Abstract
Uterine luminal epithelium (LE) is critical for establishing uterine receptivity. Microarray analysis of gestation day 3.5 (D3.5, preimplantation) and D4.5 (postimplantation) LE from natural pregnant mice identified 382 upregulated and 245 downregulated genes in the D4.5 LE. Gene Ontology annotation grouped 186 upregulated and 103 downregulated genes into 22 and 15 enriched subcategories, respectively, in regulating DNA-dependent transcription, metabolism, cell morphology, ion transport, immune response, apoptosis, signal transduction, and so on. Signaling pathway analysis revealed 99 genes in 21 significantly changed signaling pathways, with 14 of these pathways involved in metabolism. In situ hybridization confirmed the temporal expression of 12 previously uncharacterized genes, including Atp6v0a4, Atp6v0d2, F3, Ggh, Tmprss11d, Tmprss13, Anpep, Fxyd4, Naip5, Npl, Nudt19, and Tpm1 in the periimplantation uterus. This study provides a comprehensive picture of the differentially expressed genes in the periimplantation LE to help understand the molecular mechanism of LE transformation upon establishment of uterine receptivity.
Keywords: microarray analysis, uterine luminal epithelium, uterine receptivity, embryo implantation
Introduction
Uterine receptivity refers to a transient state in which the maternal endometrium is receptive for an embryo to implant.1–4 Ovarian hormones progesterone (P4) and estrogen (E2) are the master controls for the establishment of uterine receptivity in mammals.5–8 The ovarian hormone-regulated uterine receptivity is restricted and transient. Since such restriction is abolished if the uterine luminal epithelium (LE) is broken or absent, LE is considered essential for the receptive sensitivity of the uterus.9,10 In addition, LE plays a transmitter role for decidualization,11 a critical response in the stromal compartment at the implantation site upon implantation initiation. The importance of LE in the establishment of uterine receptivity is also manifested by its physical interaction with the implanting embryos during the initial stages of embryo implantation, embryo apposition to the LE, embryo adhesion to the LE, and embryo penetration through the LE.1,12–14
It has been observed that the LE ultrastructure, such as LE cell surface components, lateral adherent junctions and gap junction channels, and subepithelial extracellular matrix (ECM), changes during the establishment of uterine receptivity.15–20 The morphological transformations are likely associated with the differential gene expression in the LE, for example, upregulation of gap junction protein β-2 (GJB2/Cx26) in the LE likely contributes to the altered gap junction channels in the LE.21 Differential gene expression of many genes in the periimplantation LE has been reported, such as cytochrome P450 26A1 (Cyp26a1),22 Gjb2, 21 histidine decarboxylase (Hdc),23 leukemia inhibitory factor receptor (Lifr),24 lysophosphatidic acid receptor 3 (Lpar3),25 msh homeobox 1 (Msx1),26 myeloid differentiation primary response gene 88 (MyD88),27 ethanolamine kinase 1 (Etnk1),28 progesterone receptor (Pgr),29–31 phospholipase A2, group IVA (Pla2g4a),32 proline-rich acidic protein 1 (Prap1),33 wingless-related MMTV integration site 7B (Wnt7b),34 and so on. However, the knowledge of global molecular transformation in the LE during periimplantation is still incomplete.
Microarray analysis has been widely used to determine the gene expression profiling associated with implantation in the mouse uterus30,35,36 and in the LE.27,28,37 The LE cells represent only 5% to 10% of total uterine cells.27,37 It is possible that some significantly changed gene expression in the LE may be obscured in the whole uterus microarray.30,35 Several studies have focused on LE gene expression.27,28,37 However, the data from these studies could not reflect the gene expression profiles in the LE from natural pregnant periimplantation uteri. One study used LE from ovarian hormone P4- and E2-treated nonpregnant ovariectomized mice to replicate the window of uterine receptivity27; one study compared the gene expression profiles between LE and glandular epithelium (GE) from P4- and E2-treated ovariectomized early pregnant mice (delayed implantation model)28; and the third study compared the gene expression profiles in the postimplantation LE at implantation site and interimplantation site in delayed implantation mouse model.37 In this study, LE cells were isolated from preimplantation D3.5 and postimplantation D4.5 natural pregnant mouse uteri. The gene expression profiles in the periimplantation LE from natural pregnancy will provide us with a comprehensive picture about the molecular transformation of LE during the establishment of uterine receptivity.
Materials and Methods
Animals
C57BL6/129svj-mixed background wild-type (WT) mice were generated from a colony at the University of Georgia.25 Mice were housed in polypropylene cages with free access to regular food and water from water sip tubes in a reverse osmosis system. The animal facility is on a 12-hour light/dark cycle (6:00 am to 6:00 pm) at 23 ± 1°C with 30% to 50% relative humidity. All methods used in this study were approved by the Animal Subjects Programs of the University of Georgia and conform to National Institutes of Health guidelines and public law.
Mating, Uterine Tissue Collection, Isolation of Uterine LE, Total RNA Isolation
Young virgin females (2-4 months old) were mated naturally with WT stud males and checked for a vaginal plug the next morning. The day a vaginal plug identified was designated as gestation day 0.5 (D0.5, mating night as D0). Uterine tissues were collected from euthanized females between 11:00 and 12:00 hours on D3.5 and D4.5, respectively. About one-third of a uterine horn from each euthanized female was frozen on dry ice. The remaining uterine tissue was processed for LE isolation as previously described using 0.5% dispase enzyme and gentle scraping.38 The isolated LE sheets from D3.5 and D4.5 uteri were subjected to total RNA isolation using TRIzol (Invitrogen, Carlsbad, California). The pregnancy status was determined by the presence of blastocysts in the D3.5 uterus or implantation sites in the D4.5 uterus. At least 3 pregnant mice were included in each group.
Microarray Analysis
Microarray analysis was performed at the Emory Biomarker Service Center, Emory University using Affymetrix_Mouse Gene 1.0 ST Chip covering 28,853 genes. Three replicates from different mice were included in each group. Microarray data (GEO number: GSE44451) were analyzed using GeneSpring 12.1 GX (Agilent Technologies, Santa Clara, California).39 The negative and missing values were threshold to 1 before the log transformation. Percentile shift normalization was performed to overcome the difference among different arrays, and entries with the lowest 20 percentile of the intensity values were removed. The criteria for determining differential gene expression included a fold change of ≥2, P value of < .05, and an absolute mean difference in the intensity values of >200 between the 2 groups. Gene Ontology annotation and signaling pathway analysis were performed using DAVID Analysis and GeneSpring 12.1 GX, respectively.40
Real-time Polymerase Chain Reaction
Real-time polymerase chain reaction (PCR) was used to validate selected genes from the microarray analysis. Total RNA from D3.5 and D4.5 whole-uterine horns was isolated using TRIzol. Complementary DNA (cDNA) was reverse transcribed from 1 μg of total RNA using Superscript III reverse transcriptase with random primers (Invitrogen). Real-time PCR was performed in 384-well plates using Sybr-Green I intercalating dye on ABI 7900 (Applied Biosystems, Carlsbad, California). Primer sequences are listed in Supplemental Table 1 (Integrated DNA Technology, San Diego, California).
In Situ Hybridization
In situ hybridization was performed as described previously.31,33,38 Antisense and sense probes for Atpase, H+ transporting, lysosomal V0 subunit A4 (Atp6v0a4), Atpase, H+ transporting, lysosomal V0 subunit D2 (Atp6v0d2), coagulation factor III (F3), γ-glutamyl hydrolase (Ggh), transmembrane protease, serine 11d (Tmprss11d), transmembrane protease, serine 13 (Tmprss13), alanyl aminopeptidase (Anpep), FXYD domain-containing ion transport regulator 4 (Fxyd4), nod-like receptor family, apoptosis inhibitory protein 5 (Naip5), N-acetylneuraminate pyruvate lyase (Npl), nucleoside diphosphate linked moiety X-type motif 19 (Nudt19), tropomyosin 1 (Tpm1), tropomyosin 2 (Tpm2), tropomyosin 3 (Tpm3), and tropomyosin 4 (Tpm4) were synthesized from cDNA fragments amplified with their respective gene-specific primer pairs (Supplemental Table 1).
Statistical Analyses
Two-tail unequal variance Student t test was used to compare the messenger RNA (mRNA) expression levels. The significant level was set at P < .05.
Results
Categorization of Differentially Expressed Genes in the Periimplantation LE
Microarray analysis indicated 382 significantly upregulated genes and 245 significantly downregulated genes in the postimplantation D4.5 LE compared with that in the preimplantation D3.5 LE. The most upregulated 10 genes in the D4.5 LE were Atp6v0d2 (34.70x), Ggh, Prap1, tocopherol transfer protein (α; Ttpa), tolloid-like protein 1 (Tll1), γ-aminobutyric acid A receptor, pi (Gabrp), matrix metallopeptidase 7 (Mmp7), glutathione peroxidase 3 (Gpx3), interferon induced transmembrane protein 6 (Ifitm6), and olfactomedin 1 (Olfm1). The most downregulated 10 genes in the D4.5 LE were Npl (35.42x), calbindin 1 (Calb1), G protein-coupled receptor 128 (Gpr128), solute carrier family 2, member 3 (Slc23a), UDP-glcnac: betagal β-1,3-N-acetylglucosaminyltransferase 5 (B3gnt5), interleukin 17 receptor B (Il17rb), Fxyd4, carbonyl reductase 2 (Cbr2), transmembrane protein 158 (Tmem158), and acyl-coA thioesterase 7 (Acot7). A complete list of all significantly changed genes with functional subcategory, Gene Ontology term, gene description, accession number, and fold change is shown in Supplemental Table 2 for upregulated genes in the D4.5 LE and in Supplemental Table 3 for downregulated genes in the D4.5 LE, respectively.
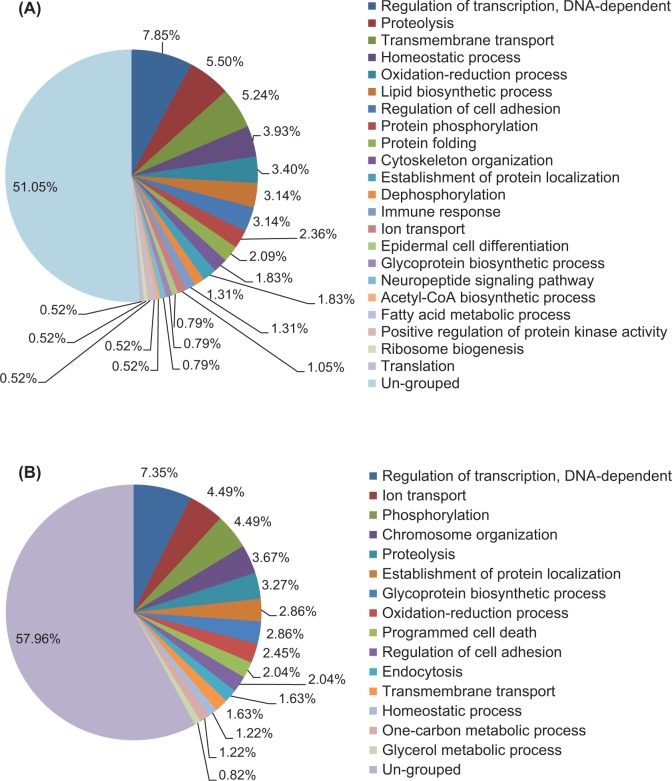
Gene Ontology annotation was conducted to categorize the differentially expressed genes based on the biological processes (Figure 1). Among the 382 significantly upregulated genes in the D4.5 LE, 187 (48.95%) genes were classified into 22 subcategories, and the remaining 195 (51.05%) genes were ungrouped. Seven largest categories with over 10 significantly upregulated genes were DNA-dependent transcription (7.85%, 30 genes), proteolysis (5.50%), transmembrane transport (5.24%), homeostatic process (3.93%), oxidation–reduction process (3.40%), lipid biosynthetic process (3.14%), and regulation of cell adhesion (3.14%; Figure 1A; Supplemental Table 2). Among the 245 downregulated genes in the D4.5 LE, 103 (42.04%) genes were grouped into 15 subcategories, including DNA-dependent transcription (7.35%, 17 genes), ion transport (4.49%), phosphorylation (4.49%), chromosome organization (3.67%), and so on, and the remaining 142 genes (57.96%) were ungrouped (Figure 1B; Supplemental Table 3). Interestingly, DNA-dependent transcription was the most enriched subcategory for both the upregulated and the downregulated genes in the D4.5 LE, in which transcription factors Arnt2 (Aryl hydrocarbon receptor nuclear translocator 2) and Myc (myelocytomatosis oncogene) were the most upregulated genes and Pgr (progesterone receptor) was the most downregulated gene (Figure 1; Supplemental Tables 2 and 3). The following 8 subcategories, proteolysis, transmembrane transport, homeostatic process, oxidation–reduction process, regulation of cell adhesion, establishment of protein localization, ion transport, and glycoprotein biosynthetic process, were shown in both the upregulated and the downregulated gene groups (Figure 1; Supplemental Tables 2 and 3).
Figure 1.
Categorization of genes whose transcript abundance is significantly changed in uterine LE upon embryo implantation via Gene Ontology Annotation. A, Pie chart of categorization (percentages) of genes significantly upregulated in the gestation day 4.5 (D4.5) LE. B, Pie chart of categorization (percentages) of genes significantly downregulated in the D4.5 LE. Only the genes with a minimal fold change of 2, P < .05, and an absolute value of mean difference greater than 200 between the 2 groups were included. N = 3 for both the groups.
Signaling Pathway Analysis of the Differentially Expressed Genes in the Periimplantation LE
In Gene Ontology annotation, each gene was grouped into a single subcategory (Figure 1; Supplemental Tables 2 and 3). However, in signaling pathway analysis, one gene could be clustered into multiple signaling pathways, for example, Cyp26a1 was shown in both metapathway biotransformation and adipogenesis pathways (Table 1). Among the 627 differentially expressed genes in the periimplantation LE, 100 genes were classified into 25 significantly changed signaling pathways. Of the top 10 most upregulated genes in the D4.5 LE, 1 gene, Gpx3, was assigned into 6 of the 25 significantly changed signaling pathways. However, Gpx3 was reported to be detected in the stromal compartment but not in LE of D4.5 uterus,41 and we confirmed this expression pattern by in situ hybridization (data not shown). The Gpx3 was thus removed from signaling pathway analysis. Without Gpx3, 99 differentially expressed genes were classified into 21 significantly altered signaling pathways in the periimplantation LE (Table 1). Fourteen (66.7%) of these signaling pathways were involved in metabolism that included the top 5 most significantly altered pathways, fatty acid biosynthesis (P = 6.11E−06), glycolysis and gluconeogenesis (P = 2.14E−04), purine metabolism (P = 2.58E−04), metapathway biotransformation (P = 2.59 E−04), and triacylglyceride synthesis (9.64E−04; Table 1). Other metabolic pathways included 1 carbon metabolism and related pathways, amino acid metabolism, prostaglandin synthesis and regulation, TCA cycle, Kennedy pathway, adipogenesis, nucleotide metabolism, fatty acid β oxidation, and urea cycle and metabolism of amino groups (Table 1). The remaining 7 pathways were involved in insulin signaling, α-6-β-4 integrin signaling pathway, complement and coagulation cascades, Wnt signaling pathway and pluripotency, micro RNA (miRNA) regulation of DNA damage response, regulation of actin cytoskeleton, and epidermal growth factor receptor 1 signaling pathway (Table 1).
Table 1.
Signaling Pathways Changed in the Periimplantation Mouse Uterine Luminal Epithelium.
| Signaling Pathways | P Value | Significantly Changed Genes | |
|---|---|---|---|
| Upregulated | Downregulated | ||
| Fattyacid biosynthesis | 6.11E−06 | Acly, Acsl4, Echs1 | Acaca, Acsl2, Fasn, Pcx |
| Glycolysis and gluconeogenesis | 2.14E−04 | Eno1, Pkm2, Slc2a1 | Pcx, Slc2a3 |
| Purine metabolism | 2.58E−04 | Entd5, Gda, Gmpr, Nt5e, Pde4d, Pkm2, Pnp, Pnp2, Polr1a, Polr1b, Prps2 | Adcy9, Nt5c2 |
| Metapathway biotransformation | 2.59E−04 | Chst12, Comt1, Cyp26a1, Fmo2, Mgst1 | Gpx1, Cyp2s1, Hs3st1 |
| Triacylglyceride synthesis | 9.64E−04 | Agpat5, Gpam | Ppap2b |
| Insulin signaling | 1.11E−03 | Eif4ebp1, Igf1r, Mapk6, Prkaa1, Prkaa2, Sgk, Xbp1 | Pik3r3, Slc2a1, |
| One carbon metabolism and related pathways | 1.55E−03 | Ahcyl1 | Chdh, Chpt1, Etnk1, Gpx1, Pcyt1b, Tyms |
| Amino acid metabolism | 1.61E−03 | Acly, Glud, Hal, Srm, Pkm2 | Adh5, Hdc, Pcx |
| α-6-β-4 integrin signaling pathway | 2.08E−03 | Dst, Eif4ebp1, Lama3, Met, Mmp7, Sfn | Pik3r3 |
| Prostaglandin synthesis and regulation | 3.88E−03 | Anxa3, Ptger2, S100a10 | Pla2g4a, Ptgdr |
| TCA cycle | 3.88E−03 | Idh2, Idh3a | Pcx |
| Complement and coagulation cascades | 4.88E−03 | C3, F3, Cfh, Kng1 | C2 |
| Kennedy pathway | 1.12E−02 | Chpt1, Etnk1, Pcyt1b | |
| Wnt signaling pathway and pluripotency | 1.68E−02 | Ccnd1, Mmp7, Myc, Ppp2r1b, Tcf4, Wnt7b | Fzd6 |
| Adipogenesis | 1.69E−02 | Cebpa, Cyp26a1, Klf7, Lifr, Mif | Gata2, Zmpste24 |
| miRNA regulation of DNA damage response | 2.43E−02 | Ccnd1, Sfn, Trp53 | Ddb2, Sesn1 |
| Nucleotide metabolism | 2.62E−02 | Prps2, Srm | Mthfd2 |
| Fatty acid β oxidation | 2.86E−02 | Acsl4, Cpt1a, Echs1 | Acss2 |
| Urea cycle and metabolism of amino groups | 3.00E−02 | Glud1, Srm | Ckb |
| Regulation of actin cytoskeleton | 3.66E−02 | Enah, Fgf9, Itga1, Mapk6, Myh10, Pip5k1b | Pik3r3, Rdx |
| EGFR1 signaling pathway | 4.19E−02 | Cblb, Cebpa, Eps8, Myc | Pik3r3, Plscr1 |
Abbreviations: EGFR1, epidermal growth factor receptor 1; miRNA, micro RNA.
Gene Expression Confirmation by Real-time PCR and In Situ Hybridization
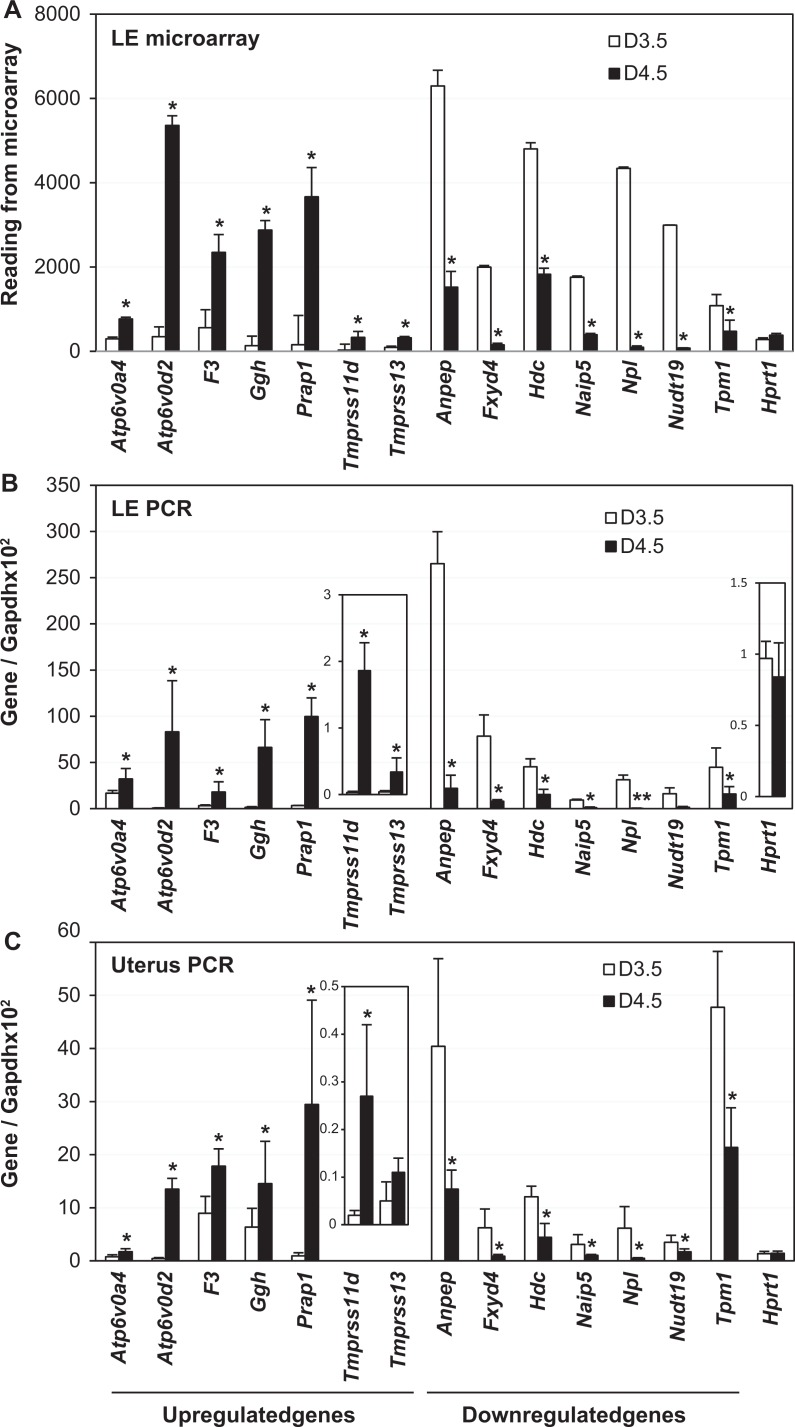
To validate microarray data, the mRNA levels of 7 upregulated and 7 downregulated genes were examined by real-time PCR and in situ hybridization. Among these 14 genes, Prap1 and Hdc served as positive controls.23,31,33 Atp6v0a4 and the most upregulated Atp6v0d2 in the D4.5 LE encode subunits for the vacuolar-type H+-ATPase (V-ATPase), which is involved in transmembrane proton translocation.42–44 F3 encodes coagulation factor III, a cell surface glycoprotein involved in initiating coagulation pathway and inflammatory signaling.45 Ggh encodes a lysosomal enzyme important for the cellular homeostasis of folate.46 Tmprss11d and Tmprss13 encode proteases.47–49 Anpep is also called CD13, encoding a membrane protein that plays a role in digesting peptides and may also play roles in regulating keratinocyte-mediated ECM remodeling and fibroblast contractile activity50 as well as immune responses.51 Fxyd4 encodes a regulator of the Na, K-ATPase.52 Naip5 is also called Birc1e, encoding an apoptosis inhibitory protein that can be upregulated by a synthetic estrogen diethylstilbestrol in the postnatal day 5 mouse LE.53 The most downregulated gene, Npl, regulates cellular sialic acid concentration.54,55 The Nudt19, also called RP2, hydrolyzes CoA esters56 and possesses mRNA decapping activity.57 The Tpm1 is 1 of the 4 genes encoding tropomyosin, an actin-binding protein involved in smooth muscle contraction and mediating actin cytoskeleton functions in nonmuscle cells.58,59
Relative readings of these genes in the microarray analysis are shown in Figure 2A. The differential expression of these genes was confirmed by real-time PCR in the LE (Figure 2B) and in the whole uterus, in which only Tmprss13 did not show significant difference (Figure 2C) but with similar trend of change as microarray data (Figure 2A) and LE real-time PCR (Figure 2B).
Figure 2.
Expression of selected upregulated and downregulated genes. A, Readings from microarray analysis. N = 3. B, Real-time PCR of gestation day 3.5 (D3.5) and D4.5 LE. N = 5 to 6. C, Real-time PCR of D3.5 and D4.5uterus. N=5 to 6. A-C: *P < .05, compared to D3.5. Error bars represent standard deviation. Atp6v0a4, Atpase, H+ transporting, lysosomal V0 subunit A4; Atp6v0d2, Atpase, H+ transporting, lysosomal V0 subunit D2; F3, coagulation factor III; Ggh, γ-glutamyl hydrolase; Prap1, proline-rich acidic protein 1; Tmprss11d, transmembrane protease, serine 11d; Tmprss13, transmembrane protease, serine 13; Anpep, alanyl aminopeptidase; Fxyd4, FXYD domain-containing ion transport regulator 4; Naip5, nod-like receptor family, apoptosis inhibitory protein 5; Npl, N-acetylneuraminate pyruvate lyase; Nudt19, nucleoside diphosphate linked moiety X-type motif 19; Tpm1, tropomyosin 1; Hprt1, hypoxanthine phosphoribosyltransferase 1, a house keeping gene; Gapdh, glyceraldehyde 3-phosphate dehydrogenase, a house keeping gene as the loading control. PCR indicates polymerase chain reaction.
Consistent with microarray data and real-time PCR results (Figure 2; Supplemental Table 2), Atp6v0a4, Atp6v0d2, F3, Ggh, Tmprss11d, and Tmprss13 were upregulated in the D4.5 LE. They were undetectable in other uterine compartments during periimplantation (Figure 3A-L). Although these 6 genes were all LE specific, there were differences in their expression patterns in the LE. Atp6v0a4, Atp6v0d2, and F3 were expressed along the entire LE (Figure 3B, D, and F); Tmprss11d was mainly detected in the LE surrounding the embryo (Figure 3J), and Ggh and Tmprss13 had higher expression levels in the LE away from the implantation site (Figure 3H and L).
Figure 3.
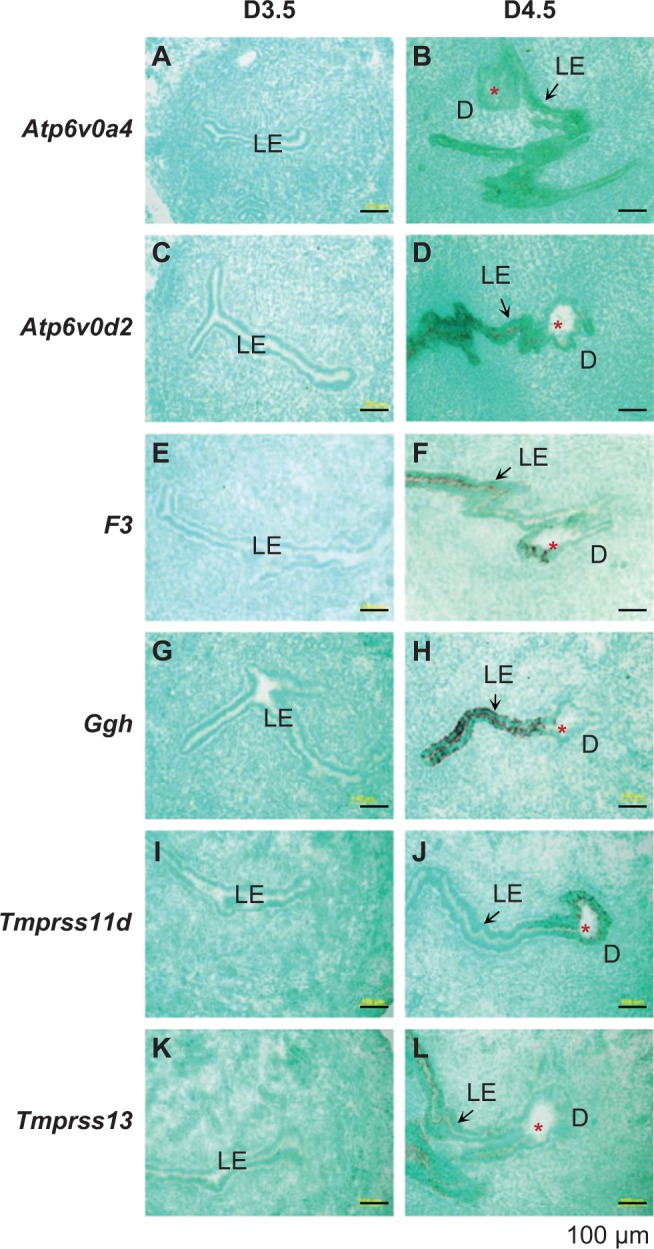
Localization of selected genes in the D3.5 and 4.5 mouse uterus by in situ hybridization using gene-specific antisense probes. These genes were shown upregulated in the D4.5 LE in microarray (Supplemental Table 2). A, Atp6v0a4, D3.5. B, Atp6v0a4, D4.5. C, Atp6v0d2, D3.5. D, Atp6v0d2, D4.5. E, F3, D3.5. F, F3, D4.5. G, Ggh, D3.5. H, Ggh, D4.5. I. Tmprss11d, D3.5. J, Tmprss11d, D4.5. K, Tmprss13, D3.5. L, Tmprss13, D4.5. D3.5, cross-sections (10 μm); D4.5, longitudinal sections (10 μm). No specific signal was detected using a sense probe (data not shown). Red star, embryo; LE, luminal epithelium; D, decidual zone; scale bar, 100 μm. N = 2 to 3.
The expression levels of Anpep, Fxyd4, Naip5, Npl, Nudt19, and Tpm1 were higher in the D3.5 LE compared to that in the D4.5 LE (Figures 4A-J and 5A and B), in which only Nudt19 remained detectable in the D4.5 LE surrounding the embryo (Figure 4J). Among these 6 genes, Fxyd4, Naip5, Npl, and Nudt19 were LE specific in the periimplantation uterus. Anpep was also abundantly expressed in the D3.5 GE and disappeared from both LE and GE in the D4.5 uterus (Figure 4A and B). The spatiotemporal expression of Tpm1 in the periimplantation uterus was unique, and it was highly expressed in the LE, GE, and myometrium of the D3.5 uterus (Figure 5A); upon embryo implantation, it disappeared from both LE and GE, remained in the myometrium, and appeared in the primary decidual zone of the D4.5 uterus (Figure 5B). Interestingly, among the 4 tropomysin isoforms TPM1-4,60 Tpm1 was the only one detected and differentially expressed in the periimplantation LE (Figure 5; Supplemental Table 3). Tpm2 was detected only in the myometrium of the D3.5 uterus (Figure 5C) and remained in the myometrium and appeared in the stromal compartment at the implantation site of the D4.5 uterus (Figure 5D). Tpm3 and Tpm4 were undetectable in the D3.5 uterus (Figure 5E and G) but detectable in the stromal compartment at the implantation site of the D4.5 uterus (Figure 5F and H).
Figure 4.
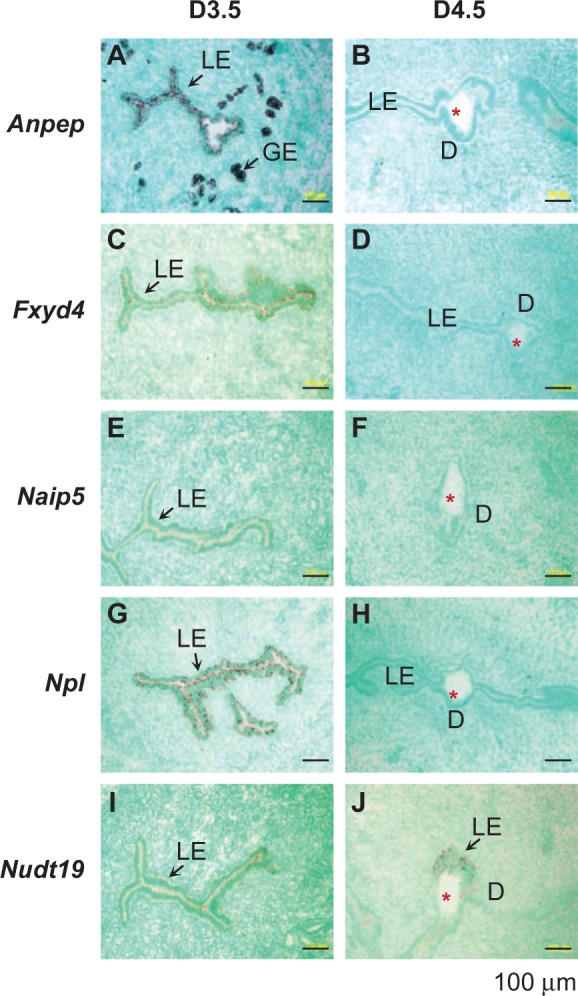
Localization of selected genes in the D3.5 and 4.5 uteri by in situ hybridization using gene-specific antisense probes. These genes were shown downregulated in the D4.5 LE in microarray (Supplemental Table 3). A, Anpep, D3.5. B, Anpep, D4.5. C, Fxyd4, D3.5. D, Fxyd4, D4.5. E, Naip5, D3.5. F, Naip5, D4.5. G, Npl, D3.5. H, Npl, D4.5. I, Nudt19, D3.5. J, Nudt19, D4.5. D3.5, cross-sections (10 μm); D4.5, longitudinal sections (10 μm). No specific signal was detected using a sense probe (data not shown). Red star, embryo; LE, luminal epithelium; GE, glandular epithelium; D, decidual zone; scale bar, 100 μm. N = 2 to 3.
Figure 5.
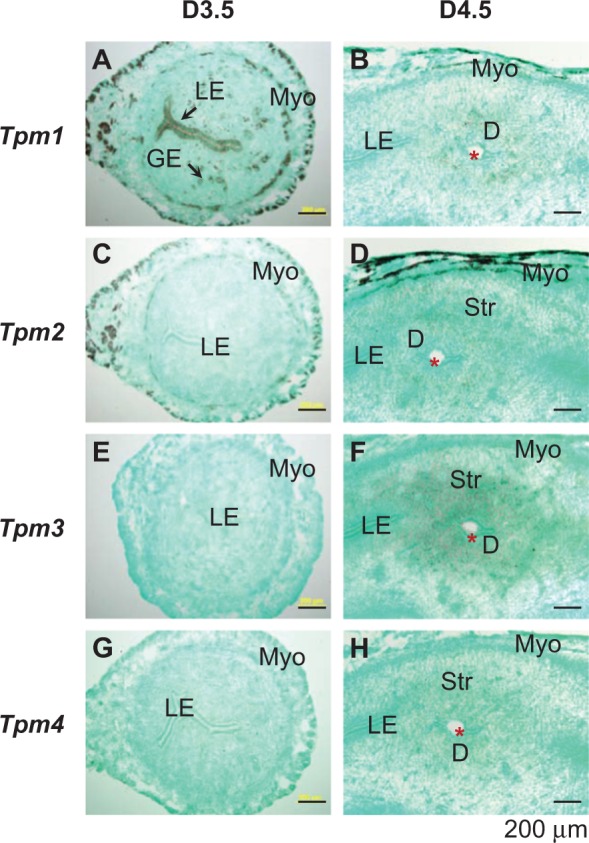
Localization of Tpm1-4 in the D3.5 and D4.5 mouse uteri by in situ hybridization using Tpm1, Tpm2, Tpm3, and Tpm4 antisense probes, respectively. A, Tpm1, D3.5. B, Tpm1, D4.5. C. Tpm2, D3.5. D. Tpm2, D4.5. E. Tpm3, D3.5. F. Tpm3, D4.5. G. Tpm4, D3.5. H, Tpm4, D4.5. D3.5, cross sections (10 μm); D4.5, longitudinal sections (10 μm). No specific signal was detected using a sense probe (data not shown). Red star, embryo; LE, luminal epithelium; GE, glandular epithelium; D, decidual zone; Myo, myometrium; scale bar, 200 μm. N = 2 to 3.
Discussion
General Discussion of the Approach
This microarray study identifies 627 differentially expressed genes in the periimplantation mouse LE, which include some well-known implantation markers in the LE but most of which have not been previously reported in the periimplantation LE. The inclusion of known implantation markers and the confirmation of previously uncharacterized genes in the LE demonstrate the effectiveness of the approach. Many stromal-specific genes upon implantation, such as Bmp2,61 Bmp7,61 Fgf2,61 Wnt4, 34,61 Wnt16,34 Gja1,62 Abp1,63,64 Hand2,65 and so on, did not show differential expression in the periimplantation LE (GEO number: GSE44451), further supporting the efficiency of the LE microarray. However, 3 genes that were reported to be mainly detected in the stromal compartment upon implantation, Gpx3,41 Angpt2,66 and Prss35,64 showed upregulation in the D4.5 LE (Supplemental Table 2). Differential expression of Gpx3 and Prss35 was confirmed by real-time PCR in D3.5 and D4.5 LE (data not shown). These observations suggest potential stromal cell contamination which would most likely occur during scraping of LE sheets with subepithelial stromal cells attached. Since many more stromal-specific genes do not show differential expression in the periimplantation LE, another explanation would be that there is true upregulation in the D4.5 LE, but the levels of upregulation are overshadowed by the much higher expression levels in the stromal compartment, and the probes used in the in situ hybridization were not sensitive enough to detect the differences in the LE but the stromal compartment.
Comparison With 3 Other Microarray Analyses Involved LE
There are three other microarray analyses involved LE.27,28,37 These reported microarray analyses are very different from our study. Regardless, each one was compared with this study to obtain the overlapped genes (Table 2). Pan et al used ovariectomized nonpregnant mice treated with E2 and E2 + P4 to replicate the window of uterine receptivity. They only reported the genes upregulated in the E2 + P4-treated LE compared with those in E2-treated LE.27 This mouse model may replicate the preimplantation uterine conditions from nonreceptive (E2-treated, ∼D0) to receptive (E2 + P4-treated, ∼D3.5 in this study), but it does not include the postimplantation D4.5 condition. There are 4 genes upregulated in both E2 + P4-treated LE and D4.5 LE, and 15 genes upregulated in E2+P4-treated LE but downregulated in D4.5 LE (Table 2). Niklaus et al compared the gene expression profiles between LE and GE from P4- and E2-treated ovariectomized early pregnant mice (delayed implantation model) before implantation initiation.28 There are 28 genes shown in both LE versus GE array and D3.5 LE versus D4.5 LE array (Table 2). Chen et al compared the gene expression profiles in the postimplantation LE at implantation site and interimplantation site in the delayed implantation mouse model.37 There are 20 genes overlapped with the results from our study. Since the study by Pan et al covers a different period from that in this study, and the studies by both Niklaus et al and Chen et al are spatial comparison but not temporal comparison as in this study (D3.5 LE vs D4.5 LE), the overlapped genes in Table 2 can only be instructive for periimplantation uterine gene expression studies.
Table 2.
Comparison of the Differentially Expressed Genes in the 3 References That Overlap With the Differentially Expressed Genes (Total 627) in This Study (D3.5 LE vs D4.5 LE).
| Reference | Reference27 | Reference28 | Reference7 |
|---|---|---|---|
| Mouse model | Ovariectomized mice treated with E2 or E2 + P4 | Delayed implantation | Delayed implantation |
| Comparison | E2-treated LE vs E2 + P4-treated LE | LE vs GE | IS LE vs inter-IS LE |
| No. of differentially expressed genes | 222 (up in E2+P4-treated LE) | 153 (up in LE) 118 (down in LE) | 136 (up in IS LE) 223 (down in IS LE) |
| No. of overlapped genes with this study | 19 | 28 | 20 |
| Detail description | Up in E2 + P4-treated LE and up in D4.5 LE (4 genes): H6pd, Igfbp3, Olfm1, and Mmp7 Up in E2 + P4-treated LE and down in D4.5 LE (15 genes): Calb1, Fxyd4, Marcks, Jam2, Lrpap1, Lrp2, Nde1, Ovgp1, Rdx, Tmod2, Cln5, Pfkfb3, Serpina1e, Hdc, and Prune | Up in LE and up in D4.5 LE (8 genes): Bcap29, Atp11a, Nt5e, Tacstd2, Atp6v0a4, Blnk, Tgfbi, and Efhd1 Up in LE & down in D4.5 LE (12 genes): Calb1, Fxyd4, Sorl1, Jam2, Nudt19, Car2, Chdh, Rdx, Cln5, Etnk1, Fasn, and Hdc Down in LE and down in D4.5 LE (2 genes): Slc2a3 and Ces3 Down in LE and up in D4.5 LE (6 genes): Lsyna1, Sult1d1, Ide, Enah, Ern1, and Manba | Up in IS LE and up in D4.5 LE (8 genes): Mgst1, Fads3, Gmpr, Hal, Pla2g7, Cfh, Wnt7b, and Ggh Down in IS LE and down in D4.5 LE (11 genes): Npl, Ank, Stx18, Lrpap1, Ckmt1, Ly75, Atp2b2, Myd88, Tmod2, Pmp22, and Plscr1 Down in IS LE and Up in D4.5 LE (1 gene): Irf1 |
Abbreviations: D3.5, gestation day 3.5, preimplantation; D4.5, gestation day 4.5, postimplantation; E2, 17β-estradiol; P4, progesterone; IS, implantation site; inter-IS, interimplantation site; LE, luminal epithelium; GE, glandular epithelium.
About the Newly Characterized Genes
The 12 newly characterized genes have potential functions in ion transport (eg, Atp6v0a4, Atp6v0d2, Fxyd4), metabolism (eg, Ggh, Npl, Nudt19), morphology (eg, Anpep, Tmprss11d, Tmprss13, Tpm1), immune responses (Anpep, F3), and apoptosis (eg, Naip5). The differential expression of these genes (Figures 2 –5) and many others revealed in the microarray analysis (Supplemental Tables 2 and 3) indicates the involvement of the above-mentioned events in the LE, which are important for the establishment of uterine receptivity.
Membrane Transport and Metabolism in Periimplantation LE
Changes in membrane transport, including ion transport and metabolism in LE, could influence the uterine histotroph, a complex mixture of enzymes, growth factors, hormones, and nutrients that are critical for the activation of conceptus–endometrium interactions, embryo development, and implantation during early pregnancy.67 Differential expression of transporters in ovine LE is involved in the altered nutrient profiles, such as glucose, amino acid, and ions in the uterine lumen during periimplantation.68–71 Although the profiles of the components in the periimplantation mouse uterus lumen is unavailable, signaling pathway analysis of our microarray data indicates that two-third of the significantly changed pathways are related to metabolism (Table 1), and a large group of genes involved in transmembrane transport are differentially expressed in the periimplantation LE (Supplemental Tables 2 and 3). These molecular changes in the LE could affect the components in the uterine lumen to influence both the endometrium and the blastocyst for embryo implantation.
Morphological Changes in Periimplantation LE
Gene Ontology annotation indicates that several groups of differentially expressed genes could be involved in the LE morphological changes, such as proteolysis, regulation of cell adhesion, cytoskeleton organization, epidermal cell differentiation, and endocytosis. Cell adhesion is thought to play an important role in the initial attachment of the embryo to the LE.72,73 Among the 15 genes in the category of regulation of cells adhesion (Supplemental Tables 2 and 3), secreted phosphoprotein 1, also called osteopontin, and its integrin receptors are among the best characterized cell adhesion molecules in the LE during embryo implantation.72,73 Integrins are cell surface glycoproteins formed by noncovalent binding of α and β subunits. Interestingly, our LE microarray analysis reveals the differential expression of integrin α1, α3, and α9 subunits in the periimplantation LE, suggesting that the integrin activity in the periimplantation LE might be regulated via α subunits (Supplemental Tables 2 and 3).
Immune Responses in Periimplantation LE
Immune responses play critical roles in embryo implantation.74 Gene Ontology annotation groups 5 genes into “immune response” subcategory, F3, Il1f6 (Interleukin 1 family, member 6), Kng1 (Kininogen 1), Mif (macrophage migration inhibitory factor), and Pla2g7 (phospholipase A2, group VII), which are all significantly upregulated in the D4.5 LE (Figure 3E and F and Supplemental Table 2). Besides the role of F3 in coagulation and proinflammation,45 IL1F6 participates in cytokine/chemokine production75; KNG1 is also involved in coagulation and proinflammation76; MIF is a cytokine with chemokine-like functions mediating host immune and inflammatory response77; and PLA2G7 metabolizes platelet-activating factor and its roles in immune responses seem to depend on the specific biological settings.78 The upregulation of these “immune response” genes indicates increased inflammatory responses in the LE during the early stages of embryo implantation.
Apoptosis in Periimplantation LE
Microarray analysis of human uterine epithelial cells between the late proliferative phase (pre-receptive) and the midsecretory phase (receptive for embryo implantation) indicates that cell cycle regulation is the most significantly enriched functional pathway in the late proliferative phase endometrial epithelium.79 Cell cycle regulation does not appear in our Gene Ontology enrichment analysis (Supplemental Tables 2 and 3). It is reasonable because the time points selected in this study have passed the cell proliferation stage in the LE, which peaks at D1.5.80 Instead, “programmed cell death” (apoptosis) is among the groups significantly downregulated in the D4.5 LE (Supplemental Table 3). Five genes in this group meet the selection criteria (fold change of ≥2, P < .05, and mean difference >200). The top 3 most differentially expressed genes in this group, Naip1, Naip5 (Figure 4E and F), and Naip7 encode apoptosis inhibitory proteins,53 the fourth gene Traf1 (TNF receptor-associated factor 1) is also a negative regulator of apoptosis.81 The fifth gene in this group, Xaf1 (XIAP associated factor 1), is an antagonist of XIAP anti-caspase activity.82 It is expected that the net result of the downregulation of these 5 genes would be increased apoptosis, which is consistent with increased LE apoptosis in rodents during embryo implantation.2,16 Caspase-3 was previously shown to be detectable in a few LE cells at the implantation site on D4.5 but undetectable in the LE of interimplantation site or D3.5 LE,80 suggesting upregulation of caspase-3 in the D4.5 LE at the implantation site. Since the average readings of caspase-3 mRNA levels in the microarray are low (<200), it is possible that the main mechanism for LE apoptosis is regulation of caspase activity instead of mRNA levels, for example, through downregulation of apoptosis-negative regulators in the LE.
Potential Mechanisms of Gene Regulation in Periimplantation LE
Gene Ontology annotation reveals that the largest group of both upregulated and downregulated genes is involved in regulation of transcription (Figure 1; Supplemental Tables 2 and 3), indicating that transcriptional regulation is most likely the main mechanism for the molecular changes in the LE during periimplantation. Interestingly, “miRNA regulation of DNA damage response” is among the significantly changed signaling pathways (Table 1), and 9 genes in “chromosome organization” category are downregulated in the D4.5 LE (Supplemental Table 3), suggesting that epigenetic mechanisms, such as miRNAs and chromosome organization, could also be involved in regulating the molecular changes in the periimplantation LE. This speculation is supported by the observation that several miRNAs and their predicated target genes are differentially expressed in the human uterine epithelium during estrous cycle.79 In addition to continuous research on understanding the LE gene network in the establishment of uterine receptivity and how LE communicates with other uterine compartments in the establishment of uterine receptivity, research on understanding the molecular mechanisms, including epigenetic mechanisms, of how the LE gene network is regulated during initial implantation stages is another important direction for deciphering uterine function in embryo implantation.
Acknowledgments
The authors thank Drs James N. Moore and Zhen Fu in the College of Veterinary Medicine, University of Georgia for access to the ABI 7900-Real-Time PCR machine and the imaging system, respectively; the Emory Biomarker Service Center for microarray analysis; and the Office of the Vice President for Research.
Footnotes
Authors’ Note: The work was done at Department of Physiology and Pharmacology, College of Veterinary Medicine, University of Georgia. SX and HD contributed equally to the work.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Interdisciplinary Toxicology Program, and Department of Physiology & Pharmacology at University of Georgia, and National Institutes of Health (NIH) R01HD065939 and NIH R15HD066301 (to XY).
Supplemental Material: The online tables are available at http://rsx.sagepub.com/supplemental.
References
- 1. Paria BC, Reese J, Das SK, Dey SK. Deciphering the cross-talk of implantation: advances and challenges. Science. 2002;296(5576):2185–2188 [DOI] [PubMed] [Google Scholar]
- 2. Wang H, Dey SK. Roadmap to embryo implantation: clues from mouse models. Nat Rev Genet. 2006;7(3):185–199 [DOI] [PubMed] [Google Scholar]
- 3. Dey SK, Lim H, Das SK, et al. Molecular cues to implantation. Endocr Rev. 2004;25(3):341–373 [DOI] [PubMed] [Google Scholar]
- 4. Sharkey AM, Smith SK. The endometrium as a cause of implantation failure. Best Pract Res Clin Obstet Gynaecol. 2003;17(2):289–307 [DOI] [PubMed] [Google Scholar]
- 5. Yochim JM, De Feo VJ. Hormonal control of the onset, magnitude and duration of uterine sensitivity in the rat by steroid hormones of the ovary. Endocrinology. 1963;72(2):317–326 [DOI] [PubMed] [Google Scholar]
- 6. Psychoyos A. Hormonal control of ovoimplantation. Vitam Horm. 1973;31:201–256 [DOI] [PubMed] [Google Scholar]
- 7. Yoshinaga K. Effect of local application of ovarian hormones on the delay in implantation in lactating rats. J Reprod Fertil. 1961;2:35–41 [DOI] [PubMed] [Google Scholar]
- 8. Finn CA, Martin L. The control of implantation. J Reprod Fertil. 1974;39(1):195–206 [DOI] [PubMed] [Google Scholar]
- 9. Denker HW. Implantation: a cell biological paradox. J Exp Zool. 1993;266(6):541–558 [DOI] [PubMed] [Google Scholar]
- 10. Cowell TP. Implantation and development of mouse eggs transferred to the uteri of non-progestational mice. J Reprod Fertil. 1969;19(2):239–245 [DOI] [PubMed] [Google Scholar]
- 11. Lejeune B, Van Hoeck J, Leroy F. Transmitter role of the luminal uterine epithelium in the induction of decidualization in rats. J Reprod Fertil. 1981;61(1):235–240 [DOI] [PubMed] [Google Scholar]
- 12. Parr EL, Tung HN, Parr MB. Apoptosis as the mode of uterine epithelial-cell death during embryo implantation in mice and rats. Biol Reprod. 1987;36(1):211–225 [DOI] [PubMed] [Google Scholar]
- 13. Enders Allen C, Schlafke S. A morphological analysis of the early implantation stages in the rat. Am J Anat. 1967;120(2):185–225 [Google Scholar]
- 14. Paria BC, Song H, Dey SK. Implantation: molecular basis of embryo—uterine dialogue. Int J Dev Biol. 2001;45(3):597–605 [PubMed] [Google Scholar]
- 15. Martel D, Frydman R, Sarantis L, Roche D, Psychoyos A. Scanning electron microscopy of the uterine luminal epithelium as a marker of the implantation window. In: Koji Y, ed. Blastocyst Implantation. Boston, MA: Adams Publishing Group; 1989:225–230 [Google Scholar]
- 16. Parr EL, Parr MB. Epithelial cell death during rodent embryo implantation. In: Yoshinaga K, ed. Blastocyst Implantation. Boston, MA: Adams Publishing Group; 1989:105–115 [Google Scholar]
- 17. Welsh AO, Enders AC. Chorioallantoic placenta formation in the rat: I. Luminal epithelial cell death and extracellular matrix modifications in the mesometrial region of implantation chambers. Am J Anat. 1991;192(3):215–231 [DOI] [PubMed] [Google Scholar]
- 18. Illingworth IM, Kiszka I, Bagley S, Ireland GW, Garrod DR, Kimber SJ. Desmosomes are reduced in the mouse uterine luminal epithelium during the preimplantation period of pregnancy: a mechanism for facilitation of implantation. Biol Reprod. 2000;63(6):1764–1773 [DOI] [PubMed] [Google Scholar]
- 19. Murphy CR. Uterine receptivity and the plasma membrane transformation. Cell Res. 2004;14(4):259–267 [DOI] [PubMed] [Google Scholar]
- 20. Aplin JD, Kimber SJ. Trophoblast-uterine interactions at implantation. Reprod Biol Endocrinol. 2004;2:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grummer R, Reuss B, Winterhager E. Expression pattern of different gap junction connexins is related to embryo implantation. Int J Dev Biol. 1996;40(1):361–367 [PubMed] [Google Scholar]
- 22. Han BC, Xia HF, Sun J, Yang Y, Peng JP. Retinoic acid-metabolizing enzyme cytochrome P450 26a1 (cyp26a1) is essential for implantation: functional study of its role in early pregnancy. J Cell Physiol. 2010;223(2):471–479 [DOI] [PubMed] [Google Scholar]
- 23. Paria BC, Das N, Das SK, Zhao X, Dileepan KN, Dey SK. Histidine decarboxylase gene in the mouse uterus is regulated by progesterone and correlates with uterine differentiation for blastocyst implantation. Endocrinology. 1998;139(9):3958–3966 [DOI] [PubMed] [Google Scholar]
- 24. Ni H, Ding NZ, Harper MJ, Yang ZM. Expression of leukemia inhibitory factor receptor and gp130 in mouse uterus during early pregnancy. Mol Reprod Dev. 2002;63(2):143–150 [DOI] [PubMed] [Google Scholar]
- 25. Ye X, Hama K, Contos JJ, et al. LPA3-mediated lysophosphatidic acid signalling in embryo implantation and spacing. Nature. 2005;435(7038):104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Daikoku T, Song H, Guo Y, et al. Uterine Msx-1 and Wnt4 signaling becomes aberrant in mice with the loss of leukemia inhibitory factor or Hoxa-10: evidence for a novel cytokine-homeobox-Wnt signaling in implantation. Mol Endocrinol. 2004;18(5):1238–1250 [DOI] [PubMed] [Google Scholar]
- 27. Pan H, Zhu L, Deng Y, Pollard JW. Microarray analysis of uterine epithelial gene expression during the implantation window in the mouse. Endocrinology. 2006;147(10):4904–4916 [DOI] [PubMed] [Google Scholar]
- 28. Niklaus AL, Pollard JW. Mining the mouse transcriptome of receptive endometrium reveals distinct molecular signatures for the luminal and glandular epithelium. Endocrinology. 2006;147(7):3375–3390 [DOI] [PubMed] [Google Scholar]
- 29. Tan J, Paria BC, Dey SK, Das SK. Differential uterine expression of estrogen and progesterone receptors correlates with uterine preparation for implantation and decidualization in the mouse. Endocrinology. 1999;140(11):5310–5321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cheon YP, Li Q, Xu X, DeMayo FJ, Bagchi IC, Bagchi MK. A genomic approach to identify novel progesterone receptor regulated pathways in the uterus during implantation. Mol Endocrinol. 2002;16(12):2853–2871 [DOI] [PubMed] [Google Scholar]
- 31. Diao H, Paria BC, Xiao S, Ye X. Temporal expression pattern of progesterone receptor in the uterine luminal epithelium suggests its requirement during early events of implantation. Fertil Steril. 2011;95(6):2087–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Song H, Lim H, Paria BC, et al. Cytosolic phospholipase A2alpha is crucial [correction of A2alpha deficiency is crucial] for 'on-time' embryo implantation that directs subsequent development. Development. 2002;129(12):2879–2889 [DOI] [PubMed] [Google Scholar]
- 33. Diao H, Xiao S, Zhao F, Ye X. Uterine luminal epithelium-specific proline-rich acidic protein 1 (PRAP1) as a marker for successful embryo implantation. Fertil Steril. 2010;94(7):2808–2811 e2801 [DOI] [PubMed] [Google Scholar]
- 34. Hayashi K, Erikson DW, Tilford SA, et al. Wnt genes in the mouse uterus: potential regulation of implantation. Biol Reprod. 2009;80(5):989–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reese J, Das SK, Paria BC, et al. Global gene expression analysis to identify molecular markers of uterine receptivity and embryo implantation. J Biol Chem. 2001;276(47):44137–44145 [DOI] [PubMed] [Google Scholar]
- 36. Yoshioka K, Matsuda F, Takakura K, et al. Determination of genes involved in the process of implantation: application of GeneChip to scan 6500 genes. Biochem Biophys Res Commun. 2000;272(2):531–538 [DOI] [PubMed] [Google Scholar]
- 37. Chen Y, Ni H, Ma XH, et al. Global analysis of differential luminal epithelial gene expression at mouse implantation sites. J Mol Endocrinol. 2006;37(1):147–161 [DOI] [PubMed] [Google Scholar]
- 38. Ye X, Herr DR, Diao H, Rivera R, Chun J. Unique uterine localization and regulation may differentiate LPA3 from other lysophospholipid receptors for its role in embryo implantation. Fertil Steril. 2011;95(6):2107–2113 e2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chu L, Scharf E, Kondo T. GeneSpringTM: tools for analyzing microarray expression data. Genome Inform. 2001;12(6):227–229 [Google Scholar]
- 40. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57 [DOI] [PubMed] [Google Scholar]
- 41. Xu X. Expression, Regulation and Function of Glutathione Peroxidase 3 in Mouse Uterus During Peri-Implantation Process [dissertation]. China: Northeast Agricultural University; 2012 [Google Scholar]
- 42. Forgac M. Structure and properties of the vacuolar (H+)-ATPases. J Biol Chem. 1999;274(19):12951–12954 [DOI] [PubMed] [Google Scholar]
- 43. Qi J, Wang YR, Forgac M. The vacuolar (H+)-ATPase: subunit arrangement and in vivo regulation. J Bioenerg Biomembr. 2007;39(5-6):423–426 [DOI] [PubMed] [Google Scholar]
- 44. Smith AN, Lovering RC, Futai M, et al. Revised nomenclature for mammalian vacuolar-type H(+)-ATPase subunit genes. Mol Cell. 2003;12(4):801–803 [DOI] [PubMed] [Google Scholar]
- 45. Chu AJ. Tissue factor, blood coagulation, and beyond: an overview. Int J Inflam. 2011;2011:367284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schneider E, Ryan TJ. Gamma-glutamyl hydrolase and drug resistance. Clin Chim Acta. 2006;374(1-2):25–32 [DOI] [PubMed] [Google Scholar]
- 47. Wysocka M, Spichalska B, Lesner A, et al. Substrate specificity and inhibitory study of human airway trypsin-like protease. Bioorg Med Chem. 2010;18(15):5504–5509 [DOI] [PubMed] [Google Scholar]
- 48. Yasuoka S, Ohnishi T, Kawano S, et al. Purification, characterization, and localization of a novel trypsin-like protease found in the human airway. Am J Respir Cell Mol Biol. 1997;16(3):300–308 [DOI] [PubMed] [Google Scholar]
- 49. Kido H, Okumura Y. Mspl/Tmprss13. Front Biosci. 2008;13(1):754–758 [DOI] [PubMed] [Google Scholar]
- 50. Lai A, Ghaffari A, Li Y, Ghahary A. Microarray-based identification of aminopeptidase N target genes in keratinocyte conditioned medium-stimulated dermal fibroblasts. J Cell Biochem. 2012;113(3):1061–1068 [DOI] [PubMed] [Google Scholar]
- 51. Ghosh M, McAuliffe B, Subramani J, Basu S, Shapiro LH. CD13 regulates dendritic cell cross-presentation and T cell responses by inhibiting receptor-mediated antigen uptake. J Immunol. 2012;188(11):5489–5499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Geering K. Function of FXYD proteins, regulators of Na, K-ATPase. J Bioenerg Biomembr. 2005;37(6):387–392 [DOI] [PubMed] [Google Scholar]
- 53. Yin Y, Huang WW, Lin C, Chen H, MacKenzie A, Ma L. Estrogen suppresses uterine epithelial apoptosis by inducing birc1 expression. Mol Endocrinol. 2008;22(1):113–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schauer R. Chemistry, metabolism, and biological functions of sialic acids. Adv Carbohydr Chem Biochem. 1982;40:131–234 [DOI] [PubMed] [Google Scholar]
- 55. Wickramasinghe S, Medrano JF. Primer on genes encoding enzymes in sialic acid metabolism in mammals. Biochimie. 2011;93(10):1641–1646 [DOI] [PubMed] [Google Scholar]
- 56. Ofman R, Speijer D, Leen R, Wanders RJ. Proteomic analysis of mouse kidney peroxisomes: identification of RP2p as a peroxisomal nudix hydrolase with acyl-CoA diphosphatase activity. Biochem J. 2006;393(pt 2):537–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Song MG, Bail S, Kiledjian M. Multiple Nudix family proteins possess mRNA decapping activity. RNA. 2013;19(3):390–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Uchida I. The role of native tropomyosin on the ATP contraction of glycerinated intestinal smooth muscle [in Japanese]. Sapporo Igaku Zasshi. 1970;37(2):123–132 [PubMed] [Google Scholar]
- 59. Lin JJ, Eppinga RD, Warren KS, McCrae KR. Human tropomyosin isoforms in the regulation of cytoskeleton functions. Adv Exp Med Biol. 2008;644:201–222 [DOI] [PubMed] [Google Scholar]
- 60. Helfman DM, Cheley S, Kuismanen E, Finn LA, Yamawaki-Kataoka Y. Nonmuscle and muscle tropomyosin isoforms are expressed from a single gene by alternative RNA splicing and polyadenylation. Mol Cell Biol. 1986;6(11):3582–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Paria BC, Ma W, Tan J, et al. Cellular and molecular responses of the uterus to embryo implantation can be elicited by locally applied growth factors. Proc Natl Acad Sci USA. 2001;98(3):1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Grummer R, Hewitt SW, Traub O, Korach KS, Winterhager E. Different regulatory pathways of endometrial connexin expression: preimplantation hormonal-mediated pathway versus embryo implantation-initiated pathway. Biol Reprod. 2004;71(1):273–281 [DOI] [PubMed] [Google Scholar]
- 63. Liang XH, Zhao ZA, Deng WB, et al. Estrogen regulates amiloride-binding protein 1 through CCAAT/enhancer-binding protein-beta in mouse uterus during embryo implantation and decidualization. Endocrinology. 2010;151(10):5007–5016 [DOI] [PubMed] [Google Scholar]
- 64. Diao H, Xiao S, Li R, Zhao F, Ye X. Distinct spatiotemporal expression of serine proteases prss23 and prss35 in periimplantation mouse uterus and dispensable function of prss35 in fertility. PLoS One. 2013;8(2):e56757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li Q, Kannan A, DeMayo FJ, et al. The antiproliferative action of progesterone in uterine epithelium is mediated by Hand2. Science. 2011;331(6019):912–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bany BM, Cross JC. Post-implantation mouse conceptuses produce paracrine signals that regulate the uterine endometrium undergoing decidualization. Dev Biol. 2006;294(2):445–456 [DOI] [PubMed] [Google Scholar]
- 67. Bazer FW, Wu G, Johnson GA, Kim J, Song G. Uterine histotroph and conceptus development: select nutrients and secreted phosphoprotein 1 affect mechanistic target of rapamycin cell signaling in ewes. Biol Reprod. 2011;85(6):1094–1107 [DOI] [PubMed] [Google Scholar]
- 68. Gao H, Wu G, Spencer TE, et al. Select nutrients in the ovine uterine lumen. I. Amino acids, glucose, and ions in uterine lumenal flushings of cyclic and pregnant ewes. Biol Reprod. 2009;80(1):86–93 [DOI] [PubMed] [Google Scholar]
- 69. Gao H, Wu G, Spencer TE, Johnson GA, Bazer FW. Select nutrients in the ovine uterine lumen. ii. glucose transporters in the uterus and peri-implantation conceptuses. Biol Reprod. 2009;80(1):94–104 [DOI] [PubMed] [Google Scholar]
- 70. Gao H, Wu G, Spencer TE, Johnson GA, Bazer FW. Select nutrients in the ovine uterine lumen. III. Cationic amino acid transporters in the ovine uterus and peri-implantation conceptuses. Biol Reprod. 2009;80(3):602–609 [DOI] [PubMed] [Google Scholar]
- 71. Gao H, Wu G, Spencer TE, Johnson GA, Bazer FW. Select nutrients in the ovine uterine lumen. IV. Expression of neutral and acidic amino acid transporters in ovine uteri and peri-implantation conceptuses. Biol Reprod. 2009;80(6):1196–1208 [DOI] [PubMed] [Google Scholar]
- 72. Lessey BA. Adhesion molecules and implantation. J Reprod Immunol. 2002;55(1-2):101–112 [DOI] [PubMed] [Google Scholar]
- 73. Singh H, Aplin JD. Adhesion molecules in endometrial epithelium: tissue integrity and embryo implantation. J Anat. 2009;215(1):3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yoshinaga K. Two concepts on the immunological aspect of blastocyst implantation. J Reprod Dev. 2012;58(2):196–203 [DOI] [PubMed] [Google Scholar]
- 75. Ichii O, Otsuka S, Sasaki N, et al. Local overexpression of interleukin-1 family, member 6 relates to the development of tubulointerstitial lesions. Lab Invest. 2010;90(3):459–475 [DOI] [PubMed] [Google Scholar]
- 76. Langhauser F, Gob E, Kraft P, et al. Kininogen deficiency protects from ischemic neurodegeneration in mice by reducing thrombosis, blood-brain barrier damage, and inflammation. Blood. 2012;120(19):4082–4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Asare Y, Schmitt M, Bernhagen J. The vascular biology of macrophage migration inhibitory factor (MIF). Expression and effects in inflammation, atherogenesis and angiogenesis. Thromb Haemost. 2013;109(3):391–398 [DOI] [PubMed] [Google Scholar]
- 78. Xu C, Reichert EC, Nakano T, et al. Deficiency of phospholipase A2 group 7 decreases intestinal polyposis and colon tumorigenesis in Apc Min/+ mice. Cancer Res. 2013;73(9):2806–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kuokkanen S, Chen B, Ojalvo L, Benard L, Santoro N, Pollard JW. Genomic profiling of microRNAs and messenger RNAs reveals hormonal regulation in microRNA expression in human endometrium. Biol Reprod. 2010;82(4):791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhang Q, Paria BC. Importance of uterine cell death, renewal, and their hormonal regulation in hamsters that show progesterone-dependent implantation. Endocrinology. 2006;147(5):2215–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Speiser DE, Lee SY, Wong B, et al. A regulatory role for TRAF1 in antigen-induced apoptosis of T cells. J Exp Med. 1997;185(10):1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liston P, Fong WG, Kelly NL, et al. Identification of XAF1 as an antagonist of XIAP anti-caspase activity. Nat Cell Biol. 2001;3(2):128–133 [DOI] [PubMed] [Google Scholar]