Abstract
Homoprotocatechuate 2,3-dioxygenase (FeHPCD) activates O2 to catalyze the aromatic ring opening of 3,4-dihydroxyphenylacetic acid (HPCA). The enzyme requires FeII for catalysis, but MnII can be substituted (MnHPCD) with essentially no change in the steady-state kinetic parameters. Near simultaneous O2 and HPCA activation has been proposed to occur through transfer of an electron(s) from HPCA to O2 through the divalent metal. In O2 reactions with MnHPCD-HPCA and the 4-nitrocatechol (4NC) complex of the His200Asn (H200N) variant of FeHPCD, this transfer has resulted in the detection of a transient MIII-O2•− species not observed during turnover of the wild type FeHPCD. The factors governing formation of the MIII-O2•− species are explored here with EPR spectroscopy using MnHPCD and nitric oxide (NO) as an O2 surrogate. Both the HPCA and dihydroxymandelic substrate complexes of MnHPCD bind NO, thus representing the first reported stable MnNO complexes of a nonheme enzyme. In contrast, the free enzyme, the MnHPCD-4NC complex, and the MnH200N and MnH200Q variants with or without HPCA bound do not bind NO. The MnHPCD-ligand complexes that bind NO are also active in normal O2-linked turnover, whereas the others are inactive. Past studies have shown that FeHPCD and the analogous variants and catecholic ligand complexes all bind NO, and are active in normal turnover. This contrasting behavior may stem from ability of the enzyme to maintain the ~0.8 V difference in the solution redox potentials of FeII and MnII. Due to the higher potential of Mn, the formation of the NO or O2 adduct requires both strong charge donation from the bound catecholic ligand and additional stabilization by interaction with the active site His200. The same non-optimal electronic and structural forces that prevent NO and O2 binding in MnHPCD variants may lead to inefficient electron transfer from the catecholic substrate to the metal center in variants of FeHPCD during O2-linked turnover. Accordingly, past studies have shown that intermediate FeIII species are observed for these mutant enzymes.
Keywords: Homoprotocatechuate Dioxygenase, Manganese, Mn(II), Mn(III), Nitric Oxide, EPR, Electron Paramagnetic Resonance
Dioxygen is a strong oxidant but requires activation by a catalytic center to overcome kinetic reaction barriers. The activation of O2 by metalloenzymes typically involves an oxy-metal complex, the formation of which is often gated by substrate binding to the enzyme to ensure that the ensuing metal-oxo intermediates specifically oxidize substrate. A combination of primary and secondary metal coordination sphere interactions may control the formation of the oxy-metal complex. The choice of metal and ligands must provide a catalytic center that can act as a reductant for O2, whereas hydrogen bonding from vicinal residues can stabilize the transfer of electron density onto O2 from the metal. The oxy-metal complex is generally formulated as superoxide bound to a trivalent metal (MIII-O2•−), and consequently, its rate of formation and the strength of the complex formed should be highly dependent on the reduction potential of the metal. The one-electron reduction of dioxygen to superoxide at neutral pH requires a moderately strong reductant, which is one rationale for the propensity of nature to utilize Fe in enzymes for O2 activation. The reduction potential for MnIII/MnII is ~0.8 V more positive than FeIII/FeII for equivalent complexes of the two metals, which implies an O2 reactivity for Mn orders of magnitude lower than Fe.
Extradiol catecholic dioxygenases catalyze the cleavage of the aromatic ring of the substrate adjacent to the vicinal OH substituents with incorporation of both oxygen atoms from O2 [1]. This reaction is a key step in the ability of nature to reclaim large quantities of carbon sequestered in aromatic compounds. The active sites of these enzymes contain a divalent metal coordinated by a 2-His-1-carboxylate facial triad, which is a common motif of nonheme enzymes that activate dioxygen [2, 3]. Homoprotocatechuate 2,3-dioxygenase (HPCD) catalyzes the ring opening of homoprotocatechuate (HPCA). As isolated from Brevibacterium fuscum, HPCD contains FeII as the native metal ion, while the enzyme from Arthrobacter globiformis contains MnII. Despite this difference, the two enzymes have approximately equal turnover numbers (kcat ≈ 10 s−1) [4-6]. Moreover, further studies have demonstrated that interchanging the metals in the two enzymes does not significantly affect the steady state kinetic parameters [7].
Given the above mentioned intrinsic differences in metal redox potential, it is surprising that the Fe and Mn forms of the HPCD display the same steady state kinetics. The crystal structures of HPCD with Fe and Mn are the same within experimental error for the entire protein, indicating no significant difference in protein interaction with the metal [7, 8], and thus the large intrinsic difference in reduction potential for Fe versus Mn is approximately maintained in the protein environment. The steady-state parameter kcat /KMO2 is particularly relevant in the context of the present study, since it incorporates all of the rate constants for the reaction cycle steps involved in O2 activation. The constant value for this parameter for the Mn and Fe forms of the enzyme suggests that the flux through this critical part of the reaction cycle is metal independent. Consequently, a mechanism has been proposed that does not invoke a kinetically significant redox event during catalysis [7].
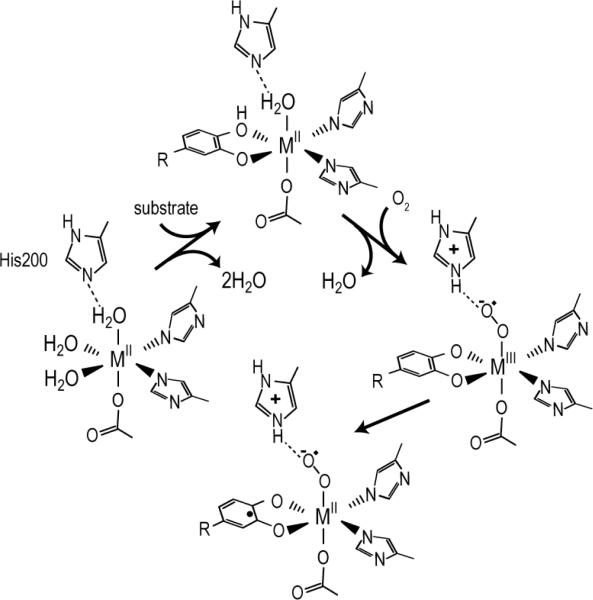
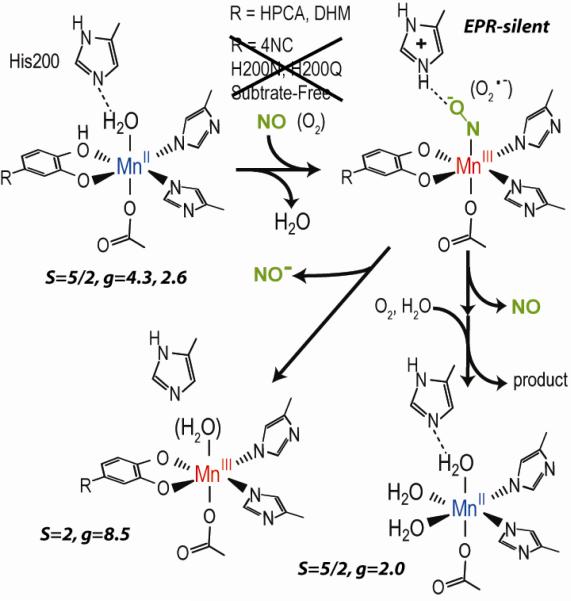
In accord with the mechanistic proposal, stopped flow (SF) and rapid freeze quench (RFQ) studies of the reaction of FeHPCD-HPCA complex with O2 reveal only FeII intermediates [9]. However, SF and RFQ studies have revealed transient MIII intermediates in MnHPCD and active site variants of FeHPCD. MnHPCD showed a short-lived intermediate upon reaction of MnHPCD-HPCA complex with O2 [10]. This intermediate was identified as a MnIII-radical species postulated to be MnIII-O2•−. It formed to a maximum of ~5% after a few milliseconds and then converted to a MnII intermediate within 30 ms. Figure 1 shows the steps of the mechanism that involve O2 activation. His200 is located adjacent to the metal active site in the secondary coordination sphere and, among other possible roles, is believed to stabilize the metal-superoxo intermediate. This intermediate has not been detected in the native form of the Fe enzyme, possibly because it is too short-lived. However, the H200N mutant of FeHPCD formed a long-lived FeIII-O2•− intermediate in nearly quantitative yield when the slow substrate 4-nitrocatechol (4NC) was used [11]. The use of 17O2 in reaction mixtures allowed measurement of the radical density on superoxide, which was found to be substantial, confirming the presence of the superoxide intermediate FeIII-O2•−. This intermediate decayed to release H2O2 and 4-nitro-o-benzoquinone rather than a ring-cleaved product, showing that the combined use of an active site His mutant and a substrate with an electron withdrawing substituent compromises the ability of FeIII-O2•− to attack the catecholic substrate. These studies show that a MIII-O2•− species can form in the active site of the enzyme, but leave open the question of whether such a species forms during normal catalysis of the Fe enzyme.
Figure 1.
Observed O2 activation steps for extradiol dioxygenases.
The O2 binding event is difficult to study as an isolated chemical step, since the ensuing reactions with substrate occur rapidly, and thus alternative probes of the reaction are desired. Nitric oxide (NO) has long been an important surrogate in studies of iron enzymes that activate O2 for several reasons [12]. NO is capable of binding to divalent metals with coordination geometries that are similar to those of O2. The reduction of a single NO by a metal is generally limited to one electron chemistry, allowing formation of a stable MIII-NO− complex [13]. NO can form hydrogen bonds with protein residues in a similar manner to O2.
Here we report for the first time NO binding to a nonheme enzyme containing Mn. This binding allows us to examine the factors governing the formation and detection of the MIII-O2•− species in all forms of HPCD. It is shown that NO, and presumably O2, binding are sensitive to both the nature of the catecholic substrate present and the nature of the active site amino acid residue at position 200. Because of the higher potential of the Mn-substituted enzyme, three factors are brought into focus: (1) the reduction potential of the active site metal, (2) the hydrogen bonding to His200, and (3) the protonation state of His200. The different catechols and enzyme variants used in this study allow these factors to be probed separately. The results provide insight into the forces that control the electron redistribution required for O2 activation and reaction with substrate in extradiol dioxygenases.
EXPERIMENTAL PROCEDURES
Reagents and Enzymes
Recombinant MnHPCD was prepared as previously described with minor changes [10]. Specifically, Mn was added to the cell cultures before and after induction of protein expression. The preparations had Mn incorporation of 0.8 equivalents per protein and no detectable Fe contamination, as determined by ICP-AES [7] and EPR spectroscopy. MnHPCD variants were produced using the over-expression vectors previously described for FeHPCD variants [14]. The cells were grown as described for MnHPCD and also purified using the same procedures. All samples were prepared with nominally 1 mM protein in 50 mM MOPS, pH 7.8 unless otherwise noted. HPCA was purchased from Sigma Aldrich and further purified with both a charcoal treatment and re-crystallization from ethyl acetate/hexane. Stock solutions of HPCA were brought to pH 7.8 with small additions of 1 M NaOH. HPCA solutions were prepared fresh daily and quantified by reacting an aliquot with MnHPCD to stoichiometrically form product, the concentration of which was determined spectrophotometrically (ε380 = 38,000 M−1 cm−1) [15]. Saturated O2-buffer was prepared by bubbling O2 from a tank into stirred buffer for no less than three hours. Anaerobic samples of MnHPCD were prepared by passing Ar(g) over the top of the sample as it was stirred at 4 °C for two hours in a sealed vial. Anaerobic buffer and substrate solutions were prepared on a Schlenk line with a minimum of five pump/purge cycles using a vacuum of 10 μTorr or better.
NO additions
Samples of MnHPCD were prepared anaerobically as described above. To these samples, NO additions were made with anaerobic solutions of diethylamine NONOate diethylammonium salts (DEA NONOates). The DEA NONOates were dissolved in rigorously degassed buffer and an aliquot was added to the protein sample shortly after dissolution, typically less than five minutes. The protein sample was then allowed to sit for twenty minutes at room-temperature to allow for the generation of NO from the dissociation of the DEA NONOates. EPR spectra were recorded before and after the addition of NO.
For the NO titration experiments, two methods were used. First, a weighed sample of DEA NONOates was dissolved in buffer and added rapidly to the protein sample as described for the normal NO addition. However, in this case, the sample was frozen immediately after addition and EPR spectra were recorded. The sample was then thawed on the Schlenk line and allowed to react at room temperature for the indicated time. The equivalents of NO were then determined based on the calculated rate of decay (half-life of 16 minutes) for the DEA NONOate solution. A second method involved dissolving DEA NONOates solution with rigorously degassed buffer and allowing this solution to sit for greater than one hour to generate an NO saturated (3 mM) solution which was then titrated into an anaerobic protein sample. In one sample, an excess of NO was added to the protein by bubbling NO gas through a base bath and then passed over the sample for a period of 20 minutes.
EPR Spectroscopy
X-band EPR spectra were recorded on a Bruker ESP 300 spectrometer equipped with an Oxford ESR 910 cryostat and a Bruker bimodal cavity for generation of microwave fields parallel and transverse to the static fields. The modulation amplitude and frequency was 1 mT at 100 kHz for all spectra. The microwave frequency was calibrated with a frequency counter and the magnetic field was calibrated with a NMR gaussmeter. The temperature was calibrated with a carbon glass resistor (Lakeshore CGR-1-1000). A CuEDTA spin standard was used to account for all relevant intensity factors. The EPR data of the mononuclear species were analyzed in terms of the standard Spin Hamiltonian (Eq. 1),
| (1) |
where all parameters have their standard meanings. The quantitative simulation of spectra use the windows software package SpinCount which allows determinations of spin concentrations.
RESULTS
Characterization of Enzyme and Enzyme-Substrate Complexes
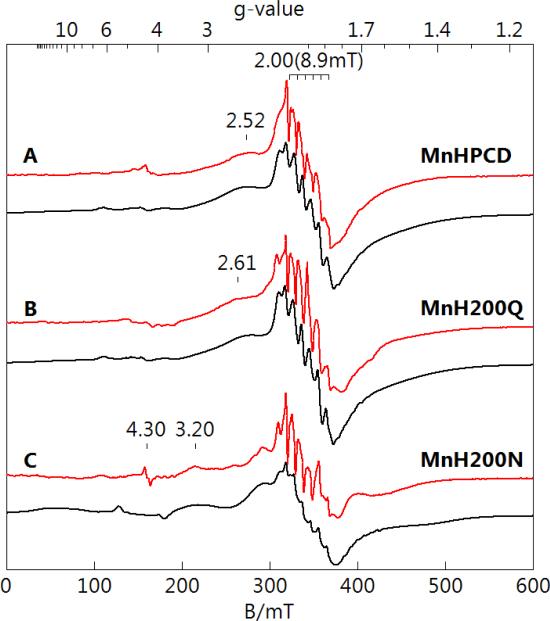
MnHPCD and variants MnH200Q and MnH200N were purified as described in Experimental Procedures. Low temperature EPR spectra (red lines) and simulations (black lines) are shown in Figure 2 for protein in 50 mM MOPS, pH 7.8. All spectra show a 6-line hyperfine pattern (a = 8.9 mT) from MnII at g = 2.00, which originates from the |±½〉 doublet of the S = 5/2 spin multiplet. These signals were clearly not from adventitious MnII as they vanished in the presence of substrate, whereas the addition of substrate to a buffered MnCl2 solution did not affect the sharp 6-line pattern of aquo-MnII. Furthermore, the addition of 0.1 eq. EDTA did not affect the intensity of the signals of Figure 2, indicating that no free MnII was available for chelation, as the EPR spectrum of MnII-EDTA did not display a 6-line pattern at g = 2.0. The spectrum of MnH200Q showed relatively small changes from that of MnHPCD. The resonance at g = 2.52 (from the |−3/2〉 to |−1/2〉 transition) shifted to g = 2.61. The spectrum of MnH200N was significantly different and consists of two species. The majority species (75%) has larger values of D and E/D (0.064 cm−1, 0.32) compared to the values of the minority species (25%), which is similar to that from MnHPCD. The larger D-value of the majority species shifted the |−3/2〉 to |−1/2〉 transition to g = 3.20. The simulations use Eq. 1 for the parameters given in Table 1, which allowed accurate determinations of the electronic symmetry parameters D and E/D of the metal environment, and the intrinsic distribution in these parameters (σ). The simulations were found to be in quantitative agreement with the Mn concentration in the samples determined from ICP-AES analysis.
Figure 2.
EPR spectra (red) and simulations (black) of (A) MnHPCD, (B) MnH200Q, and (C) MnH200N enzyme species. All spectral intensities are scaled for equal Mn concentrations, nominally 1 mM in protein, 50 mM MOPS, pH 7.8. The simulation parameters are given in Table 1. Instrumental conditions: temperature, 10 K; microwaves, 0.2 mW at 9.6 GHz.
Table 1.
Electronic parameters determined from simulation of EPR spectra, and NO binding or O2 reaction of MnHPCD and variants.
| Species | D/cm−1 (σ)a | E/D (σ) | NO/O2 reaction |
|---|---|---|---|
| MnHPCD | 0.055 (0.015) | 0.18 (0.05) | No |
| MnH200Q | 0.056 (0.015) | 0.19 (0.05) | No |
| MnH200Nb | 0.064, 0.055 | 0.32, 0.19 | No |
| MnH200N-HPCA | 0.077 (0.005) | 0.11 (0.01) | No |
| MnH200Q-HPCA | 0.082 (0.005) | 0.27 (0.03) | No |
| MnHPCD-HPCA | 0.090 (0.005) | 0.31 (0.03) | Yes |
| MnHPCD-DHM | 0.090 (0.005) | 0.31 (0.03) | Yes |
| MnHPCD-4NC | 0.125 (0.020) | 0.25 (0.03) | No |
σ value in parenthesis is width of distribution of the corresponding parameter.
parameters for two species.
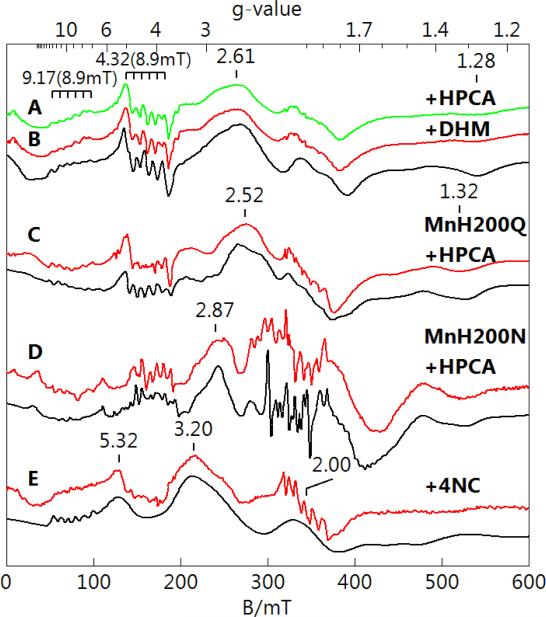
Enzyme samples were made anaerobic and 10 eq. of anaerobic substrate (or inhibitor in the case of 4NC) were added to form the enzyme-substrate complexes. The substrates and inhibitors are listed in Figure 3. EPR spectra (colored lines) and simulations (black lines) of these complexes are shown in Figure 4. The binding of substrate caused significant change to the spectra of all three species, most noticeably the loss of the g = 2 hyperfine pattern and the growth of a 6-line pattern near g = 4.3 from two transitions: . The simulations were generated with the parameters given in Table 1 and were found to be in quantitative agreement with the Mn concentration determined from ICP-AES analysis. The EPR spectrum of MnHPCD with dihydroxymandelic acid (DHM) (Figure 4B) was indistinguishable from that of MnHPCD-HPCA (Figure 4A). In contrast, the spectra of MnH200Q-HPCA (Figure 4C) and MnH200N-HPCA (Figure 4D) were notably and reproducibly different from that of MnHPCDHPCA.
Figure 3.
The substrates and inhibitors considered in this work shown in the relevant ionization state when bound to the enzyme.
Figure 4.
EPR spectra (red, green) and simulations (black) of MnHPCD and mutant enzymes after addition of substrates or inhibitors in concentrations of 10-fold in excess of protein. All spectral intensities are scaled for equal Mn concentration, nominally 1 mM in protein, 50 mM MOPS, pH 7.8. (A) MnHPCD-HPCA, (B) MnHPCD-DHM, (C) MnH200Q-HPCA, (D) MnH200N-HPCA, and (E) MnHPCD-4NC. The simulation parameters are given in Table 1. Instrumental conditions: temperature, 10 K; microwaves, 0.2 mW at 9.65 GHz.
The addition of 4NC to MnHPCD caused formation of a species (MnHPCD-4NC, Figure 4E) with EPR signals significantly different from those of the other enzyme-substrate complexes. The broad shoulder at g = 2.61 in the spectrum of MnHPCD-HPCA was shifted to g = 3.20, and the hyperfine pattern at g = 4.3 was significantly diminished. The simulation accounted for all of the observed features, with the exception of a 2% contaminating aquo-Mn(II) species at g = 2.00, and was in quantitative agreement with the Mn concentration determined by ICP-AES. MnHPCD-4NC was found to be stable upon exposure to air, consistent with 4NC being an inhibitor of MnHPCD.
Reaction of NO with enzyme complexes
Samples of MnHPCD and all the species shown in Figure 4 were prepared anaerobically and NO was added with anaerobic solutions of DEA NONOates as described in the Experimental Procedures. The addition of excess NO to MnHPCD, MnH200Q-HPCA, MnH200N–HPCA, and MnHPCD-4NC resulted in no change to the EPR signals. In contrast, NO addition to MnHPCD-HPCA and MnHPCD-DHM resulted in the loss of all EPR signals assigned to these species. Samples of these enzyme species were also assayed for reaction with O2. Table 1 lists the abilities of the various species to react with NO or O2, and as indicated, the reactivity with NO was found to parallel that of O2.
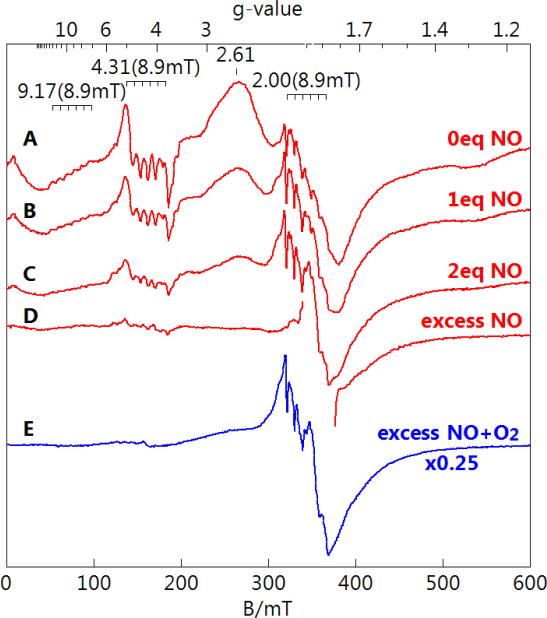
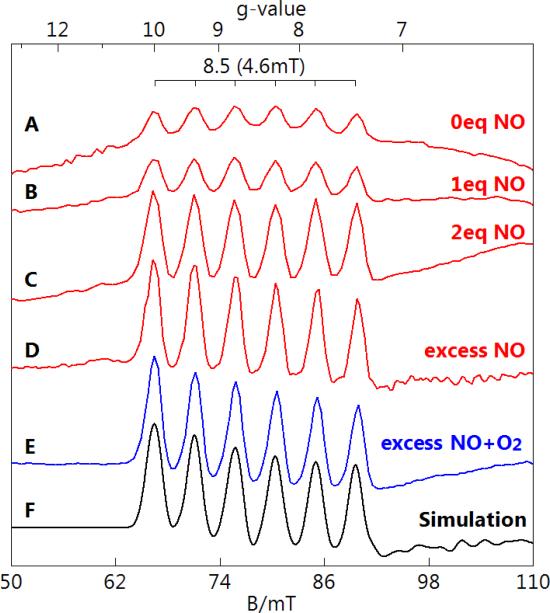
A titration of MnHPCD-HPCA with NO was performed as described in the Experimental Procedures. The addition of 0, 1 eq., 2 eq., and excess NO (NO gas bubbled over sample as described in Experimental Procedures) resulted in a decrease in the enzyme-substrate signal as shown in Figure 5, with relative signal intensities of 1, 0.57, 0.30, and 0.1, respectively. A small fraction (less than 15%) of MnHPCD without substrate bound was present in all titration points. The pH of samples was not significantly affected by the addition of NO. The spectrum with excess NO was made from a fresh protein sample. Anaerobic samples were then exposed to O2 and monitored optically for the rate of product production. The rate of product production was not affected by the exposure to NO.
Figure 5.
Perpendicular-mode EPR spectra of MnHPCD-HPCA titrated with NO. MnHPCD-HPCA (1 mM protein, 12 mM substrate) with 0 (A), 1 eq. (B), 2 eq. (C), and excess (D) NO. (E) Excess NO addition followed by aerobic incubation. All spectra are scaled for equal protein concentration. Instrumental conditions: temperature, 11 K; microwaves, 0.2 mW at 9.65 GHz.
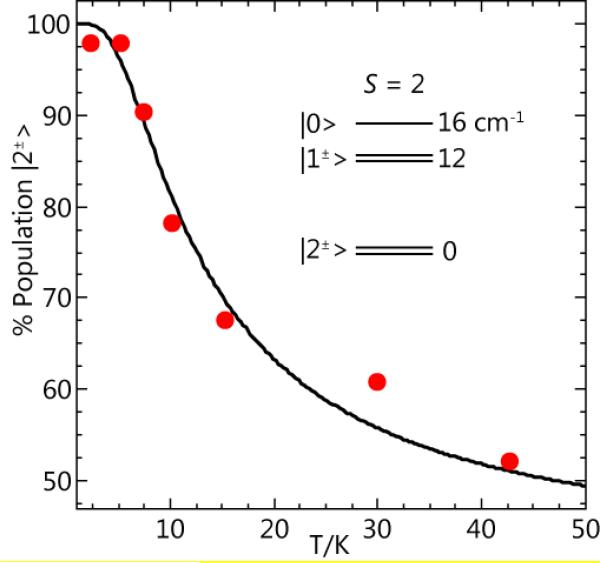
EPR spectra of the same titration samples are shown in Figure 6 with the microwave field B1 parallel to the static field B (parallel-mode). A 6-line hyperfine pattern at g = 8.5 with a = 4.6 mT was observed to grow with increasing NO. The simulation of this signal (Figure 6F) is for an S = 2 spin system using the parameters given in Figure 6. The value of D is −4(1) cm−1 as determined from the temperature dependence of the g = 8.5 signal shown in Figure 7. The electronic parameters of the species are consistent with an S = 2 MnIII ion [16]. An S = 2 system could conceivably also be observed for MnII interacting with a substrate semiquinone radical, however, the low hyperfine splitting constant of 4.6 mT rules out such a complex. The addition of more than 1 eq. NO resulted in a significant increase in the concentration of this species. The amount of MnIII determined from the simulation of the signal of the excess NO sample was between 25% and 40% of the Mn in the sample. The range in amount is due to the uncertainty in the intrinsic g-value of MnIII (1.95 to 2.03) [17-19].
Figure 6.
Parallel-mode EPR spectra of MnHPCD-HPCA titrated with NO. MnHPCD-HPCA (1 mM protein, 12 mM substrate) with 0 (A), 1 eq. (B), 2 eq. (C), and excess (D) NO. (E) Excess NO addition followed by aerobic incubation. All spectra are scaled for equal protein concentration. Simulation parameters for (F): S = 2, g = 1.95, A= 130 MHz, D = −4 cm−1, E/D = 0.10. Instrumental conditions: temperature, 5 K; microwaves, 20 mW at 9.31 GHz.
Figure 7.
The temperature dependence of the g = 8.5 EPR signal of Figure 6. The data points are determined from signal intensity times temperature. The curve is calculated for the spin population of the ground |2±〉 doublet of a S=2 system with D = −4 cm−1, as indicated by the energy level diagram.
As is evident in Figure 6A, this signal was present prior to NO addition, indicating it was not from the NO-bound species. Furthermore, the concentration of the MnIII species formed did not correspond to the amount of MnHPCD-HPCA lost. In the absence of NO, the g = 8.5 signal of Figure 6A accounts for at most 10% of the Mn in the sample. After the addition of 1 eq. NO (Figure 6B) the signal shows a small increase to at most 15% of the Mn in the sample. In contrast, the same sample shows a 40% loss of the signal intensity from the MnII center of the MnHPCD-HPCA complex (Figure 5B).
Repeated attempts to dissociate NO by nitrogen gas exchange and photolysis were unsuccessful for samples with low and high equivalents of NO, indicating high stability of the Mn-NO complex. However, the aerobic incubation of the sample resulted in product formation as evidenced by the solution changing color to bright yellow and recovery of the EPR signal (Figure 5E) from the substrate free enzyme, without a change in the amount of the MnIII species (Figure 6E). The HPCD substrate, which was 12-fold in excess over protein, was consumed during the aerobic incubation. After aerobic incubation, the amount of enzyme-free MnII species (determined from the intensity of the g = 2 signal) plus the amount of the MnIII species were found to be equal to the amount of enzyme-substrate complex before addition of NO. This indicates that O2 can displace or react with the bound NO and subsequently turn over the substrate, but the MnIII species is unreactive. In summary, the addition of NO to the MnII enzyme-substrate complex results in the formation of a majority MnNO species that is EPR-silent and a minority MnIII species. In the presence of O2, the MnNO species can undergo reactions that regenerate the enzyme-free MnII complex, whereas the MnIII species does not undergo further reaction.
DISCUSSION
The current study shows that the binding of NO to MnHPCD is regulated by both the presence and nature of the catecholic substrate ligand as well as the active site acid/base catalyst His200. Past studies with FeHPCD and the related FeII enzyme protocatechuate 4,5-dioxygenase have shown that neither a catecholic substrate nor His200 (unpublished result) is required for NO to bind, although the NO binding affinity is increased at least 40-fold when a catecholic substrate is also bound [20, 21]. By and large, the ability of these enzymes and their variants and complexes to bind NO parallels their ability to bind O2. The exception to this correlation is that substrate-free FeHPCD can bind NO but not O2, suggesting that NO can form a stronger bond. The comparison of NO and O2 binding offers insight into the mechanism of O2 activation by extradiol dioxygenases and the roles played by substrate, metal, and protein active site in this process. These aspects of the catalytic cycle are discussed here.
As summarized in Figure 8, the addition of NO to the MnHPCD-HPCA complex or the corresponding DHM complex results in the formation a new Mn-NO species. Only these two complexes are capable of reaction with NO. The resulting NO complex is found to be highly stable as evidenced by its near irreversible formation. This complex does react with O2 to form product and regenerate the substrate free MnII complex. In addition, a fraction of the Mn-NO complex may occasionally release NO−, or possibly react with NO, to form a stable MnIII enzyme complex. Presumably, the substrate remains bound to aid in stabilization of the trivalent metal state. To our knowledge, this is the first reported occurrence of the reaction between MnII and NO to form a stable Mn-NO protein species in a nonheme enzyme. Nitric oxide will form MnNO complexes in Mn-substituted heme proteins [22]. Also, nitric oxide will react with the Mn cluster of photosystem II and with the dinuclear Mn cluster of Mn-catalase [23], but for these enzymes the reaction occurs with Mn at an oxidation state higher than +2, and the addition of NO results in Mn reduction rather than oxidation as observed here.
Figure 8.
Reaction summary of NO as surrogate for O2 with MnHPCD substrate complexes. NO did not react with the substrate-free enzyme, the 4NC complex, or variants with and without substrate. The spin states and g-values of the associated EPR signals are in italics.
We are not able to assign a spectroscopic feature to the MnNO complex: it is EPR-silent. This species clearly does not give rise to the g = 8.5 signal that is assigned to a MnIII species. We have attempted to observe the MnNO species in parallel mode with low temperature Q-band EPR spectroscopy, but without success. Visible absorption measurements were also attempted on several samples, but the samples have no significant absorption. FTIR spectroscopy has been previously attempted on the well characterized FeNO complexes of HPCD, but the NO vibration could not be detected due to strong protein vibrational bands. Based on Mn model complexes [24], the expected NO vibrational frequency would be near 1700 cm−1 and again in the region of strong protein vibrational bands. Consequently, we have not attempted FTIR measurements of the present complex. We have also not attempted magnetization studies due to the presence of the variable paramagnetic MnIII species.
The MnNO complex could be formally described as either MnI-NO+, MnII-NO•, or MnIII-NO−. A reduced MnI center is generally believed to be outside the reduction potential range available in aqueous environments. An MnII-NO• species would result in S = 2 and 3 spin states from exchange coupling between the S = 5/2 MnII and S = 1/2 NO•. This species would likely be EPR detectable in parallel mode, as the zero-field energies of MnII centers are small compared to the microwave quantum of energy for X- or Q-band EPR spectroscopy. The EPR properties of such a complex would be similar (ignoring Mn hyperfine) to that of the FeIII-superoxo complex of HPCD mentioned in the introduction [11]. Thus, the absence of an EPR signal suggests an MnIII-NO− formulation. A MnIII-NO− complex could be diamagnetic [25] or have a ground S = 1 spin state from an antiferromagnetic exchange coupling between the S = 2 MnIII and S = 1 NO- ligand. The diamagnetic complex would of course be EPR-silent, and while EPR detection of S = 1 complexes is possible [26], it is unlikely due to the greater zero-field energies of MnIII ions. Paramagnetic [MnNO]6 complexes are rare. A paramagnetic MnNO complex of tropocoronand has been characterized, and similar to the present case, this MnNO complex was EPR-silent [27]. Magnetization data and DFT calculations suggested an essentially MnIII-NO− electronic structure with an S =1 spin state. This electronic structure and resulting antiferromagnetic exchange interaction is analogous to characterized FeIII-NO− complexes such as the NO adducts of FeIIHPCA or FeIIEDTA [12, 28, 29].
Effect of Metal Redox Potential
As discussed above, it is likely that the very large difference in solution reduction potentials of Fe and Mn is maintained when these metals are bound in the enzyme active site because they are located in structurally identical ligand environments [7]. The effectively identical steady state parameters for normal turnover and KmO2 indicate that this large difference in reduction potential does not substantially alter the catalytic rate and overall rate constant of the O2 binding and activation through the first irreversible step of the process. The simplest rationale for this observation is that the metal either does not change oxidation state during this process or that later steps compensate for changes in O2 binding rates. At least for the Mn enzyme, a transient MnIII state has been detected during turnover of HPCA, indicating that a metal oxidation state change is part of the overall reaction mechanism [10]. If the binding rate of O2 to Mn is significantly slower than that of the Fe enzyme, as suggested by the large potential difference of the metal sites, the steps after O2 binding must compensate such that the observed kcat and KmO2 values and their ratio are metal independent.
This same rationale can be used in a different context to explain some aspects of the difference in NO reactivity of the Fe- and MnHPCD enzymes. In contrast to the normal catalytic mechanism, NO binding generally results in an increase in metal oxidation state and spin state change. This means that the rates of complex formation and the strength of the resulting complex should be very sensitive to the reduction potential of the metal. The comparatively low potential of iron in FeHPCD is always sufficient to reduce the NO during complex formation to yield an S = 1 NO− that is antiferromagnetically coupled to the resulting S = 5/2 FeIII to give the characteristic EPR active S = 3/2 complex [12, 28]. In contrast, because of its much higher potential, the corresponding MnII center cannot transfer an electron efficiently to NO without the additional participation of both a bound catecholic substrate and the key active site His200 residue. How these latter moieties enhance NO binding has many aspects that will be addressed in turn.
Effect of the Catecholic Substrate
One likely effect of binding the anionic catecholate ligand is to tune the metal redox potential to a lower value, which presumably promotes the binding and reduction of NO (or O2). This notion has been invoked to account for the substrate regulation of O2 binding as well as the observed increase in NO affinity when substrate is bound to FeII dioxygenases [21]. The direct determination of the metal potential has not been possible for any extradiol dioxygenase due to poor coupling with redox mediator dyes and the inherently high potential of the metal center. However, the EPR characterization of MnHPCD complexes presented here provides another approach for an assessment of their relative reduction potentials.
Simulation of the EPR spectra allowed a precise determination of the electronic parameters D and E/D that characterize the symmetry of the active site as well as the distribution in these parameters due to small bonding variations in the metal coordination. The factors governing D in MnII complexes are complicated [30], and more studies would be useful, but there appears to be a correlation with reduction potential. Such a trend has been observed in Mn model complexes where an increase in the magnitude of D correlated to a decrease in the MnIII/II reduction potential [31]. In Mn superoxide dismutases, the D-value has been correlated to activity, presumably due to changes in the Mn reduction potentials [32]. The D-values for the various enzyme and substrate complexes of MnHPCD shown in Table 1 reveal three groups. The complexes lacking substrate have the smallest D-values (e.g. MnHPCD, 0.055 cm−1), whereas the substrate complexes have larger D-values (e.g. MnHPCD-HPCA, 0.090 cm−1), and the MnHPCD-4NC inhibitor complex has the largest D-value (0.125 cm−1). Because HPCA binds as a catechol monoanion [4, 33, 34] and 4NC as a dianion [35], this trend in D-value is consistent with the expected decrease in redox potential as the number of anionic ligands increases. The decrease in potential of the MnII in the MnHPCD-HPCA complex is apparently sufficient to allow NO to bind, but the lack of binding in the MnH200Q-HPCA complex and in the even lower potential MnHPCD-4NC complex cannot be rationalized by this simple redox potential argument and reveals other aspects of substrate binding that are relevant to catalysis as discussed below.
Effects of His200
It is observed here that His200 is essential for formation of the nitrosyl complex of MnHPCD in any of its complexes. His200 mutants of FeHPCD have previously been used to facilitate detection of early O2 adducts and their reaction with substrate [9, 11, 14]. This work suggested four possible roles for His200 in HPCD catalysis: (1) it may provide steric bulk to promote side-on O2 binding and the proper orientation to optimize specific reaction with the catecholic substrate, (2) it may serve as an acid-catalyst to promote the O–O bond cleavage of the alkylperoxo intermediate in the Criegee rearrangement step, (3) it may act as a hydrogen bond donor to stabilize the O2 complex, and (4) it may assume a positive charge when it acts as an active site base to deprotonate the second OH of bound HPCA. The use of NO simplifies the examination of the role His200 plays in small molecule binding because it only binds end-on, and it does not allow progression into the reaction cycle beyond the initial binding step. Thus, the first two suggested roles for His200 listed above do not come into play.
The putative role of His200 as a hydrogen bonding partner to stabilize the bound NO may contribute to the strength of the complex. This would account for the failure of the MnH200N variant to bind NO, because crystal structures show that the amide side chain is too short to be able to form an effective hydrogen bond with a bound O2 or NO [11]. In contrast, the longer side chain of H200Q would place the potential hydrogen bonding amide at a similar distance from NO as the Nε of His200, so stabilization from hydrogen bonding is likely. Consistent with the formation of a hydrogen bond for H200Q, the EPR spectra and electronic parameters of the MnH200Q variant, both with and without substrate, are close to those of the normal enzyme. This is in contrast to the spectra and parameters of the MnH200N variant, both with and without substrate, which are significantly different. In particular, the parameter E/D changes significantly for the MnH200N variant, which indicates a large symmetry change in the Mn coordination. These results are consistent with the presence of a hydrogen bond between the metal bound water and either His200 of the normal enzyme or Gln of H200Q, and the absence of the hydrogen bond to Asn of H200N.
However, the inability of the MnH200Q variant to bind NO shows that hydrogen bonding and decreased potential from the anionic catechol are together still not sufficient to form a stable NO adduct. The same arguments can be directed at the MnHPCD-4NC complex. His200 is available for hydrogen bonding for this complex, and as suggested by the trend in the D-value, the 4NC complex has an even lower potential, but again a stable NO adduct is not observed.
The final proposed role of H200 as a positively charged group to stabilize the developing charge on the NO− (or O2−) ligand can be supported by the data presented here. Neither Asn nor Gln can assume a positive charge in the physiological pH range, and thus no NO complex of H200N or H200Q with or without substrate bound would be expected to be stabilized. On the other hand, histidine is often protonated in enzymes near neutral pH, and its protonation is one step of the proposed acid/base function of His200 in the mechanism of extradiol dioxygenases [1, 36-38]. The source of the proton in this proposal is the hydroxyl group of the bound monoanionic catecholic substrate. During the course of normal catalysis, this deprotonation event serves two purposes. First, it allows the substrate to ketonize as an electron is transferred to the metal to yield a reactive substrate radical. Second, it allows the substrate to carry in the proton required to protonate the alkylperoxo intermediate resulting from oxygen attack, and thereby promote the Criegee rearrangement required for ring cleavage.
In the model presented here, HPCA or DHM could provide the proton required to create a positively charged His200 and at the same time become a dianionic catechol to further lower the potential of the Mn. The initial binding of 4NC as a dianion means that a proton is not available from the catecholic ligand, and thus His200 remains neutral and unable to stabilize NO (or O2) binding by a charge interaction, as observed.
Significance for Catalysis
It is evident from past studies of the extradiol dioxygenase mechanism by our groups and others that the structural and electronic aspects of the process must come together perfectly to allow rapid and specific catalysis [9, 11, 14, 39]. The advantage of studying the metal-replaced analogs of FeHPCD, and in particular the nitrosyl complex investigated here, is that the effects of structural or electronic perturbations are often dramatically enhanced, in some cases allowing their contributions to catalysis to be individually investigated. For example, replacement of Fe by Mn makes little change in normal catalysis, but it causes H200X mutations to convert from being quite active to being inactive, and it allows a correlation between the ability to bind NO and activity with O2 to be sharply drawn.
Given the identical active site structures, it is likely that the same interactions are present in the FeHPCD and MnHPCD active sites. For MnHPCD the interactions present a block to binding or catalysis, in contrast to FeHPCD where the interactions perturb the chemistry but are not blocking. For example, the H200N and H200Q variants of FeHPCD are both active (in contrast to the case for MnHPCD), but their activity is significantly lower than found for wild type FeHPCD [14]. Similarly, NO binds well to MnHPCD-HPCA but not at all to MnHPCD, whereas NO merely binds more weakly to the FeII containing dioxygenases in the absence of substrate.
In this context, it is very significant that a similar perturbation is not seen when FeHPCD and MnHPCD turn over HPCA (or DHM). This reinforces the idea that the very different redox potentials of Fe and Mn are of little consequence in the optimized reaction. Nevertheless, they are of significant consequence in the reactions with modified substrates and active site variants where metal oxidation state changes are seen to occur. While changes in reduction potentials do not govern reaction kinetics, they are often associated with rate changes. One way to rationalize the observations is to assume that essentially the same steps occur in both the wild type and variant enzymes, but the inefficiency incurred by altering the optimal reaction causes it to slow down at steps that are usually fast. For example, the equivalent kinetic behavior of FeHPCD and MnHPCD, as well as the failure to observe any FeIII in RFQ samples from the FeHPCD-HPCA reaction with O2 [9], supports simultaneous or nearly simultaneous transfers of electrons from the metal to O2 and from the HPCA to the metal. This would marginalize the significance of the metal redox potential even in a nonconcerted reaction because a low potential metal that could reduce O2 more rapidly would, in turn, be reduced more slowly by the substrate, resulting in compensating rates. In variants such as H200N or H200Q, the strong charge stabilization of the anionic substrate ligand suggested by the current study would be lost, and there would be less tendency for an electron to move from the catecholic substrate to the metal as a means to activate the substrate. Consequently, one might expect to observe the intermediate metal-superoxo species with a comparatively long lifetime, as has proven to be the case [11, 14]. Similar arguments could be made based on changes in the substrate that affect its protonation state or its ability to transfer an electron to the metal, and such changes have also led to the discovery of semistable intermediates with oxidized metal centers [8, 9, 11, 14, 40].
Reduction of the MIII-NO− Species
In the proposed mechanism, formation of a MIII-superoxo species would be followed by metal reduction by the catecholic substrate to generate a reactive ion pair. Since the metal is oxidized in the nitrosyl complex, this raises the question of why it is not reduced by the catecholic substrate in the substrate complex. A possible explanation is that the stability of the metal-nitrosyl complex is dependent on the metal remaining oxidized. The stability of this bond specifically in the case of the nearly irreversible MnIII-NO− complex observed here suggests that reduction would be a significantly disfavored process. However, it may occasionally occur, leading to the release of NO− and formation of a stable MnIII-HPCA complex after transfer of an electron back to the substrate semiquinone. This may account for the slow oxidation observed here for the HPCA-MnHPCD-NO complex. Alternatively, independent of substrate, a fraction of the Mn-NO complex may occasionally release NO−, or possibly react with NO, to form a stable MnIII enzyme complex.
Conclusion
The use of MnHPCD has provided the first known stable formation of a MnNO complex in a nonheme enzyme, which is probably formulated as MnIII-NO−. This complex is formed only when His200 of the wild type enzyme is present and only when a monoanionic catecholic substrate is bound to the metal. The high intrinsic reduction potential of Mn relative to Fe, which would greatly disfavor NO or O2 binding, is lowered by binding of the anionic substrate. However, this alone is not sufficient and additional stabilization from hydrogen bonds and charge interactions that are linked to substrate specificity are required to bind NO or O2. This combination is also required for ring-cleaving catalysis in the normal O2-linked reaction of MnHPCD, which affirms the importance of specific primary and secondary coordination interactions for control of oxidation specificity. The correlation between activity and ability to bind NO also applies to FeHPCD, but a much wider range of substrates and active site variants is tolerated. It is proposed that the requirement to change metal oxidation states during the formation of the NO complex makes the reaction sensitive to the redox potential of the metal and leads to the observed differences between the FeHPCD and MnHPCD NO binding reactions. It has been proposed that during normal catalysis, Fe and Mn are oxidized to activate O2 and nearly simultaneously reduced by the catecholic substrate, thereby activating it for reaction. Consequently, the metal redox potential plays a minor role. Nevertheless, the correlation between NO binding and O2-linked catalysis is consistent with the formation of a metal-superoxo species of some type during the catalytic cycle. Stabilization of that species by active site interactions as indicated by the results presented here may dictate the rate and specificity of the ensuing reaction.
Acknowledgments
Funding Sources
This work is supported by GM24689 (to J.D.L), GM33162 (to L.Q.), GM77387 (to M.P.H.)
ABBREVIATIONS
- FeHPCD
recombinant homoprotocatechuate 2,3-dioxygenase from Brevibacterium fuscum
- MnHPCD
recombinant homoprotocatechuate 2,3-dioxygenase from Brevibacterium fuscum in which the FeII is replaced by MnII
- H200N
His to Asn variant of FeHPCD at position 200
- H200Q
His to Gln variant of FeHPCD at position 200
- HPCA
homoprotocatechuate or 3,4 dihydroxyphenylacetate
- 4NC
4-nitrocatechol
- DHM
dihydroxymandelic acid
- SF
stopped flow
- RFQ
rapid freeze quench
- EPR
electron paramagnetic resonance
- ICP-AES
inductively coupled plasma atomic emission spectroscopy
References
- 1.Lipscomb JD. Curr. Opin. Struct. Biol. 2008;18:644–649. doi: 10.1016/j.sbi.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koehntop KD, Emerson JP, Que L. J. Biol. Inorg. Chem. 2005;10:87–93. doi: 10.1007/s00775-005-0624-x. [DOI] [PubMed] [Google Scholar]
- 3.Kovaleva EG, Lipscomb JD. Nat. Chem. Bio. 2008;4:186–193. doi: 10.1038/nchembio.71. DOI 10.1038/nchembio.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vetting MW, Wackett LP, Que L, Jr., Lipscomb JD, Ohlendorf DH. J. Bacteriol. 2004;186:1945–1958. doi: 10.1128/JB.186.7.1945-1958.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang YZ, Lipscomb JD. Protein Expres. Purif. 1997;10:1–9. doi: 10.1006/prep.1996.0703. DOI DOI 10.1006/prep.1996.0703. [DOI] [PubMed] [Google Scholar]
- 6.Boldt YR, Sadowsky MJ, Ellis LBM, Que L, Jr., Wackett LP. J. Bacteriol. 1995;177:1225–1232. doi: 10.1128/jb.177.5.1225-1232.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emerson JP, Kovaleva EG, Farquhar ER, Lipscomb JD, Que L., Jr. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7347–7352. doi: 10.1073/pnas.0711179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fielding AJ, Kovaleva EG, Farquhar ER, Lipscomb JD, Que L., Jr. J. Biol. Inorg. Chem. 2011;16:341–355. doi: 10.1007/s00775-010-0732-0. DOI 10.1007/s00775-010-0732-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mbughuni MM, Chakrabarti M, Hayden JA, Meier KK, Dalluge JJ, Hendrich MP, Münck E, Lipscomb JD. Biochemistry. 2011;50:10262–10274. doi: 10.1021/bi201436n. DOI 10.1021/bi201436n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunderson WA, Zatsman AI, Emerson JP, Farquhar ER, Que L, Jr., Lipscomb JD, Hendrich MP. J. Am. Chem. Soc. 2008;130:14465–14467. doi: 10.1021/ja8052255. DOI 10.1021/ja8052255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mbughuni MM, Chakrabarti M, Hayden JA, Bominaar EL, Hendrich MP, Münck E, Lipscomb JD. Proc. Natl. Acad. Sci. U. S. A. 2010;107:16788–16793. doi: 10.1073/pnas.1010015107. DOI PMID:20837547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arciero DM, Lipscomb JD, Huynh Boi H, Kent TA, Münck E. J. Biol. Chem. 1983;258:14981–14989. [PubMed] [Google Scholar]
- 13.Ford PC, Lorkovic IM. Chem. Rev. 2002;102:993–1018. doi: 10.1021/cr0000271. [DOI] [PubMed] [Google Scholar]
- 14.Groce SL, Lipscomb JD. Biochemistry. 2005;44:7175–7188. doi: 10.1021/bi050180v. DOI 10.1021/bi050180v. [DOI] [PubMed] [Google Scholar]
- 15.Whiting AK, Boldt YR, Hendrich MP, Wackett LP, Que L., Jr. Biochemistry. 1996;35:160–170. doi: 10.1021/bi951979h. [DOI] [PubMed] [Google Scholar]
- 16.Krzystek J, Ozarowski A, Telser J. Coord. Chem. Rev. 2006;250:2308–2324. DOI 10.1016/j.ccr.2006.03.016. [Google Scholar]
- 17.Zheng M, Khangulov SV, Dismukes GC, Barynin VV. Inorg. Chem. 1994;33:382–387. [Google Scholar]
- 18.Peloquin JM, Campbell KA, Randall DW, Evanchik MA, Pecoraro VL, Armstrong WH, Britt RD. J. Am. Chem. Soc. 2000;122:10926–10942. [Google Scholar]
- 19.Campbell KA, Yikilmaz E, Grant CV, Gregor W, Miller A-F, Britt RD. J. Am. Chem. Soc. 1999;121:4714–4715. [Google Scholar]
- 20.Miller MA, Lipscomb JD. J. Biol. Chem. 1996;271:5524–5535. doi: 10.1074/jbc.271.10.5524. [DOI] [PubMed] [Google Scholar]
- 21.Arciero DM, Orville AM, Lipscomb JD. J. Biol. Chem. 1985;260:14035–14044. [PubMed] [Google Scholar]
- 22.Gelb MH, Toscano WA, Jr., Sligar SG. Proc. Natl. Acad. Sci. U. S. A. 1982;79:5758–5762. doi: 10.1073/pnas.79.19.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ioannidis N, Schansker G, Barynin VV, Petrouleas V. J Biol Inorg Chem. 2000;5:354–363. doi: 10.1007/pl00010664. [DOI] [PubMed] [Google Scholar]
- 24.Franz KJ, Lippard SJ. J. Am. Chem. Soc. 1998;120:9034–9040. [Google Scholar]
- 25.Merkle AC, Fry NL, Mascharak PK, Lehnert N. Inorg. Chem. 2011;50:12192–12203. doi: 10.1021/ic201967f. DOI 10.1021/ic201967f. [DOI] [PubMed] [Google Scholar]
- 26.Taguchi T, Gupta R, Lassalle-Kaiser B, Boyce DW, Yachandra VK, Tolman WB, Yano J, Hendrich MP, Borovik AS. J. Am. Chem. Soc. 2012;134:1996–1999. doi: 10.1021/ja210957u. DOI 10.1021/ja210957u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tangen E, Conradie J, Franz K, Friedle S, Telser J, Lippard SJ, Ghosh A. Inorg. Chem. 2010;49:2701–2705. doi: 10.1021/ic901860x. [DOI] [PubMed] [Google Scholar]
- 28.Brown CA, Pavlosky MA, Westre TE, Zhang Y, Hedman B, Hodgson KO, Solomon EI. J. Am. Chem. Soc. 1995;117:715–732. [Google Scholar]
- 29.Serres RG, Grapperhaus CA, Bothe E, Bill E, Weyhermuller T, Neese F, Wieghardt K. J. Am. Chem. Soc. 2004;126:5138–5153. doi: 10.1021/ja030645+. DOI 10.1021/ja030645+ [DOI] [PubMed] [Google Scholar]
- 30.Duboc C, Collomb M-N, Neese F. Appl. Magn. Reson. 2010;37:229–245. [Google Scholar]
- 31.Gatjens J, Sjodin M, Pecoraro VL, Un S. J. Am. Chem. Soc. 2007;129:13825–13827. doi: 10.1021/ja076024o. [DOI] [PubMed] [Google Scholar]
- 32.Un S, Tabares LC, Cortez Ns, Hiraoka BY, Yamakura F. J. Am. Chem. Soc. 2004;126:2720–2726. doi: 10.1021/ja036503x. [DOI] [PubMed] [Google Scholar]
- 33.Vaillancourt FH, Barbosa CJ, Spiro TG, Bolin JT, Blades MW, Turner RF, Eltis LD. J. Am. Chem. Soc. 2002;124:2485–2496. doi: 10.1021/ja0174682. [DOI] [PubMed] [Google Scholar]
- 34.Shu L, Chiou Y-M, Orville AM, Miller MA, Lipscomb JD, Que L., Jr. Biochemistry. 1995;34:6649–6659. doi: 10.1021/bi00020a010. [DOI] [PubMed] [Google Scholar]
- 35.Reynolds MF, Costas M, Ito M, Jo D-H, Tipton AA, Whiting AK, Que L. J. Biol. Inorg. Chem. 2003;8:263–272. doi: 10.1007/s00775-002-0411-x. [DOI] [PubMed] [Google Scholar]
- 36.Bugg TD, Ramaswamy S. Curr. Opin. Chem. Biol. 2008;12:134–140. doi: 10.1016/j.cbpa.2007.12.007. DOI 10.1016/j.cbpa.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 37.Vaillancourt FH, Bolin JT, Eltis LD. Crit. Rev. Biochem. Mol. Biol. 2006;41:241–267. doi: 10.1080/10409230600817422. [DOI] [PubMed] [Google Scholar]
- 38.Costas M, Mehn MP, Jensen MP, Que L., Jr. Chem. Rev. 2004;104:939–986. doi: 10.1021/cr020628n. DOI 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 39.Boldt YR, Whiting AK, Wagner ML, Sadowsky MJ, Que L, Jr., Wackett LP. Biochemistry. 1997;36:2147–2153. doi: 10.1021/bi962362i. [DOI] [PubMed] [Google Scholar]
- 40.Fielding AJ, Lipscomb JD, Que L., Jr. J. Am. Chem. Soc. 2012;134:796–799. doi: 10.1021/ja2095365. DOI 10.1021/ja2095365. [DOI] [PMC free article] [PubMed] [Google Scholar]