Abstract
We have previously identified osteoactivin (OA), encoded by Gpnmb, as an osteogenic factor that stimulates osteoblast differentiation in vitro. To elucidate the importance of OA in osteogenesis, we characterized the skeletal phenotype of a mouse model, DBA/2J (D2J) with a loss-of-function mutation in Gpnmb. Microtomography of D2J mice showed decreased trabecular mass, compared to that in wild-type mice [DBA/2J-Gpnmb+/SjJ (D2J/Gpnmb+)]. Serum analysis showed decreases in OA and the bone-formation markers alkaline phosphatase and osteocalcin in D2J mice. Although D2J mice showed decreased osteoid and mineralization surfaces, their osteoblasts were increased in number, compared to D2J/Gpnmb+ mice. We then examined the ability of D2J osteoblasts to differentiate in culture, where their differentiation and function were decreased, as evidenced by low alkaline phosphatase activity and matrix mineralization. Quantitative RT-PCR analyses confirmed the decreased expression of differentiation markers in D2J osteoblasts. In vitro, D2J osteoblasts proliferated and survived significantly less, compared to D2J/Gpnmb+ osteoblasts. Next, we investigated whether mutant OA protein induces endoplasmic reticulum stress in D2J osteoblasts. Neither endoplasmic reticulum stress markers nor endoplasmic reticulum ultrastructure were altered in D2J osteoblasts. Finally, we assessed underlying mechanisms that might alter proliferation of D2J osteoblasts. Interestingly, TGF-β receptors and Smad-2/3 phosphorylation were up-regulated in D2J osteoblasts, suggesting that OA contributes to TGF-β signaling. These data confirm the anabolic role of OA in postnatal bone formation.
Osteoporosis is a growing public health problem, in part because of the increasing numbers of people living beyond the age of 65 years.1 It is characterized by low bone mass due to increased bone resorption by osteoclasts and decreased bone formation by osteoblasts, with significant deterioration in the bone microarchitecture leading to high bone fragility and increased fracture risk.1,2 The net effect of osteoporosis is low bone mass.1 There is an increasing demand for identifying novel bone anabolic factors with potential therapeutic benefits in treating generalized bone loss, such as osteoporosis and/or major skeletal fracture.
Osteoactivin is a novel glycoprotein first identified in natural mutant osteopetrotic rats.3 The same protein has been identified and named separately in several other species: as dendritic cell heparan sulfate proteoglycan integrin dependent ligand (DCHIL) in mouse dendritic cells,4 as transmembrane glycoprotein NMB (GPNMB) in human melanoma cell lines and melanocytes,5 and as hematopoietic growth factor inducible neurokinin (HGFIN) in human tumor cells.6 The current recommended name for the protein encoded by Gpnmb in mouse is transmembrane glycoprotein NMB (http://www.ncbi.nlm.nih.gov/protein/Q99P91.2); here, we continue to use osteoactivin (OA) for the protein and Gpnmb for the gene. OA is a type I transmembrane protein that consists of multiple domains, including an extracellular domain, transmembrane domain, and protein sorting signal sequence.7 Within the C-terminal domain, OA has an RGD motif, predicting an integrin attachment site.3,7–9
Our research group initially reported on the novel role of OA in osteoblast differentiation and function.7–10 We demonstrated that OA expression has a temporal pattern during osteoblast differentiation, being highest during matrix maturation and culture mineralization in vitro.7–11 Using loss-of–function and gain-of–function approaches in osteoblasts, we reported that OA overexpression increases osteoblast differentiation and function and that OA down-regulation decreases nodule formation, alkaline phosphatase (ALP) activity, osteocalcin (OC) production, and matrix mineralization in vitro.7 We also reported on the positive role of OA in mesenchymal stem cell (MSCs) differentiation into osteoblasts in vitro.12 In another study, we showed that recombinant OA protein induces higher osteogenic potential of fetal-derived MSCs, compared with bone marrow–derived MSCs13 and its osteogenic effects in the mouse C3H10T1/2 MSC cell line were similar to those of recombinant BMP-2.12 We also localized OA protein as associated predominately with osteoblasts lining trabecular bones in vivo,11 and showed that local injection of recombinant OA increased bone mass in a rat model.14 Moreover, in a fracture repair model OA expression increased over time, reaching a maximum 2 weeks after fracture.11 In a parallel study, recombinant OA supported bone regeneration and formation in a rat critical-size calvarial defect model.15 Others have shown that OA is highly expressed by osteoclasts in vitro, suggesting that it may regulate osteoclast formation and activity.16
There is urgent need for an animal model to fully examine the role of OA in osteogenesis. Interestingly, a natural mutation of the Gpnmb gene has been identified in the DBA/2J (D2J) mouse strain.17 These mice exhibit high-frequency hearing loss, which begins at the time of weaning and becomes severe by 2 to 3 months of age.18,19 Aged D2J mice also develop progressive eye abnormalities that closely mimic human hereditary glaucoma. The onset of disease symptoms falls roughly between 3 and 4 months of age, and disease becomes severe by 6 months of age.5,20 D2J mice are homozygous for a nonsense mutation in the Gpnmb gene sequence that induces an early stop codon, generating a truncated protein sequence of 150 amino acids (aa) instead of the full-length 562-aa OA protein.5 The control for the D2J mouse is the wild-type DBA/2J-Gpnmb+/SjJ mouse (D2J/Gpnmb+), homozygous for the wild-type Gpnmb gene.21 These Gpnmb wild-type mice do not develop glaucoma, as D2J mice do, although they exhibit mild iris stromal atrophy.21
In the present study, we used Gpnmb mutant (D2J) and Gpnmb wild-type (D2J/Gpnmb+) mice to gain insight into the role of OA in osteogenesis and in osteoblast differentiation and function. Here, we report that loss-of–function mutation of Gpnmb suppresses bone formation by directly affecting osteoblast proliferation and survival, leading to a decreased number of differentiated osteoblasts with suppressed activity in bone mineralization. Thus, our data point to OA as a novel and positive regulator of postnatal bone formation.
Materials and Methods
Mice
Gpnmb-mutant DBA/2J mice (D2J) and mice of a control strain [DBA/2J-Gpnmb+/SjJ (D2J/Gpnmb+)] with wild-type Gpnmb alleles were obtained from the Jackson Laboratory (Bar Harbor, ME). The DBA/2J-Gpnmb+/SjJ strain was generated by crossing DBA/2J-Dtnbp1sdy/J mice with modern D2J mice. DBA/2J-Dtnbp1sdy/J mice are D2J mice with a mutation for sandy coat color that appeared in 1983, before the appearance of the loss-of–function mutation in Gpnmb. The strain has the wild-type Gpnmb alleles and has been maintained as a separate strain since then. After crossing DBA/2J-Dtnbp1sdy/J mice with D2J mice, the progeny were selected for the Gpnmb wild-type alleles.21 The wild-type allele was backcrossed to D2J for a minimum of six generations. Gpnmb+ is the ancestral D2J allele, and the homozygous Gpnmb nonsense mutation is the only known genetic difference between the D2J and D2J/Gpnmb+ strains.5
All mice were housed in cages containing white pine bedding and covered with polyester filters. The environment was kept at 21°C with a 12-hour light–dark cycle. Mouse colonies were maintained at Northeast Ohio Medical University in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC), under veterinary supervision and according to the guidelines of the Institutional Animal Care and Use Committee.
Micro-CT
Femurs from 4-, 8-, and 16-week-old male D2J and D2J/Gpnmb+ mice (n ≥ 4 per group) were analyzed using a SkyScan 1172 high-resolution microtomography (micro-CT) system (Bruker, Billerica, MA). Trabecular measurements of femurs were taken 400 μm below the distal growth plate in 750 consecutive slices of 7.6-μm resolution over a distance of 5700 μm, and volumetric regions were rendered as three-dimensional arrays, using SkyScan NRecon software. Percentage of bone volume per tissue volume [(BV/TV)%], trabecular number (Tb.N; no./mm), trabecular thickness (Tb.Th; μm), and trabecular separation (Tb.Sp; μm) were measured and calculated using SkyScan CT analyzer software. Three-dimensional reconstructed images of the sagittal and axial planes of the femoral metaphysis were generated using SkyScan CTvox software version 2.4.
Biochemical Analysis
Serum samples were prepared from 8- and 16-week-old male D2J and D2J/Gpnmb+ mice (n ≥ 3 per group) that had been fasted for 12 hours. Serum ALP (BIOTANG, Waltham, MA), OC (Biomedical Technologies, Stoughton, MA) and OA (R&D Systems, Minneapolis, MN) were measured by enzyme-linked immunosorbent assay (ELISA) according to the manufacturer’s instructions. In some conditions, OA protein was quantified by ELISA in total protein isolated from the calvaria of 8-week-old mice and from primary osteoblast cultures. All ELISA data from serum, calvaria, and cultures were normalized to the total μg protein concentration per sample using a Pierce bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Waltham, MA).
Tissue Preparation and Bone Histomorphometry
At necropsy, distal femurs from 8-week-old male D2J and D2J/Gpnmb+ mice (n ≥ 3 per group) were dissected, fixed in 4% paraformaldehyde, dehydrated, and embedded undecalcified in plastic methylmethacrylate resin. Sagittal sections (5 μm thick) were stained with von Kossa–toluidine blue or Masson’s trichrome stain.22
Quantitative histomorphometry was performed in an area from 100 μm to 600 μm proximal to the growth plate, using OsteoMeasure software version 3.2.1 (Osteometrics, Decatur, GA). Images were acquired with a brightfield microscope (Olympus, Center Valley, PA; Tokyo, Japan) equipped with 10× and 20× objectives and a digital color video camera with a 0.7× reduction lens (Olympus 3CCD). Three-dimensional and two-dimensional parameters were measured and calculated in three sections per animal. The three-dimensional parameters were trabecular number [Tb.N = (4/π) × 0.5 × (B.Pm/T.Ar)] (no./mm) and trabecular separation [Tb.Sp = (1/Tb.N) × (1000 − Tb.Th)] (μm). The two-dimensional parameters were trabecular number per tissue area (Tb.N/T.Ar), bone perimeter per tissue area (B.Pm/T.Ar), osteoblast number per bone perimeter (N.Ob/B.Pm), percentage of osteoblast surface per bone surface [(Ob.S/B.S)%], percentage of osteoid area per bone area [(O.Ar/B.Ar) %], osteoid width (O.Wi μm), and osteoid maturation time [O.M.T, calculated as O.Wi/mineral apposition rate (MAR) per day].
For dynamic histomorphometry, 8-week-old male D2J and D2J/Gpnmb+ mice (n = 4 per group) were injected intraperitoneally with 10 mg/kg calcein AM (Thermo Fisher Scientific) at 7 and 2 days before sacrifice. Femurs were collected from euthanized mice, and histomorphometric analysis of undecalcified sections of the distal femur was performed with transmitted and epifluorescence microscopy using a Nikon Eclipse Ti inverted microscope (Nikon, Melville, NY; Tokyo, Japan). Single-labeled surface (sLS), double-labeled surface (dLS), mineralizing surface [MS = dLS + (sLS/2)], MAR (calculated using average dLS per day), and bone formation rate (BFR, calculated as MAR × MS) were measured and calculated.
Mouse Calvarial Osteoblast Cultures
Primary osteoblasts were isolated and cultured as described previously.7–9 In brief, the calvaria from postnatal D2J and D2J/Gpnmb+ mice at day 4 to 7 were isolated and digested with 0.25% trypsin (Thermo Fisher Scientific) and 0.1% collagenase B (Thermo Fisher Scientific) for five cycles (20 minutes/cycle) at 37°C. Only cells from digests 2 to 5 were collected. Cells were plated at a density of 2 × 104 cells/cm2 in Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Fisher Scientific) and cultured until confluent. Cells were then maintained in DMEM containing 10% fetal bovine serum (FBS) (HyClone; Thermo Fisher Scientific), 100 IU/mL penicillin, 100 μg/mL streptomycin (HyClone), and 25 ng/mL amphotericin B (HyClone). For osteoblast differentiation, cells were cultured in minimum essential medium α (HyClone) supplemented with 10% FBS, 50 μg/mL ascorbic acid, and 10 mmol/L β-glycerophosphate (Thermo Fisher Scientific). The medium was changed every 2 to 3 days.
Adult Mouse Long-Bone Osteoblast Cultures
Primary osteoblasts were isolated and cultured as described previously.23 In brief, femurs and tibias from 8-week-old D2J and D2J/Gpnmb+ mice were dissected, epiphyses were cut off, and bone marrow was flushed out. The diaphyses were cut into small pieces and digested with 0.2% collagenase B for 2 hours at 37°C. Bone pieces were transferred to 10-cm culture dishes at a density of 20 to 30 fragments per dish. Adult mouse bone cells migrate from the bone chips after approximately 5 days. Cells were trypsinized and replated at a density of 2 × 104 cells/cm2 in DMEM and cultured until confluent. Cells were then maintained in DMEM containing 10% FBS, 100 IU/mL penicillin, 100 μg/mL streptomycin, and 25 ng/mL amphotericin B. For osteoblast differentiation, cells were cultured in minimum essential medium α supplemented with 10% FBS, 50 μg/mL ascorbic acid, and 10 mmol/L β-glycerophosphate. The medium was changed every 2 to 3 days.
Proliferation–Apoptosis and Survival Assays
Primary osteoblasts from the calvaria of neonatal D2J and D2J/Gpnmb+ mice or from long bones of adult D2J and D2J/Gpnmb+ mice were cultured in 96-well plates (6 × 103 cells/cm2) in DMEM supplemented with 10% FBS in 96-well plate and total DNA content was measured after 24, 48, and 72 hours, using a CyQUANT NF cell proliferation assay kit (Life Technologies, Carlsbad, CA) according to the manufacturer’s protocol. In brief, medium was removed and replaced with 100 μL 1× dye binding solution and incubated at 37°C in for 1 hour in the dark. Sample fluorescence intensity was measured using BioTek Synergy H4 microplate reader (BioTek Instruments, Winooski, VT) with excitation at 485 nm and emission detection at 530 nm.
In parallel experiments, osteoblasts were treated with DMEM supplemented with 10% serum isolated from 8-week-old D2J or D2J/Gpnmb+ mice (n = 3 per group), and total DNA content was measured after 24 hours as described above. For cell apoptosis assay, primary osteoblasts from the calvaria of neonatal D2J and D2J/Gpnmb+ mice were cultured in 96-well plates (1 × 104 cells/cm2) in DMEM with 10% FBS; serum was then reduced to 2.5% for 24, 48, or 72 hours of culture, and cell apoptosis was measured using a Caspase-Glo 3/7 assay (Promega, Madison, WI). In brief, medium was removed and replaced with 100 μL of Caspase-Glo 3/7 reagent and cells were incubated at room temperature for 3 hours in the dark. Luciferase reaction was measured using a BioTek Synergy H4 microplate reader (BioTek Instruments) with excitation at 485 nm and emission detection at 530 nm.
For cell survival assay, primary osteoblasts were cultured in 96-well (1 × 104 cells/cm2) in DMEM with 10% FBS and then were treated with tunicamycin (5 μg/mL) (Sigma-Aldrich, St. Louis, MO) and cell survival was measured using MTT assay. In brief, medium was removed and replaced with 1.2 mmol/L MTT substrate (Sigma-Aldrich) in 1× Hanks’ balanced salt solution, and cells were then incubated at 37°C for 4 hours in the dark. MTT working reagent was removed, and formazan crystals were dissolved in 100 μL dimethyl sulfoxide. Absorbance of the colorimetric reaction was measured at 540 nm.
Immunofluorescence Staining
Primary osteoblasts from the calvaria of neonatal D2J and D2J/Gpnmb+ mice were cultured in four-well chamber slides (1 × 103 cells/cm2) for 48 hours, fixed in situ with 4% paraformaldehyde for 20 minutes, washed in PBS, permeabilized with 0.5% Triton X, and incubated with anti-mouse OA polyclonal primary antibody (R&D Systems) and/or anti-calnexin monoclonal antibody [a marker of rough endoplasmic reticulum (rER)] (BD Biosciences, San Jose, CA) overnight in a humidified chamber at 4°C. Cells were then washed and incubated with Cy2-conjugated secondary goat anti-mouse OA antibody and/or Cy3-conjugated secondary goat anti-mouse calnexin antibody (Molecular Probes; Life Technologies, Carlsbad, CA). Nuclei were labeled using Fluoroshield mounting medium with DAPI (Abcam, Cambridge, MA). Fluorescence images were captured with a Nikon Eclipse Ti inverted microscope using a 490-nm excitation filter for OA (green), a 550-nm excitation filter for calnexin (red), and a 350-nm excitation filter for DAPI (blue).
Transmission Electron Microscopy
Primary osteoblasts from calvaria of neonatal D2J and D2J/Gpnmb+ mice were cultured in DMEM (no β-glycerophosphate) in Petri dishes for 10 days, pelleted down, and fixed in situ for 1 hour at 4°C in 2% glutaraldehyde in 0.1 mol/L sodium cacodylate buffer (pH 7.4). Specimens were washed overnight with 7% sucrose buffer, postfixed for 1 hour in 2% osmium tetroxide in 0.1 mol/L sodium cacodylate buffer (pH 7.4), and stained en bloc for 30 minutes with 0.1% aqueous uranyl acetate. After dehydration, cell pellets were transferred into glass vials, washed with propylene oxide, and embedded in Spurr’s resin. For long bones, femurs from 8-week-old D2J and D2J/Gpnmb+ were fixed and embedded in Spurr’s resin. Ultrathin bone sections (2 μm) or cell pellets were stained with uranyl acetate and lead citrate before examination with a Philips CM10 transmission electron microscope (FEI, Hillsboro, OR).
Quantification of Osteoblast Matrix Formation
Matrix maturation in osteoblast cultures was determined by ALP staining and activity7,9 and matrix collagen staining. ALP staining was performed for calvarial osteoblasts at day 14, using an ALP staining kit (Sigma-Aldrich) according to the manufacturer’s instructions. ALP-positive cells were stained purple. In parallel wells, ALP activity was determined in total cell lysates using an ALP activity kit (BioAssay Systems, Hayward, CA) according to the manufacturer’s protocol. In brief, 50 μL of cell lysate was added to 150 μL of working solution, and optical density absorbance was measured at 405 nm at 0 and 4 minutes. ALP activity was calculated in enzyme units (U = μmol/L, per minute) per μg protein. For collagen, parallel culture wells were fixed in 4% paraformaldehyde and stained for collagen with Sirius Red (0.1% in saturated picric acid) for 30 minutes on a shaker at room temperature, washed twice in PBS, and air dried. Stained wells were scanned at 2400 dpi, and quantification of collagen staining was performed using ImageJ software version 1.48p (NIH, Bethesda, MD). The incorporated dye was extracted in 0.2 mol/L of sodium hydroxide in methanol (v/v) for 30 minutes with agitation, and 200 μL samples were read in a microplate reader at 490 nm. Data were normalized to the total protein concentration of each sample.
Quantification of Osteoblast Matrix Mineralization
Mineralized matrix in osteoblast cultures was determined by Alizarin Red S, calcium assay, and von Kossa staining as described previously.7,9 To visualize mineralized matrix at day 21, osteoblast cultures were stained with Alizarin Red S (40 mmol/L, pH 4.2) for 15 minutes and then were washed with distilled water. The incorporated dye was extracted in 10% (v/v) acetic acid, heated at 85°C for 10 minutes, centrifuged at 20,000 × g for 15 minutes, and neutralized with 10% (v/v) ammonium hydroxide. Optical density was then measured at 405 nm. In parallel wells, total calcium was determined in mineralized matrix lysed with 0.5 mol/L HCl using a QuantiChrom calcium assay kit (BioAssay Systems) according to the manufacturer’s instructions. In brief, 5 μL of cell lysates was added to 200 μL working solution and optical density was measured at 570 nm. Data were normalized to the total protein concentration of each sample.
To visualize mineralized nodules, parallel osteoblast cultures were stained at day 21 with von Kossa stain. In brief, cultures were stained with 5% silver nitrate solution under exposure to UV light for 20 minutes and then were washed with distilled water. Mineralized nodules were stained black for hydroxyapatite crystals and fixed with 5% sodium thiosulfate. Mineralized nodule count and size were measured using live-imaging Nikon Ti Eclipse microscopy with automated object capture using NIS-Elements software version 4.10.1.
Protein Isolation and SDS-PAGE
Total proteins were isolated from primary osteoblast cultures and subjected to SDS-PAGE electrophoresis as described previously.9 In brief, cells were lysed in radioimmunoprecipitation assay buffer (EMD Millipore, Billerica, MA), and protein concentration was measured using a Pierce bicinchoninic acid protein assay kit (Thermo Fisher Scientific). Protein samples (25 μg) were mixed with denaturing buffer, heated to 100°C, and subjected to 10% SDS-PAGE in 1 × Tris-glycine–SDS running buffer for 90 minutes. The gel was then transferred to a nitrocellulose membrane using a semidry transfer apparatus (Bio-Rad Laboratories, Hercules, CA) for 5 minutes. The blot was incubated in blocking buffer (5% nonfat milk in Tris-buffered saline) for 7 minutes using a SNAP i.d. protein detection system (EMD Millipore) and then was probed for OA (R&D Systems), GRP-78 (Thermo Fisher Scientific), TGF-β1 (Bioss, Woburn, MA), TGF-β receptor II (Bioss), and p-Smad-2 and p-Smad-3 (Cell Signaling Technology, Danvers, MA), in blocking buffer overnight at 4°C. Next, the blot was washed in 1× Tris-buffered saline/Tween 20 (TBST) and incubated with anti-rabbit IgG horseradish peroxidase–conjugated secondary antibody (R&D Systems) for 1 hour. The blot was then washed in TBST and incubated with Pierce chemiluminescent substrate (Thermo Fisher Scientific). Signals were detected using an ImageQuant LAS 4000 digital imaging system (GE Healthcare, Piscataway, NJ; Little Chalfont, UK). The blots were stripped briefly, washed in TBST, and reprobed for Smad-2, Smad-3, and actin.
RT-qPCR
For quantitative real-time RT-PCR (RT-qPCR), total RNA was extracted from calvaria, tibia, or primary osteoblasts of 8-week-old D2J and D2J/Gpnmb+ mice (n ≥ 3 per group), using TRIzol reagent (Life Technologies) and a SurePrep RNA extraction kit (Thermo Fisher Scientific). RNA quantities were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). cDNA was prepared using a high-capacity cDNA reverse transcription kit (Life Technologies). qPCR was performed with an ABI 7500 Fast real-time PCR system (Life Technologies) in duplicate 20-μL reactions, each containing 10 ng cDNA reaction mix, 100 nmol/L of each primer, and 10 μL 2× SYBR Green PCR master mix (Life Technologies). qPCR cycles consisted of an initial cycle of 50°C for 2 minutes, a second cycle of 95°C for 10 minutes, and then 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Using GAPDH as the internal control, relative gene expression among samples was calculated by comparison of CT values. A dissociation curve was checked for each qPCR run, to confirm specific amplification of target RNA. All qPCR mouse primers were purchased from Qiagen (Valencia, CA): proliferation cell nuclear antigen (PCNA), apoptosis regulator Bax, Runx2, Msx2, ALP, collagen type I (Col1), OC, OA, TGF-β1, TGF-β receptors I and II, and GAPDH.
Osteogenesis Array
Total RNA was isolated from the calvaria of 8-week-old D2J and D2J/Gpnmb+ mice using TRI Reagent (Sigma-Aldrich). After DNase treatment, RNA was further cleaned using an RNeasy mini kit (Qiagen). cDNA was synthesized using an RT2 First Strand kit (SABiosciences; Qiagen, Valencia, CA) according to the manufacturer’s instructions. Gene expression profiling was performed using a mouse osteogenesis RT2 Profiler PCR array (SABiosciences; Qiagen). This platform is designed to profile the gene expression of 84 key osteogenesis-associated genes. RT-qPCR was performed using an ABI 7500 Fast real-time PCR system (Life Technologies) according to the array manufacturer’s instructions. Relative gene expression was determined using the ΔΔCT method. Data were further analyzed with the SABiosciences PCR array data analysis web portal (http://www.SABiosciences.com/pcrarraydataanalysis.php).
Statistical Analysis
For all quantitative data generated, differences between individual groups were analyzed for statistical significance using GraphPad Prism software version 5.04 (GraphPad Software, La Jolla, CA). In most cases, when the data followed a normal distribution, one-factor or two-factor analysis of variance was performed, followed by a Bonferroni post hoc test. For comparisons between two group means, an unpaired–t-test was performed. Differences were considered statistically significant at P < 0.05. All in vitro experiments were repeated (n ≥ 3) with three replicates per experiment.
Results
Loss-of–Function Mutation in Gpnmb Reduces Bone Formation
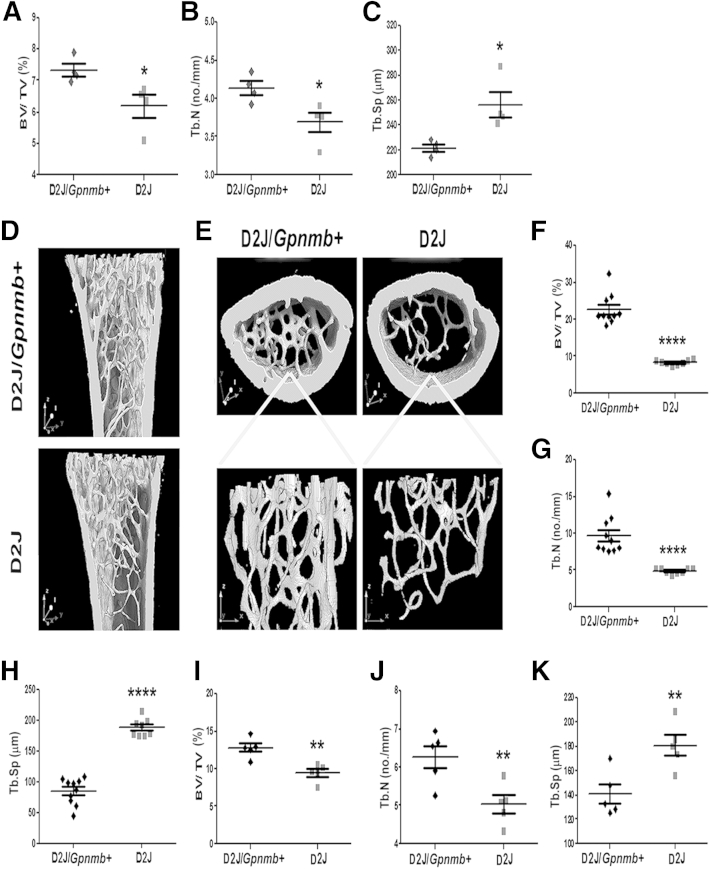
Having previously identified the role of OA in osteoblast differentiation and function in vitro,7–9,11 in the present study we evaluated the role of OA in bone formation in vivo, using a mouse model, D2J, with natural nonsense mutation of Gpnmb, compared with the D2J/Gpnmb+ wild type. Micro-CT of trabecular bone at 4 weeks of age showed a significant decrease in bone mass in D2J mice, compared to D2J/Gpnmb+ (Figure 1A). (Hereafter, if no other contrast is specified, differences reported for D2J mice are relative to D2J/Gpnmb+ mice.) Trabecular number was also significantly decreased in D2J mice, whereas trabecular separation was increased (Figure 1, B and C). At 8 weeks of age, micro-CT of the femoral metaphysis showed increased width of femoral metaphysis and consistent decrease of trabecular mass in D2J mice (Figure 1, D and E). Trabecular volume was decreased by approximately 60% in D2J mice (Figure 1F), and trabecular number was decreased by approximately 50% (Figure 1G), but trabecular separation was increased (Figure 1H).
Figure 1.
Micro-CT analysis of femur in 4-week-old (A–C), 8-week-old (F–H), and 16-week-old (I–K) D2J mutant and D2J/Gpnmb+ wild-type mice for BV/TV (A, F, and I), Tb.N (B, G, and J), and Tb.Sp (C, H, and K). D and E: Representative three-dimensional micro-CT–reconstructed image of the sagittal (D) and axial (E) planes of the femoral metaphysis, with the corresponding three-dimensional trabecular plate without cortical bone in 8-week-old mice. Data are expressed as means ± SEM. n = 4 (A–C), n = 5 (I–K), and n ≥ 8 (F–H) mice per group. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗∗P < 0.0001.
To confirm the observed decreased bone mass in D2J mice, compared to D2J/Gpnmb+ mice, we performed micro-CT of the femoral metaphysis at 16 weeks of age, when mice reach skeletal maturity. Consistent with the above data, our findings showed significant decrease in bone volume and trabecular number in D2J, while trabecular separation was increased (Figure 1, I–K). Two other micro-CT parameters, trabecular thickness and trabecular surface, were consistently decreased in D2J mice at 4, 8, and 16 weeks of age; however, trabecular surface as a percentage of bone volume was increased in D2J mice, because of marked decrease in bone volume (Table 1).
Table 1.
Micro-CT Analyses in D2J Mutant and D2J/Gpnmb+ Wild-Type Mice
| Age (wk) | Genotype | Tb.Th (μm) | B.S/TV (mm2/mm3) | B.S/BV (mm2/mm3) |
|---|---|---|---|---|
| 4 | D2J/Gpnmb+ | 17.78 ± 0.39 | 8.27 ± 0.18 | 111.7 ± 2.23 |
| D2J | 16.37 ± 0.34∗ | 7.38 ± 0.27∗ | 121 ± 2.83∗ | |
| 8 | D2J/Gpnmb+ | 23.87 ± 0.81 | 19.36 ± 1.61 | 84.75 ± 3.14 |
| D2J | 17.03 ± 0.36∗∗∗∗ | 9.8 ± 0.24∗∗∗∗ | 117.8 ± 2.51∗∗∗∗ | |
| 16 | D2J/Gpnmb+ | 21.22 ± 0.67 | 12.52 ± 0.6 | 99.07 ± 3.07 |
| D2J | 17.12 ± 1.23∗∗ | 10.07 ± 0.47∗∗ | 119.0 ± 78∗∗ |
Data are expressed as means ± SEM.
∗P < 0.05, ∗∗P < 0.01, and ∗∗∗∗P < 0.0001 versus wild type.
B.S/TV, bone surface per tissue volume; B.S/BV, bone surface per bone volume; Tb.Th, trabecular thickness.
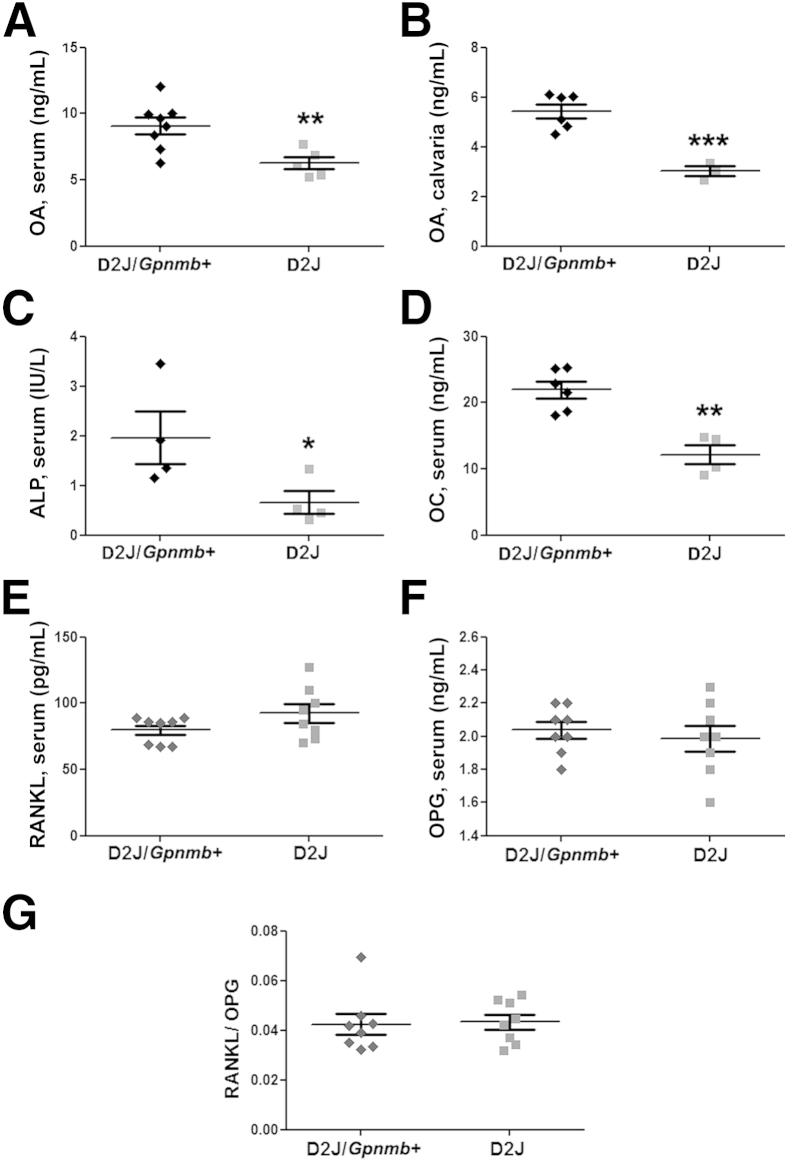
Next, we determined the levels of OA protein in 8-week-old D2J and D2J/Gpnmb+ mice. ELISA showed that OA protein in serum and calvaria of D2J mice was decreased by approximately 50% (Figure 2, A and B). To test the role of OA in osteogenesis in D2J and D2J/Gpnmb+ mice, we performed serum ELISA of the bone-formation markers ALP and OC; both markers showed a decrease of 50% or more at 8 weeks in D2J mice (Figure 2, C and D). Serum ELISA of OC showed similar results at 16 weeks (data not shown). These data suggest that OA plays a role in bone formation. Next, we examined two bone-remodeling markers, receptor activator of nuclear factor κ-B ligand (RANKL) and osteoprotegerin (OPG). RANKL is well known to stimulate osteoclastogenesis, and OPG is reported to be a decoy receptor for RANKL.24,25 Interestingly, serum ELISA revealed no significant difference in RANKL, OPG, and the RANKL/OPG ratio between D2J and D2J/Gpnmb+ mice (Figure 2, E–G).
Figure 2.
Analysis of OA and bone remodeling markers in D2J and D2J/Gpnmb+ mice. A and B: ELISA of OA protein in serum (A) and homogenized calvarial bone (B) of D2J and D2J/Gpnmb+ mice. C–G: Serum ELISA of the bone formation markers ALP (C) and OC (D) and the bone resorption markers RANKL (E) and OPG (F), along with the RANKL/OPG ratio (G). Data are expressed as means ± SEM. n ≥ 3 (A and B) and n ≥ 4 (C–G). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
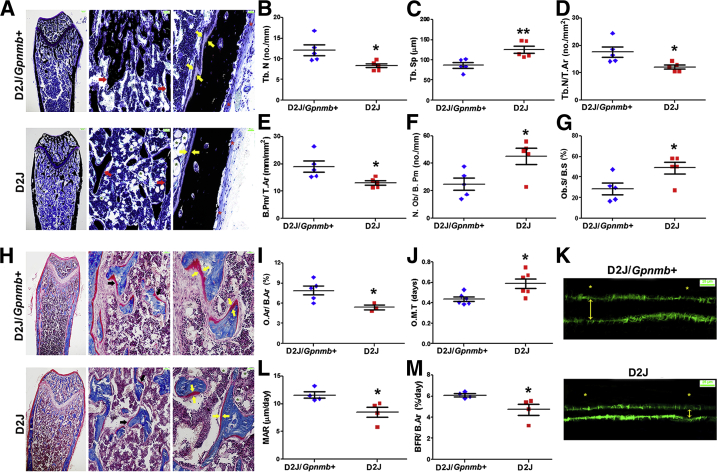
To further investigate the importance of OA in bone microstructure, we performed histomorphometric analyses of the femoral metaphysis in 8-week-old D2J and D2J/Gpnmb+ mice. The width of the femoral metaphysis was increased and trabecular bone mass was decreased in D2J mice (Figure 3A), in accord with the data reported above. Moreover, our analysis indicated an approximately 35% decrease in trabecular number (Figure 3B) and an approximately 45% increase in trabecular separation in D2J mice (Figure 3C). Furthermore, trabecular number and bone perimeter as a percentage of tissue area were also decreased in D2J mice (Figure 3, D and E). Although these findings support the positive role of OA in bone formation, we also observed a surprising increase (approximately 60%) in osteoblast number and surface in D2J mice (Figure 3, F and G).
Figure 3.
Histomorphometric analysis of femurs of D2J and D2J/Gpnmb+ mice. A: Sagittal sections with von Kossa staining for minerals (black) and toluidine blue counterstain show adipocytes (green asterisks), periosteum (red asterisks), trabecular bone (red arrows), and osteoid (yellow arrows). B–G: Histomorphometric parameters Tb.N (B), Tb.Sp (C), Tb.N/T.Ar (D), B.Pm/T.Ar (E), N.Ob/B.Pm (F), and Ob.S/B.S (G). H: Sagittal sections stained with Masson’s trichrome show osteoid (red) (black arrows, middle panel); thickness of osteoid (red) is indicated with paired yellow arrows (right panel). I and J: Histomorphometric parameters: O.Ar/B.Ar (I) and O.M.T (J). K: Calcein AM labeling for mineralized bone surface (green) shows periosteum (asterisks) and mineralized bone width (double-headed arrows). L and M: Dynamic histomorphometric parameters MAR (L) and BFR/B.Ar (M). Data are expressed as means ± SEM. n = 4 (L and M); n = 5 (B–G); n = 6 (I and J). ∗P < 0.05, ∗∗P < 0.01. Scale bars: 200 μm (A and H, left); 50 μm (A and H, middle); 25 μm (A and H, right); and 25 μm (K).
Next, we examined the importance of OA in osteoid matrix formation and mineralization in D2J and D2J/Gpnmb+ mice. Histological analysis of femoral metaphysis showed a decrease in trabecular osteoid width in D2J mice (Figure 3H). Moreover, histomorphometric analysis showed approximately 40% decrease in trabecular osteoid as a percentage of bone area in D2J mice (Figure 3I). In addition, osteoid maturation time, a parameter that detects any subtle impairment in bone mineralization, was approximately 50% prolonged in D2J mice (Figure 3J). Bone dynamic analyses showed decreased bone mineralization associated with decreased width of interlabeled surfaces in D2J mice (Figure 3K). These findings were confirmed by the significant decrease in mineral apposition and bone formation rates in D2J mice (Figure 3, L and M). Taken together, these data suggest that OA is a positive regulator of bone formation and osteoblast function in vivo.
Defective Osteoblast Differentiation in D2J Mice
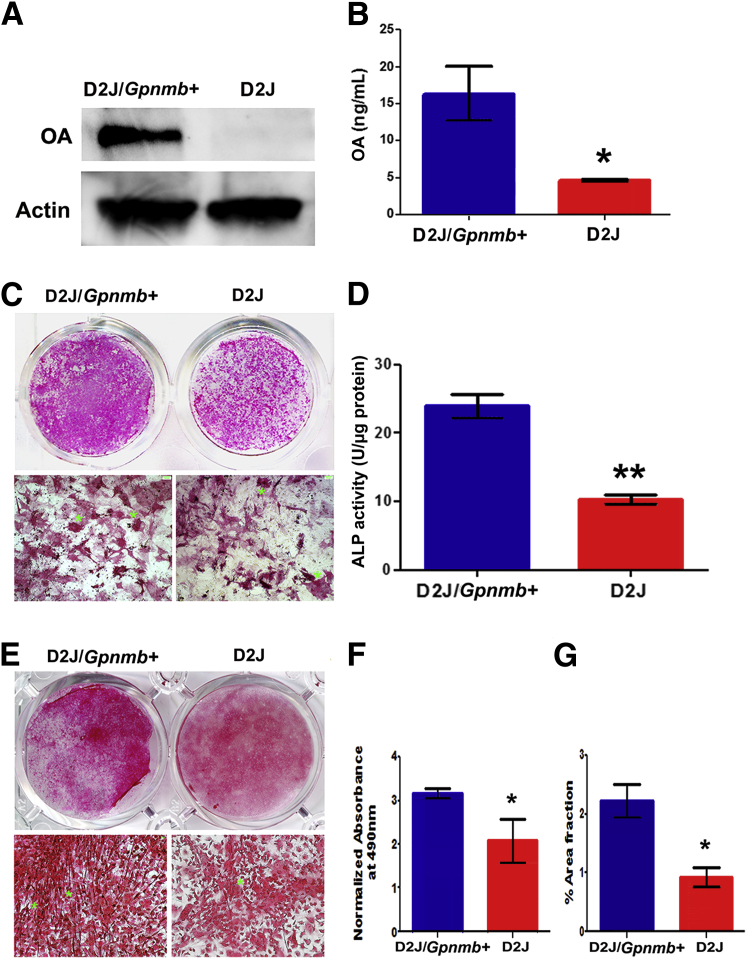
In our previous studies, using loss-of–function and gain-of–function approaches, we showed the importance of OA in osteoblast differentiation and function in vitro.7 In the present study, we characterized osteoblast differentiation in D2J and D2J/Gpnmb+ mice. First, we compared the production of the OA protein in osteoblasts. Western blotting showed OA glycoprotein present in D2J/Gpnmb+ osteoblasts, but not in D2J osteoblasts (Figure 4A). A low molecular weight protein (approximately 15 kDa) detected in D2J osteoblasts may correspond to the mutant truncated OA protein of 150 aa (see below). Quantitation of OA protein in cultured osteoblasts showed an approximately 70% decrease in production of OA protein by D2J osteoblasts (Figure 4B).
Figure 4.
Differentiation of primary calvarial osteoblasts in neonatal D2J and D2J/Gpnmb+ mice. A: Western blots of OA glycoprotein in primary osteoblasts cultured for 7 days. B: ELISA of OA protein levels in osteoblast cell lysates, normalized to total protein concentration. C: Macro- and microscopic images of 14-day differentiated osteoblasts stained for ALP (asterisks). D: ALP activity in osteoblast matrix and cell lysates. E: Macro- and microscopic images of 14-day differentiated cultures stained for collagen (asterisks) with Sirius Red in D2J and D2J/Gpnmb+. F: Quantification of Sirius Red collagen staining in D2J and D2J/Gpnmb+ cultures. G: Percentage area fraction of Sirius Red–stained matrix in D2J and D2J/Gpnmb+ cultures. Data are expressed as means ± SEM. ∗P < 0.05, ∗∗P < 0.01. Scale bar = 100 μm.
To evaluate the importance of OA in osteoblast differentiation ex vivo, we determined the expression of two early matrix markers, ALP and Col1, in differentiated D2J and D2J/Gpnmb+ osteoblasts at day 14. Both ALP staining and ALP activity were decreased in D2J osteoblasts (Figure 4, C and D). Next, we examined whether mutant OA protein has any effects on Col1 production by osteoblasts. We observed decreased staining in D2J mice, and Col1 fibers were randomly organized, in contrast to the well-oriented Col1 fibers surrounding osteoblast nodules in D2J/Gpnmb+ mice (Figure 4E). Moreover, quantification and percentage area fraction of Col1-stained matrix were significantly less in D2J mice (Figure 4, F and G).
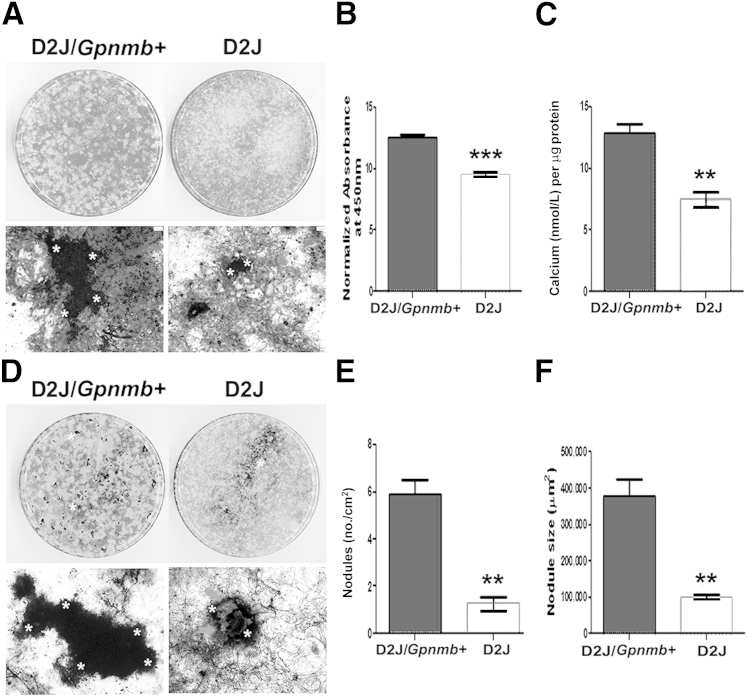
To further investigate the role of OA in later osteoblast differentiation and function, we differentiated D2J and D2J/Gpnmb+ osteoblasts and analyzed matrix mineralization markers, calcium deposition, and nodule formation at day 21. We observed decreased matrix mineralization (Figure 5A) in D2J mice, as indicated by an approximately 25% decrease in quantification of calcium-stained matrix (Figure 5B) and approximately 50% less calcium deposition (Figure 5C). Next, we examined the mineralized nodules in D2J and D2J/Gpnmb+ matrix. Nodule formation and matrix mineralization were decreased (Figure 5D), as indicated by approximately 75% decrease in number and size of mineralized nodules in D2J matrix, (Figure 5, E and F). Collectively, these data support a significant role for OA during different stages of osteoblast differentiation and matrix mineralization.
Figure 5.
Mineralization of primary calvarial osteoblasts in D2J and D2J/Gpnmb+ mice. A: Macro- and microscopic images of 21-day differentiated osteoblasts stained with Alizarin Red S for calcium (asterisks). B: Quantification of Alizarin Red S calcium staining in D2J and D2J/Gpnmb+ cultures. C: Calcium levels in total matrix and cell lysates of D2J and D2J/Gpnmb+ osteoblasts. D: Macro- and microscopic images of 21-day differentiated osteoblasts stained with von Kossa staining for hydroxyapatite (black). Asterisks in microscopic images indicate nodule border. Arrows indicate unmineralized matrix. E: Nodule number on an area basis. F: Nodule size. Data are expressed as means ± SEM. ∗∗P < 0.01, ∗∗∗P < 0.001. Scale bar = 100 μm.
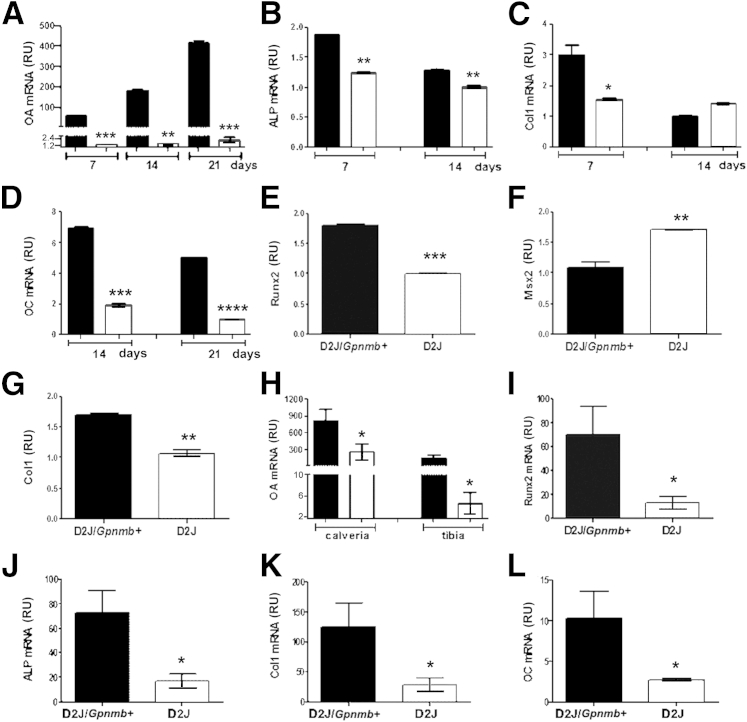
We then compared our findings for osteoblast differentiation with markers of osteogenesis. First, we used qPCR to quantitate the expression of Gpnmb encoding OA in D2J and D2J/Gpnmb+ osteoblasts differentiated for 7, 14, and 21 days. We observed a temporal increase in Gpnmb expression in osteoblasts of D2J/Gpnmb+ mice, with a maximum at day 21, but Gpnmb expression was greatly decreased in D2J osteoblasts, compared to D2J/Gpnmb+ osteoblasts, at each time point (Figure 6A). Next, we examined the osteoblast differentiation markers ALP, Col1, and OC. qPCR analysis of ALP showed decreased expression in D2J osteoblasts mice at days 7 and 14 (Figure 6B). Moreover, Col1 gene expression was also decreased in D2J osteoblasts at day 7 (Figure 6C). Interestingly, OC gene expression was approximately 80% decreased in D2J osteoblasts at days 14 and 21 (Figure 6D). We then examined early osteogenesis markers in osteoblasts of adult D2J and D2J/Gpnmb+ mice differentiated for 7 days. First, we tested Runt-related transcription factor 2 (Runx2), which is crucial for early osteoblastogenesis.26,27 Interestingly, Runx2 gene expression was down-regulated in D2J osteoblasts (Figure 6E). Next, we examined other transcription factors, including muscle segment homeobox (Msx2), which functions as a repressor for Runx2 promoter activity.28,29 Surprisingly, Msx2 expression was up-regulated in D2J osteoblasts (Figure 6F). In addition, Col1 gene expression was decreased in D2J osteoblasts, compared to D2J/Gpnmb+, at day 7 (Figure 6G).
Figure 6.
qPCR analysis of Gpnmb gene expression and bone formation markers in osteoblasts and bones of D2J and D2J/Gpnmb+ mice. A–D: Expression of OA (A) and of osteoblast differentiation markers ALP (B), Col1 (C), and OC (D) in neonatal calvarial osteoblasts differentiated for 7, 14, or 21 days. E–G: Expression of the early osteoblast differentiation markers Runx2 (E), Msx2 (F), and Col1 (G) in adult long-bone osteoblasts differentiated for 7 days. H–L: Expression of OA in calvaria and tibia (H) and expression of the bone formation markers Runx2 (I), ALP (J), Col1 (K), and OC (L) in calvaria of 8-week-old mice. Data are expressed as means ± SEM. n = 3 (A–L). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 versus wild type. Gray bars, D2J; black bars, D2J/Gpnmb+. RU, relative units.
To compare these findings with osteogenesis markers in bones, we performed qPCR analyses of OA and bone formation markers in calvaria and tibia of D2J and D2J/Gpnmb+ mice. Gpnmb expression for OA was decreased in calvaria and tibia of D2J mice (Figure 6H). qPCR analysis of other bone formation markers in calvaria showed a decrease of approximately 80% in Runx2 and ALP gene expression in D2J mice (Figure 6, I and J). Col1 and OC gene expressions were also decreased (by approximately 80%) in D2J mice (Figure 6, K and L). Taken together, these data support the importance of OA during osteoblast differentiation in vitro and osteogenesis in vivo.
Loss-of–Function Mutation in Gpnmb Decreases Proliferation and Survival of Osteoblasts
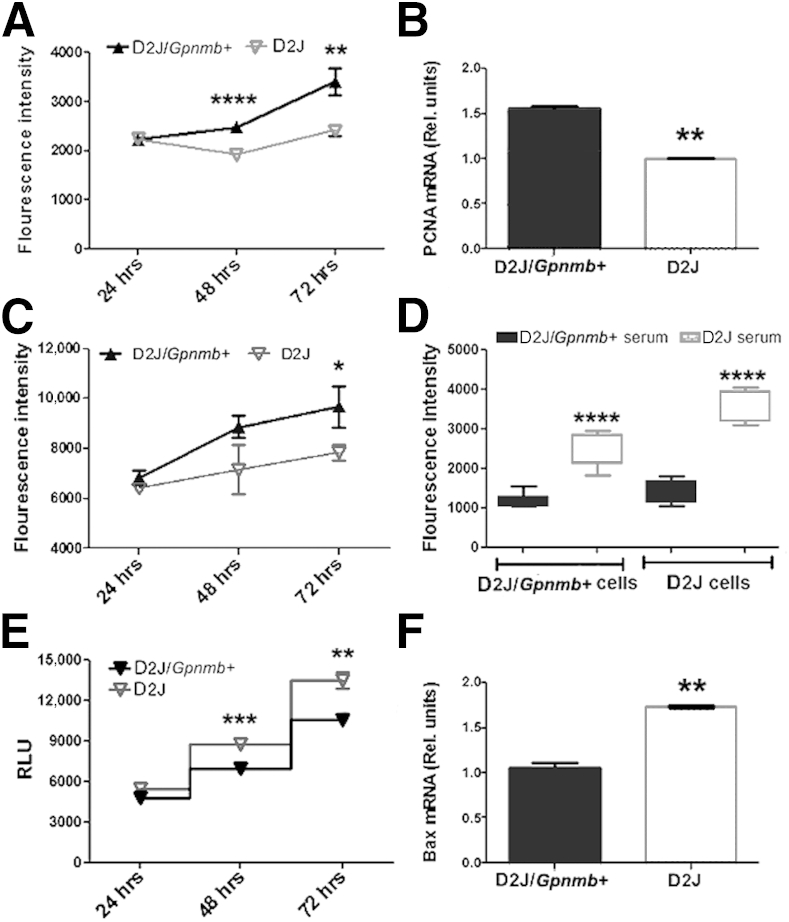
To investigate the underlying mechanism or mechanisms responsible for decreased differentiation of Gpnmb-mutant osteoblasts, we examined the proliferation and survival of osteoblasts in D2J and D2J/Gpnmb+ mice. Osteoblast proliferation was decreased in D2J mice (Figure 7A). To confirm our results, we tested the expression of proliferating cell nuclear antigen (PCNA); this proliferation marker was approximately 30% decreased in D2J osteoblasts (Figure 7B). To examine whether adult mutant osteoblasts in D2J mice are similar to neonatal calvarial mutant osteoblasts, we isolated the osteoblasts from long bones of adult D2J and D2J/Gpnmb+ mice and tested their proliferation potential in vitro. Proliferation of adult osteoblasts was decreased in D2J mice after 72 hours (Figure 7C). Given that osteoblasts from D2J mice proliferated less in vitro, and that osteoblast number was increased in vivo, we asked whether some systemic factor or factors in D2J are responsible for the high number of osteoblasts in vivo. We therefore tested the effects of serum harvested from D2J and D2J/Gpnmb+ mice on osteoblast proliferation. D2J/Gpnmb+ serum had no significant effects on either D2J/Gpnmb+ or D2J osteoblast proliferation; conversely, D2J serum increased the proliferation of both D2J and D2J/Gpnmb+ osteoblasts (Figure 7D). These findings suggest that some factor or factors in D2J serum stimulate osteoblast proliferation.
Figure 7.
Proliferation and survival of primary osteoblasts in D2J and D2J/Gpnmb+ mice. A: Proliferation assay measured by quantitation of fluorescence-labeled DNA in neonatal calvarial osteoblasts cultured in 10% FBS and measured at 24, 48, and 72 hours. B: qPCR analysis of proliferating cell nuclear antigen (PCNA) in 3-day proliferating osteoblasts. C: Proliferation assay of D2J and D2J/Gpnmb+ adult long-bone osteoblasts measured at 24, 48, and 72 hours. D: Proliferation assay of osteoblasts cultured in 10% mouse serum for 24 hours. E: Apoptosis assay measured by quantitation of caspase-3/7 luminescence of osteoblasts cultured in 2.5% FBS and measured at 24, 48, and 72 hours. F: qPCR analysis of apoptosis regulator Bax in osteoblasts cultured in 2.5% FBS serum for 3 days. Data are expressed as means ± SEM. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001. RLU, relative luminescence units.
Next, we examined whether Gpnmb mutation induces cellular apoptosis in D2J osteoblasts in vitro. Our data showed increased apoptosis of osteoblasts in D2J mice (Figure 7E). In confirmation, qPCR showed that proapoptotic marker; Bax gene expression was increased by approximately 40% in D2J mice (Figure 7F). Taken together, these data suggest a cell-autonomous defect in proliferation and survival of D2J osteoblasts.
Mutant OA Protein Does Not Induce ER Stress in Osteoblasts
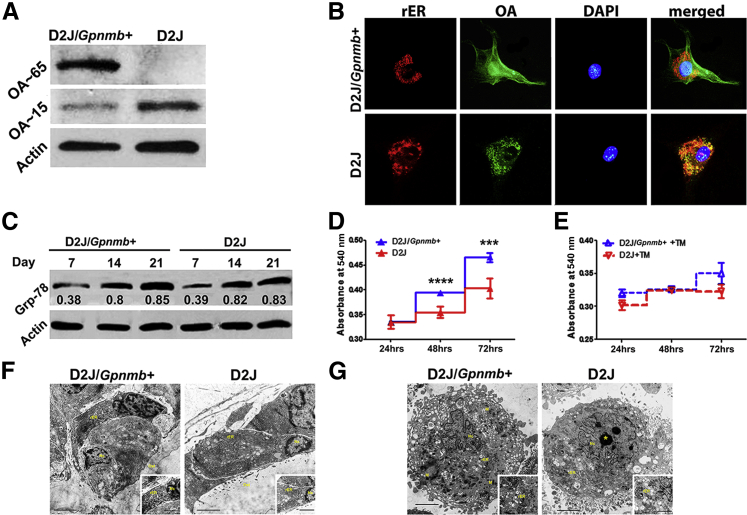
D2J mice are reported to have an early stop codon on Gpnmb gene, with a truncated protein of 150 aa.5 We therefore examined the levels of truncated OA protein by Western blotting, which showed increased levels of the truncated OA in osteoblasts of D2J mice (Figure 8A). Next, we examined the localization of OA protein in D2J osteoblasts. OA was immunodetected in a punctate, perinuclear pattern in D2J mice, compared to fine dispersion in D2J/Gpnmb+ osteoblasts (Figure 8B). Moreover, OA protein was mainly condensed and colocalized with endoplasmic reticulum (ER) in D2J osteoblasts.
Figure 8.
ER Stress in primary osteoblasts of D2J and D2J/Gpnmb+ mice. A: Western blots of native OA protein (approximately 65 kDa) and truncated OA protein (approximately 15 kDa) in primary osteoblasts cultured for 3 days. B: Immunofluorescence detection of OA colocalizing with rER) in osteoblasts. Primary osteoblasts were cultured in chamber slides, fixed, stained with anti-ER (calnexin) and anti-OA antibodies, and counterstained with DAPI for nuclei. Images were captured with a fluorescence microscope using separate excitation filters for OA (green), rER (red), and nucleus (blue), and merged images were generated. C: Western blots of the ER stress marker Grp-78 in differentiated osteoblasts at days 7, 14, and 21. The Grp-78/actin relative densitometry ratios are shown below the Grp-78 blots. D and E: MTT survival assay at 24, 48 and 72 hours in osteoblasts cultured in 10% FBS only (D) and in osteoblasts treated with 5 μg/mL tunicamycin (TM). F and G: Transmission electron microscopy images of osteoblast ultrastructure in vivo (F) and in vitro (G), showing rER, nucleus (Nc), mitochondria (M), nucleolus (asterisk), and osteoid bone (Ost). Insets: Intact rER. Data are expressed as means ± SEM. ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. Scale bars: 2000 nm (F and G); 500 nm (insets, F and G). Original magnification: ×200 (B).
Unfolded and truncated proteins are reported to increase ER stress.30 To examine whether truncated OA protein induces ER stress in D2J osteoblasts, we tested the levels of GRP-78 protein, an ER stress marker.31 We did not find any significant differences between D2J and D2J/Gpnmb+ osteoblasts differentiated for 7, 14, and 21 days (Figure 8C). We also examined ER stress response in D2J osteoblasts treated with tunicamycin, an ER stress inducer.32 Cell survival of D2J osteoblasts not treated with tunicamycin was decreased at 48 and 72 hours, compared to D2J/Gpnmb+ osteoblasts, as measured by MTT (Figure 8D); however, there was no statistical difference in cell survival between tunicamycin-treated D2J and D2J/Gpnmb+ osteoblasts at any time point (Figure 8E). These data suggest that mutant OA protein does not induce ER stress in D2J osteoblasts.
To further support our hypothesis, we examined the morphology of ER ultrastructure in D2J osteoblasts by transmission electron microscopy. For both D2J and D2J/Gpnmb+ mice, there was no evidence of morphological abnormality of ER stress in osteoblasts either in vivo (Figure 8F) or in vitro (Figure 8G). Collectively, these data showed no evidence of ER stress in D2J osteoblasts.
TGF-β1 Signaling Is Activated in D2J Osteoblasts
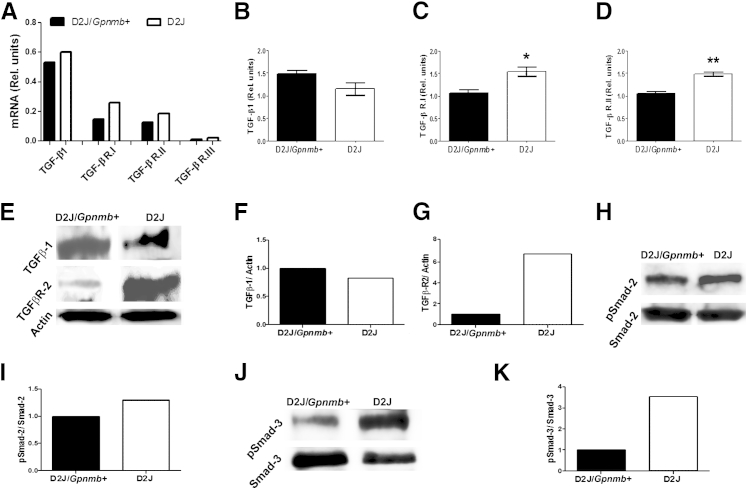
To investigate the underlying mechanism or mechanisms responsible for defective proliferation and differentiation of Gpnmb-mutant osteoblasts, we used an osteogenesis array to screen for 84 genes in D2J mice for possible differentially expressed genes. Surprisingly, TGF-β1 and TGF-β receptors I, II, and III were up-regulated in calvaria of D2J mice at 8 weeks, compared to D2J/Gpnmb+ (Figure 9A). qPCR of TGF-β1 gene expression in neonatal calvarial osteoblasts revealed no difference between D2J and D2J/Gpnmb+ mice (Figure 9B). Similar to the osteogenesis array findings, TGF-β receptors I and II were approximately 50% up-regulated in D2J osteoblasts (Figure 9, C and D). Western blotting confirmed the gene expression analysis, showing no difference in TGF-β1 protein levels (Figure 9, E and F) but a sixfold increase in TGF-β receptor II protein (Figure 9, E and G) in D2J primary osteoblasts. Next, we examined Smad activation downstream of TGF-β1 signaling in serum-deprived osteoblasts. Although phosphorylation of Smad-2 was increased by only approximately 20% in D2J osteoblasts (Figure 9, H and I), phosphorylation of Smad-3 was increased by 3.5-fold (Figure 9, J and K). Taken together, these data suggest that TGF-β receptors and p-Smad–mediated signaling are activated in D2J osteoblasts.
Figure 9.
TGF-β1 signaling in primary osteoblasts of D2J and D2J/Gpnmb+ mice. A: Osteogenesis array for TGF-β1, TGF-β receptors I, II, and III (TGF-β R.I, TGF-β R.II, and TGF-β R.III) measured in calvaria of 8-week-old mice. B–D: qPCR analyses of TGF-β1 (B), TGF-β R.I (C), and TGF-β R.II (D) in calvarial primary osteoblasts. E: Western blots of TGF-β1 and TGF-β R.II in primary osteoblasts. F and G: Corresponding densitometry for TGF-β1 (F) and TGF-β R.II (G) relative to actin. H–K: Western blots of p-Smad-2 (H) and p-Smad-3 (J), with the corresponding densitometry relative to Smad-2 (I) and Smad-3 (K). Data are expressed as means ± SEM. ∗P < 0.05, ∗∗P < 0.01. Gray bars, D2J; black bars, D2J/Gpnmb+.
Discussion
Our initial interest in the role of OA in osteogenesis emerged from our previous studies showing strong evidence of the positive effects of OA on osteoblast differentiation and function in vitro7–9 and in vivo.14,15 We sought to further characterize the anabolic role of OA in bone using a mouse model with a natural nonsense mutation in the Gpnmb gene. Because of this mutation, the D2J mouse is a unique animal model for examining the skeletal phenotype. Loss of function due to homozygous mutation of Gpnmb alleles in D2J resulted in substantial loss of bone mass, based on micro-CT findings. It is important to notice the consistent decrease in bone mass of D2J mice after 16 weeks of age. On the other hand, the sustained increase in bone mass of the wild-type mice from 4 to 8 weeks of age can be explained by the normal physiological bone growth at this age. By contrast, D2J mice showed only a slight increase in bone mass from 4 to 8 weeks, indicating a significant deficit in osteogenesis. Serum ELISA of OA and the bone-formation markers ALP and OC also were markedly decreased, despite increased number of D2J osteoblasts in vivo. Moreover, bone dynamic analyses of mineral apposition and bone formation rates supported the decreased function of D2J osteoblasts. Taken together, these findings indicate that osteoblast function is deteriorated in the D2J mouse due to absence of OA.
The fact of increased osteoblast number of D2J mice might indicate that OA negatively regulates osteoblast proliferation in vivo. Others have reported that OA (alias DC-HIL) suppresses the proliferation of cutaneous cell lymphoma through binding to syndecan-4 and trapping TGF-β1 on the cell surface33 and, moreover, OA attenuates cell proliferation and invasion of prostate carcinoma cells by dysregulating the tumor suppressor gene p53.34 These reports could explain the increased osteoblast number in D2J mice due to lack of Gpnmb. By contrast, in vitro testing showed that proliferation of D2J osteoblasts was diminished, compared to D2J/Gpnmb+, suggesting that other factors might control the cell proliferation in vivo and that these factors are absent in osteoblasts, cultured in vitro. Interestingly, proliferation of D2J osteoblasts increased significantly when treated with D2J serum in vitro. Taken together, these findings suggest the presence of systemic factors in sera of D2J mice that drive the increase in osteoblast proliferation in vivo.
In support of our hypothesis, an osteogenesis profiling array revealed an increase in gene expression of TGF-β1 and TGF-β receptors I, II, and III in D2J calvarial bone, compared to the wild type. TGF-β1 is a secreted growth factor reported to modulate proliferation and differentiation of osteoprogenitor cells into osteoblasts.35 Although osteoblasts secrete several TGF-β isoforms (TGF-β1, -β2, -β3), the TGF-β1 isoform accounts for more than 80% and up to 90% of the total TGF-β in the bone matrix.36,37 Furthermore, Smad-2 and -3 were significantly activated in D2J osteoblasts. Activation of Smad-2/3 is reported to mediate TGF-β inhibitory effects on osteoblast proliferation, which might explain in part the proliferation deficit of D2J osteoblasts in vitro38 (although it cannot yet explain the increased osteoblastogenesis in D2J mice in vivo). Interestingly, TGF-β1 has been well documented to stimulate proliferation and to promote migration of osteoprogenitors to the sites of bone remodeling through a Smad-independent PI3K/AKT pathway.39–43 Such mechanisms warrant further investigation. On the other hand, the sustained presence of TGF-β1 has been shown to inhibit osteoblast differentiation and the corresponding mineral contents,44–46 which might contribute to the decreased bone mass observed in D2J mice. Taken together, these data suggest a possible relationship between OA and TGF-β1 in modulating osteoblast proliferation and differentiation.
In terms of cell survival, Gpnmb mutation increased apoptosis and decreased survival of D2J osteoblasts through activation of caspases 3/7, which might explain the defective osteoblast differentiation in D2J. Caspases 1, 2, 3, 6, 7, and 8 regulate cell differentiation and activity,47–51 and specifically caspases 2, 3, and 8 regulate osteoblast differentiation by arresting cell proliferation at G0/G1 in the cell cycle.52 It would be interesting to test the activity of caspases 2, 3, and 8 in differentiating D2J osteoblasts lacking OA. Moreover, truncated proteins often are expected to be misfolded, and might be degraded through proteasomes or retained in ER, inducing ER stress response.53–55 In the present study, truncated OA protein did not induce ER stress in D2J osteoblasts. Moreover, ER stress markers are reported to be induced during osteoblast differentiation,56,57 that might explain the temporally increased levels of the ER stress marker GRP-78, with no significant differences in between D2J and D2J/Gpnmb+ osteoblasts. ER stress–induced osteoblast apoptosis has also been documented in a mouse model of osteogenesis imperfecta, a condition mouse characterized by decreased bone mass and multiple fractures.58 In this study D2J osteoblasts showed increased cell-apoptosis, compared to D2J/Gpnmb+, however, cell apoptosis was independent of ER stress.
The present findings of decreased osteoblast differentiation in D2J mice (mutant Gpnmb alleles) are consistent with findings from our previous studies using a loss-of–function approach of Gpnmb in MC3T3.E1 preosteoblasts, in which Gpnmb down-regulation halted osteoblast differentiation in vitro; by contrast, a gain-of-function approach of Gpnmb expedited osteoblast differentiation and matrix mineralization in vitro.7,8 Next, examining the expression of OA protein showed that truncated OA existed mostly perinuclearly and colocalized with ER in D2J osteoblasts, yet we were able to detect low levels of OA extracellularly in sera and bones of D2J mice (Figure 2, A and B). These findings suggest that D2J osteoblasts lacking the aa sequence 151-562 produce low amounts of the truncated OA protein (aa sequence: 23-150) and that is secreted extracellularly; however, the mechanism is not yet understood. Another group has reported a similar phenomenon, in which truncated mutant Fukutin-related protein (FKRP) lacking the C-terminal 185 aa because of nonsense mutation is secreted as efficiently as the normal FKRP by CHO cells.59 Decreased production of the truncated mutant OA protein by D2J osteoblasts could be explained in part by enzymatic degradation through proteasome. In a similar mechanism, mutant fibroblast growth factor receptor-2 (FGFR2), which is responsible for craniosynostosis syndrome, has increased trafficking into lysosomes and proteasomes for degradation.60 As we cannot exclude the possibility of residual function of the truncated OA, that might explain the residual osteoblast differentiation observed in D2J cultures.
Another possibility is that loss-of-function mutation of Gpnmb suppressed the osteogenic effects of other growth factors, such as bone morphogenetic proteins (BMPs), on D2J osteoblasts. We have previously shown that OA acts as a downstream mediator of BMP-2 on osteoblasts and that knocking down Gpnmb partially decreased the effects of BMP-2 on osteoblast differentiation and function.9,61 Next, we reported that OA glycoprotein consists of different domains (including extracellular, transmembrane, and cytoplasmic domains), as well as subdomains with different functions [eg, signal peptide, polycystic kidney domain (PKD), proline rich repeat domain (PRRD), RGD integrin-binding motif, and dileucine-based sorting sequence].7,62 Several reports have shown the importance of PKD in adhesive protein–protein and protein–carbohydrate interactions, and in protecting the cell from extreme environment.63,64 Other studies have elucidated the importance of RGD binding motif in adhesion of various cell types,65–70 and the role of PRRD in formation of melanosomes and melanocyte function.71
The intrinsic defect observed in differentiated D2J osteoblasts could be due to a functional domain that is missing or truncated in mutant OA protein (in contrast to normal differentiated D2J/Gpnmb+ osteoblasts, with all domains of the wild-type OA protein intact). In D2J osteoblast cultures, we observed a decreased amount of Col1 and calcium in matrix that did not match the marked decrease in osteoblast nodule size and number after 21 days. Moreover, osteoblast nodules in D2J showed incomplete mineralization. These data suggest that the mutant OA protein in D2J mice not only diminished early osteoblast differentiation but also reduced their function in vitro.
These findings also suggest the importance of other domains and subdomains of the wild-type OA protein sequence (151 to 562 aa) in osteoblast differentiation and function, in comparison to the truncated domains in mutant OA protein sequence (23 to 150 aa). In a previous study, we showed the positive role of OA C-terminal–derived peptide (538 to 556 aa), that is absent in mutant OA, in osteoblast differentiation and matrix mineralization.72 Introducing different functional domains of OA into D2J osteoblasts could help to better characterize the importance of each domain in osteoblast differentiation and function.
The highest increase in Gpnmb gene expression was detected in terminally differentiated osteoblasts of both D2J and D2J/Gpnmb+ mice, despite marked decrease in Gpnmb in D2J osteoblasts. Furthermore, Gpnmb expression was higher in calvarial bone of both D2J and D2J/Gpnmb+ mice, compared to tibia, despite lower Gpnmb in D2J bones overall. These findings may suggest a role of OA in intramembranous ossification, in which osteoprogenitor cells differentiate from MSCs.73 Other gene mutations affecting intramembranous ossification such as FGFR1, FGFR2, and FGFR3 are reported to be associated with craniosynostosis (ie, abnormal craniofacial development with premature fusions of the cranial sutures).74–76 It would be interesting to characterize any abnormality of the craniofacial development in D2J mice.
We reported previously on the defective differentiation of bone marrow–derived MSCs into osteoblasts of D2J mice, compared to D2J/Gpnmb+.7 In the present study, we observed that a periosteal thickness of D2J femur, which is the source of MSCs,77 that was thinner led to decreased bone formation overall compared to wild-type. Moreover, we noticed increased numbers of adipocytes in the marrow cavity of the D2J femur, suggesting more adipogenic and less osteogenic differentiation of MSCs. That could explain, at least in part, the resultant bone mass in D2J mice. It would be important to study the adipogenic differentiation in parallel with osteogenic differentiation of D2J MSCs. Taken together, the present findings, consistent with our previous reports,7–9,11 point to a significant role for OA in regulating osteoblast differentiation from MSCs.
Several other matrix proteins are important in osteogenesis, and loss-of–function mutations in these proteins lead to human diseases. An example is truncated Col1 resulted from genomic deletion of five to eight exons in the triple helical domain. This truncated Col1 leads to osteogenesis imperfecta, a potentially lethal disorder characterized by brittle bones, high bone deformities, high fragility, and increased fracture risk.78 Another example is mutation in the osteocalcin gene that leads to truncated protein and development of Keutel syndrome, characterized by abnormal cartilage calcification, peripheral pulmonary stenosis, and midfacial hypoplasia.79 Although our present findings strongly suggest an important role for OA in osteogenesis, based on the observed decreased bone mass and osteoblast differentiation in the Gpnmb-mutant D2J mice, we cannot exclude a role for OA in differentiation of other cell types such as osteoclasts. In one study, increased Gpnmb expression colocalized with β3 heteropolymeric complex during osteoclast differentiation.16 The same research group also showed that overexpression of Gpnmb in osteoclasts under tartrate-resistant acid phosphatase (TRAP) promoter led to increased bone resorptive activity.80
Consistent with those reports, we expect that the Gpnmb mutation would inhibit osteoclast function, which might explain the low bone turnover in D2J mice. On the other hand, the physiological role of osteoclasts in bone remodeling was marked in D2J/Gpnmb+ wild-type mice by a notable decrease in bone mass from 8 to 16 weeks old,81 consistent with similar decrease in bone mass in other mouse strains of the same age group. Interestingly, in the present study bone mass of D2J mice did not show significant changes from 8 to 16 weeks of age, suggestive of halted bone remodeling due to osteoclast functional defect. Bone resorption markers RANKL and OPG were not relevant, suggesting no difference in the total number of osteoclasts between D2J and D2J/Gpnmb+ mice in vivo.
In ongoing work with D2J mice, we are studying in depth the effect of Gpnmb mutation on osteoclast differentiation and their bone resorptive activity. Generation of other genetically engineered mouse models, such as osteoblast-specific knockout of Gpnmb or transgenic Gpnmb, may help us to understand and identify the direct effects and the possible mechanism or mechanisms of action of OA in osteoblast differentiation and their function in postnatal bone formation.
Footnotes
Supported by the NIH (National Institute of Arthritis and Musculoskeletal and Skin Diseases grant R01-AR048892-06) and by a grant from the Ohio Department of Development (F.F.S.).
Disclosures: None declared.
References
- 1.Suh T.T., Lyles K.W. Osteoporosis considerations in the frail elderly. Curr Opin Rheumatol. 2003;15:481–486. doi: 10.1097/00002281-200307000-00018. [DOI] [PubMed] [Google Scholar]
- 2.Mori S. [Contribution of bone quality to fracture risk] Clin Calcium. 2004;14:33–38. Japanese. [PubMed] [Google Scholar]
- 3.Safadi F.F., Xu J., Smock S.L., Rico M.C., Owen T.A., Popoff S.N. Cloning and characterization of osteoactivin, a novel cDNA expressed in osteoblasts. J Cell Biochem. 2001;84:12–26. doi: 10.1002/jcb.1259. [DOI] [PubMed] [Google Scholar]
- 4.Shikano S., Bonkobara M., Zukas P.K., Ariizumi K. Molecular cloning of a dendritic cell-associated transmembrane protein, DC-HIL, that promotes RGD-dependent adhesion of endothelial cells through recognition of heparan sulfate proteoglycans. J Biol Chem. 2001;276:8125–8134. doi: 10.1074/jbc.M008539200. [DOI] [PubMed] [Google Scholar]
- 5.Anderson M.G., Smith R.S., Hawes N.L., Zabaleta A., Chang B., Wiggs J.L., John S.W. Mutations in genes encoding melanosomal proteins cause pigmentary glaucoma in DBA/2J mice. Nat Genet. 2002;30:81–85. doi: 10.1038/ng794. [DOI] [PubMed] [Google Scholar]
- 6.Bandari P.S., Qian J., Yehia G., Joshi D.D., Maloof P.B., Potian J., Oh H.S., Gascon P., Harrison J.S., Rameshwar P. Hematopoietic growth factor inducible neurokinin-1 type: a transmembrane protein that is similar to neurokinin 1 interacts with substance P. Regul Pept. 2003;111:169–178. doi: 10.1016/s0167-0115(02)00288-4. [DOI] [PubMed] [Google Scholar]
- 7.Abdelmagid S.M., Barbe M.F., Rico M.C., Salihoglu S., Arango-Hisijara I., Selim A.H., Anderson M.G., Owen T.A., Popoff S.N., Safadi F.F. Osteoactivin, an anabolic factor that regulates osteoblast differentiation and function. Exp Cell Res. 2008;314:2334–2351. doi: 10.1016/j.yexcr.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Selim A.A., Abdelmagid S.M., Kanaan R.A., Smock S.L., Owen T.A., Popoff S.N., Safadi F.F. Anti-osteoactivin antibody inhibits osteoblast differentiation and function in vitro. Crit Rev Eukaryot Gene Expr. 2003;13:265–275. doi: 10.1615/critreveukaryotgeneexpr.v13.i24.180. [DOI] [PubMed] [Google Scholar]
- 9.Abdelmagid S.M., Barbe M.F., Arango-Hisijara I., Owen T.A., Popoff S.N., Safadi F.F. Osteoactivin acts as downstream mediator of BMP-2 effects on osteoblast function. J Cell Physiol. 2007;210:26–37. doi: 10.1002/jcp.20841. [DOI] [PubMed] [Google Scholar]
- 10.Owen T.A., Smock S.L., Prakash S., Pinder L., Brees D., Krull D., Castleberry T.A., Clancy Y.C., Marks S.C., Jr., Safadi F.F., Popoff S.N. Identification and characterization of the genes encoding human and mouse osteoactivin. Crit Rev Eukaryot Gene Expr. 2003;13:205–220. doi: 10.1615/critreveukaryotgeneexpr.v13.i24.130. [DOI] [PubMed] [Google Scholar]
- 11.Abdelmagid S.M., Barbe M.F., Hadjiargyrou M., Owen T.A., Razmpour R., Rehman S., Popoff S.N., Safadi F.F. Temporal and spatial expression of osteoactivin during fracture repair. J Cell Biochem. 2010;111:295–309. doi: 10.1002/jcb.22702. [DOI] [PubMed] [Google Scholar]
- 12.Arosarena O.A., Del Carpio-Cano F.E., Dela Cadena R.A., Rico M.C., Nwodim E., Safadi F.F. Comparison of bone morphogenetic protein-2 and osteoactivin for mesenchymal cell differentiation: effects of bolus and continuous administration. J Cell Physiol. 2011;226:2943–2952. doi: 10.1002/jcp.22639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raynaud C.M., Maleki M., Lis R., Ahmed B., Al-Azwani I., Malek J., Safadi F.F., Rafii A. Comprehensive characterization of mesenchymal stem cells from human placenta and fetal membrane and their response to osteoactivin stimulation. Stem Cells Int. 2012;2012:658356. doi: 10.1155/2012/658356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh M., Del Carpio-Cano F., Belcher J.Y., Crawford K., Frara N., Owen T.A., Popoff S.N., Safadi F.F. Functional roles of osteoactivin in normal and disease processes. Crit Rev Eukaryot Gene Expr. 2010;20:341–357. doi: 10.1615/critreveukargeneexpr.v20.i4.50. [DOI] [PubMed] [Google Scholar]
- 15.Bateman J.P., Safadi F.F., Susin C., Wikesjö U.M. Exploratory study on the effect of osteoactivin on bone formation in the rat critical-size calvarial defect model. J Periodontal Res. 2012;47:243–247. doi: 10.1111/j.1600-0765.2011.01428.x. [DOI] [PubMed] [Google Scholar]
- 16.Sheng M.H., Wergedal J.E., Mohan S., Lau K.H. Osteoactivin is a novel osteoclastic protein and plays a key role in osteoclast differentiation and activity. FEBS Lett. 2008;582:1451–1458. doi: 10.1016/j.febslet.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 17.Chang B., Smith R.S., Hawes N.L., Anderson M.G., Zabaleta A., Savinova O., Roderick T.H., Heckenlively J.R., Davisson M.T., John S.W. Interacting loci cause severe iris atrophy and glaucoma in DBA/2J mice. Nat Genet. 1999;21:405–409. doi: 10.1038/7741. [DOI] [PubMed] [Google Scholar]
- 18.Willott J.F., Bosch J.V., Shimizu T., Ding D.L. Effects of exposing DBA/2J mice to a high-frequency augmented acoustic environment on the cochlea and anteroventral cochlear nucleus. Hear Res. 2006;216-217:138–145. doi: 10.1016/j.heares.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Willott J.F., Bross L.S., McFadden S. Ameliorative effects of exposing DBA/2J mice to an augmented acoustic environment on histological changes in the cochlea and anteroventral cochlear nucleus. J Assoc Res Otolaryngol. 2005;6:234–243. doi: 10.1007/s10162-005-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson M.G., Smith R.S., Savinova O.V., Hawes N.L., Chang B., Zabaleta A., Wilpan R., Heckenlively J.R., Davisson M., John S.W. Genetic modification of glaucoma associated phenotypes between AKXD-28/Ty and DBA/2J mice. BMC Genet. 2001;2:1. doi: 10.1186/1471-2156-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howell G.R., Libby R.T., Marchant J.K., Wilson L.A., Cosma I.M., Smith R.S., Anderson M.G., John S.W. Absence of glaucoma in DBA/2J mice homozygous for wild-type versions of Gpnmb and Tyrp1. BMC Genet. 2007;8:45. doi: 10.1186/1471-2156-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Safadi F.F., Hermey D.C., Popoff S.N., Seifert M.F. Skeletal resistance to 1,25-dihydroxyvitamin D3 in osteopetrotic rats. Endocrine. 1999;11:309–319. doi: 10.1385/ENDO:11:3:309. [DOI] [PubMed] [Google Scholar]
- 23.Bakker A.D., Klein-Nulend J. Osteoblast isolation from murine calvaria and long bones. Methods Mol Biol. 2012;816:19–29. doi: 10.1007/978-1-61779-415-5_2. [DOI] [PubMed] [Google Scholar]
- 24.Udagawa N., Takahashi N., Jimi E., Matsuzaki K., Tsurukai T., Itoh K., Nakagawa N., Yasuda H., Goto M., Tsuda E., Higashio K., Gillespie M.T., Martin T.J., Suda T. Osteoblasts/stromal cells stimulate osteoclast activation through expression of osteoclast differentiation factor/RANKL but not macrophage colony-stimulating factor: receptor activator of NF-kappa B ligand. Bone. 1999;25:517–523. doi: 10.1016/s8756-3282(99)00210-0. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe Y., Namba A., Aida Y., Honda K., Tanaka H., Suzuki N., Matsumura H., Maeno M. IL-1beta suppresses the formation of osteoclasts by increasing OPG production via an autocrine mechanism involving celecoxib-related prostaglandins in chondrocytes. Mediators Inflamm. 2009;2009:308596. doi: 10.1155/2009/308596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franceschi R.T., Xiao G., Jiang D., Gopalakrishnan R., Yang S., Reith E. Multiple signaling pathways converge on the Cbfa1/Runx2 transcription factor to regulate osteoblast differentiation. Connect Tissue Res. 2003;44(Suppl 1):109–116. [PMC free article] [PubMed] [Google Scholar]
- 27.Karsenty G., Ducy P., Starbuck M., Priemel M., Shen J., Geoffroy V., Amling M. Cbfa1 as a regulator of osteoblast differentiation and function. Bone. 1999;25:107–108. doi: 10.1016/s8756-3282(99)00111-8. [DOI] [PubMed] [Google Scholar]
- 28.Shirakabe K., Terasawa K., Miyama K., Shibuya H., Nishida E. Regulation of the activity of the transcription factor Runx2 by two homeobox proteins, Msx2 and Dlx5. Genes Cells. 2001;6:851–856. doi: 10.1046/j.1365-2443.2001.00466.x. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi T., Kato S., Suzuki N., Kawabata N., Takagi M. Autoregulatory mechanism of Runx2 through the expression of transcription factors and bone matrix proteins in multipotential mesenchymal cell line, ROB-C26. J Oral Sci. 2005;47:199–207. doi: 10.2334/josnusd.47.199. [DOI] [PubMed] [Google Scholar]
- 30.Boot-Handford R.P., Briggs M.D. The unfolded protein response and its relevance to connective tissue diseases. Cell Tissue Res. 2010;339:197–211. doi: 10.1007/s00441-009-0877-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee A.S. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods. 2005;35:373–381. doi: 10.1016/j.ymeth.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 32.Jang W.G., Kim E.J., Koh J.T. Tunicamycin negatively regulates BMP2-induced osteoblast differentiation through CREBH expression in MC3T3E1 cells. BMB Rep. 2011;44:735–740. doi: 10.5483/BMBRep.2011.44.11.735. [DOI] [PubMed] [Google Scholar]
- 33.Chung J.S., Shiue L.H., Duvic M., Pandya A., Cruz P.D., Jr., Ariizumi K. Sezary syndrome cells overexpress syndecan-4 bearing distinct heparan sulfate moieties that suppress T-cell activation by binding DC-HIL and trapping TGF-beta on the cell surface. Blood. 2011;117:3382–3390. doi: 10.1182/blood-2010-08-302034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsui K.H., Chang Y.L., Feng T.H., Chang P.L., Juang H.H. Glycoprotein transmembrane nmb: an androgen-downregulated gene attenuates cell invasion and tumorigenesis in prostate carcinoma cells. Prostate. 2012;72:1431–1442. doi: 10.1002/pros.22494. [DOI] [PubMed] [Google Scholar]
- 35.Dallas S.L., Rosser J.L., Mundy G.R., Bonewald L.F. Proteolysis of latent transforming growth factor-beta (TGF-beta)-binding protein-1 by osteoclasts. A cellular mechanism for release of TGF-beta from bone matrix. J Biol Chem. 2002;277:21352–21360. doi: 10.1074/jbc.M111663200. [DOI] [PubMed] [Google Scholar]
- 36.Gatherer D., Ten Dijke P., Baird D.T., Akhurst R.J. Expression of TGF-beta isoforms during first trimester human embryogenesis. Development. 1990;110:445–460. doi: 10.1242/dev.110.2.445. [DOI] [PubMed] [Google Scholar]
- 37.Thorp B.H., Anderson I., Jakowlew S.B. Transforming growth factor-beta 1, -beta 2 and -beta 3 in cartilage and bone cells during endochondral ossification in the chick. Development. 1992;114:907–911. doi: 10.1242/dev.114.4.907. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y.E. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mu Y., Gudey S.K., Landström M. Non-Smad signaling pathways. Cell Tissue Res. 2012;347:11–20. doi: 10.1007/s00441-011-1201-y. [DOI] [PubMed] [Google Scholar]
- 40.Li X., Huang X., Qi Z.L., Wang W. [Transforming growth factor-beta superfamily in the regulation of new bone formation] Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2002;16:126–129. Chinese. [PubMed] [Google Scholar]
- 41.Liu P., Oyajobi B.O., Russell R.G., Scutt A. Regulation of osteogenic differentiation of human bone marrow stromal cells: interaction between transforming growth factor-beta and 1,25(OH)(2) vitamin D(3) In vitro. Calcif Tissue Int. 1999;65:173–180. doi: 10.1007/s002239900678. [DOI] [PubMed] [Google Scholar]
- 42.Kessler S., Kastler S., Mayr-Wohlfart U., Puhl W., Günther K.P. Stimulation primärer Osteoblastenkulturen mit rh-TGF-β, rh-bFGF, rh-BMP 2 und rx-BMP 4 in einem In-vitro-Modell [Stimulation of primary osteoblast cultures with rh-TGF-beta, rh-bFGF, rh-BMP 2 and rx-BMP 4 in an in vitro model] Orthopade. 2000;29:107–111. doi: 10.1007/s001320050017. [DOI] [PubMed] [Google Scholar]
- 43.Ingram R.T., Bonde S.K., Riggs B.L., Fitzpatrick L.A. Effects of transforming growth factor beta (TGF beta) and 1,25 dihydroxyvitamin D3 on the function, cytochemistry and morphology of normal human osteoblast-like cells. Differentiation. 1994;55:153–163. doi: 10.1046/j.1432-0436.1994.5520153.x. [DOI] [PubMed] [Google Scholar]
- 44.Lian N., Lin T., Liu W., Wang W., Li L., Sun S., Nyman J.S., Yang X. Transforming growth factor β suppresses osteoblast differentiation via the vimentin activating transcription factor 4 (ATF4) axis. J Biol Chem. 2012;287:35975–35984. doi: 10.1074/jbc.M112.372458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lian J.B., Stein G.S. The developmental stages of osteoblast growth and differentiation exhibit selective responses of genes to growth factors (TGF beta 1) and hormones (vitamin D and glucocorticoids) J Oral Implantol. 1993;19:95–105. discussion 36–37. [PubMed] [Google Scholar]
- 46.Alliston T., Choy L., Ducy P., Karsenty G., Derynck R. TGF-beta-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 2001;20:2254–2272. doi: 10.1093/emboj/20.9.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Basu S., Rajakaruna S., Menko A.S. Insulin-like growth factor receptor-1 and nuclear factor kappaB are crucial survival signals that regulate caspase-3-mediated lens epithelial cell differentiation initiation. J Biol Chem. 2012;287:8384–8397. doi: 10.1074/jbc.M112.341586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan G., Nogalski M.T., Yurochko A.D. Human cytomegalovirus stimulates monocyte-to-macrophage differentiation via the temporal regulation of caspase 3. J Virol. 2012;86:10714–10723. doi: 10.1128/JVI.07129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernando P., Kelly J.F., Balazsi K., Slack R.S., Megeney L.A. Caspase 3 activity is required for skeletal muscle differentiation. Proc Natl Acad Sci USA. 2002;99:11025–11030. doi: 10.1073/pnas.162172899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gabet A.S., Coulon S., Fricot A., Vandekerckhove J., Chang Y., Ribeil J.A., Lordier L., Zermati Y., Asnafi V., Belaid Z., Debili N., Vainchenker W., Varet B., Hermine O., Courtois G. Caspase-activated ROCK-1 allows erythroblast terminal maturation independently of cytokine-induced Rho signaling. Cell Death Diff. 2011;18:678–689. doi: 10.1038/cdd.2010.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rébé C., Cathelin S., Launay S., Filomenko R., Prévotat L., L’Ollivier C., Gyan E., Micheau O., Grant S., Dubart-Kupperschmitt A., Fontenay M., Solary E. Caspase-8 prevents sustained activation of NF-kappaB in monocytes undergoing macrophagic differentiation. Blood. 2007;109:1442–1450. doi: 10.1182/blood-2006-03-011585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mogi M., Togari A. Activation of caspases is required for osteoblastic differentiation. J Biol Chem. 2003;278:47477–47482. doi: 10.1074/jbc.M307055200. [DOI] [PubMed] [Google Scholar]
- 53.Ding W.X., Yin X.M. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4:141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- 54.Hiller M.M., Finger A., Schweiger M., Wolf D.H. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 55.Christis C., Fullaondo A., Schildknegt D., Mkrtchian S., Heck A.J., Braakman I. Regulated increase in folding capacity prevents unfolded protein stress in the ER. J Cell Sci. 2010;123:787–794. doi: 10.1242/jcs.041111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saito A., Ochiai K., Kondo S., Tsumagari K., Murakami T., Cavener D.R., Imaizumi K. Endoplasmic reticulum stress response mediated by the PERK-eIF2(alpha)-ATF4 pathway is involved in osteoblast differentiation induced by BMP2. J Biol Chem. 2011;286:4809–4818. doi: 10.1074/jbc.M110.152900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hamamura K., Yokota H. Stress to endoplasmic reticulum of mouse osteoblasts induces apoptosis and transcriptional activation for bone remodeling. FEBS Lett. 2007;581:1769–1774. doi: 10.1016/j.febslet.2007.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lisse T.S., Thiele F., Fuchs H., Hans W., Przemeck G.K., Abe K., Rathkolb B., Quintanilla-Martinez L., Hoelzlwimmer G., Helfrich M., Wolf E., Ralston S.H., Hrabé de Angelis M. ER stress-mediated apoptosis in a new mouse model of osteogenesis imperfecta. PLoS Genet. 2008;4:e7. doi: 10.1371/journal.pgen.0040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu P.J., Zillmer A., Wu X., Lochmuller H., Vachris J., Blake D., Chan Y.M., Lu Q.L. Mutations alter secretion of fukutin-related protein. Biochim Biophys Acta. 2010;1802:253–258. doi: 10.1016/j.bbadis.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 60.Hatch N.E., Hudson M., Seto M.L., Cunningham M.L., Bothwell M. Intracellular retention, degradation, and signaling of glycosylation-deficient FGFR2 and craniosynostosis syndrome-associated FGFR2C278F. J Biol Chem. 2006;281:27292–27305. doi: 10.1074/jbc.M600448200. [DOI] [PubMed] [Google Scholar]
- 61.Singh M., Del Carpio-Cano F.E., Monroy M.A., Popoff S.N., Safadi F.F. Homeodomain transcription factors regulate BMP-2-induced osteoactivin transcription in osteoblasts. J Cell Physiol. 2012;227:390–399. doi: 10.1002/jcp.22791. [DOI] [PubMed] [Google Scholar]
- 62.Selim A.A. Osteoactivin bioinformatic analysis: prediction of novel functions, structural features, and modes of action. Med Sci Monit. 2009;15:MT19–MT33. [PubMed] [Google Scholar]
- 63.Bycroft M., Bateman A., Clarke J., Hamill S.J., Sandford R., Thomas R.L., Chothia C. The structure of a PKD domain from polycystin-1: implications for polycystic kidney disease. EMBO J. 1999;18:297–305. doi: 10.1093/emboj/18.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jing H., Takagi J., Liu J.H., Lindgren S., Zhang R.G., Joachimiak A., Wang J.H., Springer T.A. Archaeal surface layer proteins contain beta propeller, PKD, and beta helix domains and are related to metazoan cell surface proteins. Structure. 2002;10:1453–1464. doi: 10.1016/s0969-2126(02)00840-7. [DOI] [PubMed] [Google Scholar]
- 65.Kim S.Y., Oh H.K., Ha J.M., Ahn H.Y., Shin J.C., Baek S.H., Lim S.C., Joe Y.A. RGD-peptide presents anti-adhesive effect, but not direct pro-apoptotic effect on endothelial progenitor cells. Arch Biochem Biophys. 2007;459:40–49. doi: 10.1016/j.abb.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 66.Lastres P., Bellon T., Cabanas C., Sanchez-Madrid F., Acevedo A., Gougos A., Letarte M., Bernabeu C. Regulated expression on human macrophages of endoglin, an Arg-Gly-Asp-containing surface antigen. Eur J Immunol. 1992;22:393–397. doi: 10.1002/eji.1830220216. [DOI] [PubMed] [Google Scholar]
- 67.Rezania A., Healy K.E. Integrin subunits responsible for adhesion of human osteoblast-like cells to biomimetic peptide surfaces. J Orthop Res. 1999;17:615–623. doi: 10.1002/jor.1100170423. [DOI] [PubMed] [Google Scholar]
- 68.Schofer M.D., Boudriot U., Bockelmann S., Walz A., Wendorff J.H., Greiner A., Paletta J.R., Fuchs-Winkelmann S. Effect of direct RGD incorporation in PLLA nanofibers on growth and osteogenic differentiation of human mesenchymal stem cells. J Mater Sci Mater Med. 2009;20:1535–1540. doi: 10.1007/s10856-009-3719-z. [DOI] [PubMed] [Google Scholar]
- 69.van der Pluijm G., Mouthaan H., Baas C., de Groot H., Papapoulos S., Löwik C. Integrins and osteoclastic resorption in three bone organ cultures: differential sensitivity to synthetic Arg-Gly-Asp peptides during osteoclast formation. J Bone Miner Res. 1994;9:1021–1028. doi: 10.1002/jbmr.5650090709. [DOI] [PubMed] [Google Scholar]
- 70.Wang X., Yan C., Ye K., He Y., Li Z., Ding J. Effect of RGD nanospacing on differentiation of stem cells. Biomaterials. 2013;34:2865–2874. doi: 10.1016/j.biomaterials.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 71.Hoashi T., Muller J., Vieira W.D., Rouzaud F., Kikuchi K., Tamaki K., Hearing V.J. The repeat domain of the melanosomal matrix protein PMEL17/GP100 is required for the formation of organellar fibers. J Biol Chem. 2006;281:21198–21208. doi: 10.1074/jbc.M601643200. [DOI] [PubMed] [Google Scholar]
- 72.Selim A.A., Castaneda J.L., Owen T.A., Popoff S.N., Safadi F.F. The role of osteoactivin-derived peptides in osteoblast differentiation. Med Sci Monit. 2007;13:BR259–BR270. [PubMed] [Google Scholar]
- 73.Ornitz D.M., Marie P.J. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 2002;16:1446–1465. doi: 10.1101/gad.990702. [DOI] [PubMed] [Google Scholar]
- 74.Park W.J., Meyers G.A., Li X., Theda C., Day D., Orlow S.J., Jones M.C., Jabs E.W. Novel FGFR2 mutations in Crouzon and Jackson-Weiss syndromes show allelic heterogeneity and phenotypic variability. Hum Mol Genet. 1995;4:1229–1233. doi: 10.1093/hmg/4.7.1229. [DOI] [PubMed] [Google Scholar]
- 75.Ornitz D.M. Regulation of chondrocyte growth and differentiation by fibroblast growth factor receptor 3. Novartis Found Symp. 2001;232:63–76. doi: 10.1002/0470846658.ch6. discussion 80, 272–282. [DOI] [PubMed] [Google Scholar]
- 76.Muenke M., Schell U. Fibroblast-growth-factor receptor mutations in human skeletal disorders. Trends Genet. 1995;11:308–313. doi: 10.1016/s0168-9525(00)89088-5. [DOI] [PubMed] [Google Scholar]
- 77.Mara C.S., Sartori A.R., Duarte A.S., Andrade A.L., Pedro M.A., Coimbra I.B. Periosteum as a source of mesenchymal stem cells: the effects of TGF-β3 on chondrogenesis. Clinics (Sao Paulo) 2011;66:487–492. doi: 10.1590/S1807-59322011000300022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mundlos S., Chan D., Weng Y.M., Sillence D.O., Cole W.G., Bateman J.F. Multiexon deletions in the type I collagen COL1A2 gene in osteogenesis imperfecta type IB. Molecules containing the shortened alpha2(I) chains show differential incorporation into the bone and skin extracellular matrix. J Biol Chem. 1996;271:21068–21074. doi: 10.1074/jbc.271.35.21068. [DOI] [PubMed] [Google Scholar]
- 79.Munroe P.B., Olgunturk R.O., Fryns J.P., Van Maldergem L., Ziereisen F., Yuksel B., Gardiner R.M., Chung E. Mutations in the gene encoding the human matrix Gla protein cause Keutel syndrome. Nat Genet. 1999;21:142–144. doi: 10.1038/5102. [DOI] [PubMed] [Google Scholar]
- 80.Sheng M.H., Wergedal J.E., Mohan S., Amoui M., Baylink D.J., Lau K.H. Targeted overexpression of osteoactivin in cells of osteoclastic lineage promotes osteoclastic resorption and bone loss in mice. PLoS One. 2012;7:e35280. doi: 10.1371/journal.pone.0035280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Halloran B.P., Ferguson V.L., Simske S.J., Burghardt A., Venton L.L., Majumdar S. Changes in bone structure and mass with advancing age in the male C57BL/6J mouse. J Bone Miner Res. 2002;17:1044–1050. doi: 10.1359/jbmr.2002.17.6.1044. [DOI] [PubMed] [Google Scholar]