The homing endonuclease I-CvuI from C. vulgaris has been expressed, purified and crystallized in complex with its target DNA. Preliminary X-ray diffraction analysis is reported.
Keywords: gene targeting, protein–DNA interaction, Chlorella vulgaris, homing endonuclease I-CvuI
Abstract
Homing endonucleases are highly specific DNA-cleaving enzymes that recognize long stretches of DNA. The engineering of these enzymes provides novel instruments for genome modification in a wide range of fields, including gene targeting, by inducing specific double-strand breaks. I-CvuI is a homing endonuclease from the green alga Chlorella vulgaris. This enzyme was purified after overexpression in Escherichia coli. Crystallization experiments of I-CvuI in complex with its DNA target in the presence of Mg2+ yielded crystals suitable for X-ray diffraction analysis. The crystals belonged to the orthorhombic space group P212121, with unit-cell parameters a = 62.83, b = 83.56, c = 94.40 Å. The self-rotation function and the Matthews coefficient suggested the presence of one protein–DNA complex per asymmetric unit. The crystals diffracted to a resolution limit of 1.9 Å using synchrotron radiation.
1. Introduction
Homing endonucleases (HEs), also known as meganucleases, recognize long DNA sequences, generating an accurate double-strand break that can be used to promote gene targeting through homologous recombination. They are highly sequence-specific enzymes, with recognition sites of 12–45 base pairs in length. HEs display a very low frequency of cleavage even in whole genomes. Because of this rare-cutting property, they are very powerful tools for manipulation of the genomes of mammalian and plant cells (Choulika et al., 1994 ▶, 1995 ▶; Rouet et al., 1994 ▶). However, the use of meganuclease-induced recombination has long been limited by the repertoire of natural meganucleases.
HEs are grouped into several families, among which the LAGLIDADG family is the most abundant. The family name derives from the conserved amino-acid sequence motif containing the catalytic aspartic residue. The nucleases of this family cleave their DNA target along the minor groove to generate cohesive 4 bp 3′-OH overhangs. Two classes of LAGLIDADG nucleases can be found, one of which contains a single copy of the motif and act as homodimers which bind palindromic or near-palindromic DNA target sequences. The other class contains two copies of the LAGLIDADG motif and act as monomers which recognize and cleave nonpalindromic DNA sequences (Marcaida et al., 2010 ▶). Catalysis requires the conserved acidic residue at the C-termini of the two LAGLIDADG sequences in the active site and the coordination of divalent metal cations for phosphodiester hydrolysis (Chevalier et al., 2001 ▶). Members of the LAGLIDADG family have been engineered to specifically target new DNA sequences and are emerging as a powerful tool for gene targeting. The best example is I-CreI, which has been redesigned using a combinatorial approach for binding and cleaving new specific DNA sequences (Prieto et al., 2012 ▶).
I-CvuI (GenBank accession No. L43357) is a homodimeric LAGLIDADG homing endonuclease from the chloroplast large-subunit ribosomal RNA gene of the green alga Chlorella vulgaris. This enzyme recognizes and cleaves a 22 bp DNA palindromic sequence. Here, we report the crystallization of I-CvuI in complex with its DNA target in the presence of Mg2+ as a first step towards solving its structure. The structure determination of this HE may expand the current possibilities for engineering custom specificities using I-CvuI as a new scaffold either alone or in hybrid constructs with I-CreI, I-PakI or I-MsoI proteins, taking into account that all are encoded by group I introns in site 2593 of the large subunit of the rRNA gene (Lucas et al., 2001 ▶).
2. Materials and methods
2.1. Protein expression and purification
Escherichia coli BL21 Rosetta pLysS cells were transformed with a pET-24d(+) vector containing the gene coding for the I-CvuI protein. The polypeptide chain expressed is the UniProt Q8WL02 sequence with a six-His tag at the C-terminus. Protein expression tests in Luria–Bertani (LB) medium showed high levels of protein production at 310 K. Therefore, cells were grown at this temperature in LB medium and gene expression was induced by the addition of 0.3 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 3 h at 310 K. Cells were harvested by centrifugation (at 9000g for 20 min at 277 K) and resuspended in 50 mM sodium phosphate pH 8.0, 300 mM NaCl, 5%(v/v) glycerol containing protease inhibitors (Complete EDTA-free tablets, Roche). The cells were disrupted by sonication and the cell lysate was ultracentrifuged at 12 000g for 30 min at 277 K. The protein was predominantly present in the pellet. Nevertheless, the amount of protein in the soluble fraction was sufficient to set up a purification procedure using four consecutive chromatography steps.
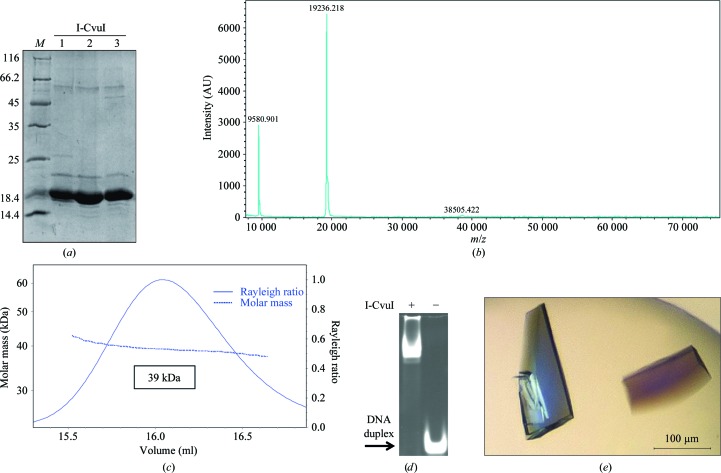
Firstly, the supernatant was applied onto a 5 ml HiTrap Chelating HP (GE Healthcare) column loaded with Co2+ ions and equilibrated in lysis buffer. The column was washed with lysis buffer [50 mM sodium phosphate pH 8.0, 300 mM NaCl, 5%(v/v) glycerol] plus 32 mM imidazole and eluted with the same buffer plus 800 mM imidazole. Enriched protein fractions were diluted twofold with 20 mM sodium phosphate pH 7.0 to reduce the imidazole and salt concentrations and were applied onto a 5 ml HiTrap Heparin HP column (GE Healthcare) equilibrated with 20 mM sodium phosphate pH 7.0, 150 mM NaCl. The protein was eluted with a linear gradient from 0 to 50% 20 mM sodium phosphate pH 7.0, 2 M NaCl in 15 column volumes. Protein-rich fractions were collected and loaded onto a HiLoad 16/60 200 Superdex column (GE Healthcare) equilibrated with 20 mM sodium phosphate pH 7.0, 500 mM NaCl (the amount of salt in the pool of protein fractions eluted from the HiTrap Heparin column was estimated from conductivity measurements). The main peak (centred at 86 ml) was diluted fivefold with 20 mM sodium phosphate pH 7.0 to reduce the salt concentration to 100 mM and was applied onto a 1 ml Mono S 5/50 GL column (GE Healthcare) equilibrated with 20 mM sodium phosphate pH 7.0, 100 mM NaCl. The protein was eluted with a linear gradient from 0 to 20% 20 mM sodium phosphate pH 7.0, 2 M NaCl in 25 column volumes. The protein eluted at a salt concentration of around 300 mM as estimated from the conductivity measurements. In some chromatograms a shoulder or even a separate peak was distinguished from the main peak and was collected separately. This minor species behaved as the major peak on SDS–PAGE. The protein peaks were concentrated (using 3 kDa cutoff Centriprep Amicon Ultra devices), flash-frozen in liquid nitrogen and stored at 193 K. The protein concentration was determined using the theoretical molar extinction coefficient at 280 nm calculated from the amino-acid composition. An overload SDS–PAGE stained with SimplyBlue SafeStain (Invitrogen) showed the protein preparation (Fig. 1 ▶ a). The molar mass measured by mass spectrometry (MALDI) indicated that the initial methionine was not present in the purified protein (Fig. 1 ▶ b). For crystallization and enzymatic activity measurements, the protein buffer was exchanged to 20 mM Tris–HCl pH 8.0, 150 mM NaCl using a PD-10 desalting column (GE Healthcare). Finally, the protein was concentrated to 24.12 mg ml−1 using an Amicon Ultra system equipped with a 10 kDa cutoff filter, aliquoted, flash-frozen in liquid nitrogen and stored at 193 K.
Figure 1.
I-CvuI expression, purification and crystallization. (a) SDS–PAGE gel showing the purified protein. Lane M contains molecular-mass marker (labelled in kDa). (b) MALDI of I-CvuI shows the molecular mass of the monomer. A small peak at 38 505 Da arising from the dimer can also be observed. (c) SEC–MALLS chromatogram of the I-CvuI protein preparation. (d) Band-shift assay. The DNA target was incubated with and without I-CvuI protein for 1 h at 298 K. The mixture was separated on a native polyacrylamide gel. The arrow indicates the band corresponding to free DNA target. (e) I-CvuI–DNA complex crystals grown in the presence of 2 mM Mg2+.
2.2. Size-exclusion chromatography–multi-angle laser light scattering
These experiments were performed at 293 K using a Superdex 200 10/300 GL column (GE Healthcare) attached to a DAWN HELEOS light-scattering detector and an Optilab rEX differential refractive-index detector (Wyatt Technology; Fig. 1 ▶ c). The column was equilibrated in 20 mM sodium phosphate pH 7.0, 500 mM NaCl, 0.03%(w/v) NaN3 and the SEC–MALLS system was calibrated with a sample of BSA at 1 g l−1 in the same buffer. A sample of 100 µl of I-CvuI at 1.7 g l−1 in the same buffer was injected and run through the column at a flow rate of 0.5 ml min−1. Data acquisition and analysis were carried out using the ASTRA software (Wyatt). Based on numerous measurements on BSA samples under the same or similar conditions, we estimate that the experimental error in the molar mass is around 5%.
2.3. I-CvuI–DNA complex formation
The I-CvuI target was purchased from Eurofins MWG Operon and consisted of one palindromic strand of sequence 5′-TCAGAACGTCGTACGACGTTCTGA-3′. The target sequence was initially chosen according to a previously reported DNA sequence (Lucas et al., 2001 ▶). This target was optimized after testing enzyme cleavage in a yeast screen assay (Smith et al., 2006 ▶). The construct forms a 24 bp blunt-end duplex after heating and cooling. The I-CvuI–DNA complex was obtained in the presence of 2 mM MgCl2 by pre-warming the meganuclease and the oligonucleotide samples at 310 K and mixing them in a 1.5:1 molar ratio (DNA:protein). The mixture was incubated for 50 min at this temperature and then spun down for 10 min to remove insoluble material. The supernatant was stored at 293 K to avoid precipitation. The final concentration of I-CvuI in the DNA–protein complex solution was 4 mg ml−1. A band-shift assay was used to test the binding of I-CvuI to the oligonucleotide (Fig. 1 ▶ d). The protein in the same buffer was incubated with the DNA duplex as previously described and loaded onto a native polyacrylamide gel.
2.4. Crystallization
Crystallization screenings were performed immediately after complex formation using a Cartesian MicroSys robot (Genomic Solutions) and the sitting-drop method in 96-well MRC plates with nanodrops of 0.1 µl protein solution plus 0.1 µl reservoir solution and a reservoir volume of 60 µl. The initial screens tested were Crystal Screen, Crystal Screen 2, Crystal Screen Cryo and Crystal Screen Lite (Hampton Research) and Wizard Cryo I and II (Emerald BioSystems). Crystals were obtained in nanodrops under several conditions (Crystal Screen 2 conditions 17, 24 and 48, Crystal Screen Lite condition 2, Wizard Cryo I conditions 3, 19, 27, 28 and 35, and Wizard Cryo II conditions 8, 11, 12, 29, 31 and 35). The small crystals grown in normal conditions were collected under Al’s oil and all crystals were tested for diffraction using synchrotron radiation. The best diffracting crystals were obtained using condition 12 of Emerald BioSystems Wizard Cryo II (40% 1,2-propanediol, 0.1 M citrate pH 5.5, 0.2 M NaCl). The initial crystals were only reproduced using the sitting-drop method with drops of 0.2 µl mixed with an equal volume of well solution, producing crystals with maximum dimensions of 0.2 × 0.1 × 0.025 mm (Fig. 1 ▶ e). These crystals, which appeared after 3 d, grew during 15 d and diffracted to 1.9 Å resolution.
2.5. DNA in vitro cleavage assay conditions
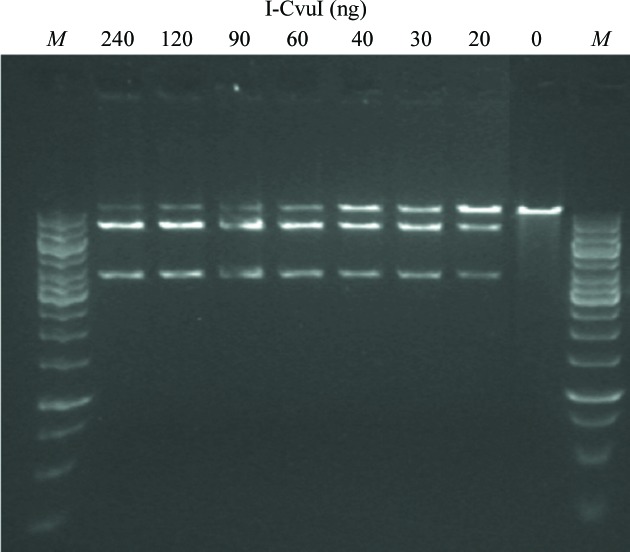
Cleavage assays were performed at 310 K in 10 mM Tris–HCl pH 8, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT. The amounts of enzyme and target were 100 ng of the BamHI-linearized DNA substrate and 0–240 ng dilutions of I-CvuI in a 25 ml final reaction volume (Fig. 2 ▶). The linearized target plasmid is 10 kb and after cleavage yields two smaller bands of 7 and 3 kb. Reactions were stopped after 1 h by adding 5 µl 45% glycerol, 95 mM EDTA pH 8, 1.5%(w/v) SDS, 1.5 mg ml−1 proteinase K, 0.048%(w/v) bromophenol blue (6× buffer stop), incubated at 310 K for 30 min and electrophoresed on a 0.6% agarose gel. The gels were stained with SYBR Safe (Invitrogen).
Figure 2.
DNA cleavage in vitro. 100 ng of the BamHI-linearized plasmid substrate containing the target was cleaved by adding increasing amounts of I-CvuI ranging from 0 to 240 ng. Lanes M contain molecular-mass marker.
2.6. Data collection
Crystals were removed from the drop and flash-cooled in liquid nitrogen. The diffraction data in Table 1 ▶ were recorded using a Pilatus 6M detector both on the PX beamline at the Swiss Light Source (SLS, Villigen, Switzerland) and on XALOC (ALBA, Barcelona, Spain). The best data set was collected using the fine ϕ-slicing method with Δϕ = 0.2° and a wavelength of 1.0 Å. Processing and scaling were accomplished using XDS (Kabsch, 2010 ▶) and SCALA from the CCP4 package (Evans, 2006 ▶; Winn et al., 2011 ▶). Data-collection statistics are summarized in Table 1 ▶.
Table 1. Data-collection statistics for the native I-CvuI–DNA crystals grown in the presence of 2 mM Mg2+ .
Values in parentheses are for the outer resolution shell.
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 62.83, b = 83.56, c = 94.40 |
| Temperature (K) | 100 |
| Wavelength (Å) | 1.00 |
| Beamline | XS06A, SLS |
| Resolution (Å) | 47.20–1.90 (2.00–1.90) |
| Total reflections | 357627 |
| Unique reflections | 39337 |
| Multiplicity | 9.1 (8.3) |
| Completeness (%) | 98.8 (95.6) |
| Mean I/σ(I) | 12.9 (3.9) |
| R merge † | 0.083 (0.42) |
R merge is defined according to XDS (Kabsch, 2010 ▶).
3. Results and discussion
The I-CvuI nuclease was expressed in E. coli, the recombinant protein was purified using four consecutive chromatography steps and the soluble fraction was subjected to HiTrap Chelating HP loaded with Co2+ and HiTrap Heparin HP affinity purification columns, a gel-filtration purification step in a HiLoad 16/60 200 Superdex column and a final polishing step using a Mono S 5/50 GL column. The recombinant I-CvuI construct used in this work is 173 residues in length, with a theoretical molecular mass of 19 392 Da and a pI of 9.21. However, mass spectrometry shows a single-charged peak of purified protein that has a measured molecular mass of 19 236 Da (Fig. 1 ▶ b), indicating that the initial methionine was processed during expression. The SEC–MALLS experiment shows that I-CvuI behaves as a homodimer in solution (Fig. 1 ▶ c), since its measured molar mass is 39 000 Da. The isolated enzyme cleaves its DNA target sequence, as shown in an in vitro cleavage assay (Fig. 2 ▶), but displays very low activity towards the DNA target of I-CreI, another LAGLIDADG meganuclease, which differs at only two bases (−9 and +10) from the known I-CvuI DNA target. In contrast, I-CreI is able to cleave the I-CvuI DNA target with high efficiency.
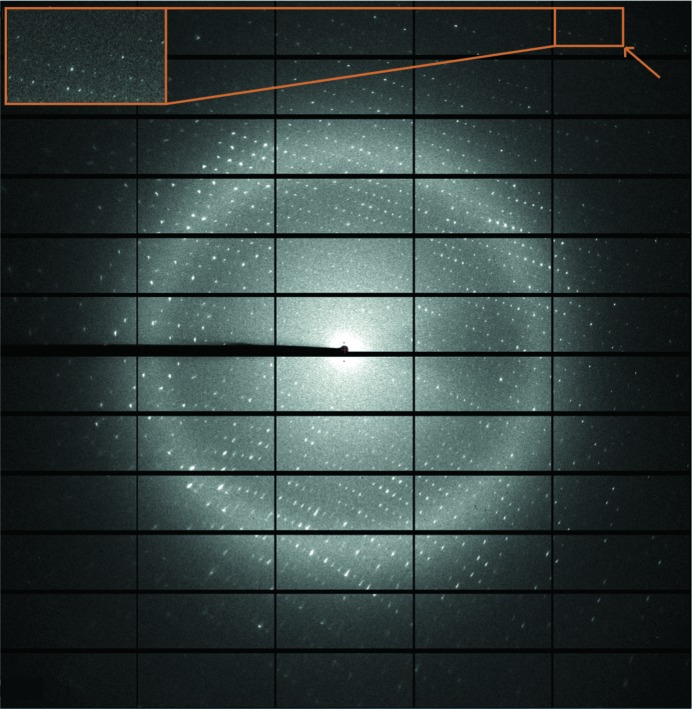
The crystal of I-CvuI in complex with its target DNA in the presence of 2 mM Mg2+ belonged to space group P212121, with unit-cell parameters a = 62.83, b = 83.56, c = 94.40 Å, α = β = γ = 90°. The Matthews coefficient (V M = 2.48 Å3 Da−1; Matthews, 1968 ▶) and the self-rotation function (data not shown) suggested the presence of one protein–DNA complex per asymmetric unit and a solvent content of 56%. The diffraction data (Fig. 3 ▶) were 98.8% complete, with a multiplicity of 9.1 and an overall mean I/σ(I) of 12.9. Detailed statistics of the data set are summarized in Table 1 ▶.
Figure 3.
X-ray diffraction pattern of I-CvuI–DNA complex crystals. The inset indicated by an arrow shows the high-resolution reflections at 1.9 Å resolution.
These are the first crystals obtained of I-CvuI in complex with DNA. We believe that these studies will help to elucidate the molecular mechanisms of DNA recognition and the cleavage mechanisms of this meganuclease. Initial attempts to solve the structure using molecular replacement are under way. These findings will facilitate the use of I-CvuI as a new scaffold that should help in the production of intelligent molecular scalpels that recognize and substitute certain DNA sequences.
Acknowledgments
We thank the Swiss Light Source and ALBA beamline staff for their support. This work was supported by Ministerio de Económia y Competitividad (JCI-2011-09308 to RM, BFU2011-23815/BMC to GM), the Fundación Ramón Areces and the Comunidad Autónoma de Madrid (CAM-S2010/BMD-2305 to GM).
References
- Chevalier, B. S., Monnat, R. J. Jr & Stoddard, B. L. (2001). Nature Struct. Biol. 8, 312–316. [DOI] [PubMed]
- Choulika, A., Perrin, A., Dujon, B. & Nicolas, J.-F. (1994). C. R. Acad. Sci. III, 317, 1013–1019. [PubMed]
- Choulika, A., Perrin, A., Dujon, B. & Nicolas, J.-F. (1995). Mol. Cell. Biol. 15, 1968–1973. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Lucas, P., Otis, C., Mercier, J.-P., Turmel, M. & Lemieux, C. (2001). Nucleic Acids Res. 29, 960–969. [DOI] [PMC free article] [PubMed]
- Marcaida, M. J., Muñoz, I. G., Blanco, F. J., Prieto, J. & Montoya, G. (2010). Cell. Mol. Life Sci. 67, 727–748. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Prieto, J., Molina, R. & Montoya, G. (2012). Crit. Rev. Biochem. Mol. Biol. 47, 207–221. [DOI] [PubMed]
- Rouet, P., Smih, F. & Jasin, M. (1994). Proc. Natl Acad. Sci. USA, 91, 6064–6068. [DOI] [PMC free article] [PubMed]
- Smith, J., Grizot, S., Arnould, S., Duclert, A., Epinat, J.-C., Chames, P., Prieto, J., Redondo, P., Blanco, F. J., Bravo, J., Montoya, G., Pâques, F. & Duchateau, P. (2006). Nucleic Acids Res. 34, e149. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.