tyr180 helps to structure the H-bond network formed by Glu202 and the catalytic waters.44 Therefore, the Y180F mutation should increase disorder within the binding cavity, resulting in less efficient, albeit still present, catalysis. Figure 5 reports the rate of isomerization for these two mutants compared to that of CRALBP. As expected, E202Q practically lost all catalytic activity, apart from a very weak basal signal (9% of the original rate), probably attributable to the weakly acidic side-chain of Tyr180. Y180F had intermediate isomerase activity, but with a significant decrease in reaction rate (52%) as compared to CRALBP (Figure 5).
Figure 5. Comparison of the reaction kinetics of CRALBP, E202Q and Y180F.
Kinetics of CRALBP:9-cis-retinal isomerization (filled circles), E202Q (open triangles) and Y180F (open squares) quantified by HPLC and plotted versus time. Rate constants (k) were determined from fitting the data to the following equation: log (A380(t)/A380(t0)) = −kt/ln(10), where ΔA(t) = A380(t = 0) − A380(t). k = 1.1×10−5s−1 (CRALBP:9-cis-retinal); k = 0.10×10−5 s−1, (E202Q), and k = 0.57×10−5 s−1 (Y180F).
General features of thermal trans/cis retinoid isomerization have been characterized in a number of protein systems. Retinal is a typical polyene with an isomerization barrier of ~48 kcal/mol making a spontaneous conversion improbable. Upon protonation, however, the retinal cation exhibits a lowered isomerization barrier in the range of 11.5 kcal/mol due to weakening of bond alternations.61 Indeed, thermal isomerization of all-trans-retinal to 13-cis-retinal has been reported to occur within bacteriorhodopsin during dark adaptation.62 In this well-studied protein system, a strong interdependence of retinal Schiff base protonation and lowering of the isomerization barrier was reported.63 The rate constant for dark adaptation in this protein was also shown to correlate positively with the fraction of the Asp85 protonated side chain64
Specifically, the mechanism of thermal all-trans to 13-cis isomerization of the chromophore in bacteriorhodopsin requires transient formation of an unprotonated Schiff base, which constitutes the reactive isomerizing species.64–66 We noted that this unprotonated Schiff base is isoelectric to the protonated cationic retinal that we observed as the isomerizing species in CRALBP. The crucial difference between bacteriorhodopsin and CRALBP is due to the chemical nature of the bound chromophore, the covalently-bound protonated Schiff base and free retinaldehyde, respectively. Therefore, in the acid/base reaction initiating isomerization in the two systems, the chromophore has to play opposite roles as either an acid or base. In the case of bacteriorhodopsin, the role of solvated excess protons in the internal cavity, represented by a Zundel cation, was shown to be critical to the transient protonation network.46 This is in agreement with the presence of under-coordinated water molecules in the CRALBP:9-cis-retinal cavity. Altogether, these findings suggest that the prototypic all-trans to 13-cis thermal isomerization of the retinal Schiff base in the dark adaptation cycle of bacteriorhodopsin critically depends on the balanced interplay of reversible deprotonation/reprotonation of an acidic amino acid side-chain, a protonated polyene and a Zundel- like cation. All three elements are presented in a different functional context in the cavity of CRALBP:9-cis-retinal suggesting a convergent evolutionary background for thermal trans/cis isomerization of retinal in a protein system.
The role of 9-cis-retinol/al in the mammalian eye has yet to be clarified. Several studies have addressed the question as to the general mechanism for slowly accumulating 9-cis-retinal in the absence of light and its physiological significance. In 1973 Futterman and Rollins provided an initial model for the non-enzymatic formation of 9-cis-retinal and 13-cis-retinal from all-trans-retinal through the action of endogenous nucleophiles.67, 68 It was noted that free dihydroriboflavin sufficed to catalyze 9-cis-retinal formation from all-trans-retinal at physiologic concentrations in vitro. More recently, an additional non-enzymatic mechanism for the in vitro and in vivo formation of 9-cis-retinol from all-trans-retinol was proposed.12 In that study 9-cis-retinol was shown to accumulate from its relative resistance compared with the other geometric cis- isomers to the non-enzymatic back-isomerization to all-trans-retinol. Accordingly, incubating non-transfected 293-F cells with all-trans-retinol for several hours evidenced accumulation of 9-cis-retinol (1.6%) besides 13-cis-retinol (5.3%) and 11-cis-retinol (0.12%).69 Overall, these observations implicate thermal uphill isomerization in the presence of nucleophiles as a possible in vitro and in vivo source of slowly accumulating 9-cis-retinol/al in membranes.
Alternatively, 9-cis-retinol may arise from the eye’s ability of endothermic reformation of 11-cis-retinol from all-trans-retinol through the visual cycle. The protein RPE65 has been shown to be essential for the function of the RPE cycle in rod-dominated vision by catalyzing the isomerization of all-trans-retinyl esters to yield 11-cis-retinol.70–72 Several lines of evidence support a carbocation mechanism for the isomerase activity of RPE65. The mechanism is consistent with a loss of bond order of the polyene substrate and the propensity of the enzyme to yield 13-cis-retinol and 11-cis-retinol in vitro.69, 73, 74 Recently a second isomerase (DES1) was discovered with a putative in vivo function in cone-dominated vision by acting directly on alltrans-retinol.6 DES1 is a transmembrane enzyme affecting the isomerization equilibrium of all-trans-retinol to cis-retinol species (including 9-cis-retinol) at inverse ratios to their free energies. CRALBP was shown by co- immunoprecipitation to physically interact with DES1 and could have a role in directing isomerization by mass action similar to that it has with RPE65.6 This indicates a high in vivo chemical selectivity for 11-cis-retinoids of the visual cycle resulting from conjunction of the involved isomerases RPE65 and DES1 with downstream factors such as CRALBP, retinol dehydrogenases and opsins.
Although it is rather unlikely that cis-retinoids other than 11-cis-retinal are relevant for the properly working eye, traces of 9-cis-retinal or 9,13-dicis-retinal would still be chaperoned by CRALBP into receptor cells to be turned over in the light reaction.75 Recently, low amounts of 9-cis-retinal (5%) have been reported in the absence of light to accumulate specifically in an animal model with disrupted 11-cis-retinal production (RPE65−/- mice). Moreover this accumulation was associated with a minimal visual response through the formation of isorhodopsin.14 The suitability of 9,13-dicis-retinal to act as surrogate for 11-cis-retinal has been demonstrated as well by its ability to reconstitute photosensitivity in bleached skate retinas by forming isorhodopsin- II.42 Overall, the fact that 9-cis-retinal is a suitable surrogate for 11-cis-retinal in vivo has led to development of synthetic 9-cis-retinoids as orally administered, pro-drug forms of 9-cis-retinal for the treatment of various forms of inherited retinal degeneration caused by defects in the retinoid cycle.13 These findings indicate that sustained delivery of 9-cis-retinal restores visual function in vivo.15
Surprisingly, no significant amounts of 9-cis-retinol/al were detected in CRALBP-knockout mice displaying heavily impaired regeneration of 11-cis-retinal and delayed dark adaptation76. However, the isomer-composition of accumulating retinyl-esters was not reported in that study. In the absence of CRALBP, mass transfer would be expected to be strongly impaired in these animals causing significant back-esterification of cis-retinols produced by RPE65.
Other than its general role as a chaperone in vision, several lines of evidence support the involvement of CRALBP in non-image- forming functions. Most importantly, its expression has been reported in the ciliary body, cornea, pineal gland, optic nerve, brain, and iris.77 In zebrafish, CRALBP expression was shown to be strongest in Müller end feet directed toward the ganglion cell layer of the inner retina and also in the pineal gland during larval development.9 A possible non- image- forming function of CRALBP could be inferred from the fact that melanopsin, the only opsin in the intrinsically photosensitive retinal ganglion cells (ipRGCs), has been shown to bind exclusively to cis-retinal species.78 CRALBP may thus chaperone the delivery of a cis-retinal species to ipRGCs found in a small subpopulation of ganglion cell neurons that primarily project to areas of the brain involved in circadian rhythm and pupillary light responses.79 In a similar manner, CRALBP may also act in the pineal gland, known to express the non-visual pineal opsin80 with implications for the production of the hormone melatonin and wake/sleep patterns. Most importantly, CRALBP has been identified as the downstream target of Pax6,81 a key transcription factor involved in the control of various aspects of embryonic development including that of the eye. Pax6 directly controls expression of CRALBP during spinal cord development indicating its putatively ancient role as a cis-retinoid chaperone in non- image-forming neuronal tissues. Therefore, the biological relevance of CRALBP’s thermal isomerase function could be associated with processes other than the visual cycle.
Conclusions
9-cis-Retinal bound to CRALBP is thermally isomerized to 9,13-dicis-retinal in the dark with reaction kinetics affected by the structure of this protein. In particular, CRALBP slowly and irreversibly accumulates the 9,13 product whereas its R234W mutant rapidly catalyzes equilibration to a ~ 2:3 ratio of the 9-cis- and 9,13-dicis isomers. Crystallographic structures of CRALBP complexes revealed a binding mode for 9-cis-retinal different from that previously reported for 11-cis-retinal. The structure of CRALBP:9-cis-retinal is characterized by the presence of buried waters in the hydrophobic pocket of the protein. By combining structural data, molecular modeling, NMR spectroscopy, and kinetic measurements, we propose a mechanism for the isomerization reaction involving transient protonation of the bound retinal. Specifically, the 13-trans-cis- isomerisation of 9-cis-retinal includes initial protontransfer from Glu202 via two water molecules onto the aldehyde oxygen prior to isomerization and subsequent recovery of bond order by re-protonation of Glu202. Kinetic data with E202Q and Y180F CRALBP mutants support the essential role of Glu202 as initial proton donor in the isomerization reaction. Our findings highlight the specific role of water molecules in both CRALBP-assisted specificity towards 9-cis-retinal and its thermal isomerase activity yielding 9,13-dicis-retinal. Other than its known role as a cis-specific retinoid binder downstream of restorative retinoid recycling, CRALBP is expressed in several tissues not directly associated with light detection functions. Therefore, the biological relevance of the newly discovered isomerase activity for CRALBP could include processes other than vision.
Supplementary Material
Acknowledgements
This research was supported by the Swiss National Science Foundation (Grants NN. PP00P2_139195 for MC, 31003A_130497 for AS). Research reported in this publication was also supported by the National Eye Institute of the National Institutes of Health (NEI/NIH) under award numbers: R01EY009339, and R01EY020551. KP is John Hord Professor of Pharmacology. We would also like to acknowledge NEI/NIH for donating 11-cis-retinal. Finally, we are grateful to Prof. Samuel Leutwyler, Dr. Hans-Martin Frey and Prof. Andrea Cannizzo for useful discussions.
Footnotes
Supporting Information
Such information includes: A Table containing X-ray data collection and refinement statistics. Figures reporting UV/VIS spectra of protein- ligand complexes, close-up view of CRALBP binding pocket, experimental 1H, 1H-1H COSY, and simulated 1H NMR spectra of different retinoids, HPLC traces of retinoid extracts of R234W:9-cisretinal. Also available are coordinates of CRALBP:9-cis-retinal and of R234W:9-cis-retinal complexes. This material is available free of charge via the Internet at http://pubs.acs.org.
Contributor Information
Michele Cascella, Email: michele.cascella@iac.unibe.ch.
Achim Stocker, Email: achim.stocker@ibc.unibe.ch.
References
- 1.Palczewski K. J. Biol. Chem. 2012;287:1612–1619. doi: 10.1074/jbc.R111.301150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palczewski K. Annu. Rev. Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Travis GH, Golczak M, Moise AR, Palczewski K. Annu. Rev. Pharmacol. Toxicol. 2007;47:469–512. doi: 10.1146/annurev.pharmtox.47.120505.105225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fleisch VC, Neuhauss SC. Prog. Ret. Eye Res. 2010;29:476–486. doi: 10.1016/j.preteyeres.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Moiseyev G, Chen Y, Takahashi Y, Wu BX, Ma JX. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12413–12418. doi: 10.1073/pnas.0503460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaylor JJ, Yuan Q, Cook J, Sarfare S, Makshanoff J, Miu A, Kim A, Kim P, Habib S, Roybal CN, Xu T, Nusinowitz S, Travis GH. Nat. Chem. Biol. 2013;9:30–36. doi: 10.1038/nchembio.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiser PD, Palczewski K. Prog. Ret. Eye Res. 2010;29:428–442. doi: 10.1016/j.preteyeres.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamb TD, Pugh EN., Jr Prog. Ret. Eye Res. 2004;23:307–380. doi: 10.1016/j.preteyeres.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Collery R, McLoughlin S, Vendrell V, Finnegan J, Crabb JW, Saari JC, Kennedy BN. Invest. Ophth. Vis. Sci. 2008;49:3812–3820. doi: 10.1167/iovs.08-1957. [DOI] [PubMed] [Google Scholar]
- 10.Golovleva I, Bhattacharya S, Wu Z, Shaw N, Yang Y, Andrabi K, West KA, Burstedt MS, Forsman K, Holmgren G, Sandgren O, Noy N, Qin J, Crabb JW. J. Biol. Chem. 2003;278:12397–12402. doi: 10.1074/jbc.M207300200. [DOI] [PubMed] [Google Scholar]
- 11.Huang J, Possin DE, Saari JC. Mol. Vis. 2009;15:223–234. [PMC free article] [PubMed] [Google Scholar]
- 12.McBee JK, Van Hooser JP, Jang GF, Palczewski K. J. Biol. Chem. 2001;276:48483–48493. doi: 10.1074/jbc.M105840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maeda T, Maeda A, Casadesus G, Palczewski K, Margaron P. Invest. Ophth. Vis. Sci. 2009;50:4368–4378. doi: 10.1167/iovs.09-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan J, Rohrer B, Moiseyev G, Ma JX, Crouch RK. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13662–13667. doi: 10.1073/pnas.2234461100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang PH, Fan J, Goletz PW, Wheless L, Crouch RK. Invest. Ophth. Vis. Sci. 2010;51:5958–5964. doi: 10.1167/iovs.10-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palczewski K. Trends Pharmacol. Sci. 2010;31:284–295. doi: 10.1016/j.tips.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rando RR. Chem. Rev. 2001;101:1881–1896. doi: 10.1021/cr960141c. [DOI] [PubMed] [Google Scholar]
- 18.Burstedt MS, Sandgren O, Holmgren G, Forsman-Semb K. Invest. Ophth. Vis. Sci. 1999;40:995–1000. [PubMed] [Google Scholar]
- 19.Humbert G, Delettre C, Senechal A, Bazalgette C, Barakat A, Bazalgette C, Arnaud B, Lenaers G, Hamel CP. Invest. Ophth. Vis. Sci. 2006;47:4719–4724. doi: 10.1167/iovs.05-1488. [DOI] [PubMed] [Google Scholar]
- 20.He X, Lobsiger J, Stocker A. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18545–18550. doi: 10.1073/pnas.0907454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams PD, Baker D, Brunger AT, Das R, Dimaio F, Read RJ, Richardson DC, Richardson JS, Terwilliger TC. Annu. Rev. Biophys. 2013 doi: 10.1146/annurev-biophys-083012-130253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Noll GN, Becker C. J. Chromatogr. A. 2000;881:183–188. doi: 10.1016/s0021-9673(00)00232-6. [DOI] [PubMed] [Google Scholar]
- 24.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. J. Comput. Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 25.Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 26.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 27.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 28.Hoover WG. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 29.Martyna GJ, Klein ML, Tuckerman M. J. Chem. Phys. 1992;97:2635–2643. [Google Scholar]
- 30.Nose S. Mol. Phys. 1984;52:255–268. [Google Scholar]
- 31.Feller SE, Zhang YH, Pastor RW, Brooks BR. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 32.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 33.Ryckaert JP, Ciccotti G, Berendsen HJC. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 34.Becke AD. Phys. Rev. A. 1988;38:3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 35.Lee CT, Yang WT, Parr RG. Phys. Rev. B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 36.Troullier N, Martins JL. Phys. Rev. B. 1991;43:1993–2006. doi: 10.1103/physrevb.43.1993. [DOI] [PubMed] [Google Scholar]
- 37.Lin IC, Coutinho-Neto MD, Felsenheimer C, von Lilienfeld OA, Tavernelli I, Rothlisberger U. Phys. Rev. B. 2007;75 doi: 10.1021/jp0750102. [DOI] [PubMed] [Google Scholar]
- 38.Laio A, VandeVondele J, Rothlisberger U. J. Phys. Chem. B. 2002;106:7300–7307. [Google Scholar]
- 39.Laio A, VandeVondele J, Rothlisberger U. J. Chem. Phys. 2002;116:6941–6947. [Google Scholar]
- 40.Tavernelli I. J. Phys. Chem. A. 2007;111:13528–13536. doi: 10.1021/jp0767056. [DOI] [PubMed] [Google Scholar]
- 41.Car R, Parrinello M. Phys. Rev. Lett. 1985;55:2471–2474. doi: 10.1103/PhysRevLett.55.2471. [DOI] [PubMed] [Google Scholar]
- 42.Crouch R, Purvin V, Nakanishi K, Ebrey T. Proc. Natl. Acad. Sci. U. S. A. 1975;72:1538–1542. doi: 10.1073/pnas.72.4.1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu ZP, Hasan A, Liu TY, Teller DC, Crabb JW. J. Biol. Chem. 2004;279:27357–27364. doi: 10.1074/jbc.M401960200. [DOI] [PubMed] [Google Scholar]
- 44.Helbling RE, Bolze CS, Golczak M, Palczewski K, Stocker A, Cascella M. J. Phys. Chem. B. 2013;117:10719–10729. doi: 10.1021/jp405410t. [DOI] [PubMed] [Google Scholar]
- 45.Raghavan NV, Das PK, Bobrowski K. J. Am. Chem. Soc. 1981;103:4569–4573. [Google Scholar]
- 46.Garczarek F, Gerwert K. Nature. 2006;439:109–112. doi: 10.1038/nature04231. [DOI] [PubMed] [Google Scholar]
- 47.Gu W, Helms V. J. Am. Chem. Soc. 2009;131:2080–2081. doi: 10.1021/ja809301w. [DOI] [PubMed] [Google Scholar]
- 48.Vendrell O, Gelabert R, Moreno M, Lluch JM. J. Am. Chem. Soc. 2006;128:3564–3574. doi: 10.1021/ja0549998. [DOI] [PubMed] [Google Scholar]
- 49.Frank RAW, Titman CM, Pratap JV, Luisi BF, Perham RN. Science. 2004;306:872–876. doi: 10.1126/science.1101030. [DOI] [PubMed] [Google Scholar]
- 50.Hummer G, Rasaiah JC, Noworyta JP. Nature. 2001;414:188–190. doi: 10.1038/35102535. [DOI] [PubMed] [Google Scholar]
- 51.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao Z, Peng YX, Yan TY, Li S, Li AL, Voth GA. J. Am. Chem. Soc. 2010;132:11395–11397. doi: 10.1021/ja1046704. [DOI] [PubMed] [Google Scholar]
- 53.Stoyanov ES, Stoyanova IV, Reed CA. J. Am. Chem. Soc. 2010;132:1484–1485. doi: 10.1021/ja9101826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saari JC, Bredberg DL. J. Biol. Chem. 1987;262:7618–7622. [PubMed] [Google Scholar]
- 55.Rando RR, Chang A. J. Am. Chem. Soc. 1983;105:2879–2882. [Google Scholar]
- 56.Zatula AS, Ryding MJ, Uggerud E. Phys. Chem. Chem. Phys. 2012;14:13907–13909. doi: 10.1039/c2cp42060k. [DOI] [PubMed] [Google Scholar]
- 57.Rini M, Magnes BZ, Pines E, Nibbering ET. Science. 2003;301:349–352. doi: 10.1126/science.1085762. [DOI] [PubMed] [Google Scholar]
- 58.Mohammed OF, Pines D, Dreyer J, Pines E, Nibbering ET. Science. 2005;310:83–86. doi: 10.1126/science.1117756. [DOI] [PubMed] [Google Scholar]
- 59.Jensen MO, Rothlisberger U, Rovira C. Biophys. J. 2005;89:1744–1759. doi: 10.1529/biophysj.104.058206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brzezinski P, Ädelroth P. Curr. Opin. Struc. Biol. 2006;16:465–472. doi: 10.1016/j.sbi.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 61.Schulten K, Dinur U, Honig B. J. Chem. Phys. 1980;73:3927–3935. [Google Scholar]
- 62.Oesterhelt D, Meentzen M, Schuhmann L. Eur. J. Biochem. 1973;40:453–463. doi: 10.1111/j.1432-1033.1973.tb03214.x. [DOI] [PubMed] [Google Scholar]
- 63.Ohno K, Takeuchi Y, Yoshida M. Biochim. Biophys. Acta. 1977;462:575–582. doi: 10.1016/0005-2728(77)90102-5. [DOI] [PubMed] [Google Scholar]
- 64.Balashov SP, Imasheva ES, Govindjee R, Sheves M, Ebrey TG. Biophys. J. 1996;71:1973–1984. doi: 10.1016/S0006-3495(96)79395-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Balashov SP. Isr. J. Chem. 1995;35:415–428. [Google Scholar]
- 66.Balashov SP, Govindjee R, Kono M, Imasheva E, Lukashev E, Ebrey TG, Crouch RK, Menick DR, Feng Y. Biochemistry. 1993;32:10331–10343. doi: 10.1021/bi00090a008. [DOI] [PubMed] [Google Scholar]
- 67.Futterman S, Rollins MH. Invest. Ophth. Visual. 1973;12:234–235. [PubMed] [Google Scholar]
- 68.Futterman S, Rollins MH. J. Biol. Chem. 1973;248:7773–7779. [PubMed] [Google Scholar]
- 69.Redmond TM, Poliakov E, Kuo S, Chander P, Gentleman S. J. Biol. Chem. 2010;285:1919–1927. doi: 10.1074/jbc.M109.027458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hamel CP, Tsilou E, Pfeffer BA, Hooks JJ, Detrick B, Redmond TM. J. Biol. Chem. 1993;268:15751–15757. [PubMed] [Google Scholar]
- 71.Redmond TM. Exp. Eye Res. 2009;88:846–847. doi: 10.1016/j.exer.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Redmond TM, Poliakov E, Yu S, Tsai JY, Lu ZJ, Gentleman S. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13658–13663. doi: 10.1073/pnas.0504167102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kiser PD, Farquhar ER, Shi W, Sui X, Chance MR, Palczewski K. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E2747–W2756. doi: 10.1073/pnas.1212025109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kiser PD, Golczak M, Lodowski DT, Chance MR, Palczewski K. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17325–17330. doi: 10.1073/pnas.0906600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cascella M, Bärfuss S, Stocker A. Arch. Biochem. Biophys. 2013;513:187–195. doi: 10.1016/j.abb.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 76.Saari JC, Nawrot M, Kennedy BN, Garwin GG, Hurley JB, Huang J, Possin DE, Crabb JW. Neuron. 2001;29:739–748. doi: 10.1016/s0896-6273(01)00248-3. [DOI] [PubMed] [Google Scholar]
- 77.Saari JC, Crabb JW. Exp. Eye Res. 2005;81:245–246. doi: 10.1016/j.exer.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 78.Sexton T, Buhr E, Van Gelder RN. J. Biol. Chem. 2012;287:1649–1656. doi: 10.1074/jbc.R111.301226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zaidi FH, Hull JT, Peirson SN, Wulff K, Aeschbach D, Gooley JJ, Brainard GC, Gregory-Evans K, Rizzo JF, 3rd, Czeisler CA, Foster RG, Moseley MJ, Lockley SW. Curr. Biol. 2007;17:2122–2128. doi: 10.1016/j.cub.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Max M, McKinnon PJ, Seidenman KJ, Barrett RK, Applebury ML, Takahashi JS, Margolskee RF. Science. 1995;267:1502–1506. doi: 10.1126/science.7878470. [DOI] [PubMed] [Google Scholar]
- 81.Boppana S, Scheglov A, Geffers R, Tarabykin V. Biochim. Biophys. Acta. 2012;1820:151–156. doi: 10.1016/j.bbagen.2011.09.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.