Abstract
We have found a previously unreported precerebellar nucleus located among the emerging fibers of the motor root of the trigeminal nerve in the mouse, which we have called the interfascicular trigeminal nucleus (IF5). This nucleus had previously been named the tensor tympani part of the motor trigeminal nucleus (5TT) in rodent brain atlases, because it was thought to be a subset of small motor neurons of the motor trigeminal nucleus innervating the tensor tympani muscle. However, following injection of retrograde tracer in the cerebellum, the labeled neurons in IF5 were found to be choline acetyltransferase (ChAT) negative, indicating that they are not motor neurons. The cells of IF5 are strongly labeled in mice from Wnt1Cre and Atoh1 CreER lineage fate mapping, in common with the major precerebellar nuclei that arise from the rhombic lip and that issue mossy fibers. Analysis of sections from mouse Hoxa3, Hoxb1, and Egr2 Cre labeled lineages shows that the neurons of IF5 arise from rhombomeres caudal to rhombomere 4, most likely from rhombomeres 6–8. We conclude that IF5 is a significant precerebellar nucleus in the mouse that shares developmental gene expression characteristics with mossy fiber precerebellar nuclei that arise from the caudal rhombic lip.
Indexing Terms: rhombic lip, tensor tympani, motor trigeminal nucleus, principal sensory trigeminal nucleus, pontine nuclei
In a study of precerebellar nuclei in the mouse using cerebellar injection of retrograde tracers, we reported a group of labeled cells located among the emerging fibers of the motor root of the trigeminal nerve in the mouse (Fu et al., 2011). These cells appeared to be coincident with an area labeled the parvicellular trigeminal nucleus (5PC) in the rat brain atlas of Paxinos and Watson (2007) as well as an area that was labeled the tensor tympani part of the motor trigeminal nucleus (5TT) in the mouse brain atlas of Franklin and Paxinos (2008). The 5TT label was adopted by the latter atlas because these cells seemed to occupy an area that contains tensor tympani motor neurons in other mammals such as cats, rabbits, guinea pigs, monkeys, and rats (Mizuno et al., 1982; Keller et al., 1983; Shaw and Baker, 1983; Takahashi et al., 1984; Friauf and Baker, 1985; Gannon and Eden, 1987; van den Berge and van der Wal, 1990). A recent retrograde tracing study of tensor tympani motor neurons in the mouse shows that these motor neurons lie ventrolateral to the main trigeminal motor nucleus (Mukerji et al., 2009). The area occupied by the mouse tensor tympani motor neurons appears to overlap the territory of the region labeled 5TT in the Franklin and Paxinos (2008) atlas, but is much smaller.
In order to break the association with the trigeminal motor neurons, we have renamed this group of precerebellar neurons on a purely topographic basis as the interfascicular trigeminal nucleus (IF5). We have attempted to answer some of the questions raised by the finding that IF5 neurons project to the cerebellum. First, is there any relationship between the IF5 precerebellar neurons and the neighboring trigeminal motor neurons (including tensor tympani motor neurons)? Second, is there a developmental relationship between IF5 neurons and other precerebellar groups in the hindbrain?
To answer the first question (is there any relationship between the IF5 precerebellar neurons and the neighboring trigeminal motor neurons?), we examined choline acetyltransferase (ChAT) expression, because ChAT is an excellent marker for motor neurons (Ichikawa et al., 1997; McHanwell and Watson, 2009). In addition, we used a combination of ChAT and neuronal labeling following cerebellar injections with retrograde tracer to specifically ask whether some neurons might project both to the cerebellum and to a muscle.
To answer the second question (is there a developmental relationship between IF5 neurons and other precerebellar groups in the hindbrain?), we examined sections of postnatal (P0) brains (Wnt1) and P12 brains (Atoh1, also called Math1), as well as sections of adult brains containing Egr2, Hoxa3, and Hoxb1 Cre labeled lineages. Wnt1 is strongly expressed throughout the rhombic lip progenitor pool in the early embryo, and the Wnt1-Cre lineage fate map reveals that almost all of the precerebellar nuclei of the hindbrain (Fu et al., 2011) arise from this germinal zone. We examined the Atoh1 fate-mapped lineage because a layer of Atoh1 expression in the rhombic lip gives rise to mossy fiber precerebellar nuclei (Landsberg et al., 2005; Ray and Dymecki, 2009). In order to determine the rhombomeric origin of the IF5 neurons, we examined the fate-mapped populations in brains of mice from two Hox-Cre transgenic lines (Hoxb1 and Hoxa3) and a Hox transcription factor, Egr2. Hoxb1 and Hoxa3 are expressed in the rhombomeres caudal to rhombomere (r) 3 and caudal to r4, respectively (Favier and Dollé, 1997). Egr2 (formerly called Krox20) is widely expressed in the forebrain, and in the hindbrain is expressed exclusively in the third and fifth rhombomeres (Farago et al., 2006). Thus, by analyzing Cre fate-mapped lineages arising in of Hoxa3, Hoxb1, and Egr2 Cre transgenic animals, we were able to narrow down the rhombomeric origin of IF5 neurons.
We also used Atoh1CreER mice to determine the birthdate of Atoh1 derivatives (Machold and Fishell, 2005), including IF5. Finally, we looked for the presence in IF5 of a number of genes commonly expressed in the hindbrain precerebellar nuclei (Fu et al., 2009), by using data from The Allen Institute for Brain Science (available at http://www.brain-map.org/) (Lein et al., 2007).
Materials and Methods
Tracer studies
Animals
The tracer experiments were carried out on either C57BL/6J mice (8-10 weeks old, male, 21-25 g, n = 19), obtained from the Animal Resources Centre (Canning Vale, WA, Australia), or on ChAT(BAC)-eGFP transgenic mice (ChAT-EGFP; 8-10 weeks old, male, 21-25 g, n = 3, animals used at 6 backcross generations and 10 inbreeding generations), obtained from The Jackson Laboratory (Bar Harbor, ME). Animal experimental procedures were in accordance with the Australian Code for the Care and Use of Animals in Research and were approved by the Animal Care and Ethics Committee of The University of New South Wales (approved ACEC no. 11/75A).
Surgery and retrograde tracing
Mice were anesthetized with a mixture of ketamine (80 mg/kg body weight, i.p.) and xylazine (5 mg/kg body weight, i.p., for C57BL6/J mice; 12 mg/kg body weight, i.p., for ChAT-EGFP mice) for surgery. The retrograde tracers horseradish peroxidase (HRP; Sigma-Aldrich, St. Louis, MO; 25% dissolved in distilled water), Fluoro-Gold (FG; Fluorochrome, Denver, CO; 5% dissolved in distilled water), and Alexa Fluor dextran conjugate (AFD; Alexa Fluor 594; 10,000 MW, no. D-22913, Invitrogen, Carlsbad, CA; dissolved in phosphate-buffered saline [PBS; pH 7.4] at 50 mg/ml) were injected either jointly or independently. The injections were made into the cerebellar vermis and/or right hemisphere at a volume of 200-300 nl for each injection, by using a stereotaxic instrument (David Kopf Instruments, Tujunga, CA). In all cases, a large region of the cerebellum of one side was filled with tracer through multiple or single injections (Fig. 1).
Figure 1.
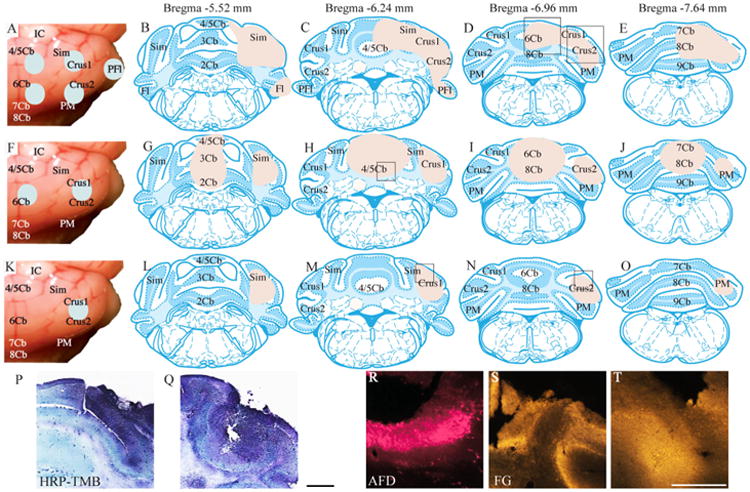
Retrograde tracer injections into the cerebellum. A: A surface view of the HRP injections. B–E: Area filled with HRP delivery is indicated in transverse section drawings taken from Franklin and Paxinos (2008). F: A surface view of AFD and FG injections. G–J: The area filled with tracer. K: A single large FG injection. L–O: The area filled with FG. P,Q: Photomicrographs of an HRP injection. The area shown in P is represented by a rectangle in the vermis in D, and the area shown in Q is represented by a rectangle in the cerebellar hemisphere in D. R: Photomicrograph of an AFD injection site (see rectangle in the H). S,T: Photomicrographs of FG sites (see rectangles in the cerebellar hemisphere in M and N, respectively). 2Cb, lobule 2 of the cerebellar vermis; 3Cb, lobule 3 of the cerebellar vermis; 4/5Cb, lobules 4 and 5 of the cerebellar vermis; 6Cb, lobule 6 of the cerebellar vermis; 7Cb, lobule 7 of the cerebellar vermis; 8Cb, lobule 8 of the cerebellar vermis; 9Cb, lobule 9 of the cerebellar vermis; AFD, Alexa Fluor dextran conjugate; Crus1, crus1 of the ansiform lobule; Crus2, crus2 of the ansiform lobule; FG, Fluoro-Gold; Fl, flocculus; HRP, horseradish peroxidase; IC, inferior colliculus; PFl, paraflocculus; PM, paramedian lobule; Sim, simple lobule; TMB, 3,3′,5,5′-tetramethylbenzidine. Scale bar = 500 μm in Q (applies to P,Q) and T (applies to R–T).
Postsurgery tissue preparation
In the cases with HRP injection (n = 5), mice were sacrificed on the second postoperative day. In cases with FG injection (n = 8, including three ChAT-EGFP mice), the mice were sacrificed on the seventh postoperative day. In cases of FG and AFD injections (n = 6), mice were sacrificed on the 18th postoperative day. The deeply anesthetized mice were perfused with 4% paraformaldehyde (4°C, pH 7.4). After cryoprotection in 30% sucrose, brains were cut with a cryostat microtome at 40 μm thickness and mounted on slides (not including the two FG injection cases used for immunofluorescence). HRP-labeled sections were reacted with 3,3′,5,5′-tetramethylbenzidine (TMB) (Fu et al., 2009, 2011) before histological examination; FG- and AFD-labeled sections were directly examined under a fluorescence microscope (Olympus BX51; Olympus, Tokyo, Japan) equipped with an AxioCam HRc digital camera (Carl Zeiss International, Oberkochen, Germany).
Immunofluorescence
FG labeling was combined with immunofluorescence to detect NeuN, a neuron-specific marker (Mullen et al., 1992). The sections from the two cases of FG injection were collected as free-floating sections and blocked in 5% serum, incubated with anti-NeuN antibody (Table 1) for 2 days at 4°C, and then incubated with Alexa Fluor 594 goat anti-mouse antibody (Invitrogen, A11032) for 2 hours at room temperature. At the end of the incubation, the sections were washed, mounted, and coverslipped with anti-fade mounting medium (Dako, Campbellfield, VIC, Australia).
Table 1. Antibodies Used in This Study.
| Antibody | Raised to | Manufacturer and cat. no. | Species raised in |
|---|---|---|---|
| NeuN | Purified cell nuclei from mouse brain | Chemicon, MAB377 | Mouse monoclonal |
| β-Gal | Raised against recombinant β-gal protein (E. coli) | Biogenesis 4600-1409 | Goat polyclonal |
| GFP | Raised against native GFP isolated from the jellyfish Aequorea victoria | Invitrogen A11122 | Rabbit polyclonal |
| Alexa Fluor 594 goat anti-mouse IgG | Prepared from affinity-purified antibodies that react with IgG heavy chains and all classes of immunoglobulin light chains from mouse | Invitrogen, A11032 | Goat highly cross-adsorbed |
| Alexa Fluor 594 donkey anti—goat IgG | Prepared from affinity-purified antibodies that react with IgG heavy chains and all classes of immunoglobulin light chains from goat | Invitrogen A11058 | Donkey |
| Alexa Fluor 488 donkey anti-rabbit IgG | Prepared from affinity-purified antibodies that react with IgG heavy chains and all classes of immunoglobulin light chains from rabbit | Invitrogen A21206 | Donkey |
Abbreviations: β-gal, β-galactosidase; GFP, green fluorescent protein; NeuN, neuron-specific neuronal protein.
Nissl stain
Brain sections were cut in the coronal or sagittal plane and directly mounted for cresyl violet staining (n = 3, C57BL/6J). Sections were photographed by using a Scan-Scope digital slide scanner (model XT, Aperio Technologies, Vista, CA). Images containing typical neurons of IF5, motor trigeminal nucleus (5N), dorsal part of the principal sensory trigeminal nucleus (Pr5-d), and ventral part of the principal sensory trigeminal nucleus (Pr5-v) were extracted for analysis.
Developmental studies using Cre transgenic mice
Wnt1-Cre, Egr2-Cre, Hoxa3-IRES-Cre, and Hoxb1-IRES-Cre
In the case of Wnt1, Egr2, Hoxa3, and Hoxb1 Cre fatemapping analyses, the mouse lines were maintained at the University of Utah, originally sourced as follows: the Wnt1-Cre line is that of Danielian et al. (1998); the Egr2-Cre line is that of Voiculescu et al. (2000); the Hoxa3-IRES-Cre line is that of Macatee et al. (2003); and the Hoxb1-IRES-Cre line is that of Arenkiel et al. (2003). Lineage analysis, in the case of Wnt1, Egr2, Hoxa3, and Hoxb1, was carried out by crossing each Cre driver line to the ROSA26R reporter line (Soriano, 1999) to generate compound transgenic animals for analysis. All Cre lines were maintained on a C57BL/6J background and the ROSA26R on a CD1 background. All mouse use at the University of Utah complied with protocols approved by the University of Utah Institutional Animal Care and Use Committee.
For β-gal staining, postnatal animals (for Wnt1) and adult animals (8–10 weeks old for Egr2, Hoxa3, and Hoxb1) were perfusion-fixed with 2% paraformaldehyde, dissected, cryosectioned, and transferred into X-gal staining solution (0.8 mg/ml X-gal, 25 mM K3Fe(CN)6, 25 mM K4Fe(CN)6-3H2O, 2 mM MgCl2, 0.01% Na deoxycholate, 0.02% NP40 in PBS [pH 7.4]).
Atoh1CreER
Atoh1CreER mice (Machold and Fishell, 2005) were crossed with Taulox-stop-lox-mGFPiresNLSLacZ reporter mice (Hippenmeyer et al., 2005) to generate compound transgenic animals. Both lines were maintained on a Swiss Webster (Taconic Farms, Germantown, NY) background as heterozygotes. The day of the observed plug was considered to be E0.5; tamoxifen (4-mg dose from a 20 mg/ml stock in corn oil; Sigma) was administered by oral gavage to pregnant dams at E9.5, E10.5, E11.5, or E12.5. Double transgenic P12 pups (Atoh1CreER; Tau reporter) identified by polymerase chain reaction (PCR) genotyping were anesthetized with pentobarbital (Nembutal; i.p. injection) and transcardially perfused with ice-cold 4% paraformaldehyde/PBS (pH 7.2), after which the brains were dissected and fixed for an additional 4 hours prior to overnight equilibration in 30% sucrose/PBS (pH 7.4). Cryoprotected brains were frozen in blocks of OCT (Tissue-Tek, VWR, Radnor, PA) on crushed dry ice, and 20-μM coronal sections were obtained by using a Leica CM3050S cryostat. Histochemistry for nuclear (β-gal as well as immunofluorescence for nuclear (β-gal and membrane-tethered EGFP (mGFP) was performed as described previously (Machold and Fishell, 2005). For immunofluorescence, goat anti-β-gal (Biogenesis, Kingston, NH) and rabbit anti- EGFP (Invitrogen) primary antibodies, and donkey anti-goat (Alexa Fluor 594 conjugated) and anti-rabbit (Alexa Fluor 488 conjugated) secondary antibodies (Invitrogen), respectively, were used. Mice were maintained and sacrificed in accordance with the protocols approved by the Institutional Animal Care and Use Committee at the New York University School of Medicine.
Antibody characterization
The NeuN antibody used in this study stains the same pattern of cellular morphology and distribution that has been demonstrated in previous publications (Mullen et al., 1992; Wolf et al., 1996; Watson and Paxinos, 2010). The GFP antibody has been used in previous studies (Allen mouse brain connectivity atlas: www.alleninstitute.org; Czupyrn et al., 2011) to identify GFP-positive neurons in mouse brains from transgenic animals expressing this reporter protein. The (β-gal antibody has been used in previous studies (Packard et al., 2011; Kim et al., 2011) to identify (β-gal-positive neurons in mouse brains from transgenic animals expressing this enzyme. For both GFP and (β-gal antibodies, no signal is detectable in brain sections collected from wild-type animals.
Image analysis
The contrast and brightness of the images were enhanced by using Adobe Photoshop CS6 (Adobe Systems, San Jose, CA). Double-labeled images were converted from red-green to magenta-green by using Adobe Photoshop CS6, and the image groups were then assembled by using Adobe Illustrator CS6.
Results
The interfascicular trigeminal nucleus defined by HRP and Nissl staining
Labeled IF5 neurons were identified in coronal sections after multiple injections of HRP into the cerebellum of one side (Fig. 1A). The labeled neurons were located in the vicinity of the emerging motor trigeminal nerve fibers from bregma −4.60 mm to bregma −5.20 mm (Fig. 2A-F). At most coronal levels, the IF5 contained three to five layers of cells arranged from medial to lateral. Except for the rostral IF5 (where 5N is not present: rostral to bregma −4.84 mm), the IF5 was always located between the 5N medially and the Pr5 laterally. In coronal sections, the IF5 neurons were clustered among the bundles of motor trigeminal fibers (m5), with the exception of the caudal IF5 neurons, which were located ventrolateral to the motor neurons of 5N (Fig. 2A-F). More HRP-labeled neurons were found in the ipsilateral IF5 than in the contralateral side (Fig. 2 shows a representative case).
Figure 2.
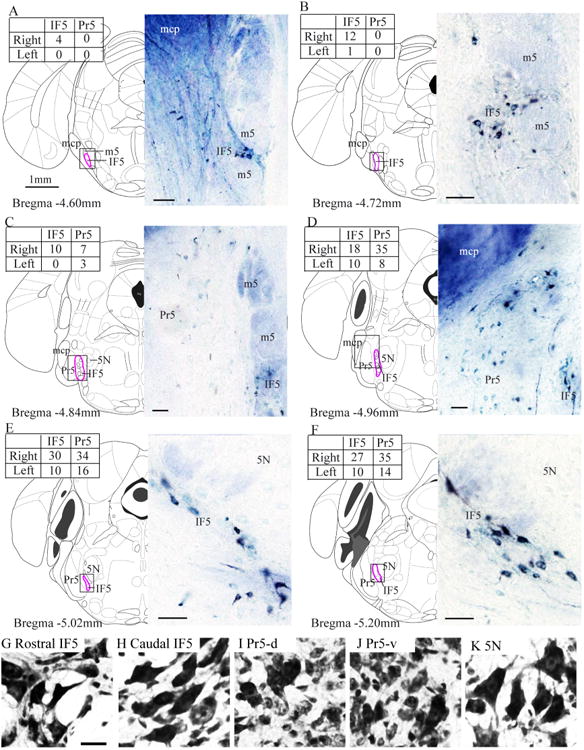
IF5 labeling after cerebellar tracer injections. A–F: Rostrocaudal series of diagrams and accompanying photographs of labeled cells following a contralateral cerebellar HRP injection (Fig. 1A). The diagrams and coordinates are taken from the atlas of Franklin and Paxinos (2008). The inset tables show the numbers of HRP-labeled cells in the IF5 and Pr5 each side for each section. G–K: Photomicrographs of neurons from each region of interest. 5N, motor trigeminal nucleus; HRP, horseradish peroxidase; IF5, interfascicular trigeminal nucleus; m5, motor root trigeminal nerve; mcp, middle cerebellar peduncle; Pr5, principal sensory trigeminal nucleus; Pr5-d, dorsal part of the principal sensory trigeminal nucleus; Pr5-v, ventral part of the principal sensory trigeminal nucleus. Scale bar = 50 μm in A–F; the scale bar in G is 20 μm, and it applies to G–K.
In cresyl violet-stained sections, the neurons in rostral IF5 were triangular, pyriform, olivary, or multipolar in shape, whereas the neurons in caudal IF5 were mostly elongated (Fig. 2G,H). The neighboring Pr5 contained mainly small round neurons mixed with a few larger olivary shaped neurons (Fig. 2I,J). The 5N mainly contained triangular or multipolar shaped neurons (Fig. 2K).
The relationship between IF5 neurons and motor trigeminal neurons
FG was injected into the cerebellum of each of three ChAT-EGFP mice (Fig. 1F). Although the injections were large and filled the cerebellar vermis and right hemisphere, we found no ChAT-EGFP cells containing FG (Fig. 3). FG-labeled IF5 cells were always located lateral or ventrolateral to the 5N motor neurons. Near the caudal pole of IF5 (bregma −4.96 mm), a few ChAT-EGFP cells were present ventrolateral to the main 5N motor neuron cluster (Fig. 3A-C, white arrows and arrowheads). We believe that these ChAT-positive cells are likely to be the tensor tympani motor neurons identified by Mukerji et al. (2009). Some of these presumed tensor tympani motor neurons were embedded in the emerging fibers of the motor trigeminal nerve and some were mingled with the IF5 neurons (Fig. 3A,B, as the white arrowheads indicate).
Figure 3.
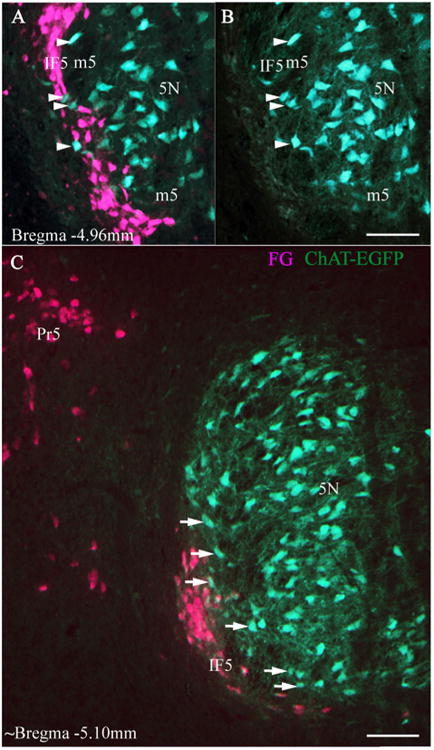
FG retrograde tracing in ChAT-EGFP mice. Photomicrographs of fluorescence images of FG-labeled IF5 neurons in sections from a ChAT-EGFP mouse brain following contralateral injections of FG in the cerebellum (Fig. 1F). A–C: Location of IF5 in relation to 5N; IF5-labeled neurons do not express ChAT. B is the single-channel view of A, showing ChAT neurons in the same area. White arrowheads in A and B indicate that ChAT-EGFP neurons with no retrograde signal are mingled with FG-labeled precerebellar neurons of IF5. White arrows in C point to the ChAT-EGFP neurons located in between 5N and IF5, which we presume to be tensor tympani motor neurons. 5N, motor trigeminal nucleus; ChAT-EGFP, choline acetyltransferase-enhanced green fluorescent protein; FG, Fluoro-Gold IF5, interfascicular trigeminal nucleus; m5, motor root trigeminal nerve; Pr5, principal sensory trigeminal nucleus. Scale bar = 100 μm in all cases.
The cerebellar target of interfascicular trigeminal nucleus
We did not attempt to define the cerebellar target of IF5, but we do have evidence that IF5 neurons project to a wide area of vermis and hemisphere. In the case in which AFD was injected in the vermis and FG was injected in the right hemisphere (Fig. 1F), few AFD-labeled cells were located in the rostrocaudal IF5, but many were located in the caudoventral IF5 (Fig. 4A), whereas the FG-labeled cells were found to be evenly distributed at rostrocaudal levels. This confirms that IF5 projects to both vermis and hemisphere. A very small number of double-labeled (AFD and FG) neurons were found (Fig. 4C-F, indicated by white arrowhead). Although we cannot completely exclude the possibility that there was some overlap of the injected regions, the fact that double-labeled neurons were very rare indicates that it is unlikely that individual IF5 neurons project to both vermis and hemisphere.
Figure 4.
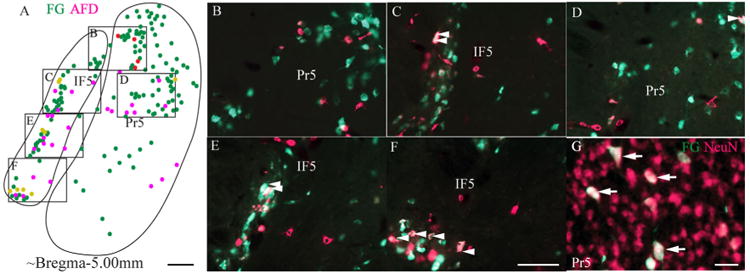
Retrograde labeling of IF5 neurons after injection of AFD and FG into the cerebellum. Location of AFD (magenta) and FG (green) labeled neurons in the IF5 after cerebellar injections (Fig. 1F). A–F: A diagram of the position of labeled cells in IF5 (A) is accompanied by five high-magnification images (B–4F). White arrowheads point to neurons located in the caudoventral IF5 that contain both AFD and FG signals, indicating that these neurons project to either the paravermal area or both the cerebellar vermis and hemisphere. G: A high-magnification image shows large FG-labeled neurons in the dorsal part of Pr5 at around bregma −4.96 mm. AFD, Alexa Fluor dextran conjugate; FG, Fluoro-Gold; IF5, interfascicular trigeminal nucleus; NeuN, neuron-specific neuronal protein; Pr5, principal sensory trigemina nucleus. Scale bar = 100 μm in A; 50 μm in F (applies to B–F); 20 μm in G.
In the case of a single FG injection in the right hemisphere (Fig. 1K), the spread of FG filled the most of the volume of the cerebellar hemisphere (crus1 of the ansiform lobule [Crus1], crus2 of the ansiform lobule [Crus2], the simple lobule [Sim], and the paramedian lobule [PM]). When this FG labeling was combined with immunofluorescence to detect NeuN, a substantial proportion of the NeuN-immunoreactive cells was found to contain FG (56.62%, n = 302), which indicates that the cerebellar hemisphere is the target of the majority of IF5 neurons. Labeled cells in Pr5 were concentrated in the dorsal part of the nucleus (Pr5-d), and these labeled cells were larger than the general population of neurons in Pr5 (Fig. 4G, as indicated by the white arrows).
Development of the mouse interfascicular trigeminal nucleus
Wnt1 Cre fate map labeling was strong in IF5, which indicates that IF5 arises from the rhombic lip. These neurons were embedded among the fibers of the emerging trigeminal motor nerve root or were located in the band of fibers that separates 5N and Pr5 (Fig. 5A).
Figure 5.
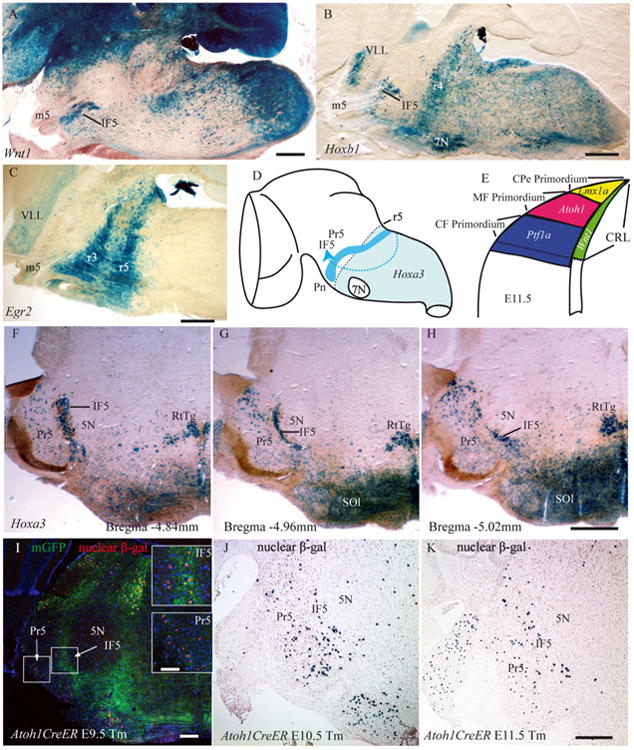
The developmental origin of IF5: evidence from gene expression. A–C: Sagittal sections of a mouse brain showing the IF5 cell groups labeled in different Cre lineages by X-Gal staining: the Wnt1-Cre lineage (A); the Hoxb1 lineage (B); and the Egr2 lineage (C). D: Drawing of the hypothesized migration stream of IF5 progenitors from the caudal rhombic lip to the rostral hindbrain. E: Diagram of a transverse section of the caudal rhombic lip showing the dorsoventral layers of gene expression that related to the development of the hindbrain precerebellar nuclei. F–H: Rostral to caudal views of Hoxa3 expression in IF5 and Pr5. I–K: Atoh1 lineage neurons are evident in IF5 and Pr5, where the Cre reporter expression (membrane-tethered GFP [mGFP] and nuclear β-gal) was induced by Tm administration at different embryonic days in the Atoh1CreER transgenic embryos. 5N, motor trigeminal nucleus; 7N, facial nucleus; β-gal, β-galactosidase; CF, climbing fiber; CPe, choroid plexus; CRL, caudal rhombic lip; IF5, interfascicular trigeminal nucleus; m5, motor root trigeminal nerve; MF, mossy fiber; mGFP, membrane tethered green fluorescent protein; Pn, pontine nuclei; Pr5, principal sensory trigeminal nucleus; r, rhombomere; RtTg, reticulotegmental nucleus of the pons; SOl, superior olive; Tm, Tamoxifen; VLL, ventral nucleus of the lateral lemniscus. Scale bar = 500 μm in A–C, H (applies to F–H), and K (applies to J,K); 200 μm in I; 100 μm in insets to I.
Hoxb1-Cre
The outstanding feature of the Hoxb1 lineage fate map was the presence of a restricted cluster of labeled cells in a dense band coincident with the fourth rhombomere (Arenkiel et al., 2004). Because the facial nucleus neurons arise in r4 and then migrate caudally to r6, they were also prominently labeled in this fate map. Although the region of Hoxb1 fate-mapped cells generally formed a sharp border at the junction of r3 and r4, labeled populations extended caudally from r4 into the spinal cord, but the caudal labeling was less intense (Fig. 5B). However, we also observed that the neurons of IF5 and the ventral nucleus of the lateral lemniscus had migrated rostrally from the main Hoxb1 expression territory. We therefore conclude that the IF5 neurons arise from the caudal hindbrain (somewhere from r4 to the junction of the hindbrain and spinal cord) and that they do not arise from r1–r3.
Egr2 (Krox20)-Cre
Consistent with many previous reports of the expression of this gene (Schneider-Maunoury et al., 1997; Voiculescu et al., 2000), we found that Egr2 lineages encompassed r3 and r5 (Fig. 5C). Because no rostral migratory groups were observed in this lineage, we conclude that the IF5 cells do not arise from r3 or r5.
Hoxa3-Cre
The main field of expression of Hoxa3 (r5 and more caudally) gives rise to a number of significant rostral migratory groups (Farago et al., 2006). Chief among these migrations were those that form the basilar pontine and reticulotegmental nuclei, but we found additional migrations into the rostral hindbrain that have not received attention in the past, including the cells of IF5 and the relatively large cerebellar projecting cells embedded in the dorsal part of Pr5 (Fig. 5F–H).
From the point of view of IF5 origin, the Hoxa3 lineage analysis reveals that the IF5 neurons do not arise from the rostral hindbrain (r1–r4), but that they arise from a region in the caudal hindbrain somewhere between r4 and the junction of the hindbrain and spinal cord. Combined with the results from Egr2-Cre (which show that IF does not arise from r5) and the logic of Farago et al. (2006), it seems likely that the IF5 neurons are generated from r6–r8.
Atoh1CreER
We examined a series of P12 brain sections from the Atoh1CreER fate-mapping experiments in which tamoxifen was administered to pregnant dams at day E9.5, E10.5, E11.5, or E12.5. We found that IF5 cells arise from Atoh1-expressing precursors, and therefore from the Atoh1 layer of the caudal rhombic lip, along with the other major mossy fiber precerebellar nuclei (Fig. 5D,E). We also found that substantial numbers of the IF5 and Pr5 precerebellar progenitors were labeled by tamoxifen administration as early as E9.5 (Fig. 5I–K) and that the Atoh1 expression in IF5 neurons persisted at least up to E12.5. There did not appear to be any substantial labeling in the pontine nuclei at stages prior to E12.5 (data not shown).
Genetic expression profile of the mouse interfascicular trigeminal nucleus
Finally, we compared the pattern of gene expression in the major precerebellar nuclei with that in IF5 and Pr5. We did this by analyzing the images available online from the Allen Brain Institute for Brain Science (Lein et al., 2007), using the AGEA tool (Table 2). The analysis confirmed that the gene expression profile of IF5 and Pr5 was similar to that seen in the mossy fiber issuing precerebellar nuclei. An exception was the gene Lgals1, which was expressed in the reticulotegmental nucleus of the pons and the lateral reticular nucleus, but not in IF5 or Pr5 (Table 2). Notably, neither IF5 nor Pr5 expressed Pou4f1, a gene that was expressed in the climbing fiber issuing precerebellar nucleus, the inferior olive.
Table 2. Gene Expression in the Major Hindbrain Precerebellar Nuclei, IF5, and Pr51.
| Major hindbrain PreCb nuclei | ||||
|---|---|---|---|---|
|
|
||||
| Gene symbol | Gene name | IF5 | Pr5 | Others |
| B2m | β-2-Microglobulin | + | + | Pn, RtTg, ECu, LRt |
| Ctsz | Cthepsin Z | + | + | Pn, RtTg, ECu, LRt, IO |
| Depdc6 | DEP domain containing 6 | + | + | Pn, RtTg, ECu, LRt |
| Dgat2 | Dacylglycerol O-acyltransferase 2 | + | + | Pn, RtTg, ECu, LRt |
| Lgals1 | Lectin, galactose binding, soluble 1 | − | − | RtTg, LRt |
| Pou4f1 | POU domain, class 4, transcription factor 1 | − | − | IO |
| Rgs4 | Regulator of G-protein signaling 4 | + | + | Pn, RtTg, ECu, LRt, IO |
| Slc17a7 | Solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 | + | + | Pn, RtTg, ECu, LRt |
| Zfp533 | Zinc finger protein 533 | + | + | Pn, RtTg, ECu, LRt, IO |
Abbreviations: ECu, external cuneate nucleus; IF5, interfascicular trigeminal nucleus; IO, inferior olive; LRt, lateral reticular nucleus; Pn, pontine nuclei; Pr5, principal sensory trigeminal nucleus; PreCb, precerebellar nuclei; RtTg, reticulotegmental nucleus of the pons
From our analysis of expression seen in ISH images in the website of the Allen Brain Institute for Brain Science (http://www.brain-map.org/), using the AGEA tool.
Discussion
The discovery of a precerebellar nucleus in the vicinity of the emerging fibers of the motor root of the trigeminal nerve is somewhat surprising. We have wondered why a similar cell group has not been reported in other mammals. Its presence in the mouse raises a number of questions as to the developmental relationships of these neurons. We have attempted to answer these questions with the use of retrograde tracing, genetic fate mapping, and gene expression profiling. To clearly distinguish the cerebellar projecting cells from the adjacent trigeminal motor neurons, we have named them the interfascicular trigeminal nucleus on account of their location.
The precerebellar neurons of the interfascicular trigeminal nucleus are distinct from the tensor tympani motor neurons
We have shown that IF5 neurons are ChAT negative and therefore cannot be motor neurons. The IF5 neurons are adjacent to the 5TT motor neurons in the mouse. The 5TT motor neurons have been identified in a number of mammals (Keller et al., 1983; Takahashi et al., 1984; Gannon and Eden, 1987; Strutz et al., 1988; van den Berge and van der Wal, 1990; Mukerji et al., 2009). In the cat and cynomolgus monkey, the 5TT motor nucleus represents a separate, ventrally located, parvicellular division of the trigeminal motor nucleus (Friauf and Baker, 1985; Gannon and Eden, 1987). In the CBA/CaJ mouse, the nucleus forms a layer ventrolateral to the motor trigeminal nucleus (Mukerji et al., 2009). On the basis of their location and shape, we consider the ChAT-EGFP neurons adjacent to IF5 in our preparations to represent the 5TT neurons identified by Mukerji et al. (2009). As noted above, although there was a small amount of overlap between the most lateral ChAT-positive neurons of 5TT and the IF5 precerebellar neurons, we found no colocalization of ChAT and retrograde cerebellar tracer. It is therefore safe to identify IF5 in the C57BL/6J mouse as a separate nucleus applied to the lateral border of the 5TT, whereas the medial border of 5TT is located close to the main body of 5N.
Generation of interfascicular trigeminal neurons
We have shown that the cells of the IF5 cell group arise from Wnt1- and Atoh1-expressing lineages. Whereas Wnt1 is expressed in all cells of the caudal rhombic lip, Atoh1 is expressed only in the caudal rhombic lip layer that gives rise to mossy fiber precerebellar nuclei (Landsberg et al., 2005; Ray and Dymecki, 2009). Furthermore, we found that IF5 contains substantial numbers of cells arising from Hoxb1 and Hoxa3 territories, which are located in the rhombomeres caudal to r3 and caudal to r4, respectively (Favier and Dollé, 1997; Creuzet et al., 2005). In addition, IF5 cells did not express Egr2, which is a marker for r3 and r5 lineages. These results therefore confirm that the IF5 progenitors are generated from the rhombic lip caudal to r5, as are the other major hindbrain precerebellar nuclei (Farago et al., 2006). An analysis of the Atoh1CreER lineage data shows that the generation of IF5 progenitors is between E9.5 and E12.5, prior to the pontine neurons.
IF5 shares gene expression patterns with major mossy fiber projecting precerebellar nuclei
IF5 neurons express many genes in common with major mossy precerebellar nuclei. IF5 cells do not express Lgals1, a gene expressed by reticulotegmental and lateral reticular nuclei, but Lgals1 is also not expressed by the pontine and external cuneate nuclei. Given that IF5 and the pontine nuclei both migrate to the rostral hindbrain from the caudal rhombic lip, we first suspected that the IF5 neurons might represent an outlying group of neurons developmentally and functionally related to the pontine nuclei. However, the Atoh1CreER birthdate study shows that pontine precursors are generated after E12.5, whereas IF5 precursors are generated as early as E9.5, indicating that the two populations have temporally different origins.
Relationship between Pr5 precerebellar neurons and IF5 neurons
Watson and Switzer (1978) showed that the principal and interpolar spinal trigeminal nuclei contained large neurons that project to the cerebellum in the rat, and the same holds true for the mouse (Fu et al., 2011). Because the cerebellar projecting neurons in Pr5 are located adjacent to IF5, we were interested in other features they may have in common. The majority of the neurons in Pr5 are somatosensory neurons that arise from the alar plate of r1 and r2 (Marin and Puelles, 1995), whereas the cerebellar projecting cells constitute a small subpopulation located mainly in the dorsal part of Pr5 (Fu et al., 2011). We have shown that the relatively large cerebellar projection neurons of Pr5 arise from the rhombic lip (i.e., arise from Wnt1 and Atoh1 lineages), and that they arise caudal to r4 (i.e., arise from Hoxa3 lineages). The precerebellar neurons in the dorsal part of Pr5 and those in IF5 both express the genes B2m, Ctsz, Depdc6, Dgat2, Rgs4, Slc17a7, and Zfp533, and they do not express Pou4f1, which characterizes climbing fiber precerebellar neurons.
Finally, both Pr5 and IF5 mainly project to the ipsilateral cerebellum, as do the external cuneate and lateral reticular nuclei, whereas the pontine nuclei project mainly to the contralateral cerebellum (Fu et al., 2011). All of these elements suggest that IF5 is more likely to be developmentally (and perhaps functionally) linked to the relatively large neurons in the dorsal part of Pr5 than to the pontine neurons. However, we cannot assume on this basis that IF5 is functionally related to the trigeminal system simply because of its position. To speculate on the functional relationships of IF5, we would require data on its afferents or electrophysiological mapping.
In summary, we have identified IF5 as a precerebellar nucleus that arises from the caudal rhombic lip. IF5 shares many developmental characteristics with the mossy fiber–projecting precerebellar nuclei, but we have no direct evidence that it gives rise to mossy fibers. IF5 lies adjacent to the motor neurons supplying the tensor tympani muscle, but the cerebellar-projecting neurons of IF5 do not express ChAT, and thus are a distinct population.
Acknowledgments
We thank Mario Capecchi of the University of Utah for generous support, and for providing the Hox Cre mouse lines.
Grant sponsor: Australian Research Council Thinking Systems Initiative; Grant number: TS0669860 (to Y.F, G.P.); Grant sponsor: National Health & Medical Research Council Australia; Grant number: fellowship grant 568605 (to G.P); Grant sponsor: National Institutes of Health; Grant number: 5R01MH068469-06 (to R.M.).
Abbreviations
- 2Cb
Lobule 2 of the cerebellar vermis
- 3Cb
Lobule 3 of the cerebellar vermis
- 4/5Cb
Lobules 4 and 5 of the cerebellar vermis
- 5N
Motor trigeminal nucleus
- 5PC
Parvicellular trigeminal nucleus
- 5TT
Motor trigeminal nucleus, tensor tympani part
- 6Cb
Lobule 6 of the cerebellar vermis
- 7Cb
Lobule 7 of the cerebellar vermis
- 7N
Facial nucleus
- 8Cb
Lobule 8 of the cerebellar vermis
- 9Cb
Lobule 9 of the cerebellar vermis
- AFD
Alexa Fluor dextran conjugate
- β-gal
β-Galactosidase
- CF
Climbing fiber
- ChAT
Choline acetyltransferase
- ChAT-EGFP
Choline acetyltransferase-enhanced green fluorescent protein
- CPe
Choroid plexus
- CRL
Caudal rhombic lip
- Crus1
Crus1 of the ansiform lobule
- Crus2
Crus2 of the ansiform lobule
- ECu
External cuneate nucleus
- FG
Fluoro-Gold
- Fl
Flocculus
- HRP
Horseradish peroxidase
- IC
Inferior colliculus
- IF5
Interfascicular trigeminal nucleus
- IO
Inferior olive
- LRt
Lateral reticular nucleus
- m5
Motor root trigeminal nerve
- mcp
Middle cerebellar peduncle
- MF
Mossy fiber
- mGFP
Membrane-tethered green fluorescent protein
- NeuN
Neuron-specific neuronal protein
- P
Postnatal
- PBS
Phosphate-buffered saline
- PFl
Paraflocculus
- PM
Paramedian lobule
- Pn
Pontine nuclei
- Pr5
Principal sensory trigeminal nucleus
- Pr5-d
Dorsal part of the principal sensory trigeminal nucleus
- Pr5-v
Ventral part of the principal sensory trigeminal nucleus
- PreCb
Precerebellar nuclei
- r
Rhombomere
- RtTg
Reticulotegmental nucleus of the pons
- Sim
Simple lobule
- SOl
Superior olive
- Tm
Tamoxifen
- TMB
3,3′,5,5′-Tetramethylbenzidine
- VLL
Ventral nucleus of the lateral lemniscus
Footnotes
Conflict of Interest Statement: The authors declare no actual or potential conflict of interest.
Role of Authors: All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: Y.F., C.W. Acquisition of data: Y.F., P.T., N.M., R.M., C.W. Analysis and interpretation of data: Y.F., P.T., N.M., R.M., G.P., C.W. Drafting of the manuscript: Y.F., C.W. Critical revision of the manuscript for important intellectual content: Y.F., P.T., N.M., R.M., G.P., C.W. Obtained funding: G.P. Administrative, technical, and material support: G.P. Study supervision: G.P., C.W.
Literature Cited
- Arenkiel BR, Gaufo GO, Capecchi MR. Hoxb1 neural crest preferentially form glia of the PNS. Dev Dyn. 2003;227:379–386. doi: 10.1002/dvdy.10323. [DOI] [PubMed] [Google Scholar]
- Arenkiel BR, Tvrdik P, Gaufo GO, Capecchi MR. Hoxb1 functions in both motoneurons and in tissues of the periphery to establish and maintain the proper neuronal circuitry. Gene Dev. 2004;18:1539–1552. doi: 10.1101/gad.1207204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creuzet S, Couly G, Le Douarin NM. Patterning the neural crest derivatives during development of the vertebrate head: insights from avian studies. J Anat. 2005;207:447–459. doi: 10.1111/j.1469-7580.2005.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czupryn A, Zhou YD, Chen X, McNay D, Anderson MP, Flier JS, Macklis JD. Transplanted hypothalamic neurons restore leptin signaling and ameliorate obesity in db/db mice. Science. 2011;334:1133–1137. doi: 10.1126/science.1209870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Farago AF, Awatramani RB, Dymecki SM. Assembly of the brainstem cochlear nuclear complex is revealed by intersectional and subtractive genetic fate maps. Neuron. 2006;50:205–218. doi: 10.1016/j.neuron.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Favier B, Dollé P. Developmental functions of mammalian Hox genes. Mol Hum Reprod. 1997;3:115–131. doi: 10.1093/molehr/3.2.115. [DOI] [PubMed] [Google Scholar]
- Franklin KBJ, Paxinos G. The mouse brain in stereotaxic coordinates. San Diego: Elsevier Academic Press; 2008. [Google Scholar]
- Friauf E, Baker R. An intracellular HRP-study of cat tensor tympani motoneurons. Exp Brain Res. 1985;57:499–511. doi: 10.1007/BF00237837. [DOI] [PubMed] [Google Scholar]
- Fu Y, Tvrdik P, Makki N, Palombi O, Machold R, Paxinos G, Watson C. The precerebellar linear nucleus in the mouse defined by connections, immunohistochemistry, and gene expression. Brain Res. 2009;1271:49–59. doi: 10.1016/j.brainres.2009.02.068. [DOI] [PubMed] [Google Scholar]
- Fu Y, Tvrdik P, Makki N, Paxinos G, Watson C. Precerebellar cell groups in the hindbrain of the mouse defined by retrograde tracing and correlated with cumulative Wnt1-cre genetic labeling. Cerebellum. 2011;10:570–584. doi: 10.1007/s12311-011-0266-1. [DOI] [PubMed] [Google Scholar]
- Gannon PJ, Eden AR. A specialized innervation of the tensor tympani muscle in Macaca fascicularis. Brain Res. 1987;404:257–262. doi: 10.1016/0006-8993(87)91376-x. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa T, Ajiki K, Matsuura J, Misawa H. Localization of two cholinergic markers, choline acetyltransferase and vesicular acetylcholine transporter in the central nervous system of the rat: in situ hybridization histochemistry and immunohistochemistry. J Chem Neuroanat. 1997;13:23–39. doi: 10.1016/s0891-0618(97)00021-5. [DOI] [PubMed] [Google Scholar]
- Keller JT, Saunders MC, Ongkiko CM, Johnson J, Frank E, Van Loveren H, Tew JM., Jr Identification of motoneurons innervating the tensor tympani and tensor veli palatini muscles in the cat. Brain Res. 1983;270:209–215. doi: 10.1016/0006-8993(83)90594-2. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Hori K, Wyckoff A, Dickel LK, Koundakjian EJ, Goodrich LV, Johnson JE. Spatiotemporal fate map of neurogenin1 (Neurog1) lineages in the mouse central nervous system. J Comp Neurol. 2011;519:1355–1370. doi: 10.1002/cne.22574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsberg RL, Awatramani RB, Hunter NL, Farago AF, DiPietrantonio HJ, Rodriguez CI, Dymecki SM. Hindbrain rhombic lip is comprised of discrete progenitor cell populations allocated by Pax6. Neuron. 2005;48:933–947. doi: 10.1016/j.neuron.2005.11.031. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Macatee TL, Hammond BP, Arenkiel BR, Francis L, Frank DU, Moon AM. Ablation of specific expression domains reveals discrete functions of ectoderm- and endoderm-derived FGF8 during cardiovascular and pharyngeal Development. 2003;130:6361–6374. doi: 10.1242/dev.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Marin F, Puelles L. Morphological fate of rhombomeres in quail/chick chimeras: a segmental analysis of hindbrain nuclei. Eur J Neurosci. 1995;7:1714–1738. doi: 10.1111/j.1460-9568.1995.tb00693.x. [DOI] [PubMed] [Google Scholar]
- McHanwell S, Watson C. Localization of motoneurons in the spinal cord. In: Watson C, Paxinos G, Kayalioglu G, editors. Spinal cord: A Christopher and Dana Reeve Foundation text and atlas. San Diego: Elsevier Academic Press; 2009. pp. 94–114. [Google Scholar]
- Mizuno N, Nomura S, Konishi A, Uemura-Sumi M, Takahashi O, Yasui Y, Takada M, Matsushima R. Localization of motoneurons innervating the tensor tympani muscles: a horseradish peroxidase study in the guinea pig and cat. Neurosci Lett. 1982;31:205–208. doi: 10.1016/0304-3940(82)90020-9. [DOI] [PubMed] [Google Scholar]
- Mukerji S, Brown MC, Lee DJ. A morphologic study of Fluorogold labeled tensor tympani motoneurons in mice. Brain Res. 2009;1278:59–65. doi: 10.1016/j.brainres.2009.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RJ, Buck CR, Smith AM. NeuN, a neuronal specific nuclear protein in vertebrates. Development. 1992;116:201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- Packard A, Giel-Moloney M, Leiter A, Schwob JE. Progenitor cell capacity of NeuroD1-expressing globose basal cells in the mouse olfactory epithelium. J Comp Neurol. 2011;519:3580–3596. doi: 10.1002/cne.22726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The rat brain in stereotaxic coordinates. San Diego: Elsevier Academic Press; 2007. [Google Scholar]
- Ray RS, Dymecki SM. Rautenlippe redux—toward a unified view of the precerebellar rhombic lip. Curr Opin Cell Biol. 2009;21:741–747. doi: 10.1016/j.ceb.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Maunoury S, Seitanidou T, Charnay P, Lumsden A. Segmental and neuronal architecture of the hindbrain of Krox-20 mouse mutants. Development. 1997;124:1215–1226. doi: 10.1242/dev.124.6.1215. [DOI] [PubMed] [Google Scholar]
- Shaw MD, Baker R. The locations of stapedius and tensor tympani motoneurons in the cat. J Comp Neurol. 1983;216:10–19. doi: 10.1002/cne.902160103. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Strutz J, Munker G, Zollner C. The motor innervation of the tympanic muscles in the guinea pig. Arch Otorhinolaryngol. 1988;245:108–111. doi: 10.1007/BF00481446. [DOI] [PubMed] [Google Scholar]
- Takahashi O, Mizuno N, Mitani A, Takeuchi Y, Matsushima R. Identification of motoneurons innervating the tensor tympani muscle in the rabbit: a retrograde horseradish peroxidase study. Neurosci Lett. 1984;49:19–23. doi: 10.1016/0304-3940(84)90129-0. [DOI] [PubMed] [Google Scholar]
- van den Berge H, van der Wal JC. The innervation of the middle ear muscles of the rat. J Anat. 1990;170:99–109. [PMC free article] [PubMed] [Google Scholar]
- Voiculescu O, Charnay P, Schneider-Maunoury S. Expression pattern of a Krox-20/Cre knock-in allele in the developing hindbrain, bones, and peripheral nervous system. Genesis. 2000;26:123–126. doi: 10.1002/(sici)1526-968x(200002)26:2<123::aid-gene7>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Watson C, Paxinos G. Chemoarchitectonic atlas of the mouse brain. San Diego: Elsevier Academic Press; 2010. [Google Scholar]
- Watson C, Switzer RC. Trigeminal projections to cerebellar tactile areas in the rat–origin mainly from n. interpolaris and n. principalis. Neurosci Lett. 1978;10:77–82. doi: 10.1016/0304-3940(78)90015-0. [DOI] [PubMed] [Google Scholar]
- Wolf HK, Buslei R, Schmidt-Kastner R, Schmidt-Kastner PK, Pietsch T, Wiestler OD, Blumcke I. NeuN: a useful neuronal marker for diagnostic histopathology. J Histochem Cytochem. 1996;44:1167–1171. doi: 10.1177/44.10.8813082. [DOI] [PubMed] [Google Scholar]


