Abstract
Regional expression of Wingless/Int (Wnt) genes plays a central role in regulating intestinal development and homeostasis. However, our knowledge of such regional Wnt proteins in the colon remains limited. To understand further the effect of Wnt signaling components in controlling intestinal epithelial homeostasis, we investigated whether the physiological heterogeneity of the proximal and distal colon can be explained by differential Wnt signaling. With the use of a Wnt signaling-specific PCR array, expression of 84 Wnt-mediated signal transduction genes was analyzed, and a differential signature of Wnt-related genes in the proximal versus distal murine colon was identified. Several Wnt agonists (Wnt5a, Wnt8b, and Wnt11), the Wnt receptor frizzled family receptor 3, and the Wnt inhibitory factor 1 were differentially expressed along the colon length. These Wnt signatures were associated with differential epithelial cell proliferation and migration in the proximal versus distal colon. Furthermore, reduced Wnt/β-catenin activity and decreased Wnt5a and Wnt11 expression were observed in mice lacking commensal bacteria, an effect that was reversed by conventionalization of germ-free mice. Interestingly, myeloid differentiation primary response gene 88 knockout mice showed decreased Wnt5a levels, indicating a role for Toll-like receptor signaling in regulating Wnt5a expression. Our results suggest that the morphological and physiological heterogeneity within the colon is in part facilitated by the differential expression of Wnt signaling components and influenced by colonization with bacteria.
One of the fundamental aspects in the development of the gastrointestinal tract is the spatiotemporal expression of signaling molecules that regulate cell fate and differentiation. Previous studies have highlighted a central role of the evolutionarily conserved Wingless/Int (Wnt) signaling pathways as key regulators of embryonic development and epithelial homeostasis in the gut.1–3 In development, local expression patterns of Wnt signaling components play an important role in organogenesis.4,5 Wnt signals control important biological processes required for cell proliferation, differentiation, polarity, and movement, depending on the target cell and the cellular environment.
Recent reports have highlighted the importance of understanding the role of Wnt signaling in the intestinal tract. The intestinal epithelium is highly dynamic and, depending on the species and location, is actively turned over in <1 week.6 Wnt/β-catenin signaling regulates intestinal epithelial cell (IEC) homeostasis and proliferation by increasing β-catenin stability in crypt epithelial cells, whereas IEC migration and differentiation are believed to be in part facilitated through noncanonical (Wnt) signaling pathways independent of β-catenin.6,7 The renewal of intestinal epithelia requires a delicate balance of signaling proteins to control epithelial cell proliferation and migration that in turn is vital for maintaining mucosal homeostasis. Interestingly, regional differences in Wnt gene expression in small versus large intestine are observed in adult mice, suggesting the importance of differential local Wnt expression in regulation of intestinal mucosal homeostasis.7
Although the entire colon exhibits considerable morphological and physiological heterogeneity along its length,8–11 the expression pattern of Wnt signaling components in the different regions of the adult colon remains poorly understood. Embryologically, the cecum, ascending colon, and the proximal two-thirds of the transverse colon are derived from the midgut, whereas the distal colon originates from the hindgut. Such distinct origins of the colonic segment support specific biological characteristics and suggest that distinct regulatory factors are likely to control epithelial homeostasis in the proximal versus distal colon. In addition, important contributing factors that influence Wnt/β-catenin signaling and intestinal epithelial proliferation might be microbial communities that localize in the intestine in distinct regions.6,12 Such a delicate physiological balance of Wnt signaling and intestinal epithelial homeostasis is further perturbed in mucosal inflammatory and neoplastic diseases,3,13 which also indicate regional differences in the proximal versus distal colonic segments.14–17
In the present study, we investigate the regional heterogeneity of Wnt genes in the proximal versus distal colon. Given the importance of luminal microbiota in influencing intestinal epithelial homeostasis18 and to determine whether the Wnt signatures are influenced by microflora colonization, we examined expression of Wnt proteins in the colonic segments of mice raised under germ-free (GF) conditions.
Materials and Methods
Animal Experiments
All procedures that used animals were reviewed and approved by the Emory University Institutional Animal Care and Use Committee and were performed according to the criteria outlined by the NIH. Animals are maintained on a 12-hour light/12-hour dark cycle under specific pathogen-free (SPF) conditions. The mice have ad libitum access to a standard diet and water until reaching the desired age (8 to 10 weeks) and/or weight (20 to 25 g).
Measurement of Crypt Length
Colon was dissected from C57BL/6 mice, rolled into Swiss rolls, and processed for H&E staining. Crypt length was determined by bottom-to-surface measurement of at least 50 correctly aligned crypts of three mice per group. Analysis and measurements were done with Aperio ImageScope version 11.1.2.760 (Vista, CA).
Assessment of IEC Migration in Vivo
In vivo labeling of migrating IECs was achieved by intraperitoneal injection of 100 μg of 5-ethynyl-2-deoxyuridine (EdU; Invitrogen; Carlsbad, CA) per mouse. Colon was harvested from mice and flushed with ice-cold PBS, cut longitudinally, formed into Swiss rolls, and fresh frozen in OCT and dry ice. After freezing, the tissue blocks were sectioned into 8-μm sections for histological analysis. EdU incorporation was determined by using a Click-iT EdU Cell Proliferation Kit (Invitrogen), according to the supplier’s instructions. IEC migration along the crypt luminal axis was assessed with an EdU pulse chase approach. Therefore, a labeling index distribution curve was established to accurately measure the cutoff position between proliferating and migrating cells. Thus, a flash label (2 hours) was used to designate a baseline position of crypt proliferating cells. A pulse chase was then performed to evaluate migration of epithelial cells in the crypt-luminal axis. We calculated the distance of migrated cells from the baseline at 4, 8, and 12 hours after the flash label.
Real-Time Quantitative PCR
RNA was extracted from Isol-RNA–embedded tissues by phenol-chloroform extraction with the use of standard protocols. Purification of RNA was then performed with the RNeasy Mini Kit (Qiagen, Germantown, MD). The Fermentas first-strand kit (Fermentas, Waltham, MA) was used for cDNA synthesis of 1 μg of total RNA. The cDNA was then diluted 1:10, and 2 μL of cDNA was used with primers and SYBR Green Master Mix (SA Biosciences, Frederick, MD) according to the manufacturer’s instructions. The following primer sets were used: Axin2 (forward, 5′-GGGGGAAAACACAGCTTACA-3′; reverse, 5′-TTGACTGGGTCGCTTCTCTT-3′), wingless-type mouse mammary tumor virus (MMTV) integration site family member 5A (Wnt5a) (forward, 5′-GCAGGACTTTCTCAAGGACA-3′; reverse, 5′-CCCTGCCAAAGACAGAAGTA-3′), wingless-type MMTV integration site family member 8b (Wnt8b) (forward, 5′-GGGTAAGAGGTAACCCCAGA-3′; reverse, 5′-GCCAACCTGCCTACTACAGA-3′), wingless-type MMTV integration site family member 11 (Wnt11) (forward, 5′-ACAGAGCTCCCCTGACTTCT-3′; reverse, 5′-TGGTCTCACTTGCAGACGTA-3′), frizzled family receptor 3 (Fzd3) (forward, 5′-TCTTGCCAAAGCCATTAGAC-3′; reverse, 5′-TCGATCACCATTTTGGACTT-3′), and WNT inhibitory factor 1 (WIF1) (forward, 5′-CGGCAGACACTGCAATAAGA-3′; reverse, 5′-GCATTTGAACATCCAACACG-3′). Murine glyceraldehyde-3-phosphate dehydrogenase was used as an internal control in each reaction. The relative abundance of mRNA was determined by using the ΔΔCT method.19
RT2 Profiler PCR Array
The murine RT2 Profiler PCR array specific for the Wnt signaling pathway (PAMM-243Z) was purchased from SA Biosciences and was used according to the manufacturer’s instructions.
cDNA was synthesized from 1 μg of RNA by using the RT2 First-Strand cDNA synthesis kit (SA Biosciences). Amplification was performed with iCycler (Bio-Rad, Hercules, CA) for 40 cycles, with each cycle consisting of a 15-second denaturation at 95.0°C followed by 1 minute of annealing at 60.0°C.
Conventionalization of GF Mice
For experiments that used GF mice, wild-type (WT) C57BL/6 mice were born and raised in GF isolators (Gnotobiotic Rodent Resource Center at University of North Carolina, Chapel Hill). For tissue analysis GF mice were sacrificed in the GF facility according to protocol via carbon dioxide and cervical dislocation. Colon tissue was collected, then Swiss-rolled and fixed in formalin for paraffin embedding and H&E staining or collected for RNA isolation by using the method described (see Real-Time Quantitative PCR). A cohort of GF mice was transferred to the SPF facility (housed two to four mice per cage) for 4 weeks to allow the acquisition of conventional microbiota as described previously.20 The colon tissue from these conventionalized mice was harvested in the animal facility as described for GF mice.
In Situ Hybridization
For analysis of Wnt gene expression frozen blocks of large intestinal Swiss rolls were sectioned at a thickness of 10 μm. In brief tissue was fixed for 10 minutes with 4% para-formaldehyde, washed in PBS, and treated with proteinase K (10 μg/mL) for 10 minutes. After another washing and fixation step (10 minutes with 4% paraformaldehyde) tissue was incubated in acetic anhydride for 10 minutes before a 2-hour prehybridization step (prehybridization buffer: 50% formamide, 5× standard saline citrate, pH 4.5, 50 μg/mL yeast tRNA, 1% SDS, and 50 μg/mL heparin). Sections were then incubated overnight in hybridization buffer [50% formamide, 5× standard saline citrate, 5× Denhardt’s, 250 mg/mL Baker’s yeast RNA (Sigma R6750; Sigma-Aldrich, St. Louis, MO), 500 ng/mL herring sperm] that contained the digoxigenin-labeled ribo-probes at a concentration of 500 ng/mL. Hybridization and washes after hybridization were performed at 60°C. After hybridization the sections were treated with RNase A, and the samples were then incubated with anti-digoxigenin alkaline phosphatase at 4°C overnight. Signals were detected with BM purple. After being fixed with 4% paraformaldehyde/0.1% glutaraldehyde, samples were cleared with a graded series of glycerol/PBS and stored in 80% glycerol/PBS at 4°C.
Statistical Analysis
Results are expressed as means ± SEM. Statistical comparisons were performed by Student’s two-tailed t-tests. A value of P < 0.05 was considered significant.
Results
Differential Expression of Wnt Genes and Epithelial Crypt Architecture in Proximal Versus Distal Colon
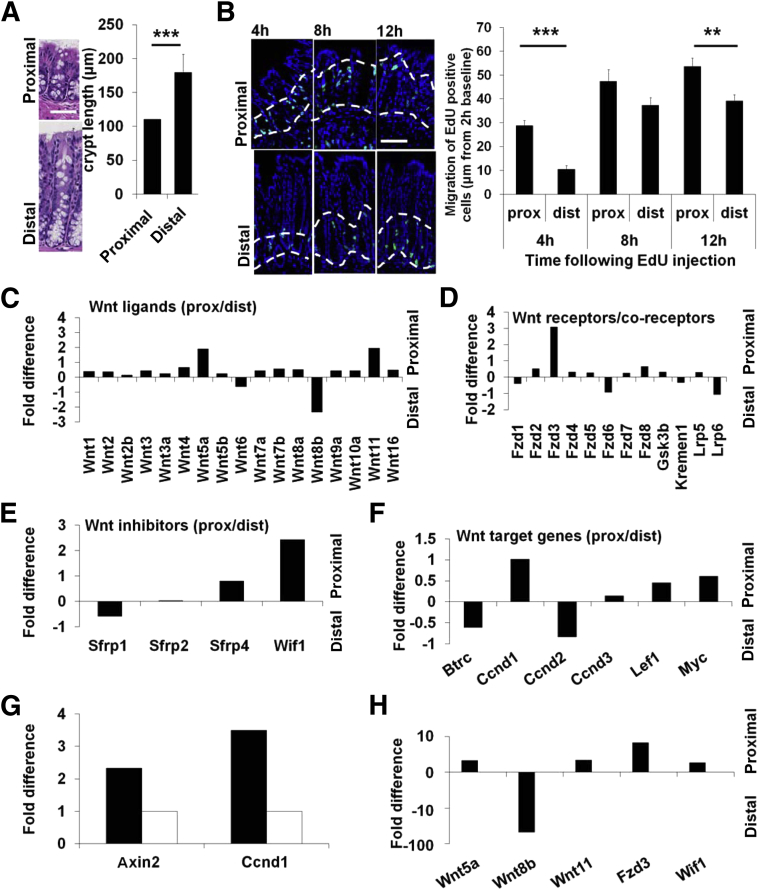
The proximal and distal colon exhibit considerable physiological differences. Measurement of colonic crypt length in the proximal and distal colon found a mean crypt length of 106.3 ± 1.78 μm in the proximal colon compared with 203.2 ± 2.63 μm in the distal colon (P < 0.0001) (Figure 1A). In addition, migration of IECs along the luminal crypt axis differs in the proximal versus distal colon. EdU pulse chase experiments found significantly increased IEC migration along the crypt-luminal axis in the proximal colon compared with the distal colon (Figure 1B). After 4 hours of EdU incorporation, cells that retained the EdU label migrated along the crypt-luminal axis with a mean of 28.8 ± 2.3 μm in the proximal colon compared with 10.5 ± 1.6 μm in the distal colon (P < 0.0001). Eight hours after flash labeling, IECs migrated 47.4 ± 5 μm in the proximal colon and 37.3 ± 3.3 μm in the distal colon (P < 0.0001). Finally, 12 hours after EdU administration, IECs migrated 53.7 ± 3.5 μm in the proximal colon compared with 39.2 ± 2.7 μm in the distal colon (P = 0.0025).
Figure 1.
Heterogeneity of the proximal and distal colon is accompanied by distinct Wnt signaling expression patterns. A: Morphology of proximal and distal colonic crypts exhibit notable histomorphologic differences (representative H&E image in left panel). Quantification of crypt length shows significant differences between the proximal and distal colonic crypts with a mean crypt length of 106.3 ± 1.78 μm in the proximal colon and 203.2 ± 2.63 μm in the distal colon (P < 0.0001). B: IEC migration along the crypt-luminal axis was measured with an EdU pulse-chase experiment [EdU-positive cells in green; nuclear staining (TO-PRO-3)]; migration distance was measured from 2-hour baseline in proximal and distal colonic crypts at 4 hours (proximal, 28.8 ± 2.3 μm; distal, 10.5 ± 1.6 μm; P < 0.0001), 8 hours (proximal, 47.4 ± 5 μm; distal, 37.3 ± 3.3 μm), and 12 hours (proximal, 53.7 ± 3.5 μm; distal, 39.2 ± 2.7 μm; P = 0.0025) after flash label with EdU. C–F: A qPCR array was used to analyze Wnt signaling-related gene expression in the proximal colon compared with the distal colon. Wnt5a, Wnt8b, Wnt11, Fzd3, and WIF1 showed significantly different expression patterns in the proximal colon compared with the distal colon (positive fold differences indicate higher expression in the proximal colon; negative fold differences indicate higher expression in the distal colon). G: Wnt target gene expression (Axin2, Ccnd1) indicates higher Wnt/β-catenin activity in the proximal (black bars) colon than in the distal (white bars) colon (difference: Axin2, 2.33-fold; Ccnd1 3.5-fold). H: qPCR verification of the array results. Data are expressed as means ± SEM and represent three experiments with three mice per group. ∗∗P < 0.01 and ∗∗∗P < 0.001. Scale bars: 50 μm (A and B). Original magnification: ×20 (A and B). Btrc, β-transducin repeat containing protein; Dist, distal; Gsk3b, glycogen synthase kinase 3 β; h, hour; Kremen1, kringle containing transmembrane protein 1; Lef1, lymphoid enhancer binding factor 1; Lrp, low-density lipoprotein receptor-related protein; Myc, (alias c-myc) myelocytomatous oncogene; prox, proximal; Sfrp, secreted frizzled-related protein.
Recent studies have highlighted a role of the evolutionarily conserved Wnt signaling pathway as a key regulator of embryologic development and epithelial homeostasis in the gut. To determine whether the morphological and functional differences in the proximal versus distal colon were reflected by differences in gene expression of the Wnt signaling pathway components, expression of 84 Wnt signaling-related genes was analyzed in the proximal and distal colon of adult C57BL/6 mice with a real-time PCR–based Wnt Signaling Pathway array. Genes with differential expression were then ordered according to their role in Wnt signaling: as Wnt ligands, Wnt receptors, or Wnt-inhibitory and modulatory factors.
The gene array study found significantly increased expression of Wnt5a (difference, 3.74-fold; P < 0.0001) and Wnt11 (difference, 3.89-fold; P < 0.01) in the proximal colon compared with the distal colon. On the converse, Wnt8b expression was significantly higher in the distal colon compared with the proximal colon (difference, 5.15-fold; P < 0.01) (Figure 1C).
Analysis of Wnt receptors found increased expression of Fzd3 (difference, 8.52-fold; P < 0.001) in the proximal colon versus the distal colon. All other analyzed receptors (Fzd1, Fzd2, Fzd4, Fzd5, Fzd6, Fzd7, Fzd8) did not show notable gene expression differences in the two colonic segments (Figure 1D).
Wnt signaling is negatively controlled by proteins that inhibit this signaling pathway at different levels. Analysis of these regulatory genes found a significantly higher expression of WIF1 in the proximal colon compared with the distal colon (difference, 5.43-fold; P < 0.0001) (Figure 1E). In addition, study of the Wnt target genes Axin2 and Cyclin D1 (Ccnd1) found increased expression in the proximal versus distal colon (difference: Axin2, 2.33-fold; Ccnd1, 3.5-fold), supporting increased Wnt activity in this region of the gut (Figure 1, F and G). The results of the array were confirmed with real-time quantitative PCR (qPCR; Figure 1H).
Wnt5a has been reported to be expressed by mesenchymal cells in the lamina propria.21 To detect expression of Wnt8b and Wnt11 mRNA in the intestinal mucosa we performed in situ hybridization. We observed that canonical Wnt8b is expressed by IECs with increased expression in the base of crypts in the distal colon (Supplemental Figure S1A). In contrast, noncanonical Wnt11 was expressed by surface/luminal epithelial cells of the proximal colon (Supplemental Figure S1B). These findings are in keeping with results reported by Ouko et al.22 Taken together Wnt ligands Wnt8b and Wnt11 indicate distinct gradients in the crypt-luminal axis as well as differential expression in the proximal versus distal colonic mucosa.
Wnt Gene Expression in the Colon of Mice Raised in GF Conditions
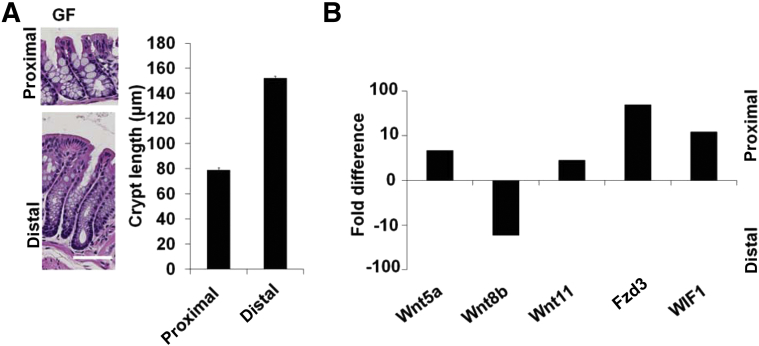
Because luminal microbiota of the colon are known to interact and influence intestinal epithelial homeostasis,23 we next investigated whether the heterogeneity of the proximal versus the distal colon could in part be explained by bacterial colonization. The morphological differences in terms of colonic crypt length and shape observed in SPF mice can also be found in mice raised in GF conditions with a mean crypt length of 79.08 ± 1.72 μm in the proximal colon and 152.10 ± 2.57 μm in the distal colon (Figure 2A). In addition the Wnt gene expression pattern in GF mice resembled the expression profile observed in SPF mice. Thus, increased expression of Wnt5a (difference, 4.65-fold), Wnt11 (difference, 2.82-fold), Fzd3 (difference, 49.51-fold), and WIF1 (difference, 12.17-fold) in the proximal versus distal colon of GF mice was observed (Figure 2B). Conversely, as in the SPF mice, expression of Wnt8b was higher in the distal colon than in the proximal colon (difference, 16.95-fold).
Figure 2.
GF mice show differences in crypt morphology in the proximal colon compared with the distal colon but resemble Wnt gene expression patterns of SPF mice. A: Crypt morphology in the proximal (79.08 ± 1.72 μm) versus the distal (152.10 ± 2.57 μm) colon of GF mice. B: Results of qPCR for Wnt signaling genes in GF mice were Wnt5a (difference, 4.65-fold), Wnt11 (difference, 2.82-fold), Fzd3 (difference, 49.51-fold), and WIF1 (difference, 12.17-fold) in the proximal colon and Wnt8b (difference, 16.95-fold) in the distal colon. Data are expressed as means ± SEM. Scale bar = 50 μm (A).
Comparison of Wnt Expression Levels in GF Mice versus SPF Mice Shows Decreased Gene Expression of Wnt11 and Wnt5a in GF Mice
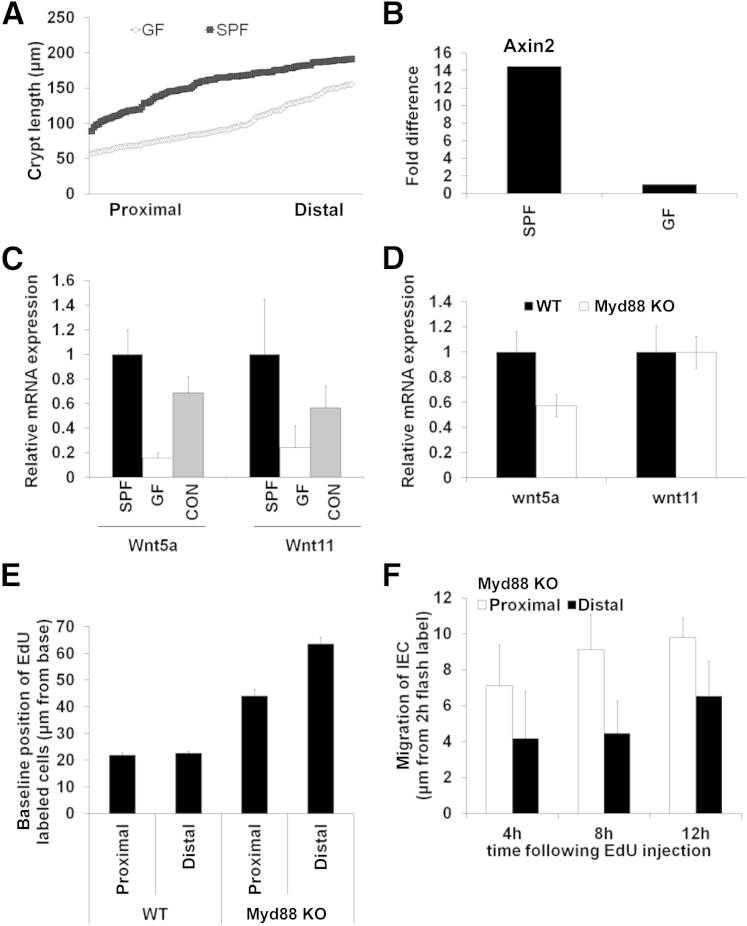
Mice raised in GF conditions showed decreased length of colonic crypts along the length of the colon (Figure 3A). In addition, decreased Axin2 expression was observed, suggesting reduced β-catenin activity in GF mice compared with SPF mice (Figure 3B). Although the expression pattern of the analyzed Wnt genes was comparable in GF and SPF mice, distinct differences in expression levels in GF mice were observed. Direct quantitative comparison with the use of qPCR found differences of Wnt gene expression with decreased mRNA levels of Wnt5a (−6.41-fold) and Wnt11 (−4.18-fold) in GF mice versus SPF mice. To analyze the effect of colonization with commensal bacteria, GF mice were conventionalized for 4 weeks before Wnt gene expression was analyzed. Conventionalization resulted in a significant increase in Wnt5a (difference, 4.4-fold) and Wnt11 (difference, 2.36-fold) expression levels (Figure 3C).
Figure 3.
Mice raised in GF conditions exhibit shorter intestinal crypt length and altered Wnt gene expression levels compared with SPF mice. A: Total crypt length from proximal to distal colon of mice raised in GF conditions compared with SPF mice. B: Axin2 mRNA expression was used as a surrogate marker for active Wnt/β-catenin signal transduction. Axin2 expression (difference, 14.46-fold) was higher in SPF mice than in GF mice. C: Gene expression levels of the Wnt ligands were Wnt5a (difference, −6.41-fold) and Wnt11 (difference, −4.18-fold) compared with WT controls. Conventionalization of GF mice for 4 weeks induces expression of Wnt5a (difference, 4.4-fold) and Wnt11 (difference, 2.36-fold). D: Gene expression of Wnt5a (difference, −1.75-fold) but not Wnt11 is markedly decreased in Myd88 KO mice. E: Detection of EdU-positive cells in crypt-luminal axis in the baseline state (2 hours after flash label) was 44.2 ± 2.2 μm proximal and 63.5 ± 2.6 μm distal in Myd88 KO mice compared with 21.9 ± 0.9 μm proximal and 22.7 ± 0.6 μm distal in WT controls, measured from crypt base. F: IEC migration in Myd88 KO mice, measured as distance from 2-hour baseline position, at 4 hours was 7.1 ± 2 μm proximal and 4.2 ± 1.8 μm distal, at 8 hours was 9.2 ± 1.1 μm proximal and 4.5 ± 1.9 μm distal, and at 12 hours was 9.8 ± 1.5 μm proximal and 6.5 ± 1.8 μm distal after the EdU flash label. Data are expressed as means ± SEM and represent three experiments with three mice per group. CON, conventionalized; h, hour.
Bacterial epithelial crosstalk in the intestine is mediated by different receptors and signaling pathways. Toll-like receptors have been described to play a major role in mediating the interaction of the intestinal microbiome and the intestinal mucosa.24 Specifically, the myeloid differentiation primary response gene 88 (MYD88) has been linked to Toll-like receptor–related signal transduction.25 Therefore, Wnt gene expression was analyzed in Myd88 knockout (KO) mice and compared with WT controls. Here, reduced expression of Wnt5a but not Wnt11 was observed, indicating the differential importance of this pathway in regulating Wnt5a expression levels (Figure 3D). Taken together these data showed distinct differences in Wnt gene expression, specifically of the Wnt ligands Wnt5a and Wnt11. Furthermore, we report that Wnt5a expression but not Wnt11 expression depended on MYD88-mediated signaling during intestinal homeostasis.
To evaluate IEC migration after KO of Myd88, EdU pulse chase experiments were performed in Myd88 KO mice. As shown in Figure 3E, in Myd88 KO mice EdU-positive cells were detected at higher positions in crypt-luminal axis in the baseline state (2-hour flash label) compared with WT mice. This observation is consistent with the original report of proliferation in Myd88 KO mice.26 However, migration of epithelial cells from the baseline toward the luminal surface is decreased in Myd88 KO mice (Figure 3F). At the 4-hour position IECs migrated 7.1 ± 2 μm in the proximal colon and 4.2 ± 1.8 μm in the distal colon. At 8 hours after flash label we observed migration of 9.2 ± 1.1 μm in the proximal colon and 4.5 ± 1.9 μm in the distal colon. Finally, 12 hours after flash label IEC migration was 9.8 ± 1.5 μm in the proximal colon and 6.5 ± 1.8 μm in the distal colon.
Discussion
The Wnt signaling pathway is a critical mediator of intestinal epithelial and mucosal homeostasis. Although it is well established that canonical Wnt/β-catenin signaling promotes proliferation of intestinal stem cells in the stem cell niche, knowledge about regional patterns of Wnt signaling activity and its components is limited.2 In this report we demonstrate a distinct profile of Wnt target gene expression in the proximal versus distal colon of mice.
Our findings indicate increased expression of Wnt/Wnt target genes that correspond with increased canonical Wnt/β-catenin activity (Axin2, Ccnd1) in the proximal colon compared with the distal colon. Furthermore, we observed increased mRNA expression of the noncanonical Wnt ligands Wnt5a and Wnt11 in the proximal colon compared with the distal colon. All together our results suggest that the elevated activity of canonical Wnt/β-catenin signaling in the proximal colon is accompanied by increased expression of noncanonical signaling mediators and by increased cell migration along the crypt-luminal axis in the proximal compared with the distal colonic crypts. The heterogeneity of the proximal and distal colon might, therefore, be partly connected to regional differences in Wnt gene expression and Wnt/β-catenin activity.
This is particularly interesting, because specific distribution of tumors is observed in patients with colon cancer, including hereditary nonpolyposis colorectal cancer. Adenocarcinomas in the proximal colon have a higher likelihood of microsatellite instability and are associated with older age and poorer differentiation compared with tumors localized in the distal colon. Sporadic cancers independent of family history are more frequently seen in the distal colon.27,28 At this point we can only speculate whether this might be pathophysiologically connected to increased cell turnover and different Wnt gene expression as reported in this study. However, there appears to be a relationship between physiological epithelial homeostatic events and dysregulation of the respective pathways in development of diseases. Specifically, mucosal inflammation and ulceration as seen in inflammatory bowel disease has been associated with altered Wnt gene expression.13 Thus, distinct regional expression of Wnt signaling proteins could attribute to differences in pathological involvement of the colon in these diseases. Future studies that address these questions will be of value in further understanding of the molecular mechanisms associated with inflammation-associated cancer such as observed in inflammatory bowel disease.
The bacterial ecosystem in the mammalian gut consists of a high number of microorganisms that reside in homeostasis with the host’s immune system. The effect of commensal microbiota on the host’s biology ranges from morphological integrity of the IECs to metabolic, absorptive, and endocrine functions.29,30 Comparative gene expression studies of GF and SPF mice have shown direct host responses to commensal bacteria that induce changes at a molecular level.31–34 Sun et al35 have investigated the effect of the nonpathogenic Salmonella strain PhoP(c) on IECs and reported that this crosstalk enhances β-catenin signaling through blocking β-catenin degradation. In addition, Cheesman et al18 have shown that in zebrafish the stability of β-catenin in IECs can be enhanced by resident intestinal bacteria that thereby induce cell proliferation of the IECs. Concordant with the previous reports, we saw significantly higher Wnt/β-catenin activity in SPF mice than in GF mice. Microbiota have also been associated with development of intestinal disease such as inflammatory bowel disease, inflammatory bowel disease–associated carcinoma, and hereditary colon carcinoma.36 However, little is known about the effect of commensal bacteria on upstream effectors of β-catenin signaling. Here, we report differential expression of Wnt signaling components in GF mice compared with colonized animals. With the use of comparative qPCR, we found that the colon of mice raised in GF conditions exhibits decreased expression levels of the ligands Wnt5a and Wnt11 compared with SFP mice. Conventionalization of GF mice with commensal bacteria for 4 weeks is sufficient to notably increase gene expression levels of Wnt5a and Wnt11. In this context Wnt5a could function as an important regulator of molecular responses to bacterial epithelial crosstalk. Secretion of Wnt5a has been reported to alter production of cytokines such as IL-10 and IL-6 by dendritic cells and therefore play a role in regulation of the balance between tolerance and inflammation.37 By contrast, Wnt5a has been described to be of central importance in mediating mucosal wound repair in the intestine.21 Therefore, Wnt5a not only plays a role during morphogenesis and embryonic intestinal development4 but might also contribute to the regulation of homeostasis in the adult intestine. However, it has to be pointed out that intestinal homeostasis is a complex process with a variety of factors that influence this process. These factors include an influence of luminal bacteria and bacterial products that modulate intestinal epithelial homeostasis. Interestingly, compared with WT controls, Myd88 KO mice showed decreased mRNA expression of the Wnt ligand Wnt5a but not Wnt11. The mechanisms that might be involved in regulating Wnt11 expression still need further investigation. Thus, molecular pathways that lead to induction of Wnt ligands in the intestinal mucosa are not yet well established, and different signal mediators, such as transforming growth factor-β, have been associated with the induction of Wnt11 expression.38 Interestingly, the proximal promoter region of Wnt11 contains Tcf/LEF binding sites, suggesting that Wnt/β-catenin signals may increase expression of Wnt11.39 Here, we report reduced expression of Wnt11 and Wnt/β-catenin activity in GF mice compared with SPF mice. At this point, we can only speculate whether this is a causative or associated mechanism, and future investigations are needed to further understand these mechanisms.
Taken together our results indicate distinct regional expression patterns of the Wnt signaling pathway in the proximal versus distal colon. Furthermore, our studies suggest that the commensal bacteria in the intestine influence Wnt signaling pathways that in turn contribute to the regional differences in intestinal epithelial homeostasis.
Acknowledgments
We thank Dr. Alyssa Bushey Long (Emory University) and Dr. Jing Yu for their help and suggestions with in situ hybridization of Wnt ligands in the colon.
Footnotes
Supported by the NIH grants R01 DK055679 and DK059888 (A.N.) and R01HD059122 (P.D.) and the German Research Foundation (DFG) grant NE 1834/1-1 (P.A.N.).
Disclosures: None declared.
Supplemental Data
In situ hybridization was performed to detect Wnt8b and Wnt11 mRNA expression in the intestinal mucosa. A: Canonical Wnt8b was expressed in epithelial cells in the base of crypts (arrow) with increased expression in the distal colon. B: Noncanonical Wnt11 was predominantly expressed in surface epithelial cells (arrow) in the proximal colon. Scale bar = 50μm.
References
- 1.Nusse R. Wnt signaling in disease and in development. Cell Res. 2005;15:28–32. doi: 10.1038/sj.cr.7290260. [DOI] [PubMed] [Google Scholar]
- 2.Pinto D., Gregorieff A., Begthel H., Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nava P., Koch S., Laukoetter M.G., Lee W.Y., Kolegraff K., Capaldo C.T., Beeman N., Addis C., Gerner-Smidt K., Neumaier I., Skerra A., Li L., Parkos C.A., Nusrat A. Interferon-gamma regulates intestinal epithelial homeostasis through converging beta-catenin signaling pathways. Immunity. 2010;32:392–402. doi: 10.1016/j.immuni.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lickert H., Kispert A., Kutsch S., Kemler R. Expression patterns of Wnt genes in mouse gut development. Mech Dev. 2001;105:181–184. doi: 10.1016/s0925-4773(01)00390-2. [DOI] [PubMed] [Google Scholar]
- 5.Tai C.C., Sala F.G., Ford H.R., Wang K.S., Li C., Minoo P., Grikscheit T.C., Bellusci S. Wnt5a knock-out mouse as a new model of anorectal malformation. J Surg Res. 2009;156:278–282. doi: 10.1016/j.jss.2009.03.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinto D., Clevers H. Wnt control of stem cells and differentiation in the intestinal epithelium. Exp Cell Res. 2005;306:357–363. doi: 10.1016/j.yexcr.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 7.Gregorieff A., Pinto D., Begthel H., Destree O., Kielman M., Clevers H. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129:626–638. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Sellin J.H., DeSoignie R. Rabbit proximal colon: a distinct transport epithelium. Am J Physiol. 1984;246:G603–G610. doi: 10.1152/ajpgi.1984.246.5.G603. [DOI] [PubMed] [Google Scholar]
- 9.Potten C.S., Kellett M., Roberts S.A., Rew D.A., Wilson G.D. Measurement of in vivo proliferation in human colorectal mucosa using bromodeoxyuridine. Gut. 1992;33:71–78. doi: 10.1136/gut.33.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finkel Y., Larsson L. Morphometric and functional studies of proximal and distal colon in young and adult rats. J Pediatr Gastroenterol Nutr. 1987;6:454–459. [PubMed] [Google Scholar]
- 11.Clauss W., Hornicke H. Segmental differences in K-transport across rabbit proximal and distal colon in vivo and in vitro. Comp Biochem Physiol A Comp Physiol. 1984;79:267–269. doi: 10.1016/0300-9629(84)90427-4. [DOI] [PubMed] [Google Scholar]
- 12.Nava G.M., Friedrichsen H.J., Stappenbeck T.S. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 2011;5:627–638. doi: 10.1038/ismej.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.You J., Nguyen A.V., Albers C.G., Lin F., Holcombe R.F. Wnt pathway-related gene expression in inflammatory bowel disease. Dig Dis Sci. 2008;53:1013–1019. doi: 10.1007/s10620-007-9973-3. [DOI] [PubMed] [Google Scholar]
- 14.Christie M., Jorissen R.N., Mouradov D., Sakthianandeswaren A., Li S., Day F., Tsui C., Lipton L., Desai J., Jones I.T., McLaughlin S., Ward R.L., Hawkins N.J., Ruszkiewicz A.R., Moore J., Burgess A.W., Busam D., Zhao Q., Strausberg R.L., Simpson A.J., Tomlinson I.P., Gibbs P., Sieber O.M. Different APC genotypes in proximal and distal sporadic colorectal cancers suggest distinct WNT/β-catenin signalling thresholds for tumourigenesis. Oncogene. 2003;32:4675–4682. doi: 10.1038/onc.2012.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bara J., Nardelli J., Gadenne C., Prade M., Burtin P. Differences in the expression of mucus-associated antigens between proximal and distal human colon adenocarcinomas. Br J Cancer. 1984;49:495–501. doi: 10.1038/bjc.1984.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bufill J.A. Colorectal cancer: evidence for distinct genetic categories based on proximal or distal tumor location. Ann Intern Med. 1990;113:779–788. doi: 10.7326/0003-4819-113-10-779. [DOI] [PubMed] [Google Scholar]
- 17.Engel M.A., Khalil M., Mueller-Tribbensee S.M., Becker C., Neuhuber W.L., Neurath M.F., Reeh P.W. The proximodistal aggravation of colitis depends on substance P released from TRPV1-expressing sensory neurons. J Gastroenterol. 2012;47:256–265. doi: 10.1007/s00535-011-0495-6. [DOI] [PubMed] [Google Scholar]
- 18.Cheesman S.E., Neal J.T., Mittge E., Seredick B.M., Guillemin K. Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4570–4577. doi: 10.1073/pnas.1000072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Arthur J.C., Perez-Chanona E., Muhlbauer M., Tomkovich S., Uronis J.M., Fan T.J., Campbell B.J., Abujamel T., Dogan B., Rogers A.B., Rhodes J.M., Stintzi A., Simpson K.W., Hansen J.J., Keku T.O., Fodor A.A., Jobin C. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyoshi H., Ajima R., Luo C.T., Yamaguchi T.P., Stappenbeck T.S. Wnt5a potentiates TGF-beta signaling to promote colonic crypt regeneration after tissue injury. Science. 2012;338:108–113. doi: 10.1126/science.1223821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ouko L., Ziegler T.R., Gu L.H., Eisenberg L.M., Yang V.W. Wnt11 signaling promotes proliferation, transformation, and migration of IEC6 intestinal epithelial cells. J Biol Chem. 2004;279:26707–26715. doi: 10.1074/jbc.M402877200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chow J., Lee S.M., Shen Y., Khosravi A., Mazmanian S.K. Host-bacterial symbiosis in health and disease. Adv Immunol. 2010;107:243–274. doi: 10.1016/B978-0-12-381300-8.00008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marques R., Boneca I.G. Expression and functional importance of innate immune receptors by intestinal epithelial cells. Cell Mol Life Sci. 2011;68:3661–3673. doi: 10.1007/s00018-011-0829-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wells J.M., Rossi O., Meijerink M., van Baarlen P. Epithelial crosstalk at the microbiota-mucosal interface. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4607–4614. doi: 10.1073/pnas.1000092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rakoff-Nahoum S., Paglino J., Eslami-Varzaneh F., Edberg S., Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Andrieu N., Launoy G., Guillois R., Ory-Paoletti C., Gignoux M. Estimation of the familial relative risk of cancer by site from a French population based family study on colorectal cancer (CCREF study) Gut. 2004;53:1322–1328. doi: 10.1136/gut.2003.036376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinicrope F.A., Rego R.L., Foster N., Sargent D.J., Windschitl H.E., Burgart L.J., Witzig T.E., Thibodeau S.N. Microsatellite instability accounts for tumor site-related differences in clinicopathologic variables and prognosis in human colon cancers. Am J Gastroenterol. 2006;101:2818–2825. doi: 10.1111/j.1572-0241.2006.00845.x. [DOI] [PubMed] [Google Scholar]
- 29.Round J.L., Mazmanian S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith K., McCoy K.D., Macpherson A.J. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Backhed F., Ding H., Wang T., Hooper L.V., Koh G.Y., Nagy A., Semenkovich C.F., Gordon J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hooper L.V., Wong M.H., Thelin A., Hansson L., Falk P.G., Gordon J.I. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 33.Salzman N.H., Underwood M.A., Bevins C.L. Paneth cells, defensins, and the commensal microbiota: a hypothesis on intimate interplay at the intestinal mucosa. Semin Immunol. 2007;19:70–83. doi: 10.1016/j.smim.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 34.Stappenbeck T.S., Hooper L.V., Gordon J.I. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci U S A. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun J., Hobert M.E., Rao A.S., Neish A.S., Madara J.L. Bacterial activation of beta-catenin signaling in human epithelia. Am J Physiol Gastrointest Liver Physiol. 2004;287:G220–G227. doi: 10.1152/ajpgi.00498.2003. [DOI] [PubMed] [Google Scholar]
- 36.Arthur J.C., Jobin C. The struggle within: microbial influences on colorectal cancer. Inflamm Bowel Dis. 2011;17:396–409. doi: 10.1002/ibd.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oderup C., LaJevic M., Butcher E.C. Canonical and noncanonical Wnt proteins program dendritic cell responses for tolerance. J Immunol. 2013;190:6126–6134. doi: 10.4049/jimmunol.1203002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang P., Cai Y., Soofi A., Dressler G.R. Activation of Wnt11 by transforming growth factor-beta drives mesenchymal gene expression through non-canonical Wnt protein signaling in renal epithelial cells. J Biol Chem. 2012;287:21290–21302. doi: 10.1074/jbc.M112.357202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uysal-Onganer P., Kypta R.M. Wnt11 in 2011-the regulation and function of a non-canonical Wnt. Acta Physiol (Oxf) 2012;204:52–64. doi: 10.1111/j.1748-1716.2011.02297.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In situ hybridization was performed to detect Wnt8b and Wnt11 mRNA expression in the intestinal mucosa. A: Canonical Wnt8b was expressed in epithelial cells in the base of crypts (arrow) with increased expression in the distal colon. B: Noncanonical Wnt11 was predominantly expressed in surface epithelial cells (arrow) in the proximal colon. Scale bar = 50μm.