Abstract
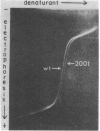
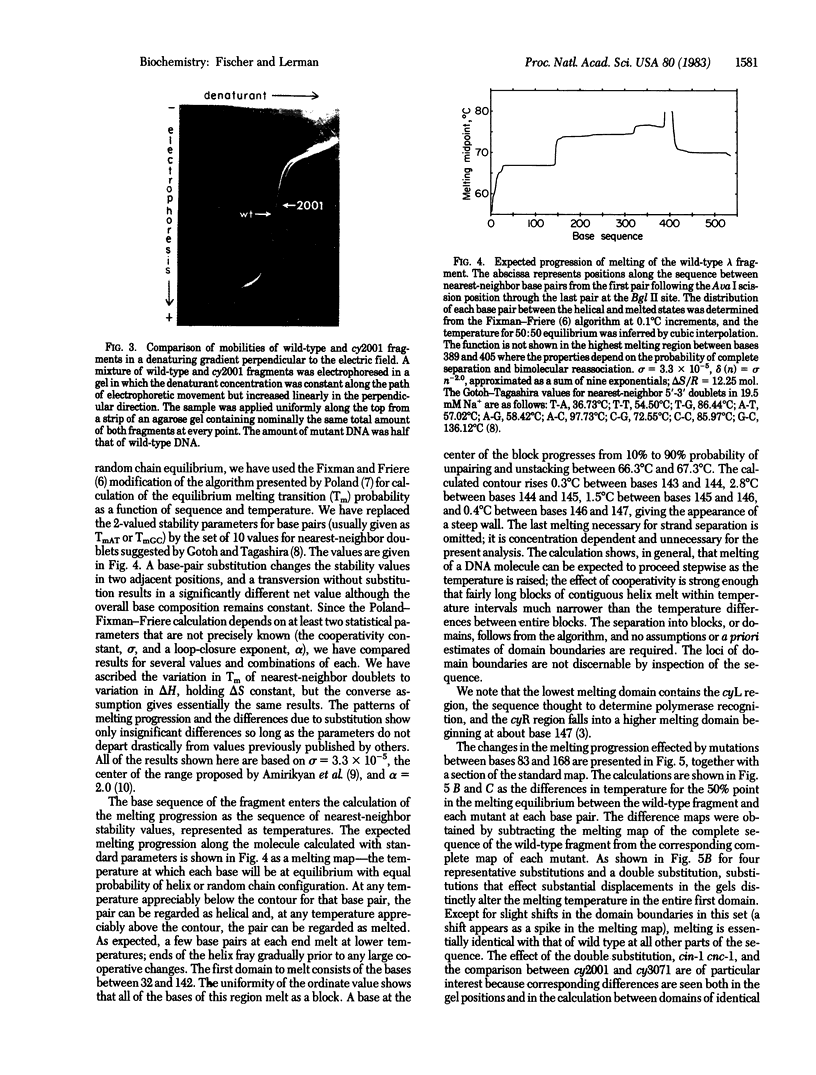
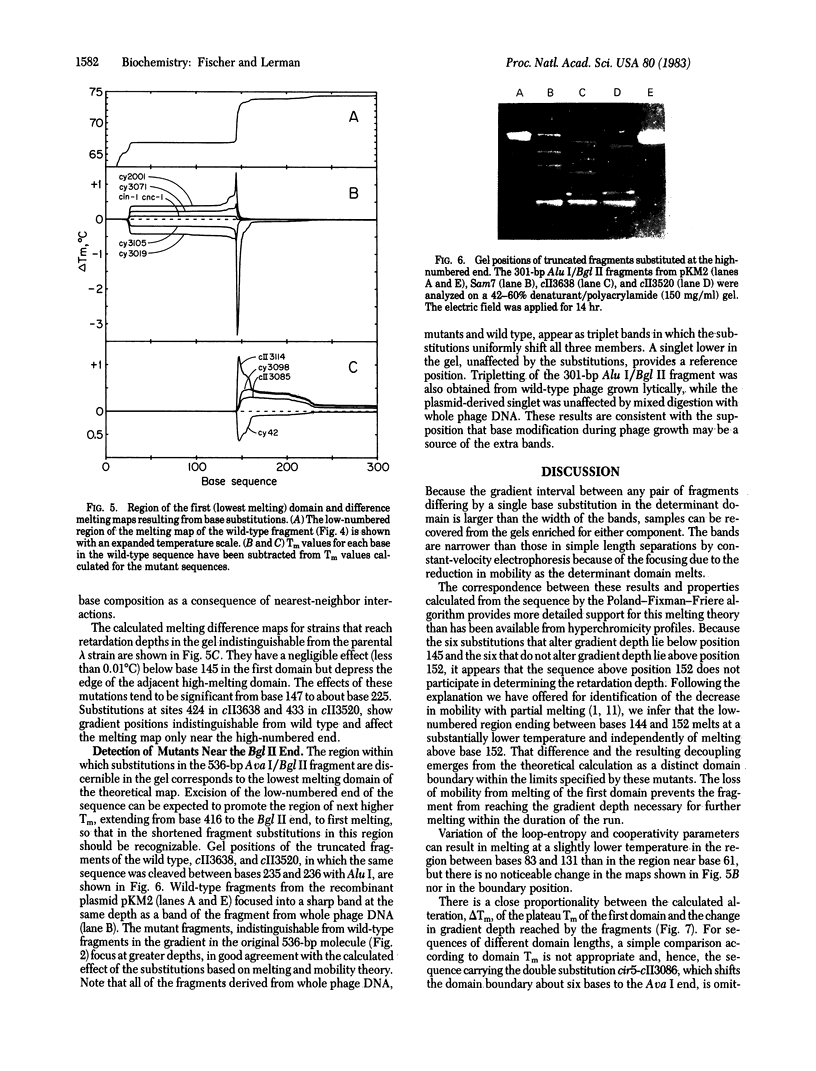
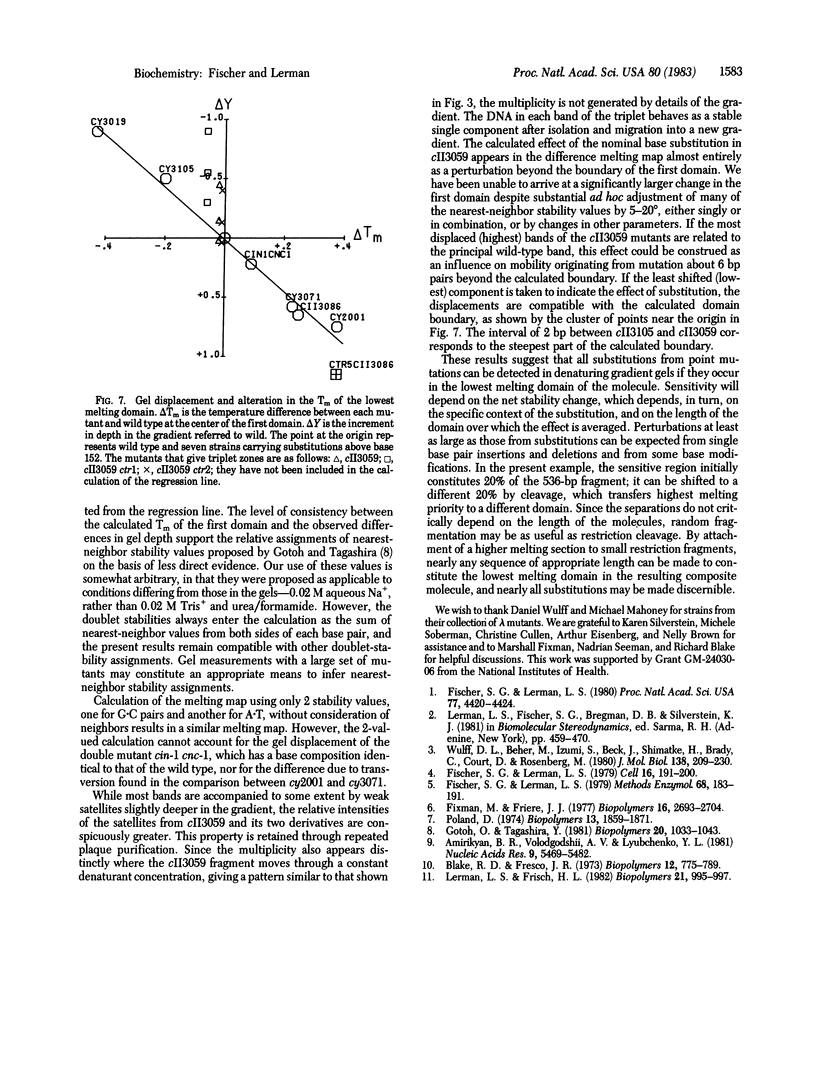
DNA fragments 536 base pairs long differing by single base-pair substitutions were clearly separated in denaturing gradient gel electrophoresis. Transversions as well as transitions were detected. The correspondence between the gradient gel measurements and the sequence-specific statistical mechanical theory of melting shows that mutations affecting final gradient penetration lie within the first cooperatively melting sequence. Fragments carrying substitutions in domains melting at a higher temperature reach final gel positions indistinguishable from wild type. The gradient data and the sites of substitution bracket the boundary between the first domain and its neighboring higher-melting domain within eight base pairs or fewer, in agreement with the calculated boundary. The correspondence between the gradient displacement of the mutants and the calculated change in helix stability permits substantial inference as to the type of substitution. Excision of the lowest melting domain allows recognition of mutants in the next ranking domain.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amirikyan B. R., Vologodskii A. V., Lyubchenko YuL Determination of DNA cooperativity factor. Nucleic Acids Res. 1981 Oct 24;9(20):5469–5482. doi: 10.1093/nar/9.20.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake R. D., Fresco J. R. Polynucleotides. XI. Thermodynamics of (A) N -2(U) infinity from the dependence of T m N on oligomer length. Biopolymers. 1973 Apr;12(4):775–786. doi: 10.1002/bip.1973.360120407. [DOI] [PubMed] [Google Scholar]
- Fischer S. G., Lerman L. S. Length-independent separation of DNA restriction fragments in two-dimensional gel electrophoresis. Cell. 1979 Jan;16(1):191–200. doi: 10.1016/0092-8674(79)90200-9. [DOI] [PubMed] [Google Scholar]
- Fischer S. G., Lerman L. S. Separation of random fragments of DNA according to properties of their sequences. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4420–4424. doi: 10.1073/pnas.77.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S. G., Lerman L. S. Two-dimensional electrophoretic separation of restriction enzyme fragments of DNA. Methods Enzymol. 1979;68:183–191. doi: 10.1016/0076-6879(79)68013-8. [DOI] [PubMed] [Google Scholar]
- Fixman M., Freire J. J. Theory of DNA melting curves. Biopolymers. 1977 Dec;16(12):2693–2704. doi: 10.1002/bip.1977.360161209. [DOI] [PubMed] [Google Scholar]
- Gotoh O., Tagashira Y. Locations of frequently opening regions on natural DNAs and their relation to functional loci. Biopolymers. 1981 May;20(5):1043–1058. doi: 10.1002/bip.1981.360200514. [DOI] [PubMed] [Google Scholar]
- Lerman L. S., Frisch H. L. Why does the electrophoretic mobility of DNA in gels vary with the length of the molecule? Biopolymers. 1982 May;21(5):995–997. doi: 10.1002/bip.360210511. [DOI] [PubMed] [Google Scholar]
- Poland D. Recursion relation generation of probability profiles for specific-sequence macromolecules with long-range correlations. Biopolymers. 1974;13(9):1859–1871. doi: 10.1002/bip.1974.360130916. [DOI] [PubMed] [Google Scholar]
- Wulff D. L., Beher M., Izumi S., Beck J., Mahoney M., Shimatake H., Brady C., Court D., Rosenberg M. Structure and function of the cy control region of bacteriophage lambda. J Mol Biol. 1980 Apr;138(2):209–230. doi: 10.1016/0022-2836(80)90284-3. [DOI] [PubMed] [Google Scholar]