Abstract
Most insects harbor two paralogous circadian genes, namely timeout and timeless. However, in the Hymenoptera only timeout is present. It remains unclear whether both genes, especially timeout in hymenopteran insects, have distinct evolutionary patterns. In this study, we examine the molecular evolution of both genes in 25 arthropod species, for which whole genome data are available, with addition of the daily expression of the timeout gene in a pollinating fig wasp, Ceratosolen solmsi (Hymenoptera: Chalcidoidea: Agaonidae). Timeless is under stronger purifying selection than timeout, and timeout has positively selected sites in insects, especially in the Hymenoptera. Within the Hymenoptera, the function of timeout may be conserved in bees and ants, but still evolving rapidly in some wasps such as the chalcids. In fig wasps, timeout is rhythmically expressed only in females when outside of the fig syconium but arrhythmically in male and female wasps inside the syconium. These plastic gene expressions reflect adaptive differences of males and females to their environment.
Timeless1 and timeout2 are paralogous genes in animals. Phylogenetic analyses suggest that timeless originated as a duplication of timeout around the time of the Cambrian explosion3. In insects, both timeless and timeout occur in fruit flies, mosquitoes, butterflies, moths, Tribolium beetles and pea aphids, whereas hymenopteran insects have only timeout4. Timeless is reported to function as a canonical circadian clock gene in Drosophila and some other insects5,6,7. In contrast, timeout is a multifunctional gene, which plays an essential role in the maintenance of chromosome integrity, light entrainment of the circadian clock, embryonic development, and regulation of DNA replication8,9,10. Recent studies have provided evidence that timeout also plays a critical role in the circadian clock11. This functional divergence suggests that timeout and timeless may have distinctive evolutionary patterns. In addition, hymenopterans have lineage-specific loss of timeless, and to compensate for the function of timeless, natural selection may have driven the evolution of timeout.
In this study, we used gene structure analysis on timeout, and evolutionary selection analysis on the timeout/timeless family, in 25 species of arthropods to gain further insights into the evolutionary history of this gene family. We also characterized the daily expression of timeout in a pollinating fig wasp, Ceratosolen solmsi (Hymenoptera: Agaonidae), to examine in detail plasticity of gene expression according to the ecology of this species.
Results
Gene structure and evolutionary analyses on timeout
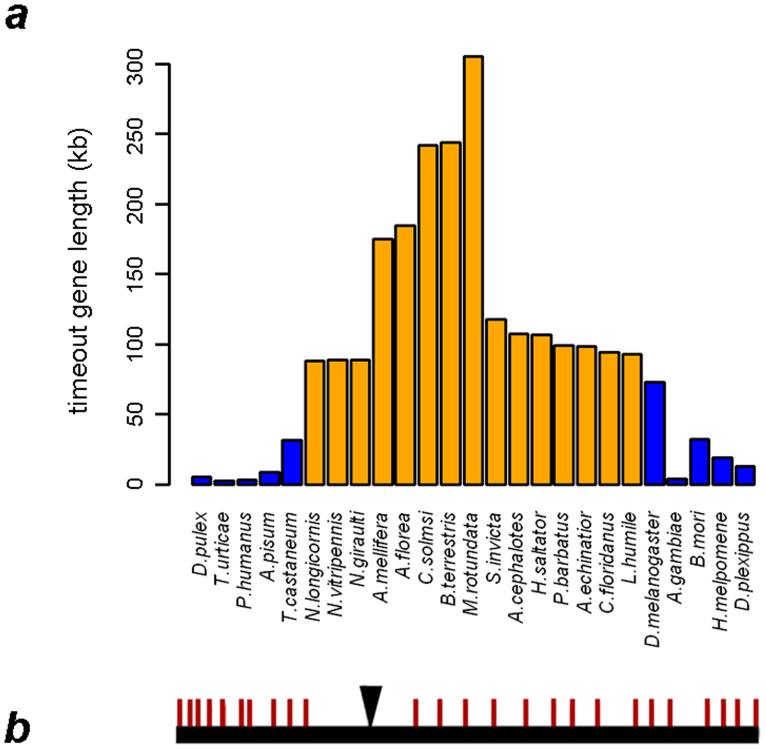
We obtained orthologous data for the timeout gene from 25 arthropod species which have had their entire genomes sequenced. Our gene structure analysis detected that timeout has varied gene lengths, with the maximum length of 305.6 kb in Megachile rotundata (Hymenoptera: Apoidea: Megachilidae) and a minimum length of 3.09 kb in Tetranychus urticae (Arachnida: Trombidiformes: Tetranychidae). Hymenopteran insects tended to have longer timeout genes than non-hymenopteran insects (Fig. 1a, orange bar vs. blue bar). This divergence was due not to exon length (Table S1), but longer introns (average total-intron-length-of-timeout of 141,932 bp) in hymenopterans compared with all other arthropods (average total-intron-length-of-timeout of 16,395 bp). We also detected one actively transcribing gene “nested” in the intron of timeout in all hymenopterans as well as in Tribolium and Drosophila (species in red branches, Fig. 2). The gene structure of timeout of C. solmsi was shown in Fig. 1b. This spans 241.73 kb in the scaffold and comprises of 25 exons and 24 introns, with the tenth intron harboring one actively transcribing gene.
Figure 1. Gene structure analyses on timeout.
(a) Variation of gene lengths among arthropods. The orange columns represent hymenopteran species, and the blue columns represent non-hymenopteran species. (b) The genomic structure of timeout in Ceratosolen solmsi. The red bars represent exons, and the black reversed triangle denotes the nested gene, putatively encoding an insulin growing banding protein.
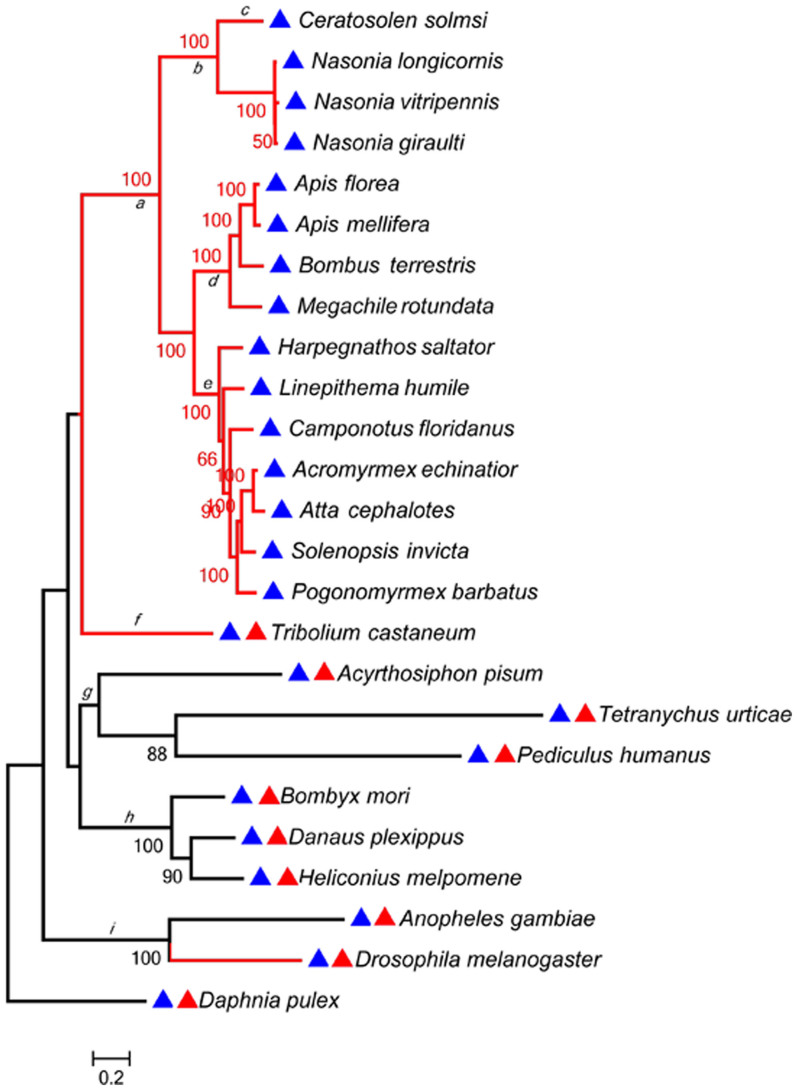
Figure 2. The phylogeny of timeout in 25 species of arthropod.
A red triangle before each species denotes the presence of timeless in a species, with a blue triangle denoting the existence of timeout. The branches a to i are the lineages tested for positive selection by a branch-site model using PAML software. The red colored branches denote the species in which a “nested” transcribed gene was detected in the intron of timeout.
A phylogeny was constructed based on timeout sequences for all 25 species. The hymenopterans, which have no timeless gene, clustered into a single clade (Fig. 2, branch a). Based on this gene tree, we tested if timeout/timeless had distinct evolutionary patterns by using site- and branch-site models nested in PAML (Table 1)12. Potential positive selection was tested based on the ratio (ω) of non-synonymous (Ka) to synonymous (Ks) substitutions rates (ω = Ka/Ks). Generally, if ω = 1 amino acid substitutions were assumed to be largely neutral; ω > 1 is evidence of positive selection, ω < 1 was consistent with purifying selection. Five models were used to test for positively selected sites: M1, M2, M7, M8, and M8a. We employed site models M7 vs. M8 to test for positive selection of specific codons. The results showed that both genes were under purifying selection, but selection pressure on timeless was stronger than that on timeout (one-ratio ω value of 0.00925 for timeless, and 0.08332 for timeout, table S2). The results from the site-model (M7 vs. M8) indicated that timeout had one positively selected site (242 V, ω = 1.10056) (Table S2). Our branch-site model results further demonstrated that timeout had positively selected sites on almost all branches (Fig. 2, branches a, b, c, f, g, h, and i) except those of ants and bees (Fig. 2, branches d and e). In general, more significant positive sites were present in the Hymenoptera than other lineages (Table S2). For example, the fig wasp C. solmsi had one significant positively selected site (45 S, Table S2).
Table 1. Tests for positive selection using site- and branch-site models.
| Model | Models Compared | -2lnΔL | df | p value |
|---|---|---|---|---|
| site model | M7 versus M8 | 19. 568712 | 2 | p < 0.001 |
| branch-site model | Branch-site a (Hymenoptera) | 54.141658 | 2 | p < 0.001 |
| Branch-site b (wasp) | 9.816718 | 2 | p = 0.007 | |
| Branch-site c (C. solmsi) | 10.708974 | 2 | p = 0.0047 | |
| Branch-site d (bee) | 13.471594 | 2 | p < 0.001 | |
| Branch-site e (ant) | 2.762636 | 2 | p = 0.25 | |
| Branch-site f (T. castaneum) | 35.287362 | 2 | p < 0.001 | |
| Branch-site g (Hemiptera) | 7.42465 | 2 | p = 0.012 | |
| Branch-site h (Lepidoptera) | 40.693252 | 2 | p < 0.001 | |
| Branch-site i (Diptera) | 31.38715 | 2 | p < 0.001 |
Daily expression of timeout in the fig wasp C. solmsi
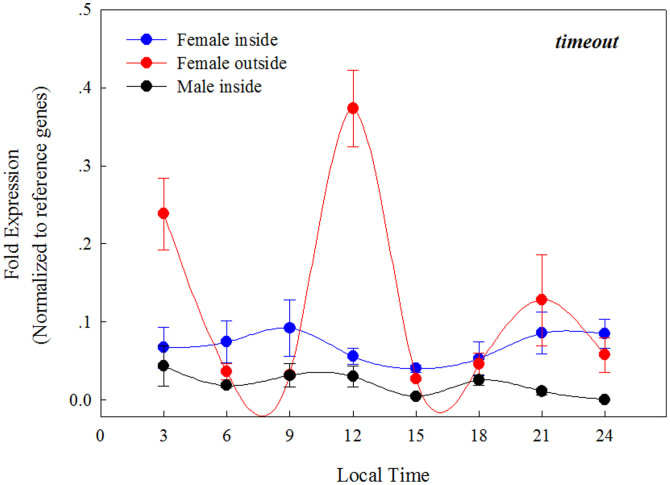
We measured daily levels of timeout mRNA in both female and male fig wasps using real time qPCR. At the early stage of development, both males and females develop within the fig syconium, and after mating, only the females leave the fig syconium. Taking the ecological microcosm of the fig syconium into consideration, three sample groups were allocated: 1) female wasps collected from within the syconium cavity; 2) male wasps collected in the same manner, and 3) female wasps that had successfully emerged from their natal syconium, and had been exposed to natural light for at least three hours. Repeated measures ANOVAs were employed to show the expression values of timeout fluctuate over time (Fig. 3). In addition, we performed cosinor analysis13 (http://www.circadian.org/softwar.html) to reveal if timeout is rhythmically expressed. The parameters used for these tests on cosinor software were: start at period 8 h, and end at period 34 h, with incremental steps being set at 0.1. The results showed that timeout is only rhythmically expressed in females that had successfully dispersed from their syconium (F2, 5 = 6.35, P = 0.043), but arrhythmic in both males (F2, 5 = 5.77, p = 0.051) and females (F2, 5 = 5.66, p = 0.050) still within the syconium.
Figure 3. The expression of timeout in the fig wasp Ceratosolen solmsi over a 24 h period in males (that do not leave their syconium), females still within their syconium, and females that had naturally dispersed from their syconium.
Discussion
Phylogenetic analysis for the timeless/timeout family suggest that these two paralogous genes evolved from gene duplication in early animals3. Therefore, possession of only one of these genes in some insects represents lineage-specific gene loss events. Among the arthropods we examined, only hymenopterans, including ants, bees and wasps, possessed timeout but not timeless. It is likely that the loss of timeless occurred in their common ancestor. Moreover, we discover that timeout is significantly longer in the Hymenoptera compared to the other insects mainly due to long introns. These long introns may harbor undetected information in regulating the expression of this gene14.
Our analyses of the timeless/timeout family in arthropods reveals distinct evolutionary patterns of each gene. Timeless is under stronger purifying selection than timeout, and both site- and branch-site models provide evidence of positive selection sites for timeout. The strong purifying selection for timeless is predictable, because the function of timeless as a component of the canonical circadian clock is clearly conservative. However, considering its multifunctional nature, timeout should be subject to stronger positive selection to maintain its variable function in different lineages. In the Hymenoptera, and in the absence of the function conferred by timeless, timeout may have been subject to stronger positive selection to compensate for the absence of timeless. The stronger signal of positive selection in this lineage supports this possibility. Previous studies have also supported this hypothesis for another circadian gene, cryptochrome15. Although in general hymenopterans had more positively selected sites than did other lineages, evidence of positive selection in taxa that diverged from the common ancestor of ants and bees was absent. We suggest that the function of TIMEOUT has been conserved in these lineages. However, for parasitoid fig wasps and Nasonia, timeout may have evolved rapidly under positive selection for adaptations associated with their distinct ecological conditions16. For example, we detected strong signals of positive selection at one site in C. solmsi. We further examined the functional role of the only positive selection site, Serine (S), in C. solmsi by mapping it onto the predicted secondary structure of TIMEOUT (Fig. S1). In this amino acid position, compared with the amino acids Leucine (L), Isoleucine (I), Tyrosine (Y) or Asparagine (N), which occur in all the other arthropods, Serine is a polar amino acid, which readily forms a random coil structure17. This random coil structure usually includes the active sites of enzyme and functional sites of proteins18,19. All of our results suggest that timeout may show plastic expressions associated with adaptations to ecology or life-history, especially in wasps, all of which lack timeless but have the positively selected timeout.
Fig trees (Ficus: Moraceae) have an obligate mutualism with their pollinating wasps (Hymenoptera: Agaonidae)20. Wasps gall individual flowers and develop within the characteristic enclosed inflorescences (syconia) of the trees. Male wasps do not leave their natal syconium. Only females disperse from the syconium upon maturation, through exit tunnels chewed by males21. The ecological and life-history differences between the sexes may thus explain the expression patterns of circadian genes. Our results show that in both males and females still inside the syconium cavity arrhythmically expressed timeout. However, females that have successfully dispersed from their natal syconium show rhythmic expression of timeout. This may reflect differences in exposure to light because light is absent within a syconium. Previous studies of other insects have shown that the expression of timeout is light-dependent. For example, timeout is needed for circadian photoreception in Drosophila8 and has circadian expression in the mammalian retina22,23. We suggest that the expression of timeout may thus affect the light input pathway in C. solmsi, which is adaptive due to light changes between the inside and the outside of the syconium. However, another factor, temperature also exerts an important influence on the expression of circadian genes24. Thus, differences in temperature within and outside the syconium may also affect the expression of timeout in C. solmsi. Contrary to our previous studies on the expression of opsin genes25, the clear shift from arrhythmic to rhythmic expression of timeout in female fig wasps as they move from the syconium cavity to the outside of the syconium, suggests an environmental determinant of timeout expression. In addition, our closer examination of timeout expression in C. solmsi provides a good model system to test in the future if evolutionary compensation affects the loss of timeless. Further studies of additional hymenopteran species are needed to clarify the role timeout plays in the absence of timeless.
Methods
Phylogenetic analysis
We searched public genomic DNA databases at NCBI26, FlyBase27, Bombyxmori genome28 and BeeBase (http://racerx00.tamu.edu/bee_resources.html) for genes encoding homologs of known timeout/timeless proteins using TBLASTN29. The FGENESH+ gene predictor (http://linux1.softberry.com/berry.phtml) was occasionally used to improve the translation. The searched and selected gene sequences (all the sequences used in this study are shown in Table S3) were aligned using ClustalW implemented in MEGA 5.030. We improved the alignment based on the translated protein sequences. We translated the codon sequences into protein sequences, and performed an alignment based on these protein sequences. We then re-translated the aligned protein sequences into codon sequences, which were used to instruct the improvement of the real alignment of the codon sequences. The alignment was manually edited using BioEdit31. (For the manually edited sequences before and after deleting gap, see supplementary information). A maximum likelihood (ML) tree was constructed using PhyML3.032, with the best-fit model of nucleotide substitutions of GTR + G, according to the Akaike Information Criterion in jModeltest33. One thousand ML bootstrap replicates were obtained to assess clade robustness. Timeout data from a crustacean species, Daphnia pulex, were set as the outgroup.
Selective pressure analysis
Maximum likelihood methods were used to explore the selective pressure acting on timeout, and all tests were conducted using the CodeML in PAML 412. Potential positive selection was tested based on the ratio (ω) of non-synonymous (Ka) to synonymous (Ks) substitutions rates (ω = Ka/Ks). To evaluate positive selection on timeout across the examined arthropod species, we first used the site-specific models, M7 restricted sites with ω ≤ 1, whereas models M8 included a class of sites with ω > 1. The sites with a posterior probability >0.9 were considered as candidates for selection. Positive selection was further detected with the improved branch-site likelihood method34. Test 1 (M1a vs. Branch site model) and test 2 (Branch site null model vs. Branch site model), which could differentiate positive selection from the relaxation of selective constraints, were used. The branch leading to the Hymenoptera (a), parasitoid wasps (b), Ceratosolen solmsi (c), bees (d), ants (e), Tribolium castaneum (f), Hemiptera (g), Lepidoptera (h), and Diptera (i), were labeled as the foreground lineage to test whether positive selection occurred along these branches. For each model, the ratio of ω was estimated, and likelihood ratio tests (LRT) were performed to compare pairs of nested models. We calculated twice the difference in log-likelihood values between the two models against a chi-square distribution. When the LRT was significant, a Bayes Empirical Bayes (BEB) analysis was used to identify positively selected sites. In addition, we obtained the secondary structure of TIMEOUT in C. solmsi by using the SOPMA Server (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html)35. This positively selected site was mapped onto the resulting figure (Fig. S1).
Sample collections, RNA isolation, cDNA synthesis and real time qPCR expression analysis of the fig pollinator wasp, Ceratosolen solmsi
We made field collections of the fig wasp C. solmsi, the pollinator of Ficus hispida, from Danzhou (19°30′ N, 109°29′ E), Hainan province, China between July and August 2012. Adult females and males were collected from naturally growing syconia when the loose female wasps were yet to emerge. (Male wasps do not emerge from the syconium.) We also collected adult females from different syconia. These females were exposed to natural light for at least three additional hours prior to processing.
After collection, all wasps were flash frozen in liquid nitrogen every 3 hr over a 24 h period. We then isolated total RNA from each wasp, with the first cDNA strand synthesized using TransScript II First Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). Ultimately, we used a Real Time-qPCR (RT-qPCR) technique to obtain daily transcript levels of timeout. RPL13a and UBC were selected as the reference genes for normalizing the RT-qPCR data15. The detailed methods of samples collection, RNA isolation and cDNA synthesis, and RT-qPCR expression analysis are provided as supplement material.
Author Contributions
H.F.G., J.H.X. and D.W.H. designed the work and wrote the main manuscript text. H.F.G., L.M.N. and G.C.M. collected the sample. H.F.G. demonstrated the molecular experiments. H.F.G., J.H.X. and B.W. analyzed the data. D.W.D. participated in the writing and revisions. All the authors have reviewed and approved the manuscript.
Supplementary Material
supplement_materials
Acknowledgments
This project was supported by the National Natural Science Foundation of China (NSFC grant nos 31090253, 31172072, 31210103912), partially by the Major Innovation Program of Chinese Academy of Sciences (KSCX2-EW-Z-2), a grant (O529YX5105) from the Key Laboratory of the Zoological Systematics and Evolution of the Chinese Academy of Sciences, and the National Science Fund for Fostering Talents in Basic Research (Special Subjects in Animal Taxonomy, NSFC-J0930004). We thank Dr. Wen Xin and TransGen Biotech for providing most of the reagents used for the study.
References
- Myers M. P., Wager-Smith K., Wesley C. S., Young M. W. & Sehgal A. Positional cloning and sequence analysis of the Drosophila clock gene, timeless. Science 270, 805–808, 10.1126/science.270.5237.805 (1995). [DOI] [PubMed] [Google Scholar]
- Benna C. et al. A second timeless gene in Drosophila shares greater sequence similarity with mammalian tim. Curr. Biol. 10, R512–R513, 10.1016/S0960-9822(00)00594-7 (2000). [DOI] [PubMed] [Google Scholar]
- Rubin E. B. et al. Molecular and phylogenetic analyses reveal mammalian-like clockwork in the honey bee (Apis mellifera) and shed new light on the molecular evolution of the circadian clock. Genome Res. 16, 1352–1365, 10.1101/gr.5094806 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S., Merlin C., Boore J. L. & Reppert S. M. The monarch butterfly genome yields insights into long-distance migration. Cell 147, 1171–1185, 10.1016/j.cell.2011.09.052 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X. & Sehgal A. Probing the relative importance of molecular oscillations in the circadian clock. Genetics 178, 1147–1155, 10.1534/genetics.107.088658 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai S., Fukui Y., Fujiwara Y. & Takeda M. Structure and expressions of two circadian clock genes, period and timeless in the commercial silkmoth, Bombyx mori. J. Insect Physiol. 52, 625–637, 10.1016/j.jinsphys.2006.03.001 (2006). [DOI] [PubMed] [Google Scholar]
- Zhu H. et al. Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS. Biol. 6, e4, 10.1371/journal.pbio.0060004 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benna C. et al. Drosophila timeless2 is required for chromosome stability and circadian photoreception. Curr. Biol. 20, 346–352, 10.1016/j.cub.2009.12.048 (2010). [DOI] [PubMed] [Google Scholar]
- Gotter A. L. et al. A time-less function for mouse timeless. Nat. Neurosci. 3, 755–756, 10.1038/77653 (2000). [DOI] [PubMed] [Google Scholar]
- Gotter A. L., Suppa C. & Emanuel B. S. Mammalian TIMELESS and Tipin are evolutionarily conserved replication fork-associated factors. J. Mol. Biol. 366, 36–52, 10.1016/j.jmb.2006.10.097 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes J. W. et al. Requirement of mammalian timeless for circadian rhythmicity. Science 302, 439–442, 10.1126/science.1086593 (2003). [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591, 10.1093/molbev/msm088 (2007). [DOI] [PubMed] [Google Scholar]
- Nelson W., Tong Y. L., Lee J. K. & Halberg F. Methods for cosinor-rhythmometry. Chronobiologia 6, 305–323 (1979). [PubMed] [Google Scholar]
- Swinburne I. A., Miguez D. G., Landgraf D. & Silver P. A. Intron length increases oscillatory periods of gene expression in animal cells. Genes Dev. 22, 2342–2346, 10.1101/gad.1696108 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Xiao J. H., Bian S. N., Gu H. F. & Huang D. W. Adaptive evolution of vertebrate-type cryptochrome in the ancestors of Hymenoptera. Biol. Lett. 9, 20120958, 10.1098/rsbl.2012.0958 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodnitskii D. Adaptation to flapping flight in different holometabolic Insects. Entomol. Rev. 76, 1209–1216 (1996). [Google Scholar]
- Brazill D. T. et al. A protein containing a serine-rich domain with vesicle fusing properties mediates cell cycle-dependent cytosolic pH regulation. J. Bio.Chem. 275, 19231–19240, 10.1074/jbc.M000900200 (2000). [DOI] [PubMed] [Google Scholar]
- Ferraroni M. et al. Crystal structure of 4-chlorocatechol 1, 2-dioxygenase from the chlorophenol-utilizing gram-positive Rhodococcus opacus 1CP. J. Bio. Chem. 279, 27646–27655, 10.1074/jbc.M401692200 (2004). [DOI] [PubMed] [Google Scholar]
- Gurevich V. V. & Gurevich E. V. How and why do GPCRs dimerize? Trends Pharmacol. Sci. 29, 234–240, 10.1016/j.tips.2008.02.004 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J. M. & Rasplus J.-Y. Mutualists with attitude: coevolving fig wasps and figs. Trends Ecol. Evol. 18, 241–248, 10.1016/S0169-5347(03)00062-4 (2003). [Google Scholar]
- Suleman N., Raja S. & Compton S. G. Only pollinator fig wasps have males that collaborate to release their females from figs of an Asian fig tree. Biol. Lett. 8, 344–346, 10.1098/rsbl.2011.1016 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangoram A. M. et al. Mammalian circadian autoregulatory loop: a timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription. Neuron 21, 1101–1113, 10.1016/S0896-6273(00)80627-3 (1998). [DOI] [PubMed] [Google Scholar]
- Takumi T. et al. A mammalian ortholog of Drosophila timeless, highly expressed in SCN and retina, forms a complex with mPER1. Genes Cells 4, 67–75, 10.1046/j.1365-2443.1999.00238.x (1999). [DOI] [PubMed] [Google Scholar]
- Buhr E. D., Yoo S. H. & Takahashi J. S. Temperature as a universal resetting cue for mammalian circadian oscillators. Sci. Signal. 330, 379–385, 10.1126/science.1195262 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. et al. Evolution and expression plasticity of opsin genes in a fig pollinator, Ceratosolen solmsi. PLoS One 8, e53907, 10.1371/journal.pone.0053907 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J. & Sayers E. W. GenBank. Nucleic Acids Res. 38, D46–D51, 10.1093/nar/gkp1024 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale R. A. & Crosby M. A. FlyBase: genes and gene models. Nucleic Acids Res. 33, D390–D395, 10.1093/nar/gki046 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Q. et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 306, 1937–1940, 10.1126/science.1102210 (2004). [DOI] [PubMed] [Google Scholar]
- Altschul S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402, 10.1093/nar/25.17.3389 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739, 10.1093/molbev/msr121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Series 41, 95–98 (1999). [Google Scholar]
- Guindon S. P. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321, 10.1093/sysbio/syq010 (2010). [DOI] [PubMed] [Google Scholar]
- Darriba D., Taboada G. L., Doallo R. & Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772–772, 10.1038/nmeth.2109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Nielsen R. & Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22, 2472–2479, 10.1093/molbev/msi237 (2005). [DOI] [PubMed] [Google Scholar]
- Geourjon C. & Deleage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 11, 681–684, 10.1016/j.tips.2008.02.004 (1995). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplement_materials