Abstract
Proteasomes are self-compartmentalized energy-dependent proteolytic machines found in Archaea, Actinobacteria species of bacteria and eukaryotes. Proteasomes consist of two separate protein complexes, the core particle that hydrolyzes peptide bonds and an AAA+ ATPase domain responsible for the binding, unfolding and translocation of protein substrates into the core particle for degradation. Similarly to eukaryotes, proteasomes play a central role in protein degradation and can be essential in Archaea. Core particles associate with and utilize a variety of ATPase complexes to carry out protein degradation in Archaea. In actinobacterial species, such as Mycobacterium tuberculosis, proteasome-mediated degradation is associated with pathogenesis and does not appear to be essential. Interestingly, both actinobacterial species and Archaea use small proteins to covalently modify proteins, prokaryotic ubiquitin-like proteins (Pup) in Actinobacteria and ubiquitin-like small archaeal modifier proteins (SAMP) in Archaea. These modifications may play a role in proteasome targeting similar to the ubiquitin-proteasome system in eukaryotes.
Keywords: Prokaryotic ubiquitin-like protein, Pupylation, Small archaeal modifier proteins, Sampylation
Introduction
Proteasomes are found in Archaea, actinobacterial families of bacteria and eukaryotes and function in protein quality control through the degradation of misfolded or otherwise damaged proteins [Volker and Lupas, 2002]. Proteasomes can also play a regulatory role through the timed and targeted degradation of proteins responsible for DNA replication, cell division, transcription and a variety of other central cellular processes [Bader and Steller, 2009; Bengtson and Joazeiro, 2010; Kirkland et al., 2007; Kirkland and Maupin-Furlow, 2009; Mehnert et al., 2010; Poulsen et al., 2010]. All ATP-dependent proteases with solved structures exist in an autoinhibited state [Sauer and Baker, 2011]. The overall three-dimensional structure of these complexes results in the sequestration of the active site residues responsible for protein degradation within a small nanochamber. This feature results in simultaneously increasing the concentration of the active site residues within the degradation chamber and protecting the proteins in the cytosol from random proteolysis or degradation by the protease. Prokaryotic 20S proteasomes are no exception to this rule [Löwe et al., 1995]. The overall architectures of 20S proteasomes are conserved from the different domains of life and consist of a stack of heptameric rings of α-type and β-type subunits in the arrangement α7β7β7α7. The active site residues are found in the β-rings. The α-rings are needed for β subunit assembly and serve as a gate into the inner chambers [Humbard et al., 2009; Religa et al., 2010; Stadtmueller and Hill, 2011], only to be opened while interacting with a regulatory AAA+ ATPase complex.
The AAA+ ATPase portion of the proteasome is responsible for the binding, delivery, unfolding and translocation of substrates into the core particle for degradation. Prokaryotic organisms use a variety of hexameric ATPase rings for this function. In Archaea, at least three different ATPases function in concert with the core particle for protein degradation, proteasome-activating nucleotidase (Pan), AMA (AAA protein of methanogenic archaea and Archaeoglobus) and Cdc48 (or VAT, a p97 homologue) [Barthelme and Sauer, 2012, 2013; Forouzan et al., 2012; Wilson et al., 2000; Zwickl et al., 1999]. While core particle and Cdc48 homologues are universally conserved in Archaea, the Pan and AMA proteins are not. Many Pan, AMA and Cdc48 homologues have a conserved C-terminal motif (HbYX) that functions in proteasome activation (where Hb is a hydrophobic residue and X is any C-terminal amino acid residue) [Stadtmueller and Hill, 2011; Yu et al., 2010]. Proteasomal core particles of Actinobacteria also utilize hexameric ATPases for degradation of substrate proteins. In Mycobacterium, mycobacterial proteasome ATPase (Mpa) is required for the degradation of certain proteasomal substrates as well as pathogenesis and protection from nitric oxide stress [Darwin et al., 2005; Pearce et al., 2006]. AAA+ ATPase forming a ring-shaped complex (Arc) is another ATPase found in Rhodococcus species of Actinobacteria that is closely related to Mpa (82% identity) and likely involved in the activation of core particles [Wolf et al., 1998], based on analogy to its close relatives.
Even though the overall structure, enzymology and biochemistry of prokaryotic and eukaryotic proteasomes are closely related, the mechanism of how proteins are targeted for degradation by proteasomes was presumed to be quite different between these two groups. This difference was due to the apparent lack of a prokaryotic equivalent to ubiquitin, a small protein that can covalently modify the lysine residues of proteins and target them for degradation by proteasomes in eukaryotic cells. The first prokaryotic ubiquitin-like protein was discovered in Mycobacterium tuberculosis and was termed Pup [Pearce et al., 2008]. Pup is covalently attached to hundreds of proteins [Poulsen et al., 2010; Watrous et al., 2010] with dozens of these modified proteins confirmed to be degraded by the mycobacterial proteasome [Burns et al., 2009, 2010b; Striebel et al., 2010]. In the case of M. tuberculosis, protein modification by Pup (pupylation) and proteasome activity are connected to the pathogenesis of the organism [Festa et al., 2010; Pearce et al., 2006]. More recently, protein modifiers related to ubiquitin (termed ubiquitin-like small archaeal modifier proteins or SAMPs), along with structural homologues of deubiquitinating enzymes and E1-activating enzymes, were identified in the halophilic archaeon Haloferax volcanii [Hepowit et al., 2012; Humbard et al., 2010a; Miranda et al., 2011]. SAMP homologues are widespread in Archaea with a single archaeal genome typically encoding multiple homlogs that are predicted to each form covalent attachments, with not all of these protein modifications directly related to proteolysis [Humbard et al., 2010a; Makarova and Koonin, 2010]. In this review, we will describe recent advances in the understanding of the structure and function of prokaryotic proteasome complexes. Also, we will discuss the roles ubiquitin-like proteins (Pup and SAMP) may play in protein targeting and proteasome-mediated degradation in prokaryotic cells.
Proteasome Structure, Subunit Composition and Activity
Proteasomes consist of two separate protein complexes, the 20S core particle that catalyzes the hydrolysis of peptide bonds and an ATPase component. These two complexes come together to form an energy-dependent protease. The makeup and composition of the ATPase component can vary from organism to organism, but the 20S core particle is highly conserved across Bacteria, Archaea and Eukarya domains [Dahlmann et al., 1989]. The overall architecture of the core particle is a barrel-shaped stack of four heptameric rings with an overall dimension of 15 nm in length and a diameter of about 12 nm [Löwe et al., 1995]. The rings stack to form a central channel and are made up of two different types of structurally related subunits termed α- and β-type [Baumeister et al., 1998]. The outer rings of the core particle complex are composed exclusively of α-type subunits and the inner rings are of β-type subunits, resulting in an overall organization of α7β7β7α7 [Grziwa et al., 1991; Lupas et al., 1994] (fig. 1a). The active sites of the proteolytic domain formed by the β-subunits are located on the interior of the 20S cylindrical structure. Generally, in prokaryotic proteasomes, the complexity of α- and β-type subunits is low, with usually only one to two members of each subunit group present per organism, with some exceptions. By contrast, eukaryotic proteasomes such as Saccharomyces cerevisiae have 7 related but distinct members of each group with 7 members of the α-type and 7 members of the β-type in each 20S proteasome [Groll et al., 1997; Löwe et al., 1995].
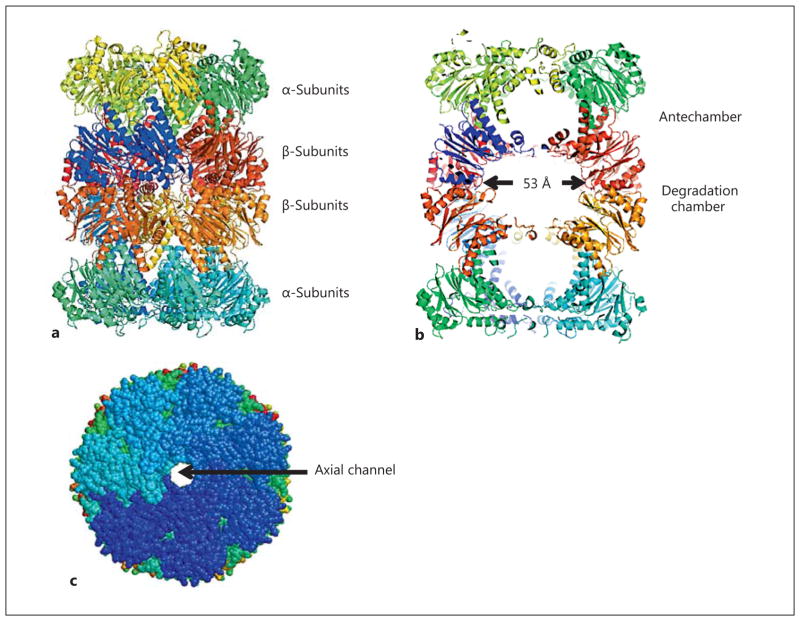
Fig. 1.
20S core particle: three views and dimensions. a 20S core particle from T. acidophilium (PDB 1PMA) consisting of a stack of heptameric rings of α- and β-type subunits arranged as α7β7β7α7. b Cross-sectional view of T. acidophilium 20S core particle displaying antechambers and degradation chambers within the structure. c Axial view of 20S core particle in ‘closed’ conformation with an obstructed axial channel leading into the antechamber (PDB 3C92).
Despite the variance in the subunit complexity between 20S proteasomes, the three-dimensional structure of core particles is quite conserved. The stacking of the four heptameric rings results in the formation of three small interior chambers, two antechambers of 60 nm 3 and a central chamber of 84 nm3, with the innermost chamber containing the active site residues (fig. 1b) [Groll et al., 1997; Hegerl et al., 1991; Löwe et al., 1995]. These chambers are formed at the interior interfaces of the heptameric rings, with the two antechambers at the interface of the α- and β-subunit rings and the central chamber at the interface of the two β-subunit rings. The 6–14 (number depends on the organism) active site residues that hydrolyze the peptide bonds are found lining the innermost chamber. A central channel runs through the core particle connecting each of the three chambers and connects pores of up to 2.2 nm in diameter that are located on each of the cylinder. The functional role of the antechambers is not entirely understood, especially since not all energy-dependent proteases have these antechambers [Sauer and Baker, 2011]. The antechambers are speculated to aid in holding proteins in an unfolded state prior to degradation [Ruschak et al., 2010; Sharon et al., 2006] or be a consequence of the α-type subunit gating function that sequesters the proteolytic active site residues from nonsubstrate proteins [Baumeister et al., 1998; Bochtler et al., 1999].
The 20S core particles exist in an autoinhibited state. Structural elements in the N-terminal domains of α-type subunits form an axial ‘gate’ that blocks the central channel and, thus, prevents substrates from accessing the central chamber and the active site residues within this structure [Stadtmueller and Hill, 2011] ( fig. 1c). This axial gate can be visualized in the crystal structure of the 20S core particle of yeast (S. cerevisiae) where the N-terminal tails of α2, α3, and α4 subunits form the gate [Groll et al., 1997]. In the case of the yeast proteasome, the gating can be abolished by the addition of low levels of anionic detergent (sodium dodecyl sulfate), introduction of a D to N substitution in the conserved YDR motif of the α3 subunit or a mutation that prevents the N-terminal acetylation of certain α-type subunits in the proteasome [Forster and Hill, 2003; Kimura et al., 2000]. In the original crystal structure of 20S core particles of the archaeon Thermo-plasma acidophilum [Löwe et al., 1995], the N-terminal residues of the α-subunit were disordered implying that this domain is unstructured or highly variable. Biochemical data further demonstrated that small peptides could gain access to the interior chamber of archaeal proteasomes in the absence of an ATPase activator and that this activity did not require sodium dodecyl sulfate or heat treatment [Akopian et al., 1997; Maupin-Furlow et al., 1998]. Aside from these early findings that suggested prokaryotic proteasomes are not gated, the residues necessary for gate formation in the eukaryotic proteasomes, which included the conserved N-terminal helix of α-subunits with a YDR motif, are strictly conserved in Archaea and partially conserved in Actinobacteria [Lin et al., 2006]. Deletion of this N-terminal helix results in a substantial increase in the rate and size of peptides hydrolyzed by the core particles of proteasomes from diverse prokaryotes including the archaea T. acidophilium and Hfx. volcanii, as well as M. tuberculosis [Benaroudj et al., 2003; Humbard et al., 2009; Lin et al., 2006]. Additionally, visualization of core particles from Mycobacterium and archaeal species by transmission electron microscopy and cryoelectron microscopy revealed a closed-gate conformation [Hu et al., 2006; Li et al., 2010; Lin et al., 2006; Rabl et al., 2008; Yu et al., 2010]. Measurements of the mobility of the archaeal core particle using TROSY NMR further suggested that the N-terminal tails of α-subunits function as gates and that these gates switch between closed and open conformations that are correlated with rates of peptide hydrolysis [Religa et al., 2010]. The N-terminal domains of the α-subunits from the actinobacterial structure can adopt three different conformations that establish the gating required for the basal inhibition [Li et al., 2010]. Together, these data suggest a gating mechanism is present in eukaryotic and prokaryotic proteasomes that limits substrate access to the core particle active sites and, at least in Archaea, is dynamic [Religa et al., 2010].
As mentioned above, the complexity of the α- and β-type subunits is considerably less in prokaryotic than eukaryotic proteasomes. Many prokaryotic 20S core particles are composed of only one α- and one β-type subunit in α7β7β7α7 configuration such as those of the archaea T. acidophilum, Archaeoglobus fuldigus, Methanocaldococcus jannaschii (formerly Methanococcus jannaschii), and Methanosarcina thermophila [Maupin-Furlow et al., 2000] as well as the Actinobacteria M. tuberculosis, Streptomyces coelicolor, and many Rhodococcus species [De Mot et al., 1999; Lin et al., 2006]. Several examples also exist of prokaryotes that encode two or more α- and/or two or more β-type subunits associated as one or more different proteasomal core particles in α7β7β7α7 configuration. The actinobacterium Rhodococcus erythropolis N186/21 synthesizes two α-type and two β-type subunits as a single core particle [Tamura et al., 1995; Zühl et al., 1997]. Many archaeal organisms are predicted to encode 3 or more different proteasome proteins (e.g. most haloarchaea, Sulfolobus and Pyrococcus spp.) [Madding et al., 2007; Maupin-Furlow et al., 2000]. Hfx. volcanii synthesizes a single β-type subunit and two α-type subunits (α1 and α2) [Kaczowka and Maupin-Furlow, 2003; Wilson et al., 1999] that form 20S core particles of α1β, α2β and α1β α2 subunit composition that are functional in the hydrolysis of short peptides [Kaczowka and Maupin-Furlow, 2003; Karadzic et al., 2012; Wilson et al., 1999]. In Hfx. volcanii, α1 and α2 are not likely to be functionally redundant due to their differential regulation in the cell with respect to expression. The α2 subunit is upregulated in stationary phase, while the α1 subunit is expressed throughout growth [Kaczowka and Maupin-Furlow, 2003]. Despite this difference in expression patterns, the two α-subunits can complement each other for some functions since neither is essential but rather the double knockout is lethal [Zhou et al., 2008]. However, the deletion of psmA (α1) renders the cells more sensitive to osmotic stress, and this phenotype is not rescued by the overexpression of psmC (α2) [Zhou et al., 2008]. The finding that psmC (α2) can rescue the lethality of a psmA psmC (α1 α2) double mutant but not complement the phenotype of the psmA (α1) single mutant during osmotic stress provides evidence that the two α-subunits are not redundant for all functions of the cell. Pyrococcus furiosus also exemplifies the potential for multiple proteasomal subtypes based on its genome sequence, which predicts two different β-subunits (β1 and β2) and one α-subunit. Transcriptional profiling reveals the transcript for β1 increases immediately following heat shock [Madding et al., 2007]. Consistent with this finding, proteasome core particles assembled in vitro with recombinantly expressed subunits at heat shock temperatures have a higher proportion of β1 and are more thermally stable than proteasomes formed at standard growth temperatures [Madding et al., 2007]. The combination of β1- and α-subunits alone (in the absence of β2) fails to yield properly assembled or functional core particles, even though β1 has a conserved active site (Thr1) residue [Madding et al., 2007]. By contrast, core particles composed of αβ2 and αβ1β2 hydrolyze peptides [Madding et al., 2007]. These few examples, where multiple subunits of α- or β-type subfamilies are in the same organism, support the notion of specialized subpopulations of core particles with redundant as well as unique physiological roles which appear advantageous in certain prokaryotes.
Several steps occur during the assembly of α- and β-type subunits into active proteasomal core particles. Although archaea are predicted to encode homologues of the PAC2 family of chaperones required for proper proteasomal core particle assembly in eukaryotes [Le Tallec et al., 2007], it is currently unclear whether any factors are required for core particle assembly in prokaryotes. In the Archaea, α-subunits spontaneously assemble to form heptameric rings that act as a scaffold for the formation of the heptameric rings of β-subunits [Groll et al., 2003]. In the Actinobacteria, α- and β-subunits form heterodimers (αβ) that oligomerize to form the half proteasome [Kwon et al., 2004; Li et al., 2010]. These intermediate ‘half proteasomes’ or 11S proteasomes of α7β7 configuration are inactive due to the presence of propeptides on the β-subunits. Many β-type subunits belong to the amino-terminal hydrolase family of proteases (Ntn family) [Brannigan et al., 1995; Löwe et al., 1995]. Active β-subunits are synthesized with an N-terminal propeptide that upon autocatalytic removal exposes an N-terminal threonine that functions as a proteolytic-active site as the proteasome halves come together. Initial evidence that proteasomal core particles were Ntn-hydrolases came from the close proximity of the peptide aldehyde inhibitor, acetyl-leu-leu-norleucinal, to the Thr1 of the β-subunits exposed within the innermost chamber of core particle crystal structures derived from T. acidophilum [Löwe et al., 1995]. Guided by this structure, a Thr1Ala substitution of the β-subunit was shown to abolish the peptidase activity of core particles [Seemuller et al., 1995].
Due to the simplified composition of subunits that form a prokaryotic proteasome, activities with respect to peptide hydrolysis with 7-amino-4-methylcoumarin-linked substrates are typically less diverse than eukaryotic proteasomes. Most prokaryotic core particles catalyze chymotrypsin-like peptidase activity [Dahlmann et al., 1992; Nagy et al., 1998; Tamura et al., 1995; Wilson et al., 1999]. A few exceptional proteasomes have both chymotrypsin- and caspase-like activity, specifically, core particles from M. thermophila and M. jannaschii [Maupin-Furlow et al., 1998; Wilson et al., 2000]. Some archaeal proteasomes also exhibit low levels of trypsin-like activity [Akopian et al., 1997; Maupin-Furlow et al., 1998; Wilson et al., 2000]. The lack of diversity of β-subunits in the prokaryotic core particle has another implication. The active sites in the central chamber are more numerous than is typical for eukaryotic proteasomes. As discussed earlier, eukaryotic proteasomes have 7 different β-subunits, but only 3 of those are active [Groll et al., 1997], resulting in 6 active sites in the central chamber (3 per heptameric ring). However, prokaryotes such as T. acidophilum, M. tuberculosis, Hfx. volcanii and methanogenic archaea have only a single type of β-subunit that assembles into the core particle to form a total of 14 identical active sites estimated at a concentration of 50 mM based on dimensions of the central chamber [Löwe et al., 1995; Maupin-Furlow et al., 1998; Wilson et al., 2000]. Proteasome-mediated degradation of proteins into short peptides is typically processive [Akopian et al., 1997], which means once proteolysis is initiated the protein is completely digested into peptides before the next protein substrate is hydrolyzed. Multiple factors likely influence product (peptide) size including substrate sequence, structural elements within substrates, direction of translocation and influences by the ATPase associated with the core particle. Initial analysis of non-ATP dependent substrates using core particles from T. acidophilum revealed products between 7 and 9 amino acids in length [Wenzel et al., 1994]. Another study with saturating substrate demonstrated a wider distribution of product lengths between 3 and 30 amino acids with no apparent preference of cleavage site [Kisselev et al., 1998]. Surprisingly, mammalian proteasomes, which contain only 6 active sites per core particle, generate peptides with a similar size distribution as do archaeal proteasomes, which have 14 identical sites [Kisselev et al., 1999]. Furthermore, inactivation of two of the six active sites of the mammalian proteasomes influences the pattern but does not affect the size distribution of the peptide products [Kisselev et al., 1999]. The direction of translocation was recently found to have an effect on the length of peptide products for some substrates [Berko et al., 2012]. The rates of degradation of certain protein substrates is also slower when translocated in a C-to-N direction by both eukaryotic and archaeal proteasomes [Berko et al., 2012]. This slower degradation rate is correlated with an increase in product size, and this altered product size is believed to be caused by a partial refolding, at least reestablishment of secondary structure, of certain substrates that may prevent interaction with the hydrophobic patches in the antechamber [Ruschak et al., 2010]. The localization of active sites within the central chamber and the processive degradation of protein substrates that are translocated into the chamber make proteasome core particles efficient peptidases. Selection of substrates and the ATP-dependent unfolding and translocation are the rate-limiting step for degradation by proteasome complexes.
Prokaryotic Proteasome Activators
Proteasomes typically require interaction with AAA+ (ATPase associated with various cellular activities) ATPase complexes to activate the degradation of folded or aggregated proteins [for a review of AAA+ ATPase classification, see Snider et al., 2008]. As discussed in the previous section, structural conformations of the N-terminal domains of α-subunit rings in 20S core particles form an ‘axial gate’ and restrict substrate entry into the degradation chamber except for peptides and small unfolded proteins. In a sense, AAA+ ATPase complexes function as reverse chaperones by binding, unfolding, and translocating substrates through α-subunit rings into the interior chambers of the 20S core particles [Medalia et al., 2009; Smith et al., 2007, 2005]. In eukaryotes, the 19S cap is the major ATPase associated with core particles and is made up of several different proteins. The 19S cap can be divided into two domains, the base and the lid. The base of the complex consists of 9 separate protein products, six of which are ATPases (Rpt1–6) that form a hexameric ring [Tomko et al., 2010]. This ring of ATPases is the portion of the complex that interacts with the α-subunit rings of the core particle [Tomko and Hochstrasser, 2011]. In bacteria and archaea, several different ATPase complexes are proposed to function as regulatory particles for 20S proteasomes. In archaea, proteins of the AAA ATPase family including Pan, Cdc48 and AMA can activate protein degradation by the proteasome [Forouzan et al., 2012]. In the Actinobacteria, the divergent group of AAA+ ATPases called Arc is thought to be responsible for proteasome-mediated protein degradation in Rhodococcus spp. [Wolf et al., 1998], and the related Mpa can interact with the Mycobacterium proteasome for energy-dependent proteolysis [Striebel et al., 2010; Wang et al. 2009] (fig. 2).
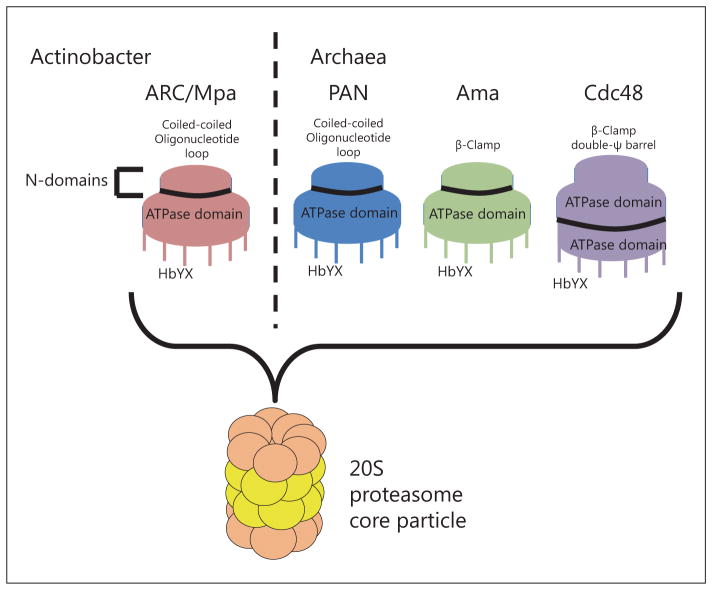
Fig. 2.
Regulatory ATPases. Hexameric ATPase complexes activate protease activity of 20S proteasomes through interaction with the outer α-rings of the proteasome and the HbYX C-terminal motif on the ATPase. Variation in the N-domains between the different ATPases may influence substrate binding of those complexes.
Completion of the first archaeal (M. jannaschii) genome sequence revealed a prokaryotic gene homologue closely related to the six Rpt subunits in the eukaryotic 19S cap [Zwickl et al., 1999]. This archaeal AAA ATPase homologue (Pan) was 41–45% similar to the Rpt subunits, higher in amino acid similarity than between the individual Rpt proteins. Pan could be expressed and purified from recombinant Escherichia coli, and this form of Pan was shown to hydrolyze ATP and to increase the rates of degradation of several proteins in an ATP-dependent manner by 20S proteasomes of M. jannaschii and T. acidophilium [Smith et al., 2005; Wilson et al., 2000]. Pan was also demonstrated to associate in electron micrographs with proteasomes of M. jannaschii [Wilson et al., 2000]. Pan was purified as a 550-kDa complex corresponding to the molecular weight of a dodecamer [Wilson et al., 2000]. Other closely related ATPases are known to adopt a do-decameric structure, but the likely complex that interacts with proteasomes in vivo is a hexamer [Reuter et al., 2004; Smith et al., 2005; Yu et al., 2010]. Electron micrographs of hexameric Pan of M. jannaschii revealed that these complexes consist of what appeared to be a stack of two rings, a proximal and a distal ring. The proximal ring was 120 Å in diameter and 80 Å high, approximately the diameter of the core particle, and the distal ring was 75 Å in diameter and 30 Å high [Smith et al., 2005]. Similar micrographs of the Mpa ATPase from M. tuberculosis were obtained, showing a hexameric structure in complex with a proteasome core particle [Wang et al., 2009]. The outer ring consists of two domains, a coiled-coil and oligonucleotide binding fold domain. The coiled-coil domain is made of the N-terminal extensions of Pan subunits required for substrate binding [Djuranovic et al., 2009]. The coiled-coil domains of the archaeal and actinobacterial ATPases appear to be solvent exposed and likely make direct contact with substrates [Djuranovic et al., 2009; Wang et al., 2010; Zhang et al., 2009a]. The N-terminal coiled-coil domains are arrayed around a central channel in the ATPase ring formed by an oligonucleotide-binding fold, common to other proteins [Murzin, 1993], which holds the ring together and may be responsible for the initial unfolding of substrates on the surface of the ring [Benaroudj et al., 2003; Navon and Goldberg, 2001; Sauer and Baker, 2011; Zhang et al., 2009a].
In a hexameric ring, Pan subunits can exist in three different states, high and low nucleotide affinity states and an ADP-bound state [Smith and Goldberg, 2011; Bar-Nun and Glickman, 2012]. The current model, based on ATP hydrolysis by the ClpX ATPase from E. coli [Sauer and Baker, 2011] and Pan of M. jannaschii [Smith et al., 2011], is that opposite subunits in the ATPase ring adopt the same conformation. ATP binding is thought to cause a conformational change in the neighboring subunit; upon hydrolysis, ADP is released from one of the neighboring subunits (counterclockwise to the ATP-bound subunit) and the other neighboring partner binds ATP. This process proceeds around the ring. The structural perturbations that occur as a result of ATP binding and hydrolysis are thought to provide the mechanical energy required for substrate unfolding on the surface of the ring [Navon and Goldberg, 2001; Zhang et al., 2009b]. Similarly to other AAA+ ATPases, a highly conserved hydrophobic loop (Ar-φ loop) is found at the narrowed portion of the substrate channel of Pan [Zhang et al., 2009a]. This loop is absolutely required for the unfolding action of the ATPase ring and is thought to hold onto hydrophobic residues of substrate proteins in the channel and pull proteins into and ultimately through the channel through successive rounds of ATP hydrolysis based on analysis of ClpX [Sauer and Baker, 2011]. As the substrate protein is unfolded and passed through this central channel, the protein is translocated through the axial channel of the 20S core particle for degradation.
Degradation of unfolded substrates presented to 20S core particles by ATPases, such as Pan, requires the opening of the axial gates that keep proteasomes in an autoinhibited state. Several recent studies have demonstrated that a conserved C-terminal motif, hydrophobic-tyrosine-X (HbYX), is required for ATP-dependent proteasome activation [Gillette et al., 2008; Smith et al., 2007]. This motif is present in some archaeal Pan proteins and three of the six Rpt subunits from eukaryotic 19S cap [Yu et al., 2010]. Peptides with the C-terminal sequence of Pan bind to the intersubunit pocket between adjacent α-subunits in the outer ring of the core particle. This binding induces a rotation in the α-subunits and stabilizes the ‘open gate’ conformation in the proteasome [Rabl et al., 2008; Yu et al., 2010]. The binding of ATP to subunits of the ATPase complex is thought to cause a conformational change in the C-terminal domains that may facilitate the binding of this tail into the intersubunit space in the α-ring [Smith et al., 2005]. While the HbYX motif is conserved, it is not universally so. Several Pan proteins do not contain this exact motif. Hfx. volcanii synthesizes two Pan proteins (PanA and PanB), both suspected of interacting with the proteasome to activate proteolysis in vivo, but only PanA has a C-terminal tail (-AFA) similar to the HbYX motif [Reuter et al., 2004]. C-terminal fusions to PanA fail to complement the osmolarity sensitivity of a panA knockout in Hfx. volcanii while expression of wild-type panA did complement [Zhou et al., 2008], indicating that the C-terminus was important for in vivo activity of PanA. PanB, however, does not contain the conserved HbYX motif (instead it has a C-terminal tail of -YQY). PanB can associate with PanA [Humbard et al., 2010b]. This structure of mixed subunits, some with and some without the conserved HbYX motif, is analogous to the eukaryotic 19S cap where every other subunit in the ring has the HbYX motif [Tomko et al., 2010]. While the subunit arrangement of the mixed PanA/PanB complex from Hfx. volcanii is not currently known, it is possible that an alternating structure could still facilitate gate opening in the core particle in a similar fashion to the eukaryotic structure.
Much is known about the structures of ATPase complexes and the biochemical evidence that ATPase complexes interact with and therefore activate 20S core particles is convincing; however, showing native in vivo interaction between a 20S core particle and an intact ATPase complex in prokaryotes has been difficult [Forster and Hill, 2003; Ruepp et al., 2001; Smith et al., 2005]. The symmetry mismatch between the heptameric proteasome and the hexameric ATPases or coexpression of highly variable ATPases are likely culprits for the difficulties. At present, no crystal structure of the 20S core particle and ATPase in complex or of a Pan hexamer complex alone is available. Recently, structures of the 26S proteasomes from the yeast Schizosaccharomyces pombe at 9.1 Å [Bohn et al., 2010] and S. cerevisiae at 9.1 Å [Beck et al., 2012] were solved by cryoelectron microscopy and single particle analysis. Coupled with chemical crosslinking, mass spectrometry and molecular dynamics-based flexible fitting, many protein-protein contacts were elucidated, giving to date the best view of an ATPase structure and core particle in complex [Bohn et al., 2010]. Even though no crystal structures of the complete complexes from prokaryotes are available, several structures of the domains of Pan and Arc/Mpa have been solved that give specific insight into the various roles these complexes can play in concert with core particles and the domain organization within the complexes [Djuranovic et al., 2009; Wang et al., 2009, 2010; Zhang et al., 2009a]. Two separate structures of domains from the Pan complex of M. jannaschii, including the ATPase ring and the coiled-coil N-domain ring were obtained after limited proteolysis removed a variable loop that connects these two domains [Zhang et al., 2009a]. When modeled into a likely association based on protein sequence of the subunits, the structure appears as two stacked rings. This structure is in good agreement with the two-ring structure seen previously in cryoelectron microscopy studies [Smith et al., 2005]. The presence of the oligonucleotide-binding fold in the N-domain portion of the structure revealed that the distal ring of the Pan complex forms a constriction that is too narrow to allow folded proteins to pass through the ATPase ring [Zhang et al., 2009a]. This structural model provides evidence that the Ar-φ loop of the ATPase domain interacts with unfolded regions of substrates. Additional crystallization and analysis of the chaperone activity of the N-domains of both Pan (of Archeoglobus fulgidus) and Arc (of R. erythropolis) reveal a nearly identical ring structure of the oligonucleotide-binding domain despite highly divergent protein sequences [Djuranovic et al., 2009]. The energy-dependent chaperone activity was restricted to the N-domains with the coiled-coil structures and ATPase active sites. These ATPase subdomain structures and function studies support the hypothesis that protein unfolding occurs as a result of movements in the N-domain stimulated by ATP hydrolysis mediated by the ATPase itself. Also, the dimensions of the tunnel through the oligonucleotide-binding domain support the previous notion that substrates are unfolded prior to translocation through the channel [Djuranovic et al., 2009].
Several species of Archaea encode one or more Pan proteins, but some archaea have no homologue to Rpt or Pan including T. acidophilium [Barthelme and Sauer, 2012; Smith et al., 2005]. In addition, while the proteasome 20S core particle is essential in Hfx. volcanii, and presumably in other archaea, a strain with double deletion of the genes encoding the two Pan proteins, panA and panB, remains viable [Zhou et al., 2008]. These observations raise the possibility of additional ATPases that could interact with the proteasome and play a role in protein degradation. Proteins in the Cdc48/p97/VAT family, double-ring AAA+ ATPases involved in a variety of cellular processes [Meyer et al., 2012], are widespread among the archaea and some members contain the C-terminal HbYX motif required for proteasome activation [Barthelme and Sauer, 2012; Forouzan et al., 2012]. Cdc48 from T. acidophilum can interact with 20S proteasomes and stimulate proteasome-mediated degradation of nonapeptides in an ATP-dependent manner. 20S core particle subunits (α and β) can be pulled down when purified Cdc48 is added to cell extracts [Barthelme and Sauer, 2012]. Cdc48 proteins from eukaryotes can interact with the 20S core particle through both the HbYX motif and through a second loop that protrudes from the D2 domain of the Cdc48 and can stimulate the degradation of GFP-SsrA by the 20S proteasome in a regulatory manner in the same fashion of the Rpt subunits [Barthelme and Sauer, 2013]. Thus, Cdc48 is the first two-domain ATPase involved in proteasome-mediated degradation of substrates [Barthelme and Sauer, 2013; Meyer et al., 2012]. The physiological role Cdc48 plays in protein degradation in archaea remains to be elucidated. In addition to Pan and Cdc48, proteins from the AMA AAA+ ATPase family can activate proteasomes for degradation [Djuranovic et al., 2006; Forouzan et al., 2012; Summer et al., 2006]. AMA proteins are related to Cdc48 but diverge in the conserved fold of the N-domain. Examination of the difference between these complexes will likely reveal varying affinities for different classes of substrates. For example, the Cdc48 N-domain folds into a double-ψ barrel and a β-clam fold, which has been implicated in ubiquitin binding in eukaryotes [Park et al., 2005], while Pan N-domains consist of a coiled-coil and oligonucleotide-binding domain [Bar-Nun and Glickman, 2012; Forouzan et al., 2012]. AMA proteins have the β-clam fold but lack the double- ψ β-barrel of the Cdc48 N-domain [Djuranovic et al., 2006]. Much like the variation in the core particle subunits seen in some prokaryotes, the finding that different ATPase complexes are capable of proteasome activation open the possibility of subpopulations of complexes with redundant and unique sets of substrates for degradation. M. mazei is predicted to encode five different AAA+ proteins (two Pan, two Cdc48 proteins and one AMA protein) that activate proteasomal degradation [Forouzan et al., 2012]. While all five complexes from recombinant E. coli stimulated the degradation of casein by proteasomal core particles, only one of the Pan proteins (PanA) showed an energy-dependent increase in degradation of GFP-SsrA, a folded substrate [Forouzan et al., 2012]. Further work is needed to demonstrate direct interactions of these complexes with 20S proteasomes and to establish unique substrate ranges for each of the complexes.
Targeting Proteins for Degradation: Bacterial and Archaeal Ubiquitin-Like Proteins
In eukaryotic systems, proteins are targeted for degradation by the proteasome through a variety of mechanisms [Asher et al., 2005; Baugh et al., 2009]. Often, proteins destined for degradation by proteasomes will be covalently modified by addition of ubiquitin, a 76-aminoacid protein [Kerscher et al., 2006]. Ubiquitin is conjugated to primary amine groups on target proteins through an isopeptide bond with its C-terminal carboxyl group [Hochstrasser, 2009]. Ubiquitin conjugation is carried out by a cascade of enzymes and can be repeated to form a polyubiquitin chain that can ultimately serve as a signal for degradation by proteasomes. Lacking an obvious homologue to ubiquitin in the prokaryotic systems, it was unclear until recently how proteins were targeted for degradation by prokaryotic proteasomes. In the past few years, ubiquitin-like proteins were identified first in the M. tuberculosis system [Pearce et al., 2008] followed by the haloarchaea [Humbard et al., 2010a] (fig. 3). Through bioinformatic efforts, additional homologues of these prokaryotic counterparts to ubiquitin have since been identified in various phyla of Bacteria and Archaea [Burroughs et al., 2011; Nunoura et al., 2011]. Thus, along with proteasomes, conjugation to small protein modifiers is found conserved across all three domains of life.
Fig. 3.
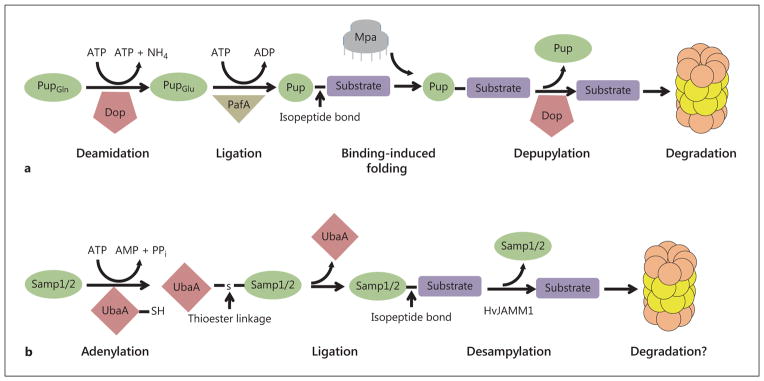
Pupylation and sampylation pathways. a Pupylation pathway in M. tuberculosis for the deamidation, ligation and depupylation of proteasome substrates for degradation. b Sampylation pathway for the adenylation (activation), ligation to substrates and desampylation for possible degradation by proteasomes.
The initial discovery of proteins that fulfill the role of ubiquitin in prokaryotes was in the actinobacterial species M. tuberculosis [Pearce et al., 2008]. A small protein, encoded by a gene closely linked to the proteasome subunit genes, termed Pup, forms covalent attachments to target proteins, and these conjugated forms are degraded by proteasomes [Burns et al., 2009; Pearce et al., 2008] (fig. 3a). While Pup is a functional equivalent of ubiquitin with respect to targeting proteins for degradation, there are several important features that make Pup unique. First, while ubiquitin and ubiquitin-like proteins in eukaryotic organisms share a β-grasp fold [Burroughs et al., 2007], Pup is an intrinsically disordered protein [Chen et al., 2009; Liao et al., 2009]. In addition, the conjugation to ubiquitin takes place between a free primary amine at either the N-terminus or the ε-amino group of lysine residues of a substrate protein and the C-terminal carboxyl group of an ubiquitin molecule. During pupylation, the C-terminal residue (glutamine 64) is deamidated to glutamate enzymatically by Dop (deaminase of Pup) [Imkamp et al. 2010a; Striebel et al., 2009]. The resulting γ-carboxylate is attached to the primary amine of lysine residues by PafA [Striebel et al., 2009]. Dop and PafA are homologous to glutamate synthetase proteins but share no clear structural or primary sequence relationship to the ubiquitin-activating E1 enzymes of eukaryotes [Striebel et al., 2009]. Interestingly, Dop can remove Pup from substrate proteins, suggesting Dop plays a similar role in regulating proteasome-mediated degradation of pupylated substrates that deubiquitinating enzymes play in eukaryotic systems [Burns et al., 2010a, 2012; Imkamp et al., 2010b]. While Pup is a disordered protein without definitive structure in solution, upon binding to the N-terminal coiled-coil domains of the ATPase Mpa, a portion of the C-terminal region of Pup forms an α-helix [Wang et al., 2010]. This binding-induced folding mechanism is believed to play a role in stabilizing the interaction between the pupylated substrate and Mpa or the translocation of the substrate through Mpa into the proteasome for degradation [Wang et al., 2010].
While the proteasome system in M. tuberculosis is not essential outside of a host [Darwin et al., 2003], several hundred substrates of the Pup system have been identified to date. The substrates are from a variety of pathways including respiration, intermediate metabolism, virulence and detoxification [Poulsen et al., 2010]. Pupylation is also used to regulate the activity of Mpa through modification of the C-terminal tails of this ATPase [Delley et al., 2012]. Covalent attachment of Pup prevents Mpa from interacting with the proteasome and disassembles the complex from the hexameric form into monomers. The pupylation of Mpa is reversible by Dop, thus, reactivating Mpa for energy-dependent degradation of pupylated substrates by proteasomes [Delley et al., 2012]. It is currently unclear whether there are additional factors other than PafA and Dop in the pupylation pathway. The system can be reconstituted in E. coli by the expression of PafA and Pup alone [Cerda-Maira et al., 2011]. However, the broad importance of pupylation and the proteasome in the physiology of M. tuberculosis and the survival within the host organisms is clear [Gandotra et al., 2007; Lamichhane et al., 2006].
A protein conjugation system in Archaea, similar to the ubiquitin pathway in eukaryotes, was initially described in Hfx. volcanii [Humbard et al., 2010a] and termed SAMP (small archaeal modifier proteins). Sampylation is more homologous to eukaryotic ubiquitination than the previously described pupylation pathway in a few ways. SAMPs are covalently attached to target proteins through C-terminal carboxyl groups to ε-amine groups of internal lysine residues of substrate proteins [Hepowit et al., 2012; Humbard et al., 2010a; Miranda et al., 2011], conjugation is carried out by a homologue of an ubiquitin-activating E1 enzyme termed UbaA [Miranda et al., 2011], Hfx. volcanii encodes two different JAMM-MPN deubiqutinating enzyme homologues (one of which is demonstrated to cleave SAMP conjugates; fig. 3b) [Hepowit et al., 2012], and SAMPs share the structural β-grasp fold of the ubiquitin-like protein family and the C-terminal diglycine motif found in many members of that family [Jeong et al., 2011; Ranjan et al., 2011]. Although there are similarities between ubiquitination and sampylation, the pathways are not identical. Sampylation does not require the activity of E2-conjugating enzyme or E3 protein ligase homologues for the attachment of SAMPs to target proteins [Burroughs et al., 2011; Humbard et al., 2010a; Miranda et al., 2011]. Also, in addition to protein conjugation, SAMPs function as sulfur-carrier proteins for the synthesis of molybdenum cofactors (MoCo) and the thiolation of tRNA [Miranda et al., 2011].
Hfx. volcanii encodes multiple SAMP proteins. SAMP1 and SAMP2 form protein conjugates in a UbaA-dependent manner [Humbard et al., 2010a; Miranda et al., 2011]. SAMP1 and SAMP2 also act as sulfur carriers for MoCo biosynthesis and thiolation of tRNA LysUUU, respectively [Miranda et al., 2011]. This dual role of SAMPs is analogous to the ubiquitin-like protein Urm1, which is responsible for thiolation of tRNA and also acts as a protein conjugation partner under oxidative stress in yeast and humans [Leidel et al., 2009; Van der Veen et al., 2011]. Each SAMP appears to modify a unique set of proteins [Humbard et al., 2010a]. Conjugation targets were identified through affinity purification of SAMPs and mass spectrometry, and sites of conjugation have been mapped to specific lysine residues [Hepowit et al., 2012; Humbard et al., 2010a]. Several dozen SAMP conjugation substrates have been identified to date. Only three proteins were commonly associated with SAMP1 and SAMP2, including homologues of E1 ubiquitin-activating enzymes (UbaA), thiosulfate sulfurtransferase (TssA) and methionine-S-sulfoxide reductase (MsrA). SAMP2 was identified as conjugated to itself, perhaps forming a poly-SAMP chain [Humbard et al., 2010a]. Alterations in proteasome composition also altered the conjugation patterns for the two SAMPs. Hfx. volcanii strains with knockouts panA (Pan ATPase) and psmA (α1 subunit) increased the levels of SAMP1 conjugates in cells and decreased the level of SAMP2 conjugates [Humbard et al., 2010a]. Since the proteasome is essential [Zhou et al., 2008], only one of the two α-type subunits could be removed. Whether a complete inhibition of the proteasome would cause further accumulation or an alteration in the pattern of SAMP conjugation remains to be seen.
Although the sampylation has only been demonstrated in Hfx. volcanii to date, homologues to SAMP and the conjugation machinery are predicted for all archaea [Humbard et al., 2010a; Makarova and Koonin, 2010]. Examination of the components of the sampylation pathway in Methanocarcina acetivorans revealed that the UbaA (E1) homologue, Elsa, can adenylate the C-terminus of SAMP from M. acetivorans in the presence of ATP [Ranjan et al., 2011], this is consistent with the observation that UbaA is required for the detection of conjugates of both SAMP1 and SAMP2 in Hfx. volcanii [Miranda et al., 2011]. In addition, the solution structures of SAMPs from M. acetivorans and P. furiosus are solved with NMR [Ranjan et al., 2011; Valafar et al., 2004]. In combination with a crystal structure of SAMP1 from Hfx. volcanii and a three-dimensional model of SAMP2, the predicted β-grasp fold of this family of ubiquitin-like proteins was confirmed [Jeong et al., 2011; Ranjan et al., 2011]. The structure of Hfx. volcanii SAMP1 had additional α-helices that make it more closely related, structurally, to the ubiquitin-like proteins MoaD and Urm1, compared to SAMP2 which is more similar to Ub [Jeong et al., 2011; Ranjan et al., 2011].
While the conjugation of SAMP1 and SAMP2 in Hfx. volcanii is demonstrated by mass spectrometry and the levels of conjugation are modulated by environmental conditions or knockouts of different proteasome genes, it remains to be seen whether the modification of proteins by SAMP1 or SAMP2 directly leads to degradation by the proteasome. Interestingly, while the proteasome itself is essential, SAMP1, SAMP2 and UbaA (E1) are not essential [Miranda et al., 2011]. This observation implies that if SAMP1 and/or SAMP2 target proteins for degradation by the proteasome, there must be additional or redundant pathways for degradation that are not clearly understood at this time. Several proteins in Hfx. volcanii seem to be degraded by the proteasome independent of SAMP conjugation. For example, proteomic experiments on cells treated with a proteasome inhibitor revealed several dozen proteins that did not appear sampylated that accumulated in the cell [Kirkland et al., 2007], the DNA clamp-loading protein PCNA accumulates in a panA mutant [Kirkland and Maupin-Furlow, 2009] as well as phosphorylated proteins [Kirkland et al., 2008]. Thus, additional targeting mechanisms (such as phosphorylation) possibly lead to degradation of proteins by proteasomes in archaea.
Conclusions
A significant amount of new information concerning the overall structure, role and targeting of proteins to prokaryotic proteasomes has been revealed in the past few years. Prokaryotic proteasomes remain an interesting and important thrust of research into the overall field of energy-dependent proteolysis with new developments in the overall structure of the larger complexes and new targeting mechanisms through ubiquitin-like proteins in Mycobacteria and Archaea. Many basic questions remain concerning this central proteolytic system such as, how does the regulatory ATPase interact with the core particle? What ATPase and non-ATPase structures can interact with core particles? Under what conditions and why would one ATPase be favored over another? How are the ubiquitin-like proteins SAMP and Pup recognized as signals for degradation? Are these Ubl proteins removed prior to degradation? Do they form poly-Ubl chains as is the case with ubiquitin? What are the other signals for degradation that do not involve Ubl proteins? Future studies of prokaryotic proteasomes and ubiquitin-like modification pathways are anticipated to answer these key questions.
Acknowledgments
This work was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Md., USA to M.A.H. and grants awarded to J.M.-F. through the National Institutes of Health (R01 GM57498) and the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of the US Department of Energy (Grant DE-FG02-05ER15650).
References
- Akopian TN, Kisselev AF, Goldberg AL. Processive degradation of proteins and other catalytic properties of the proteasome from Thermoplasma acidophilum. J Biol Chem. 1997;272:1791–1798. doi: 10.1074/jbc.272.3.1791. [DOI] [PubMed] [Google Scholar]
- Asher G, Tsvetkov P, Kahana C, Shaul Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005;19:316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader M, Steller H. Regulation of cell death by the ubiquitin-proteasome system. Curr Opin Cell Biol. 2009;21:878–884. doi: 10.1016/j.ceb.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nun S, Glickman MH. Proteasomal AAA-ATPases: structure and function. Biochim Biophys Acta. 2012;1823:67–82. doi: 10.1016/j.bbamcr.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Barthelme D, Sauer RT. Identification of the Cdc48*20S proteasome as an ancient AAA+ proteolytic machine. Science. 2012;337:843–846. doi: 10.1126/science.1224352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelme D, Sauer RT. Bipartite determinants mediate an evolutionarily conserved interaction between Cdc48 and the 20S peptidase. Proc Natl Acad Sci USA. 2013;110:3327–3332. doi: 10.1073/pnas.1300408110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh JM, Viktorova EG, Pilipenko EV. Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. J Mol Biol. 2009;386:814–827. doi: 10.1016/j.jmb.2008.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, Sakata E, Nickell S, Plitzko JM, Villa E, Baumeister W, Forster F. Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci USA. 2012;109:14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaroudj N, Zwickl P, Seemuller E, Baumeister W, Goldberg AL. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol Cell. 2003;11:69–78. doi: 10.1016/s1097-2765(02)00775-x. [DOI] [PubMed] [Google Scholar]
- Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berko D, Tabachnick-Cherny S, Shental-Bechor D, Cascio P, Mioletti S, Levy Y, Admon A, Ziv T, Tirosh B, Goldberg AL, Navon A. The direction of protein entry into the proteasome determines the variety of products and depends on the force needed to unfold its two termini. Mol Cell. 2012;48:601–611. doi: 10.1016/j.molcel.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochtler M, Ditzel L, Groll M, Hartmann C, Huber R. The proteasome. Annu Rev Biophys Biomol Struct. 1999;28:295–317. doi: 10.1146/annurev.biophys.28.1.295. [DOI] [PubMed] [Google Scholar]
- Bohn S, Beck F, Sakata E, Walzthoeni T, Beck M, Aebersold R, Forster F, Baumeister W, Nickell S. Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc Natl Acad Sci USA. 2010;107:20992–20997. doi: 10.1073/pnas.1015530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannigan JA, Dodson G, Duggleby HJ, Moody PC, Smith JL, Tomchick DR, Murzin AG. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature. 1995;378:416–419. doi: 10.1038/378416a0. [DOI] [PubMed] [Google Scholar]
- Burns KE, Cerda-Maira FA, Wang T, Li H, Bishai WR, Darwin KH. ‘Depupylation’ of prokaryotic ubiquitin-like protein from mycobacterial proteasome substrates. Mol Cell. 2010a;39:821–827. doi: 10.1016/j.molcel.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KE, Liu WT, Boshoff HI, Dorrestein PC, Barry CE., 3rd Proteasomal protein degradation in Mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J Biol Chem. 2009;284:3069–3075. doi: 10.1074/jbc.M808032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KE, McAllister FE, Schwerdtfeger C, Mintseris J, Cerda-Maira F, Noens EE, Wilmanns M, Hubbard SR, Melandri F, Ovaa H, Gygi SP, Darwin KH. Mycobacterium tuberculosis prokaryotic ubiquitin-like protein-deconjugating enzyme is an unusual aspartate amidase. J Biol Chem. 2012;287:37522–37529. doi: 10.1074/jbc.M112.384784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KE, Pearce MJ, Darwin KH. Prokaryotic ubiquitin-like protein provides a two-part degron to Mycobacterium proteasome substrates. J Bacteriol. 2010b;192:2933–2935. doi: 10.1128/JB.01639-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Balaji S, Iyer LM, Aravind L. Small but versatile: the extraordinary functional and structural diversity of the beta-grasp fold. Biol Direct. 2007;2:18. doi: 10.1186/1745-6150-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Iyer LM, Aravind L. Functional diversification of the RING finger and other binuclear treble clef domains in prokaryotes and the early evolution of the ubiquitin system. Mol Biosyst. 2011;7:2261–2277. doi: 10.1039/c1mb05061c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda-Maira FA, McAllister F, Bode NJ, Burns KE, Gygi SP, Darwin KH. Reconstitution of the Mycobacterium tuberculosis pupylation pathway in Escherichia coli. EMBO Rep. 2011;12:863–870. doi: 10.1038/embor.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Solomon WC, Kang Y, Cerda-Maira F, Darwin KH, Walters KJ. Prokaryotic ubiquitin-like protein pup is intrinsically disordered. J Mol Biol. 2009;392:208–217. doi: 10.1016/j.jmb.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlmann B, Kopp F, Kuehn L, Niedel B, Pfeifer G, Hegerl R, Baumeister W. The multicatalytic proteinase (prosome) is ubiquitous from eukaryotes to archaebacteria. FEBS Lett. 1989;251:125–131. doi: 10.1016/0014-5793(89)81441-3. [DOI] [PubMed] [Google Scholar]
- Dahlmann B, Kuehn L, Grziwa A, Zwickl P, Baumeister W. Biochemical properties of the proteasome from Thermoplasma acidophilum. Eur J Biochem. 1992;208:789–797. doi: 10.1111/j.1432-1033.1992.tb17249.x. [DOI] [PubMed] [Google Scholar]
- Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- Darwin KH, Lin G, Chen Z, Li H, Nathan CF. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol Microbiol. 2005;55:561–571. doi: 10.1111/j.1365-2958.2004.04403.x. [DOI] [PubMed] [Google Scholar]
- De Mot R, Nagy I, Walz J, Baumeister W. Proteasomes and other self-compartmentalizing proteases in prokaryotes. Trends Microbiol. 1999;7:88–92. doi: 10.1016/s0966-842x(98)01432-2. [DOI] [PubMed] [Google Scholar]
- Delley CL, Striebel F, Heydenreich FM, Ozcelik D, Weber-Ban E. Activity of the mycobacterial proteasomal ATPase Mpa is reversibly regulated by pupylation. J Biol Chem. 2012;287:7907–7914. doi: 10.1074/jbc.M111.331124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Hartmann MD, Habeck M, Ursinus A, Zwickl P, Martin J, Lupas AN, Zeth K. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol Cell. 2009;34:580–590. doi: 10.1016/j.molcel.2009.04.030. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Rockel B, Lupas AN, Martin J. Characterization of AMA, a new AAA protein from Archaeoglobus and methanogenic archaea. J Struct Biol. 2006;156:130–138. doi: 10.1016/j.jsb.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Festa RA, McAllister F, Pearce MJ, Mintseris J, Burns KE, Gygi SP, Darwin KH. Prokaryotic ubiquitin-like protein (Pup) proteome of Mycobacterium tuberculosis. PLoS One. 2010;5:e8589. doi: 10.1371/journal.pone.0008589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forouzan D, Ammelburg M, Hobel CF, Stroh LJ, Sessler N, Martin J, Lupas AN. The archaeal proteasome is regulated by a network of AAA ATPases. J Biol Chem. 2012;287:39254–39262. doi: 10.1074/jbc.M112.386458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A, Hill CP. Proteasome degradation: enter the substrate. Trends Cell Biol. 2003;13:550–553. doi: 10.1016/j.tcb.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S. In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat Med. 2007;13:1515–1520. doi: 10.1038/nm1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette TG, Kumar B, Thompson D, Slaughter CA, DeMartino GN. Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J Biol Chem. 2008;283:31813–31822. doi: 10.1074/jbc.M805935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M, Brandstetter H, Bartunik H, Bourenkow G, Huber R. Investigations on the maturation and regulation of archaebacterial proteasomes. J Mol Biol. 2003;327:75–83. doi: 10.1016/s0022-2836(03)00080-9. [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Grziwa A, Baumeister W, Dahlmann B, Kopp F. Localization of subunits in proteasomes from Thermoplasma acidophilum by immunoelectron microscopy. FEBS Lett. 1991;290:186–190. doi: 10.1016/0014-5793(91)81256-8. [DOI] [PubMed] [Google Scholar]
- Hegerl R, Pfeifer G, Puhler G, Dahlmann B, Baumeister W. The three-dimensional structure of proteasomes from Thermoplasma acidophilum as determined by electron microscopy using random conical tilting. FEBS Lett. 1991;283:117–121. doi: 10.1016/0014-5793(91)80567-m. [DOI] [PubMed] [Google Scholar]
- Hepowit NL, Uthandi S, Miranda HV, Toniutti M, Prunetti L, Olivarez O, De Vera IM, Fanucci GE, Chen S, Maupin-Furlow JA. Archaeal JAB1/MPN/MOV34 metalloenzyme (HvJAMM1) cleaves ubiquitin-like small archaeal modifier proteins (SAMPs) from protein-conjugates. Mol Microbiol. 2012;86:971–987. doi: 10.1111/mmi.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Lin G, Wang M, Dick L, Xu RM, Nathan C, Li H. Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate. Mol Microbiol. 2006;59:1417–1428. doi: 10.1111/j.1365-2958.2005.05036.x. [DOI] [PubMed] [Google Scholar]
- Humbard MA, Miranda HV, Lim JM, Krause DJ, Pritz JR, Zhou G, Chen S, Wells L, Maupin-Furlow JA. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature. 2010a;463:54–60. doi: 10.1038/nature08659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbard MA, Reuter CJ, Zuobi-Hasona K, Zhou G, Maupin-Furlow JA. Phosphorylation and methylation of proteasomal proteins of the haloarcheon Haloferax volcanii. Archaea. 2010b;2010:481725. doi: 10.1155/2010/481725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbard MA, Zhou G, Maupin-Furlow JA. The N-terminal penultimate residue of 20S proteasome α1 influences its Nα acetylation and protein levels as well as growth rate and stress responses of Haloferax volcanii. J Bacteriol. 2009;191:3794–3803. doi: 10.1128/JB.00090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imkamp F, Rosenberger T, Striebel F, Keller PM, Amstutz B, Sander P, Weber-Ban E. Deletion of dop in Mycobacterium smegmatis abolishes pupylation of protein substrates in vivo. Mol Microbiol. 2010a;75:744–754. doi: 10.1111/j.1365-2958.2009.07013.x. [DOI] [PubMed] [Google Scholar]
- Imkamp F, Striebel F, Sutter M, Ozcelik D, Zimmermann N, Sander P, Weber-Ban E. Dop functions as a depupylase in the prokaryotic ubiquitin-like modification pathway. EMBO Rep. 2010b;11:791–797. doi: 10.1038/embor.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong YJ, Jeong BC, Song HK. Crystal structure of ubiquitin-like small archaeal modifier protein 1 (SAMP1) from Haloferax volcanii. Biochem Biophys Res Commun. 2011;405:112–117. doi: 10.1016/j.bbrc.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Kaczowka SJ, Maupin-Furlow JA. Subunit topology of two 20S proteasomes from Haloferax volcanii. J Bacteriol. 2003;185:165–174. doi: 10.1128/JB.185.1.165-174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karadzic I, Maupin-Furlow J, Humbard M, Prunetti L, Singh P, Goodlett DR. Chemical cross-linking, mass spectrometry, and in sili-co modeling of proteasomal 20S core particles of the haloarchaeon Haloferax volcanii. Proteomics. 2012;12:1806–1814. doi: 10.1002/pmic.201100260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Takaoka M, Tanaka S, Sassa H, Tanaka K, Polevoda B, Sherman F, Hirano H. Nα-acetylation and proteolytic activity of the yeast 20 S proteasome. J Biol Chem. 2000;275:4635–4639. doi: 10.1074/jbc.275.7.4635. [DOI] [PubMed] [Google Scholar]
- Kirkland PA, Gil MA, Karadzic IM, Maupin-Furlow JA. Genetic and proteomic analyses of a proteasome-activating nucleotidase A mutant of the haloarchaeon Haloferax volcanii. J Bacteriol. 2008;190:193–205. doi: 10.1128/JB.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland PA, Maupin-Furlow JA. Stabilization of an archaeal DNA-sliding clamp protein, PCNA, by proteasome-activating nucleotidase gene knockout in Haloferax volcanii. FEMS Microbiol Lett. 2009;294:32–36. doi: 10.1111/j.1574-6968.2009.01547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland PA, Reuter CJ, Maupin-Furlow JA. Effect of proteasome inhibitor clastolactacystin-β-lactone on the proteome of the haloarchaeon Haloferax volcanii. Microbiology. 2007;153:2271–2280. doi: 10.1099/mic.0.2007/005769-0. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Akopian TN, Goldberg AL. Range of sizes of peptide products generated during degradation of different proteins by archaeal proteasomes. J Biol Chem. 1998;273:1982–1989. doi: 10.1074/jbc.273.4.1982. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Akopian TN, Woo KM, Goldberg AL. The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes: implications for understanding the degradative mechanism and antigen presentation. J Biol Chem. 1999;274:3363–3371. doi: 10.1074/jbc.274.6.3363. [DOI] [PubMed] [Google Scholar]
- Kwon YD, Nagy I, Adams PD, Baumeister W, Jap BK. Crystal structures of the Rhodococcus proteasome with and without its pro-peptides: implications for the role of the pro-peptide in proteasome assembly. J Mol Biol. 2004;335:233–245. doi: 10.1016/j.jmb.2003.08.029. [DOI] [PubMed] [Google Scholar]
- Lamichhane G, Raghunand TR, Morrison NE, Woolwine SC, Tyagi S, Kandavelou K, Bishai WR. Deletion of a Mycobacterium tuberculosis proteasomal ATPase homologue gene produces a slow-growing strain that persists in host tissues. J Infect Dis. 2006;194:1233–1240. doi: 10.1086/508288. [DOI] [PubMed] [Google Scholar]
- Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458:228–232. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- Le Tallec B, Barrault MB, Courbeyrette R, Guerois R, Marsolier-Kergoat MC, Peyroche A. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol Cell. 2007;27:660–674. doi: 10.1016/j.molcel.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Li D, Li H, Wang T, Pan H, Lin G. Structural basis for the assembly and gate closure mechanisms of the Mycobacterium tuberculosis 20S proteasome. Embo J. 2010;29:2037–2047. doi: 10.1038/emboj.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Shang Q, Zhang X, Zhang J, Xu C, Tu X. Pup, a prokaryotic ubiquitin-like protein, is an intrinsically disordered protein. Biochem J. 2009;422:207–215. doi: 10.1042/BJ20090738. [DOI] [PubMed] [Google Scholar]
- Lin G, Hu G, Tsu C, Kunes YZ, Li H, Dick L, Parsons T, Li P, Chen Z, Zwickl P, Weich N, Nathan C. Mycobacterium tuberculosis prcBA genes encode a gated proteasome with broad oligopeptide specificity. Mol Microbiol. 2006;59:1405–1416. doi: 10.1111/j.1365-2958.2005.05035.x. [DOI] [PubMed] [Google Scholar]
- Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- Lupas A, Koster AJ, Walz J, Baumeister W. Predicted secondary structure of the 20 S proteasome and model structure of the putative peptide channel. FEBS Lett. 1994;354:45–49. doi: 10.1016/0014-5793(94)01082-x. [DOI] [PubMed] [Google Scholar]
- Madding LS, Michel JK, Shockley KR, Conners SB, Epting KL, Johnson MR, Kelly RM. Role of the β1 subunit in the function and stability of the 20S proteasome in the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 2007;189:583–590. doi: 10.1128/JB.01382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV. Archaeal ubiquitin-like proteins: functional versatility and putative ancestral involvement in tRNA modification revealed by comparative genomic analysis. Archaea. 2010;2010:710303. doi: 10.1155/2010/710303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin-Furlow JA, Aldrich HC, Ferry JG. Biochemical characterization of the 20S proteasome from the methanoarchaeon Methanosarcina thermophila. J Bacteriol. 1998;180:1480–1487. doi: 10.1128/jb.180.6.1480-1487.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin-Furlow JA, Wilson HL, Kaczowka SJ, Ou MS. Proteasomes in the Archaea: from structure to function. Front Biosci. 2000;5:D837–D865. doi: 10.2741/furlow. [DOI] [PubMed] [Google Scholar]
- Medalia N, Beer A, Zwickl P, Mihalache O, Beck M, Medalia O, Navon A. Architecture and molecular mechanism of PAN, the archaeal proteasome regulatory ATPase. J Biol Chem. 2009;284:22952–22960. doi: 10.1074/jbc.M809643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehnert M, Sommer T, Jarosch E. ERAD ubiquitin ligases: multifunctional tools for protein quality control and waste disposal in the endoplasmic reticulum. Bioessays. 2010;32:905–913. doi: 10.1002/bies.201000046. [DOI] [PubMed] [Google Scholar]
- Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- Miranda HV, Nembhard N, Su D, Hepowit N, Krause DJ, Pritz JR, Phillips C, Soll D, Maupin-Furlow JA. E1- and ubiquitin-like proteins provide a direct link between protein conjugation and sulfur transfer in Archaea. Proc Natl Acad Sci USA. 2011;108:4417–4422. doi: 10.1073/pnas.1018151108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. Embo J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy I, Tamura T, Vanderleyden J, Baumeister W, De Mot R. The 20S proteasome of Streptomyces coelicolor. J Bacteriol. 1998;180:5448–5453. doi: 10.1128/jb.180.20.5448-5453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navon A, Goldberg AL. Proteins are unfolded on the surface of the ATPase ring before transport into the proteasome. Mol Cell. 2001;8:1339–1349. doi: 10.1016/s1097-2765(01)00407-5. [DOI] [PubMed] [Google Scholar]
- Nunoura T, Takaki Y, Kakuta J, Nishi S, Sugahara J, Kazama H, Chee GJ, Hattori M, Kanai A, Atomi H, Takai K, Takami H. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2011;39:3204–3223. doi: 10.1093/nar/gkq1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Isaacson R, Kim HT, Silver PA, Wagner G. Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites. Structure. 2005;13:995–1005. doi: 10.1016/j.str.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Pearce MJ, Arora P, Festa RA, Butler-Wu SM, Gokhale RS, Darwin KH. Identification of substrates of the Mycobacterium tuberculosis proteasome. Embo J. 2006;25:5423–5432. doi: 10.1038/sj.emboj.7601405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen C, Akhter Y, Jeon AH, Schmitt-Ulms G, Meyer HE, Stefanski A, Stuhler K, Wilmanns M, Song YH. Proteome-wide identification of mycobacterial pupylation targets. Mol Syst Biol. 2010;6:386. doi: 10.1038/msb.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan N, Damberger FF, Sutter M, Allain FH, Weber-Ban E. Solution structure and activation mechanism of ubiquitin-like small archaeal modifier proteins. J Mol Biol. 2011;405:1040–1055. doi: 10.1016/j.jmb.2010.11.040. [DOI] [PubMed] [Google Scholar]
- Religa TL, Sprangers R, Kay LE. Dynamic regulation of archaeal proteasome gate opening as studied by TROSY NMR. Science. 2010;328:98–102. doi: 10.1126/science.1184991. [DOI] [PubMed] [Google Scholar]
- Reuter CJ, Kaczowka SJ, Maupin-Furlow JA. Differential regulation of the PanA and PanB proteasome-activating nucleotidase and 20S proteasomal proteins of the haloarchaeon Haloferax volcanii. J Bacteriol. 2004;186:7763–7772. doi: 10.1128/JB.186.22.7763-7772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A, Rockel B, Gutsche I, Baumeister W, Lupas AN. The chaperones of the archaeon Thermoplasma acidophilum. J Struct Biol. 2001;135:126–138. doi: 10.1006/jsbi.2001.4402. [DOI] [PubMed] [Google Scholar]
- Ruschak AM, Religa TL, Breuer S, Witt S, Kay LE. The proteasome antechamber maintains substrates in an unfolded state. Nature. 2010;467:868–871. doi: 10.1038/nature09444. [DOI] [PubMed] [Google Scholar]
- Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- Seemuller E, Lupas A, Stock D, Lowe J, Huber R, Baumeister W. Proteasome from Thermoplasma acidophilum: a threonine protease. Science. 1995;268:579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- Sharon M, Witt S, Felderer K, Rockel B, Baumeister W, Robinson CV. 20S proteasomes have the potential to keep substrates in store for continual degradation. J Biol Chem. 2006;281:9569–9575. doi: 10.1074/jbc.M511951200. [DOI] [PubMed] [Google Scholar]
- Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s α ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell. 2011;144:526–538. doi: 10.1016/j.cell.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Kafri G, Cheng Y, Ng D, Walz T, Goldberg AL. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Snider J, Thibault G, Houry WA. The AAA+ superfamily of functionally diverse proteins. Genome Biol. 2008;9:216. doi: 10.1186/gb-2008-9-4-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtmueller BM, Hill CP. Proteasome activators. Mol Cell. 2011;41:8–19. doi: 10.1016/j.molcel.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striebel F, Hunkeler M, Summer H, Weber-Ban E. The mycobacterial Mpa-proteasome unfolds and degrades pupylated substrates by engaging Pup’s N-terminus. Embo J. 2010;29:1262–1271. doi: 10.1038/emboj.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striebel F, Imkamp F, Sutter M, Steiner M, Mamedov A, Weber-Ban E. Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat Struct Mol Biol. 2009;16:647–651. doi: 10.1038/nsmb.1597. [DOI] [PubMed] [Google Scholar]
- Summer H, Bruderer R, Weber-Ban E. Characterization of a new AAA+ protein from Archaea. J Struct Biol. 2006;156:120–129. doi: 10.1016/j.jsb.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Tamura T, Nagy I, Lupas A, Lottspeich F, Cejka Z, Schoofs G, Tanaka K, De Mot R, Baumeister W. The first characterization of a eubacterial proteasome: the 20S complex of Rhodococcus. Curr Biol. 1995;5:766–774. doi: 10.1016/s0960-9822(95)00153-9. [DOI] [PubMed] [Google Scholar]
- Tomko RJ, Jr, Funakoshi M, Schneider K, Wang J, Hochstrasser M. Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: implications for proteasome structure and assembly. Mol Cell. 2010;38:393–403. doi: 10.1016/j.molcel.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko RJ, Jr, Hochstrasser M. Order of the proteasomal ATPases and eukaryotic proteasome assembly. Cell Biochem Biophys. 2011;60:13–20. doi: 10.1007/s12013-011-9178-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valafar H, Mayer KL, Bougault CM, LeBlond PD, Jenney FE, Jr, Brereton PS, Adams MW, Prestegard JH. Backbone solution structures of proteins using residual dipolar couplings: application to a novel structural genomics target. J Struct Funct Genomics. 2004;5:241–254. doi: 10.1007/s10969-005-4899-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Veen AG, Schorpp K, Schlieker C, Buti L, Damon JR, Spooner E, Ploegh HL, Jentsch S. Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proc Natl Acad Sci USA. 2011;108:1763–1770. doi: 10.1073/pnas.1014402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker C, Lupas AN. Molecular evolution of proteasomes. Curr Top Microbiol Immunol. 2002;268:1–22. doi: 10.1007/978-3-642-59414-4_1. [DOI] [PubMed] [Google Scholar]
- Wang T, Darwin KH, Li H. Binding-induced folding of prokaryotic ubiquitin-like protein on the Mycobacterium proteasomal ATPase targets substrates for degradation. Nat Struct Mol Biol. 2010;17:1352–1357. doi: 10.1038/nsmb.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Li H, Lin G, Tang C, Li D, Nathan C, Darwin KH. Structural insights on the Mycobacterium tuberculosis proteasomal ATPase Mpa. Structure. 2009;17:1377–1385. doi: 10.1016/j.str.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous J, Burns K, Liu WT, Patel A, Hook V, Bafna V, Barry CE, 3rd, Bark S, Dorrestein PC. Expansion of the mycobacterial ‘PUPylome’. Mol Biosyst. 2010;6:376–385. doi: 10.1039/b916104j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel T, Eckerskorn C, Lottspeich F, Baumeister W. Existence of a molecular ruler in proteasomes suggested by analysis of degradation products. FEBS Lett. 1994;349:205–209. doi: 10.1016/0014-5793(94)00665-2. [DOI] [PubMed] [Google Scholar]
- Wilson HL, Aldrich HC, Maupin-Furlow J. Halophilic 20S proteasomes of the archaeon Haloferax volcanii: purification, characterization, and gene sequence analysis. J Bacteriol. 1999;181:5814–5824. doi: 10.1128/jb.181.18.5814-5824.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson HL, Ou MS, Aldrich HC, Maupin-Furlow J. Biochemical and physical properties of the Methanococcus jannaschii 20S proteasome and PAN, a homolog of the ATPase (Rpt) subunits of the eucaryal 26S proteasome. J Bacteriol. 2000;182:1680–1692. doi: 10.1128/jb.182.6.1680-1692.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S, Nagy I, Lupas A, Pfeifer G, Cejka Z, Muller SA, Engel A, De Mot R, Baumeister W. Characterization of ARC, a divergent member of the AAA ATPase family from Rhodococcus erythropolis. J Mol Biol. 1998;277:13–25. doi: 10.1006/jmbi.1997.1589. [DOI] [PubMed] [Google Scholar]
- Yu Y, Smith DM, Kim HM, Rodriguez V, Goldberg AL, Cheng Y. Interactions of PAN’s C-termini with archaeal 20S proteasome and implications for the eukaryotic proteasome-ATPase interactions. Embo J. 2010;29:692–702. doi: 10.1038/emboj.2009.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Hu M, Tian G, Zhang P, Finley D, Jeffrey PD, Shi Y. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009a;34:473–484. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wu Z, Zhang P, Tian G, Finley D, Shi Y. Mechanism of substrate unfolding and translocation by the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009b;34:485–496. doi: 10.1016/j.molcel.2009.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Kowalczyk D, Humbard MA, Rohatgi S, Maupin-Furlow JA. Proteasomal components required for cell growth and stress responses in the haloarchaeon Haloferax volcanii. J Bacteriol. 2008;190:8096–8105. doi: 10.1128/JB.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zühl F, Seemuller E, Golbik R, Baumeister W. Dissecting the assembly pathway of the 20S proteasome. FEBS Lett. 1997;418:189–194. doi: 10.1016/s0014-5793(97)01370-7. [DOI] [PubMed] [Google Scholar]
- Zwickl P, Ng D, Woo KM, Klenk HP, Goldberg AL. An archaebacterial ATPase, homologous to ATPases in the eukaryotic 26 S proteasome, activates protein breakdown by 20 S proteasomes. J Biol Chem. 1999;274:26008–26014. doi: 10.1074/jbc.274.37.26008. [DOI] [PubMed] [Google Scholar]