SUMMARY
Lysine acetylation regulates transcription by targeting histones and nonhistone proteins. Here we report that the central regulator of transcription, RNA polymerase II, is subject to acetylation in mammalian cells. Acetylation occurs at eight lysines within the C-terminal domain (CTD) of the largest polymerase subunit and is mediated by p300/KAT3B. CTD acetylation is specifically enriched downstream of the transcription start sites of polymerase-occupied genes genome-wide, indicating a role in early stages of transcription initiation or elongation. Mutation of lysines or p300 inhibitor treatment causes the loss of epidermal growth-factor-induced expression of c-Fos and Egr2, immediate-early genes with promoter-proximally paused polymerases, but does not affect expression or polymerase occupancy at housekeeping genes. Our studies identify acetylation as a new modification of the mammalian RNA polymerase II required for the induction of growth factor response genes.
INTRODUCTION
Acetylation of lysines has emerged as an important posttranslational modification affecting more than 1,700 cellular proteins (Choudhary et al., 2009). The process is reversible and involves balanced activities of lysine acetyltransferases (KATs) and histone deacetylases (HDACs). We examined the effects of acetylation on a key regulatory element of the RNA polymerase II complex, the C-terminal domain (CTD).
The CTD is a highly repetitive stretch of amino acid heptad repeats within the largest polymerase subunit (RPB1). While the consensus sequence of heptad repeats (Y1S2P3T4S5P6S7) is conserved from yeast to mammals, the overall number of repeats expanded from 26–29 in yeast to 52 in mammals, and most of the newly acquired repeats in mammals diverge from the consensus sequence (Chapman et al., 2008). Heptad repeats undergo a sequence of characteristic posttranslational modifications during transcription, most notably phosphorylation of S5 and S2 residues during transcription initiation and elongation, respectively. S5 phosphorylation tightly colocalizes with polymerases that have initiated transcription but “pause” during early elongation through the promoter-proximal region (20–70 nt downstream of the transcription start site [TSS]) (Adelman and Lis, 2012), an important regulatory step in mammalian gene transcription, which is widespread in embryonic stem cells (ESCs) and other cell types (Core et al., 2008; Guenther et al., 2007; Kim et al., 2005; Muse et al., 2007; Rahl et al., 2010; Zeitlinger et al., 2007). Overall, polymerase pausing is associated with high transcriptional activity and is enriched at genes in signal-responsive pathways (Min et al., 2011).
Other CTD modifications include phosphorylation of Y1, which prevents premature transcription termination (Mayer et al., 2012), and modifications of S7, T4, and R1810, which have important functional relevance at subsets of genes (Baillat et al., 2005; Egloff et al., 2007; Hsin et al., 2011; Sims et al., 2011).
We examined acetylation in eight of the distal heptad repeats in the mammalian CTD, which contain a lysine at position 7 (K7). K7-containing repeats are only sporadically found in fungi (none in Saccharomyces cerevisiae and Saccharomyces pombe, one in Yarrowia lipolytica) but are frequently found in the extended CTD of higher eukaryotes or of pathogens that coevolved with eukaryotic hosts (e.g., nine in Plasmodium falciparum) (Liu et al., 2010). The function and modification status of these “non-consensus” repeats are unknown. Notably, distal, nonconserved repeats are particularly important in activator-dependent transcription, suggesting a role at specific gene classes (Chapman et al., 2005; Gerber et al., 1995). We report here that K7-containing repeats are acetylated by p300/KAT3B. Our data indicate that K7 acetylation is a mammalian CTD modification detected at the 5′ ends of most polymerase-occupied genes with a critical role in growth-factor-dependent transcription activation.
RESULTS
K7 Residues Are Acetylated by p300
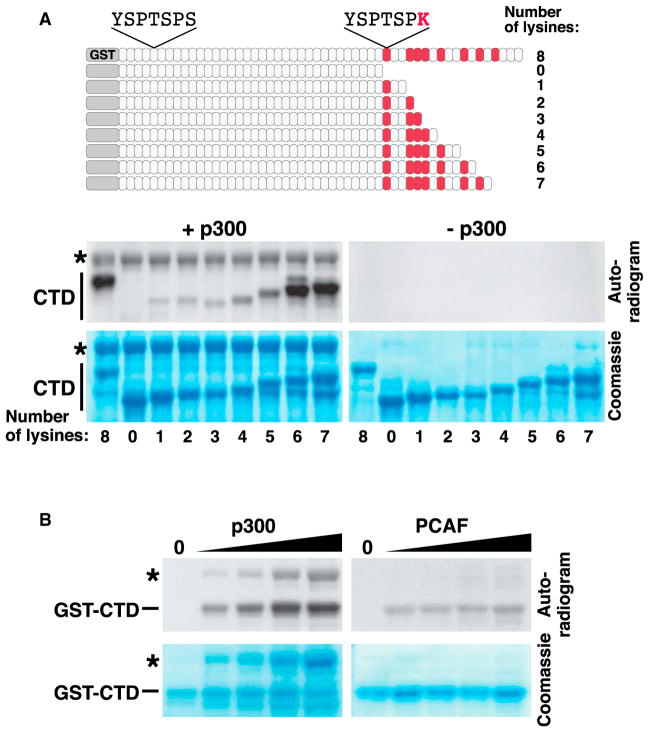
To determine if K7-containing repeats are acetylated, we performed in vitro acetylation assays with purified murine GST-CTD and the recombinant acetyltransferase domain of p300/KAT3B. Acetylation occurred in the full-length CTD, but not in a deletion mutant lacking all K7-containing repeats (Figure 1A, +p300). Progressive addition of K7-containing repeats restored acetylation. Maximal levels were reached when six K7-containing repeats were present. No acetylation was observed in the absence of p300/KAT3B (Figure 1A, −p300). PCAF/KAT2B was markedly less efficient in acetylating the CTD, although both p300 and PCAF effectively acetylated histones in vitro (Figure 1B and see Figure S1 available online). These findings demonstrate that K7 residues are in vitro targets of p300/KAT3B, but not PCAF/KAT2B.
Figure 1. In Vitro Acetylation of K7 Repeats by p300.
(A) Scheme of the GST-CTD wild-type construct and a series of GST-CTD deletion mutants (upper panel) and the in vitro acetylation of these constructs with the purified acetyltransferase domain of p300 and [14C] acetyl-CoA (lower panel). Asterisk marks autoacetylation of p300.
(B) In vitro acetylation of the murine GST-CTD with p300 or PCAF. Reactions were analyzed by autoradiography. p300, but not PCAF, acetylates the GST-CTD construct in a dose-dependent manner. Asterisk marks the autoacetylation band of p300.
See also Figure S1.
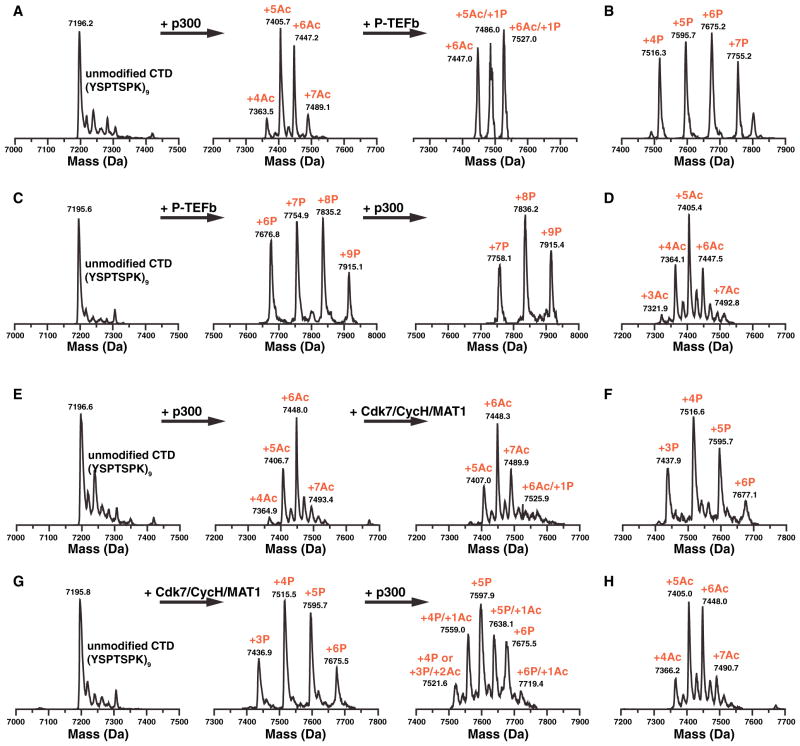
We confirmed these data by electrospray ionization-mass spectrometry of a synthetic CTD substrate composed of nine K7-containing consensus repeats (Figure S2). Recombinant p300/KAT3B acetylated the peptide in a time-dependent manner, with eight of the nine lysines acetylated after 2 hr incubation (Figure S2). We also tested the interplay between phosphorylation and acetylation using this technique. Acetylation by p300/KAT3B specifically suppressed subsequent phosphorylation by P-TEFb (Figures 2A and 2B), an important CTD kinase that regulates transcription elongation (Marshall et al., 1996; Marshall and Price, 1995). Only two phosphate groups were detected at a peptide carrying six acetyl groups, while seven phosphate groups were added to a nonmodified peptide in a parallel control reaction (Figures 2A and 2B). Vice versa, prior phosphorylation by P-TEFb inhibited acetylation by p300/KAT3B (Figures 2C and 2D). Similar results were obtained with CDK7/Cyclin H/MAT1, a CTD kinase associated with transcription initiation (Lu et al., 1992) (Figures 2E–2H). These data indicate that in vitro acetylation and phosphorylation events are mutually exclusive in K7-containing repeats.
Figure 2. In Vitro Acetylation/Phosphorylation of a Synthetic CTD Peptide.
(A) Sequential in vitro acetylation/phosphorylation of a synthetic peptide containing nine K7-containing repeats (YSPTSPK)9 with p300 and P-TEFb followed by ESI mass spectrometry.
(B) Control phosphorylation of nonacetylated (YSPTSPK)9 by P-TEFb.
(C) Sequential in vitro phosphorylation/acetylation of (YSPTSPK)9.
(D) Control acetylation of nonphosphorylated (YSPTSPK)9 by p300.
(E) Sequential in vitro acetylation/phosphorylation of a synthetic peptide containing nine K7-containing repeats (YSPTSPK)9 with p300 and CDK7/CyclinH/MAT1 followed by ESI mass spectrometry.
(F) Control phosphorylation of nonacetylated (YSPTSPK)9 by CDK7/CyclinH/MAT1.
(G) Sequential in vitro phosphorylation/acetylation of (YSPTSPK)9.
(H) Control acetylation of nonphosphorylated (YSPTSPK)9 by p300.
See also Figure S2.
RNA Polymerase II Is Acetylated in Mammalian Cells
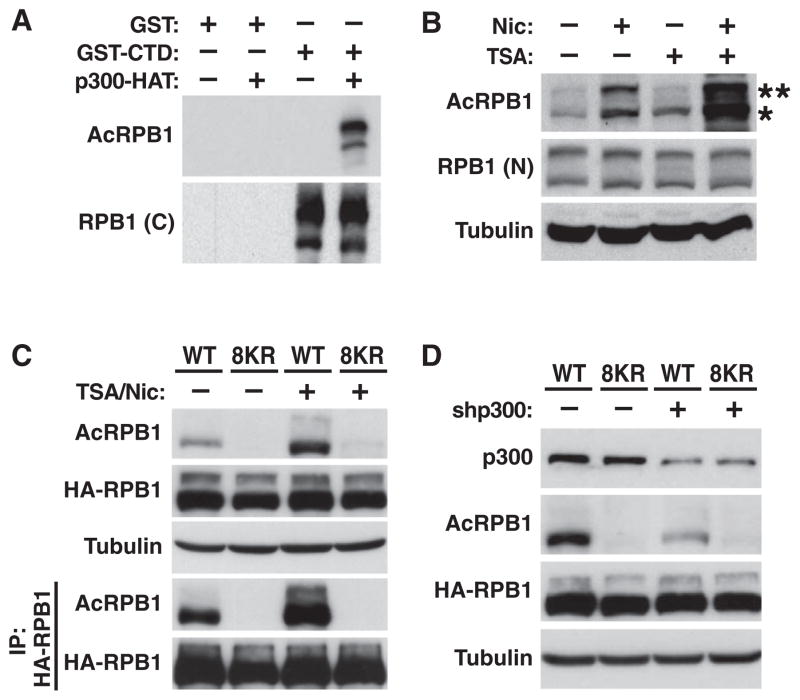
To study CTD acetylation in cells, we generated acetylation-specific polyclonal antibodies against a mix of three CTD peptides covering six of the eight K7-containing repeats (AcRPB1) (Figures S3A–S3C). In western blotting experiments, the AcRPB1 antibodies reacted with the recombinant murine GST-CTD protein only when acetylated by p300/KAT3B (Figure 3A). In human cell lysates, the antibodies recognized the two bands representing endogenous RPB1 with a preference for the faster-migrating, hypophosphorylated form (Figure 3B). The signal was enhanced when cells were treated with HDAC inhibitors trichostatin A (TSA) or nicotinamide, and hyperacetylation of both RPB1 isoforms was strongly induced after treatment with both (Figure 3B). Thus, K7 residues in the CTD are dynamically acetylated and deacetylated in cells.
Figure 3. Reversible K7 Acetylation in Human Cells.
(A) Recognition of in vitro acetylated GST-CTD by AcRPB1 antibodies by western blotting. The RPB1 antibody 8WG16 was used to control for total polymerase.
(B) Representative western blot of endogenous RNA polymerase II in 293T cells treated with deacetylase inhibitors nicotinamide (Nic) and/or trichostatin A (TSA). *, hypophosphorylated RPB1; **, hyperphosphorylated RPB1. The RPB1 antibody N-20 was used to control for total polymerase.
(C) Representative western blot of wild-type or 8KR mutant HA-RPB1 over-expressed in 293T cells before and after immunoprecipitation with HA antibodies.
(D) Representative western blot of wild-type or 8KR HA-RPB1 in 293T cells after transfection of p300 shRNAs or scrambled control shRNAs.
See also Figure S3.
Next, we generated mutant murine HA-tagged RPB1, in which all K7 residues were substituted by arginines (8KR), a conservative mutation that preserves the positive charge at the K7 position and so mimics nonacetylated lysines. Wild-type and mutant proteins contained an additional point mutation that renders HA-RPB1 resistant to α-amanitin, an inhibitor of transcription elongation that triggers degradation of endogenous RPB1 (Bartolomei and Corden, 1987; Lee et al., 2003). After overexpression in 293T cells and culture in medium containing α-amanitin, wild-type and 8KR HA-RPB1 were expressed at equivalent levels, but acetylation was detected in wild-type, but not mutant, HA-RPB1 (Figure 3C).
To confirm the role of p300/KAT3B in CTD acetylation, we introduced a specific shRNA targeting p300/KAT3B and observed an ~70% decrease in expression that correlated with a similar decrease in the acetylation of wild-type HA-RPB1 (Figure 3D). Likewise, treatment with the p300/CBP inhibitor C646, but not the inactive C37 control compound (Bowers et al., 2010), suppressed HA-RPB1 acetylation (Figure S3D). Moreover, overexpressing p300/KAT3B, but not PCAF/KAT2B, enhanced wild-type HA-RPB1 acetylation, underscoring the role of p300/KAT3B as a CTD acetyltransferase in human cells (Figure S3E).
Acetylated RPB1 Associates with 5′ Ends of Genes
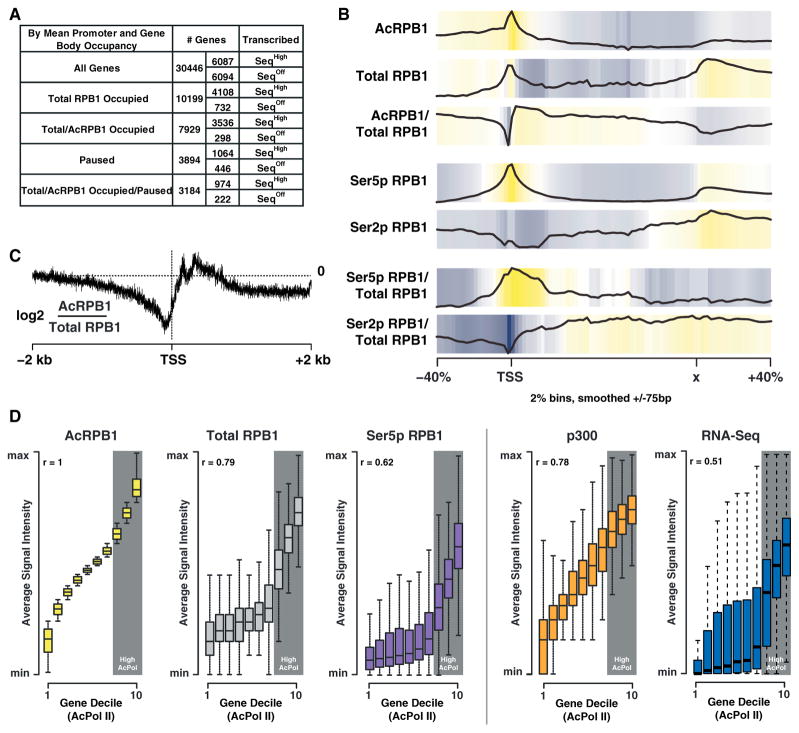
To gain insight into the function of K7 acetylation, we used ChIP-Seq with antibodies against acetylated and total RPB1 in murine ESCs. Using a cutoff of 2-fold over input, we found one-third of all genes occupied by total RPB1. Of these genes, ~78% were also occupied by acetylated RPB1, the majority of which were transcriptionally active (Figure 4A). In a meta-analysis of all occupied genes, RPB1 acetylation peaked at 5′ gene ends, with a steady decline observed within gene bodies and at 3′ gene ends (Figure 4B). When normalized to total RPB1, the occupancy profile of acetylated RPB1 showed a broad enrichment of K7 acetylation downstream of the TSS (Figures 4B and 4C), with relatively lower signal immediately upstream of the TSS. This distribution was unique. The S5-phosphorylated form displayed a tight colocalization with total promoter-proximal polymerases, consistent with widespread polymerase pausing in ESCs (Min et al., 2011; Rahl et al., 2010). The S2 form was barely detectable in this region, in accordance with its role during transcription elongation (Figure 4B).
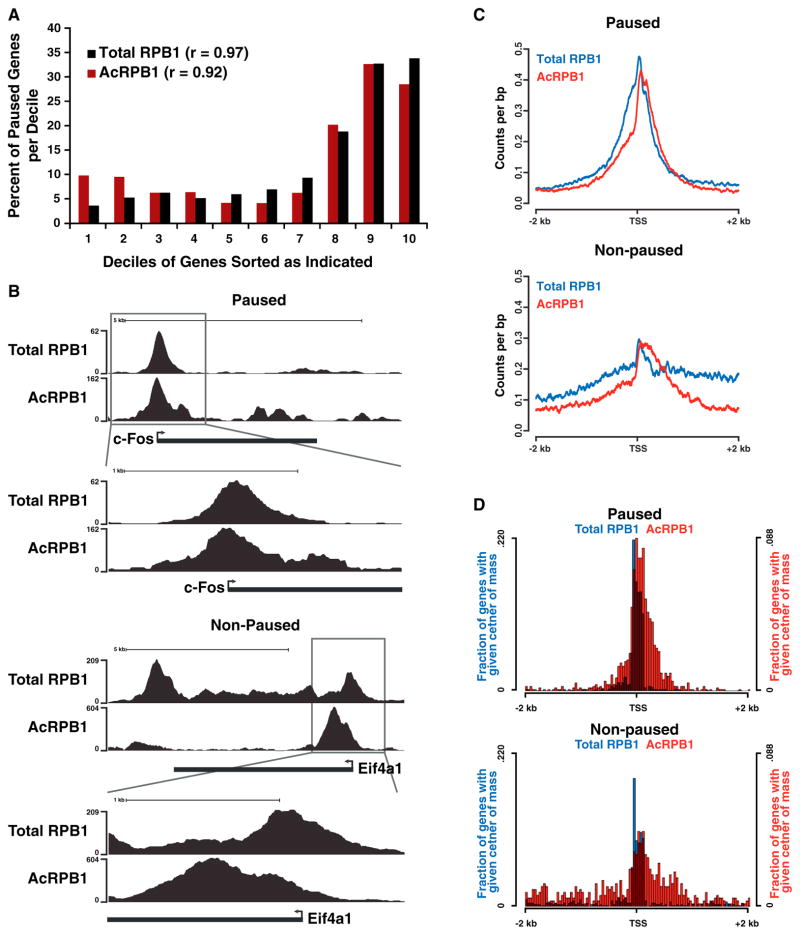
Figure 4. K7 Acetylation Is Enriched Downstream of the Transcription Start Site.
(A) Table showing the number of genes occupied by total and acetylated RPB1 as well as the pausing and expression state of the occupied genes (pausing index cutoff = 3).
(B) Input normalized average gene occupancies of the indicated RPB1 isoforms as well as their relative enrichment over total RPB1 plotted from −40% to +40% of the length normalized genes. The locations of the TSS as well as the termination site (x) are indicated.
(C) Plot of the relative acetylated/total RPB1 occupancy anchored at the TSS. Acetylated RPB1 enrichment is shown for a range of ± 2 kb of the TSS in log2 space at a single base resolution.
(D) Plots of gene deciles ranked (1–10) by acetylated RPB1 levels (left) within ±1 kb of the TSS. The occupancy levels for total RPB1, Ser5p RPB1, p300 (GEO accession number GSM918750), and RNA-Seq (GEO accession number GSM929718) ±1 kb of the TSS for each gene decile, as well as the genome-wide correlation (r) of these marks with acetylated RPB1, are shown on the right. The ranges are calculated by the default boxplot function in R, with the box describing the interquartile range (IQR) of the data, which contains the central 50% of data points. The whiskers extend to the furthest outlier from the edge of the box or 1.5 times the width of the IQR.
When all genes were binned into gene deciles according to the average intensity of acetylated RPB1 occupancy at the TSS, acetylation intensities correlated well with total polymerase (r = 0.79) and p300/KAT3B (r = 0.78) occupancy, underscoring the importance of p300/KAT3B as a cellular CTD acetyltransferase (Figure 4D). Highest acetylated RPB1 occupancy was found at genes with the highest level of total promoter-proximal RPB1 occupancy, suggesting that levels of CTD acetylation could correlate with the pausing state of genes (Figure 4D, deciles 8–10).
This was confirmed when we formally determined the pausing index of genes with high acetylated and total RPB1 occupancies. This index calculates the ratio of promoter-proximal versus promoter-distal polymerase occupancy (Muse et al., 2007), and only genes with at least a 3-fold enrichment of promoter-proximal polymerase signals were considered paused. Notably, most paused genes were found in the three deciles of genes harboring the highest levels of acetylated and total RPB1, and the pausing index of these genes correlated well with acetylated and total RPB1 occupancy levels (r > 0.9), supporting the model that CTD acetylation is linked to polymerase pausing (Figure 5A, deciles 8–10).
Figure 5. K7 Acetylation Marks Paused Polymerases.
(A) Graph showing the percentage of paused (pausing index cutoff = 3) genes per gene decile. Genes were sorted into deciles according to their occupancy by total or acetylated RPB1, with the lowest occupied genes in decile one and genes with the highest occupancies in decile ten.
(B) UCSC browser tracks (http://genome.ucsc.edu) for acetylated and total RPB1 at the entire c-Fos and Eif4a1 genes as well as just the TSS.
(C) Plots showing the acetylated and total RPB1 occupancy in counts per base pair at the TSS for paused and nonpaused genes.
(D) Plots showing the acetylated and total RPB1 center of mass distribution at the TSS for paused and nonpaused genes.
Peak Positions of Acetylated Polymerases Are Different in Paused versus Nonpaused Genes
Next, we analyzed the acetylation status of TSS-associated polymerases at paused versus nonpaused genes in the high-total/high-acetylated RPB1 occupancy category. Although TSS-associated polymerases were acetylated in both gene types, the distributions of acetylated RPB1 signals were distinctly different (Figure 5B). For example, at the immediate-early gene c-Fos, a “classical” paused gene (Plet et al., 1995), the peak for acetylated RPB1 occupancy was centered close to the TSS, mirroring the peak of total polymerase occupancy. This suggests that the bulk of acetylation occurs on the promoter-paused polymerase, with lower levels of acetylated Pol II observed escaping into the gene body. In contrast, at the nonpaused eukaryotic translation initiation factor gene Eif4a1, the main peak of acetylated RPB1 occupancy was shifted into the gene body relative to total RPB1 occupancy (Figure 5B). The differences of acetylated polymerase distribution were evident in the analysis across all genes in the paused and nonpaused categories (Figure 5C) and were independently confirmed in a “center of mass” analysis of total and acetylated RPB1 occupancy (Figure 5D). At paused genes, both total and acetylated RPB1 occupancies were centered in the promoter-proximal region, with acetylation occurring predominantly near the pause site. At nonpaused genes, the center of acetylated RPB1 occupancy was significantly less tightly associated with the TSS (p < 0.001) and was shifted overall into the gene body (Figure 5D). These findings identify a distinct difference in K7 acetylation between genes carrying paused and nonpaused polymerases and indicate that CTD acetylation occurs early in the transcription cycle, shortly after the polymerase initiates synthesis and begins elongation. Our data further suggest that the long lifetime of promoter-proximal polymerases at paused genes may allow them to be efficiently marked by K7 acetylation, whereas at nonpaused genes the rapid transition of the polymerase into productive elongation may cause CTD acetylation to occur at less defined positions within gene bodies.
Acetylation of K7 Residues Is Required for EGF-Induced Expression of Immediate-Early Genes
To study how CTD acetylation affects gene expression, we stably expressed wild-type or 8KR HA-RPB1 in murine fibroblasts and examined signal-induced gene expression of c-Fos and Egr2, both immediate-early genes with paused polymerases (Byun et al., 2012; Plet et al., 1995). Wild-type and mutant HA-RPB1-expressing cell cultures proliferated well when selected with α-amanitin; cells expressing 8KR HA-RPB1 grew slightly faster than cells expressing wild-type RPB1 (Figure S4A). When both cell cultures were stimulated with epidermal growth factor (EGF) to induce immediate-early gene expression, induction of c-Fos and Egr2 transcription was suppressed in 8KR-expressing cells as measured by quantitative RT-PCR (Figure 6A). In contrast, the expression of the housekeeping gene Eif4a1 (Figure 6A) was unchanged.
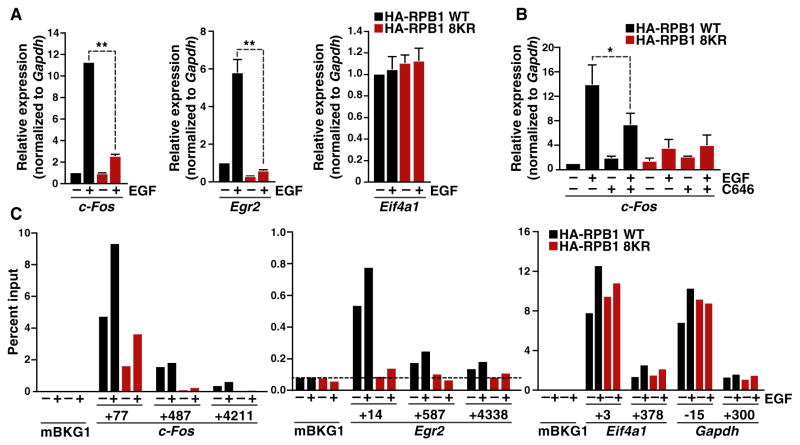
Figure 6. K7 Acetylation Regulates Growth-Factor-Induced Gene Expression.
(A) Real-time RT-PCR analysis of c-Fos, Egr2, and Eif41a gene expression in NIH/3T3 cells stably expressing WT or 8KR HA-RPB1 treated with epidermal growth factor (EGF) for 30 min. Results are shown as fold expression over untreated WT HA-RPB1 normalized to Gapdh (mean ± SD, n = 6, **p < 0.01).
(B) Real-time RT-PCR analysis of c-Fos gene expression in NIH/3T3 cells stably expressing WT or 8KR HA-RPB1 treated with the p300/CBP-specific inhibitor C646 for 1 hr followed by stimulation with EGF for 30 min. Results are shown as fold expression over untreated WT HA-RPB1 normalized to Gapdh (mean ± SD, n = 4, *p < 0.05).
(C) ChIP analysis of HA-RPB1 followed by real-time PCR analysis of the c-Fos and Egr2 genes at three different locations and Eif4a1 and Gapdh at two different locations shown relative to the TSS, and an intergenic background control (mBKG1). Data are expressed relative to input PCR values and represent the average of two ChIPs and duplicate PCRs. The dotted line marks the limit of detection of PCR method.
See also Figure S4.
To determine if acetylation of K7 residues is necessary for immediate-early gene transcription, we analyzed the induction of c-Fos and Egr2 expression in the presence of the p300/ KAT3B inhibitor C646 in wild-type or mutant RPB1-expressing cells. Treatment with C646, but not the solvent control, significantly decreased induction of c-Fos and Egr2 gene expression in wild-type cells, while no effect was observed in 8KR-expressing cells, confirming that EGF-induced transcription of immediate-early genes requires K7 acetylation (Figures 6B and S4B). Of note, a recent study also highlighted the importance of p300/KAT3B in EGF-induced c-Fos transcription but attributed this effect to the acetylation of histone H3K9 in the c-Fos promoter region (Crump et al., 2011).
Next, we performed ChIP experiments in these cell lines with HA-specific antibodies to measure total levels of wild-type and 8KR HA-RPB1 at c-Fos and Egr2 genes before and after EGF stimulation (Figure 6C). These studies confirmed the presence of polymerases downstream of the TSS before EGF treatment in cells expressing wild-type RPB1, consistent with polymerase pausing (Figure 6C). This occupancy was markedly lower in 8KR-expressing cells, before and during EGF treatment, indicating that K7 residues are involved in the recruitment of RNA polymerase II to the TSS, or in the establishment of stable promoter-associated polymerase complexes at these immediate-early genes (Figure 6C).
Notably, the 8KR mutation nearly abolished productive elongation at EGF-stimulated genes, with only background levels of 8KR RPB1 detected within the gene bodies (Figure 6C). This was not observed at two control genes, Eif4a1 and Gapdh, where polymerase occupancy was unchanged regardless of K7 status or EGF treatment (Figures 6C and S4C). These data underscore the importance of K7 residues in growth-factor-induced immediate-early gene transcription and are consistent with the concept that CTD acetylation is involved in initiation and/or early elongation steps.
DISCUSSION
Our data identify the RNA polymerase II CTD as an important new target for acetylation. Acetylation occurs at K7 residues located in the distal part of the CTD and is required for the regulation of growth-factor-induced gene expression. By using shRNAs and a small-molecule inhibitor, we demonstrate that p300/KAT3B is an important CTD acetyltransferase in vivo, but we do not exclude the possibility that additional acetyltransferases target the CTD in cells. CTD acetylation is reversible and likely involves the action of multiple HDACs of different classes, as differential hyperacetylation of polymerase isoforms is observed after treatment with TSA and nicotinamide.
Acetylation sites in human cells are frequently located in regions with ordered secondary structure (Choudhary et al., 2009; Kim et al., 2006), but the free CTD is considered largely flexible, with only some residual structure (Meinhart et al., 2005). A direct interplay with phosphorylation has previously been described in acetylated proteins, most notably histones (Cheung et al., 2000; Edmondson et al., 2002). We find that CTD phosphorylation suppresses acetylation of neighboring lysine residues and vice versa, an effect that is confined to the distal CTD where K7-containing repeats are located. Although it is difficult to determine how many and which repeats are phosphorylated at a given time during the transcription cycle in cells, our findings point to a mutual local control mechanism between acetylation and phosphorylation, i.e., by acetylation preventing aberrant or premature phosphorylation within the distal CTD and vice versa. While these modifications are mutually exclusive in K7-containing repeats, phosphorylation in the proximal part of the CTD likely remains undisturbed by acetylation, explaining why acetylation of both RPB1 isoforms, the hypo- and the hyper-phosphorylated forms, is detected by western blotting.
As distal CTD repeats and K7 residues are lacking in most fungi, this control mechanism appears unique to mammals and possibly other higher eukaryotes. This is in accordance with our finding that K7 acetylation is required for growth-factor-induced transcription, as EGF-induced growth regulation only evolved in higher eukaryotes (Schneider and Wolf, 2009). Polymerase pausing also evolved in higher eukaryotes as a regulatory mechanism, slowing the pace of the RNA polymerase II complex after initiation to ensure fidelity, allow assembly of accessory complexes, and enable accurate responses to extracellular signals such as EGF (Adelman and Lis, 2012). Drosophila melanogaster and Caenorhabditis elegans contain three and one K7-containing repeat, respectively. For both, polymerase pausing is well-studied in connection with external stimuli such as heat shock and food deprivation (Baugh et al., 2009; Rougvie and Lis, 1988). This—together with our data on EGF stimulation—provides a possible link between the presence of K7-containing repeats, polymerase pausing, and the transcriptional response to certain external stimuli in different higher eukaryotes.
It is important to note that in our study, levels of acetylation are comparable at paused and nonpaused genes; instead, we observe a distinct and statistically significant difference in the distribution of acetylation between both gene types. Notably, the (high) promoter-proximal total polymerase peak at paused genes overlaps well with a (high) peak of CTD acetylation at that position, indicating that the paused polymerase complex is primarily acetylated at the pause site. In contrast, the relative discordance of the total and acetylated CTD peaks at non-paused genes indicates that the polymerase is efficiently acetylated as it enters productive elongation. This finding, coupled with the dramatic decrease in promoter-proximal polymerase occupancy observed upon mutation of K7 residues, suggests that CTD acetylation might play a role in recruiting or stabilizing the paused complex. Indeed, p300/KAT3B is known to have a role in polymerase recruitment and transcription initiation (Eckner et al., 1994; Kraus et al., 1999), but important functions of the enzyme in transcription elongation have recently been described (Guermah et al., 2006). Likewise, we envision that CTD acetylation could impact several steps in the transcription cycle, especially at immediate-early genes, whose accurate activation requires both efficient elongation and rapid reinitiation of newly recruited RNA polymerase II.
Collectively, our data identify CTD acetylation as an important mechanism of transcription regulation in mammalian cells. CTD acetylation is implicated specifically during the induction of immediate-early genes, in agreement with previous reports showing that the nonconsensus distal CTD repeats are particularly important for activator-induced gene transcription (Chapman et al., 2005; Gerber et al., 1995). As acetylated lysines provide specific interaction interfaces with bromodomains proteins (Dhalluin et al., 1999), acetylated K7 residues may recruit bromodomain-containing coactivators to the elongating polymerase complex. Future experiments will focus on defining which proteins are involved and will determine the contribution of acetylated K7 residues to transcription initiation and elongation steps during the transcription cycle.
EXPERIMENTAL PROCEDURES
Plasmids
The GST-p300-KAT construct used in this study is described in Gu and Roeder (1997), and the HA-tagged PCAF plasmid is described in Dorr et al. (2002). The HA-tagged wild-type RNA polymerase II (HA-RPB1) construct carrying an α-amanitin resistance-inducing point mutation was a kind gift from Jeffrey L. Corden and has been described (Gerber et al., 1995), and the GST-CTD construct was a kind gift from Andrew P. Rice (Peterson et al., 1992). To generate the GST-tagged CTD truncation constructs, the codons for the eight C-terminal lysine residues were replaced by a STOP codon. Primer sequences are available upon request.
The mutant 8KR RNA polymerase II construct was generated by replacing all C-terminal lysine residues with arginines as described in Supplemental Experimental Procedures.
Cell Lines and Culture Conditions
Mouse ESCs were cultured in standard ES-DMEM: DMEM (GIBCO), 2 mM GlutaMax (GIBCO), 0.1 mM 2-mercaptoethanol (Sigma), 0.1 mM nonessential amino acids (GIBCO), 1 mM sodium pyruvate (GIBCO), 15% FBS (ESC tested, Hyclone catalog number SH30071.03), and 106 U/L leukemia inhibitory factor (ESGRO LIF #ESG1107). Cells were maintained in a 10 cm dish until confluency. HEK293T and NIH/3T3 cells were obtained from the American Type Culture Collection and grown under standard cell-culture conditions. Cells were transfected using the FuGENE 6 Transfection Reagent (Roche) according to the supplier’s instructions. Generation of stable wild-type and mutant RPB1-expressing cells was done as follows: 293T cells were transfected with linearized α-amanitin-resistant constructs expressing HA-tagged wild-type or 8KR RNA polymerase II. Forty-eight hours after transfection, 10 μg/ml α-amanitin (AppliChem) was added to the cell culture medium to select for HA-RPB1 expression. After 4 days, the selection pressure was removed, and the cells were allowed to recover for 1 week. The cells were then reselected in 5 μg/ml α-amanitin for 1 week. HA-RPB1 expression was confirmed by western blotting. NIH/3T3 cells were transfected with the respective linearized α-amanitin-resistant constructs together with a puromycin-resistance gene. Forty-eight hours after transfection, 2 μg/ml puromycin was added to the cell-culture medium to select for HA-RPB1 expression. After 3 days, the selection pressure was removed, and the cells were allowed to recover for 4 days. The cells were then selected in 5 μg/ml α-amanitin for 3 days, allowed to recover for 4 days, and then reselected in 5 μg/ml α-amanitin for 1 week. Growth rates of cells were determined by counting total cell numbers.
Antibodies and Reagents
The following antibodies and reagents were obtained commercially: anti-α-tubulin antibody (mouse, Sigma, T6074), anti-RPB1 antibody (N-20, rabbit, Santa Cruz Biotechnology, Inc., sc-899), anti-RPB1 antibody (Santa Cruz Biotechnology, sc-9001), anti-KAT3B/p300 antibody (NM11, mouse, Abcam, ab3164), anti-p300 antibody (Santa Cruz Biotechnology, sc-585), anti-acetylated-lysine antibody (rabbit, Cell Signaling Technology, 9441L), anti-RPB1 antibody (8WG16, mouse, Covance, MMS-126R), anti-RPB1 phosphoserine 2 antibody (Abcam, Ab5095), anti-c-Myc antibody (9E10, mouse, Santa Cruz Biotechnology, sc-40), anti-HA antibody (3F10, rat, Roche Applied Science, 1 867 423), and endothelial growth factor (EGF, Cell Signaling, 8916SC).
Enzymes for molecular cloning were purchased from New England Biolabs, cell culture reagents from Mediatech, fetal bovine serum from Gemini BIO-Products, and fine chemicals, if not noted otherwise, from Sigma.
Immunoprecipitation and Western Blotting
For western blotting, cells were lysed in p300 lysis buffer (7.6 mM NaH2PO4, 12.4 mM Na2HPO4, 250 mM NaCl, 30 mM NaPPi, 5 mM EDTA, 10 mM NaF, and 0.1% NP-40 [pH 7.0], and protease inhibitor cocktail [Sigma]). Immunoprecipitations were carried out using anti-HA-agarose (Sigma) and rProtein G and A agarose (Invitrogen); for details, see Supplemental Experimental Procedures.
Antibody Generation
Polyclonal AcRPB1 antibodies were generated in a rabbit immunized with chemically synthesized acetylated RNA polymerase II CTD peptides (PTSPK [Ac]YTPTSC, PTSPK[Ac]YSPTSC, PTTPK[Ac]YSPTSC, supplied by Dr. Hans-Richard Rackwitz, Peptide Specialty Laboratories GmbH, Heidelberg, Germany) in a 118-day rabbit peptide protocol (Covance) and purified on affinity columns loaded with the same immunogen (Affi-Gel Hz Immunoaffinity Kit, BioRad) as described previously (Pagans et al., 2011).
Expression and Purification of Recombinant Proteins
GST-fusion proteins were expressed in, and isolated from, Escherichia coli; see Supplemental Experimental Procedures. Recombinant His-Cdk9 was expressed in, and isolated from, baculovirus-infected Sf21 cells; see Supplemental Experimental Procedures.
In Vitro Acetylation Assay of GST-CTD
For in vitro acetylation assays, GST-tagged substrates were added to the reaction bound to 10 μl of a 50% glutathione Sepharose 4B slurry. Total calf thymus histones (20 μg, Sigma) were used as a control target for the acetylation reactions.
Reactions were all carried out in acetylation buffer with 200 nCi [14C] acetyl-coenzyme A (Amersham) for 1 hr at 37°C and were stopped by adding Laemmli buffer to the samples.
After SDS-PAGE, gels were fixed for 15 min in 10% acetic acid/50% methanol, stained in Coomassie blue staining solution (10% acetic acid, 50% methanol, 0.05% Coomassie Brilliant Blue G-250, Bio-Rad), and destained in destaining buffer (10% acetic acid, 7.5% methanol). Gels were then incubated for 30 min with Amplify Fluorographic Reagent (GE Healthcare), dried for 2 hr at 60°C under vacuum, and exposed to AmershamHyperfilm (GE Healthcare) at −70°C.
In Vitro Acetylation Assay of CTD Peptides
For in vitro acetylation or phosphorylation reactions of a CTD substrate with Ser7 mutated to lysine (CTD-K7), 50 μM of a CTD-K7 substrate comprising nine repeats (YSPTSPK)9 was incubated with 10 μM GST-p300 or 0.2 μM P-TEFb or 0.1 μM CDK7/CyclinH/MAT1 (ProQuinase, # 0366-0360-4) in the presence of acetylation buffer (25 mM HEPES [pH 8.0], 5% glycerol, 5 mM Na butyrate, 0.5 mM DTT), 7 mM MgCl2, and 1 mM acetyl-CoA (Sigma Aldrich) as coenzyme and/or ATP (Carl Roth). The reaction was incubated at 37°C for a defined reaction time (15–480 min) during mild rocking and was stopped by flash freezing (in case of acetylation) or addition of 50 mM EDTA (phosphorylation). The modification state of the CTD substrate was analyzed by electrospray ionization-mass spectroscopy with a Finnigan LCQ Advantage MAX ESI (Thermo Fisher Scientific) before desalting by a C4 column (Grace). For serial acetylation and phosphorylation reactions or vice versa, the first modification reaction was carried out for 4 hr at 37°C, followed by addition of the second enzyme (kept on ice). To ensure intact enzyme activity, control reactions were started at the same time when the second enzyme was added.
RNA Purification and Real-Time RT-PCR
Total RNA was purified from cells with the RNA STAT-60 reagent (TEL-TEST) according to the supplier’s specifications, followed by TURBO DNase (Ambion) treatment. The RNA was extracted with phenol-chloroform, precipitated with ethanol, and resuspended in an appropriate volume of water. For real-time RT-PCR analysis, the RNA was reverse transcribed with SuperScript III Reverse Transcriptase (Invitrogen) according to the supplier’s instructions, using Oligo(dT)18 primer (Fermentas) and 5 μg total RNA as a template. The real-time PCR reactions were carried out using a 2× HotSybr Real-time PCR Kit (McLAB) and a 7900HT Fast Real-Time PCR System (Applied Biosystems). Fold changes were calculated with the 2−ΔΔCt method (Livak and Schmittgen, 2001) and normalized for Gapdh. Primer sequences are available upon request.
Chromatin Immunoprecipitation Assay
ChIP-Seq in Embryonic Stem Cells
The ChIP material was prepared and processed, with slight modifications, as described by the Farnham lab protocol (O’Geen et al., 2011) using the anti-RNA polymerase II CTD repeat YSPTSPS antibody [4H8] (ab5408, Abcam), which recognizes both the phosphorylated and the non-phosphorylated CTD, for pan-RNA polymerase II and the polyclonal AcRPB1 antibody described above.
Data for S2-phosphorylated RPB1 (Rahl et al., 2010) as well as S5-phosphorylated RPB1 (Wamstad et al., 2012) and p300 (GEO accession number GSM918750) were analyzed in conjunction with the ChIP-seq analyses performed here. Tags were mapped to the mm9 genome using Bowtie2 (Lang-mead and Salzberg, 2012), and 10 million unique tags were sampled for each data set. Tag densities for browser tracks were calculated as the number of tags mapping within 75 bp of each 20 bp genomic bin, with each tag shifted 100 bp in polarity with read direction to account for fragment size. Custom tracks were then uploaded to the UCSC genome browser (Kent et al., 2002). Data sets were log2 normalized to their matching input by multiplying a normalization factor calculated by taking the ratio of average tag densities across intergenic regions (Xu et al., 2010). For the polymerase data sets, the log2 fold change for total RPB1 was then subtracted from the RPB1 modification variant input fold change to get the levels of the variant relative to total RPB1. To create gene-level plots, the genomic region around each transcript, from +40% of gene length upstream through −40% of gene length downstream, was divided into 2% sized bins according to total length including flanks. Then the log2 fold change for each data set relative to total RPB1 across all equivalent gene-level bins was calculated and plotted in R (R Core Team, 2012).
The pausing index was determined for total RPB1-occupied genes as follows: pausing index = (total RPB1 promoter/input promoter)/(total RPB1 gene body/input gene body). Only genes with a RPB1 promoter/Input promoter ratio of ≥2 were considered to be occupied by total RPB1.
The region within 2 kb of each TSS was divided into 1 bp bins, and the number of 10 mUUM tags from each data set that map to each bin was counted. To generate position-specific distributions across all TSS, the counts were processed as follows: (1) the counts were oriented by strandedness of the gene in question, and (2) the counts for each gene were normalized such that the bin with the highest number of tags for each gene within 2 kb has the value of 1 divided by the number of genes in the subset being examined.
The center of mass for a particular ChIP signal and for a particular gene was defined as the position relative to TSS that equally divides the area under the position-specific signal distribution within 2 kb of the TSS, as calculated above. Histograms of the center of mass were assembled for genes in high or low pausing groups.
ChIP in NIH/3T3 Cells Expressing Wild-Type or 8KR Mutant RPB1
ChIP material was prepared from cells ± EGF stimulation by crosslinking for 25 min with 1% formaldehyde essentially as described (Gilchrist et al., 2009). Immunoprecipitations were performed on 7.5 × 106 cells with 7 μl of antibody against HA (Abcam ab9110). Real-time PCR was performed on replicate samples, and the error represents the range. Primer sequences are available upon request.
Supplementary Material
Acknowledgments
We thank Jeffrey Corden and Andrew Rice for sharing reagents; Alexander Williams, Kirsten Eilertson, Brian Egan, Pao-Chen Li, and Ginger Muse for advice and assistance with experiments; and Gary Howard and Anna Lisa Lucido for editorial assistance. We gratefully acknowledge funds from Gladstone, the NIH (AI083139-02 [M.O.], GM62437 [P.A.C.], GM82901 [K.S.P. and J.A.C.], U01HL098179 [K.S.P. and B.G.B.]), the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (K.A., Z01 ES101987), the Deutsche Forschungsgemeinschaft (M.G., GE 976/5), the Boehringer Ingelheim Fonds (S.S.), the Human Frontiers Science Program and an E.G.G. fellowship (E.H.), and the PhRMA Foundation (J.A.C.).
Footnotes
ACCESSION NUMBERS
The ChIP-Seq data have been deposited at the Sequence Read Archive (SRA) under accession number SRX338012.
Supplemental Information includes four figures and Supplemental Experimental Procedures and can be found with this article at http://dx.doi.org/10.1016/j.molcel.2013.10.009.
References
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Näär AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Corden JL. Localization of an alpha-amanitin resistance mutation in the gene encoding the largest subunit of mouse RNA polymerase II. Mol Cell Biol. 1987;7:586–594. doi: 10.1128/mcb.7.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Demodena J, Sternberg PW. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science. 2009;324:92–94. doi: 10.1126/science.1169628. [DOI] [PubMed] [Google Scholar]
- Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H, et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem Biol. 2010;17:471–482. doi: 10.1016/j.chembiol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun JS, Fufa TD, Wakano C, Fernandez A, Haggerty CM, Sung MH, Gardner K. ELL facilitates RNA polymerase II pause site entry and release. Nat Commun. 2012;3:633. doi: 10.1038/ncomms1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RD, Conrad M, Eick D. Role of the mammalian RNA polymerase II C-terminal domain (CTD) nonconsensus repeats in CTD stability and cell proliferation. Mol Cell Biol. 2005;25:7665–7674. doi: 10.1128/MCB.25.17.7665-7674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Hintermair C, Eick D. Molecular evolution of the RNA polymerase II CTD. Trends Genet. 2008;24:289–296. doi: 10.1016/j.tig.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump NT, Hazzalin CA, Bowers EM, Alani RM, Cole PA, Mahadevan LC. Dynamic acetylation of all lysine-4 trimethylated histone H3 is evolutionarily conserved and mediated by p300/CBP. Proc Natl Acad Sci USA. 2011;108:7814–7819. doi: 10.1073/pnas.1100099108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Dorr A, Kiermer V, Pedal A, Rackwitz HR, Henklein P, Schubert U, Zhou MM, Verdin E, Ott M. Transcriptional synergy between Tat and PCAF is dependent on the binding of acetylated Tat to the PCAF bromodomain. EMBO J. 2002;21:2715–2723. doi: 10.1093/emboj/21.11.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingston DM. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- Edmondson DG, Davie JK, Zhou J, Mirnikjoo B, Tatchell K, Dent SYR. Site-specific loss of acetylation upon phosphorylation of histone H3. J Biol Chem. 2002;277:29496–29502. doi: 10.1074/jbc.M200651200. [DOI] [PubMed] [Google Scholar]
- Egloff S, O’Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber HP, Hagmann M, Seipel K, Georgiev O, West MAL, Litingtung Y, Schaffner W, Corden JL. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature. 1995;374:660–662. doi: 10.1038/374660a0. [DOI] [PubMed] [Google Scholar]
- Gilchrist DA, Fargo DC, Adelman K. Using ChIP-chip and ChIP-seq to study the regulation of gene expression: genome-wide localization studies reveal widespread regulation of transcription elongation. Methods. 2009;48:398–408. doi: 10.1016/j.ymeth.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guermah M, Palhan VB, Tackett AJ, Chait BT, Roeder RG. Synergistic functions of SII and p300 in productive activator-dependent transcription of chromatin templates. Cell. 2006;125:275–286. doi: 10.1016/j.cell.2006.01.055. [DOI] [PubMed] [Google Scholar]
- Hsin JP, Sheth A, Manley JL. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science. 2011;334:683–686. doi: 10.1126/science.1206034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Kraus WL, Manning ET, Kadonaga JT. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol Cell Biol. 1999;19:8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KC, Li J, Cole PA, Wong J, Kraus WL. Transcriptional activation by thyroid hormone receptor-beta involves chromatin remodeling, histone acetylation, and synergistic stimulation by p300 and steroid receptor coactivators. Mol Endocrinol. 2003;17:908–922. doi: 10.1210/me.2002-0308. [DOI] [PubMed] [Google Scholar]
- Liu P, Kenney JM, Stiller JW, Greenleaf AL. Genetic organization, length conservation, and evolution of RNA polymerase II carboxyl-terminal domain. Mol Biol Evol. 2010;27:2628–2641. doi: 10.1093/molbev/msq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu H, Zawel L, Fisher L, Egly JM, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- Mayer A, Heidemann M, Lidschreiber M, Schreieck A, Sun M, Hintermair C, Kremmer E, Eick D, Cramer P. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science. 2012;336:1723–1725. doi: 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. A structural perspective of CTD function. Genes Dev. 2005;19:1401–1415. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Geen H, Echipare L, Farnham PJ. Using ChIP-seq technology to generate high-resolution profiles of histone modifications. Methods Mol Biol. 2011;791:265–286. doi: 10.1007/978-1-61779-316-5_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagans S, Sakane N, Schnölzer M, Ott M. Characterization of HIV Tat modifications using novel methyl-lysine-specific antibodies. Methods. 2011;53:91–96. doi: 10.1016/j.ymeth.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SR, Dvir A, Anderson CW, Dynan WS. DNA binding provides a signal for phosphorylation of the RNA polymerase II heptapeptide repeats. Genes Dev. 1992;6:426–438. doi: 10.1101/gad.6.3.426. [DOI] [PubMed] [Google Scholar]
- Plet A, Eick D, Blanchard JM. Elongation and premature termination of transcripts initiated from c-fos and c-myc promoters show dissimilar patterns. Oncogene. 1995;10:319–328. [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. 2012 http://www.r-project.org/
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Schneider MR, Wolf E. The epidermal growth factor receptor ligands at a glance. J Cell Physiol. 2009;218:460–466. doi: 10.1002/jcp.21635. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Rojas LA, Beck D, Bonasio R, Schüller R, Drury WJ, 3rd, Eick D, Reinberg D. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science. 2011;332:99–103. doi: 10.1126/science.1202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Handoko L, Wei X, Ye C, Sheng J, Wei CL, Lin F, Sung WK. A signal-noise model for significance analysis of ChIP-seq with negative control. Bioinformatics. 2010;26:1199–1204. doi: 10.1093/bioinformatics/btq128. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.