Abstract
The insulin-sensitizing hormone, adiponectin, belongs to the expanding C1q/TNF (tumour necrosis factor) family of proteins. We recently identified a family of adiponectin paralogues designated as CTRP (C1q/TNF-related protein) 1–7, and in the present study describe CTRP10. In the present study, we show that CTRP1, CTRP2, CTRP3, CTRP5 and CTRP7 transcripts are expressed predominantly by adipose tissue. In contrast, placenta and eye expressed the highest levels of CTRP6 and CTRP10 transcripts respectively. Expression levels of CTRP1, CTRP2, CTRP3, CTRP6 and CTRP7 transcripts are up-regulated in 8-week-old obese (ob/ob)mice relative to lean controls. Treatment of mice with a PPAR-γ (peroxisome-proliferator-activated receptor-γ) agonist, rosiglitazone, increased the expression of CTRP1 and decreased CTRP6 transcript levels. All CTRPs are secreted glycoproteins when expressed in mammalian cells. CTRP1, CTRP2, CTRP3, CTRP5 and CTRP6 circulate in the blood and are potential endocrine hormones; their serum levels vary according to the sex and genetic background of mice. Importantly, serum levels of CTRP1 and CTRP6 are increased in adiponectin-null mice. Like adiponectin, all secreted CTRP proteins form trimers as their basic structural units. CTRP3, CTRP5, CTRP6 and CTRP10 trimers are further assembled into higher-order oligomeric complexes via disulfide bonding mediated by their N-terminal cysteine residues. Besides forming homo-oligomers, CTRP1/CTRP6, CTRP2/CTRP7 and adiponectin/CTRP2 are secreted as heterotrimers, thus providing a mechanism to potentially generate functionally distinct ligands. Functional characterization of one such family member, CTRP1, showed that it specifically activates Akt and p44/42-MAPK (mitogen-activated protein kinase) signalling pathways in differentiated mouse myotubes. Moreover, injection of recombinant CTRP1 into mice significantly reduced their serum glucose levels. Thus at least CTRP1 may be considered a novel adipokine. In summary, these molecular, biochemical and functional data provide an important framework to further address the physiological functions and mechanisms of the action of this family of secreted glycoproteins in normal and disease states.
Keywords: adipokine, adipose tissue, C1q/TNF family, C1q/TNF-related protein (CTRP), diabetes, obesity
INTRODUCTION
Adipose tissue is an active endocrine organ that expresses and secretes a variety of proteins that include adiponectin, resistin, leptin, IL (interleukin)-1β, IL-6, TNFα (tumour necrosis factor α), MCP-1 (monocyte chemoattractant protein-1), adipsin and PAI-1 (plasminogen-activator inhibitor-1) [1]. Collectively, these adipocyte-secreted proteins are termed adipokines. In the context of metabolism, these adipokines play major roles in regulating whole-body glucose and lipid metabolism by affecting directly or indirectly (via modulating inflammatory responses) insulin sensitivity in the liver, muscle and adipose tissue [1]. The hypothalamus region of the brain is also a target organ for leptin and adiponectin [2].
One of the most intensely studied adipokines is Acrp30/adiponectin whose expression is almost exclusively restricted to adipocytes [3–5]. The transcript and protein levels of adiponectin are paradoxically down-regulated in conditions of obesity and diabetes, both in animal models and in humans [4,6]. Molecular mechanisms that underlie reduced adiponectin expression despite increased adiposity remain largely unknown, although TNFα secreted by adipocytes and the infiltrated macrophages are a likely cause for adiponectin down-regulation [7]. Reduction in serum levels of adiponectin is associated with insulin resistance [8], whereas increased levels of serum adiponectin due to weight loss, calorie restriction or TZD (thiazolidinedione) treatment correlates with increased insulin sensitivity [9,10]. In vitro and in vivo studies using recombinant proteins, adenovirus-mediated overexpression, transgenic and knockout mice collectively demonstrate that the insulin-sensitizing effects of adiponectin are due to its ability to increase glucose uptake and fatty acid oxidation in muscle [11,12], while potently suppressing glucose production in liver [13].
We previously described seven highly conserved paralogues of adiponectin designated as CTRP (C1q/TNF-related protein) 1–7 [14]. All of these proteins share the same modular organization as adiponectin: a signal peptide, a short variable region, a collagen domain and a globular C1q domain. The crystal structure of the adiponectin globular domain strikingly resembles the three-dimensional structure of TNFα [15]. Thus all proteins with the signature C1q domain (~135 amino acids) are now classified within the expanding C1q/TNF protein family [16]. Fruebis and co-workers [17] recently showed that CTRP1 is expressed by vascular smooth muscle cells and recombinant CTRP1 significantly prevents platelet thrombosis in vivo by inhibiting vWF (von Willebrand factor) binding to collagen, thereby blocking collagen-induced platelet aggregation. Similarly to adiponectin, recombinant CTRP2 activates AMPK (AMP-activated protein kinase) in muscle cells, resulting in increased glycogen deposition and enhanced fatty acid oxidation [14]. CTRP3, also known as CORS26/cartducin, is expressed by chrondrocytes among other tissues and recombinant CTRP3 stimulates proliferation of chondrogenic precursor cells by activating ERK1/2 (extracellular-signal-regulated kinase 1/2) and Akt signalling pathways [18]. Among the tissues that express CTRP5 is the retinal pigment epithelium. Mutations in this gene cause late-onset retinal macular degeneration in humans [19]. In a screen to discover host genes that can limit ASFV (African Swine Fever virus) that causes acute haemorrhagic fever in domestic pigs, six cDNA clones, one of which is CTRP6/C1qTNF6, were identified that are required for the replication of ASFV in HeLa andHT144 cells [20]. The mechanism by which CTRP6 promotes ASFV replication is not known. The natural target cells of all the CTRPs and their functions are currently under investigation.
Several groups have recently generated adiponectin-null mice [21–23]. With one exception [21], all adiponectin-null mice developed insulin resistance when maintained under a high-fat diet [22,23]; however, these mice have mild or no detectable metabolic abnormality when maintained under a normal chow diet, suggesting that under this condition other proteins, such as CTRPs, may compensate for the absence of adiponectin. This prompted us to investigate the relative abundance of CTRP transcripts in adipose tissue and whether these are serum proteins and hence may function as endocrine hormones. In the present study, we show that CTRP1, CTRP2, CTRP3, CTRP5 and CTRP7 transcripts are expressed predominantly by adipose tissue. Factors such as gender, age, genetic backgrounds of mice and TZD drug treatment affect the relative expression levels of CTRP transcripts. Most CTRPs circulate in the blood with levels varying according to the sex and genetic background of mice, thus they are potential endocrine hormones. We found that the serum levels of CTRP1 and CTRP6 are increased in adiponectin-null mice. All CTRPs form trimers as their basic structural units and some are further assembled into higher-order oligomeric complexes involving N-terminal cysteine residues. Remarkably, CTRP1/CTRP6, CTRP2/CTRP7 and adiponectin/CTRP2 can also be secreted as heterotrimers in co-transfected cells. Functional characterization of one such family member, CTRP1, showed that it activates Akt and p44/42 MAPK (mitogen-activated protein kinase) signalling pathways in differentiated myotubes and significantly lowered serum glucose levels when injected into mice. Collectively, these molecular, biochemical and functional data provide us with a framework to further evaluate the physiological functions and mechanisms of action of CTRPs using molecular, cellular and in vivo approaches.
MATERIALS AND METHODS
Identification and cloning of CTRP10
A search for adiponectin-like proteins in the NCBI GenBank® databases identified another novel protein different from the seven recently identified adiponectin paralogues designated as CTRP1–7 [14]. We designated our novel adiponectin paralogue as CTRP10. The GenBank® accession numbers for mouse and human CTRP10 are AAY21934 and EAW95208 respectively. Based on the sequences of overlapping EST (expressed sequence tag) clones corresponding to CTRP10, a PCR strategy was used to clone the entire coding region of CTRP10.
DNA constructs
C-terminal FLAG- and HA (haemagglutinin)-tagged full-length adiponectin, CTRP1, CTRP2, CTRP3, CTRP5, CTRP6, CTRP7 and CTRP10 were generated by PCR and cloned into the mammalian expression vector pCDNA3.1 (Invitrogen). All constructs were verified by DNA sequencing.
A site-directed mutagenesis kit from Stratagene was used to mutate cysteine residues to alanine found at the N-termini of each CTRP preceding their respective globular C1q domain (refer to Figure 6C). AdiponectinΔCys refers to the C39A mutant; CTRP1ΔCys refers to the C73A/C76A/C77A/C141A mutant; CTRP2ΔCys refers to the C36A/C141A/C143A mutant; CTRP3ΔCys refers to the C39A/C42A/C43A mutant; CTRP5ΔCys refers to the C28A/C98A mutant; CTRP6ΔCys mutant refers to the C44A/C47A/C48A/C125A mutant; CTRP7ΔCys mutant refers to the C34A/C139A/C141A mutant; CTRP10ΔCys refers to the C29A/C33A mutant. Each of the cysteine residues was mutated sequentially or in tandem to alanine using a PCR-based site-directed mutagenesis kit (Stratagene). DNA templates used in the PCR mutagenesis were C-terminal FLAG-tagged adiponectin, CTRP1, CTRP3, CTRP5, CTRP6 and CTRP10 cloned into the pCRII TOPO vector (Invitrogen). Once the mutagenesis had been verified by DNA sequencing, adiponectin, CTRP1ΔCys, CTRP2ΔCys, CTRP3ΔCys, CTRP5ΔCys, CTRP6ΔCys, CTRP7ΔCys and CTRP10ΔCys inserts were excised with EcoRI from the pCRII TOPO vector and cloned into the EcoRI site of the mammalian expression vector pCDNA3.1 (Invitrogen). Expression of untagged, or FLAG- or HA-tagged CTRP2 and CTRP7 constructs in HEK (human embryonic kidney)-293T cells did not always result in consistently robust secretion of the expressed proteins compared with other CTRP constructs. Supernatants from transfected HEK-293T cells containing CTRP2 or CTRP7 proteins generally need to be concentrated. To circumvent this issue, we also generated FLAG- and HA-tagged CTRP2 and CTRP7 where we replaced their native leader sequences with that of adiponectin. Results shown in Figures 7(C) and 8 were obtained using these modified CTRP2 and CTRP7 constructs.
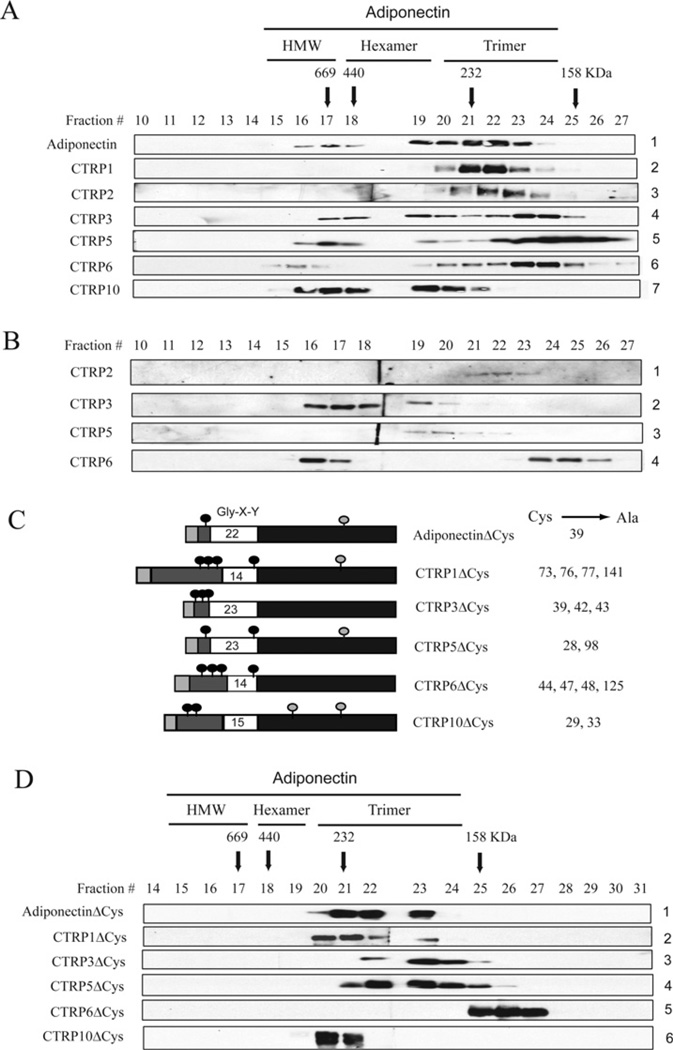
Figure 6. CTRP1, CTRP2, CTRP3, CTRP5, CTRP6 and CTRP10 proteins are secreted in multiple oligomeric forms.
(A) Gel-filtration chromatographic analyses of FLAG-tagged adiponectin and CTRPs. Fractions 10–27 were analysed by immunoblot analyses using an anti-FLAG antibody. Arrows with the molecular mass markers of 669, 440, 232 and 158 kDa correspond to the peak elution fraction of molecular standards thyroglobulin, ferritin, catalase and aldolase respectively. FPLC fractions 16–17, 18–19 and 20–23 that correspond to adiponectin trimers, hexamers and HMW oligomers respectively are indicated along the top of the Figure. (B) Gel-filtration analyses of serum derived from an 8-week-old C57BL/6 female mouse. Fractions 10–27 were analysed by immunoblot analyses using CTRP-specific antibodies. (C) A schematic diagram of adiponectin, CTRP1, CTRP3, CTRP5, CTRP6 and CTRP10. The light grey box represents the signal peptide, the dark grey box represents the N-terminal region, the white box represents the collagen domain with the number of Gly-X-Y repeats indicated, and the black box represents the globular C1q domain. Light grey circles represent cysteine residues found within the globular domains. Black circles represent cysteine residues found in the N-terminus. Numbers in the right-hand column refer to the position of the cysteine residues (indicated by black circles) that were mutated to alanine. (D) Gel-filtration analyses of FLAG-tagged adiponectinΔCys, CTRP1ΔCys, CTRP3ΔCys, CTRP5ΔCys, CTRP6ΔCys and CTRP10ΔCys proteins. Fractions 10–27 were analysed by immunoblot analyses using an anti-FLAG antibody.
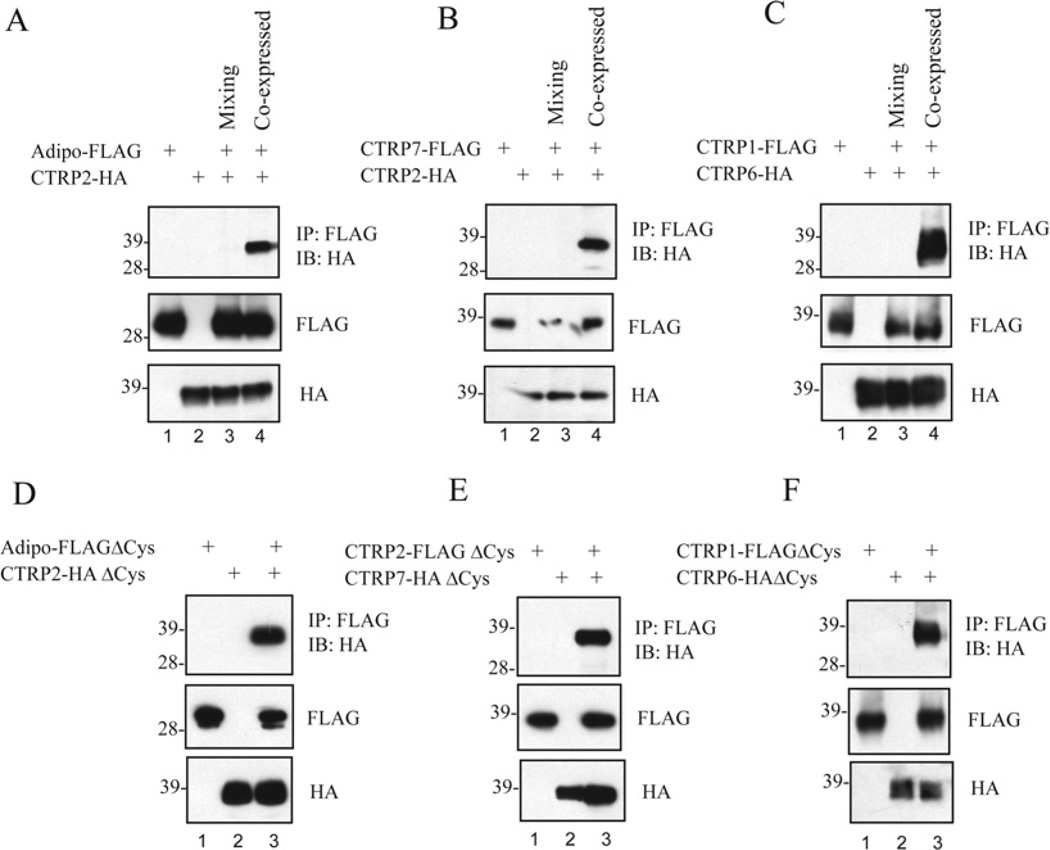
Figure 7. Formation of hetero-oligomers between different CTRPs.
(A–D) FLAG- and HA-tagged adiponectin and CTRPs were co-expressed in different combinations and the supernatants were subjected to co-immunoprecipitations using an anti-FLAG affinity gel and immunoblotted with an anti-HA antibody (top panel). The middle and bottom panels indicate the presence of FLAG- and HA-tagged input proteins in the supernatants. The molecular mass in kDa is indicated on the left-hand side of each gel. IP, immunoprecipitation; IB, immunoblot. (E) Summary of 72 co-immunoprecipitation results (from A–D; Supplementary Figure S3 at http://www.BiochemJ.org/bj/416/bj4160161add.htm). Grey boxes represent homo-oligomerization of adiponectin and each of the CTRPs. Black boxes represent hetero-oligomerization between secreted adiponectin/CTRP2, adiponectin/CTRP9, CTRP1/CTRP6 and CTRP2/CTRP7.
Figure 8. CTRP1/CTRP6, CTRP2/CTRP7 and CTRP2/adiponectin hetero-oligomers formed during biosynthesis and do not involve their N-terminal cysteine residues.
(A–C) FLAG- and HA-tagged adiponectin, CTRP1, CTRP2, CTRP6 and CTRP7 were expressed individually or in combination and the supernatants were subjected to co-immunoprecipitations. For the mixing experiments, individually expressed adiponectin–FLAG and CTRP2–HA, CTRP7–FLAG and CTRP2–HA, and CTRP1–FLAG and CTRP6–HA were mixed for 4 h prior to co-immunoprecipitation with an anti-FLAG affinity gel followed by immunoblotting with an anti-HA antibody (top panels). The middle and bottom panels indicate the presence of FLAG- and HA-tagged input proteins in the supernatants. (D–F) FLAG- and HA-tagged adiponectin, CTRP1, CTRP2, CTRP6 and CTRP7 that lack their N-terminal cysteine residues (ΔCys, refer to the schematic diagram in Figure 6C) were expressed individually or in combination, and the supernatants were subjected to co-immunoprecipitations using an anti-FLAG affinity gel followed by immunoblotting with an anti-HA antibody (top panels). The middle and bottom panels are immunoblots that show the presence of FLAG- and HA-tagged input proteins in the supernatants. The molecular mass in kDa is indicated on the left-hand side of each gel. IP, immunoprecipitation; IB, immunoblot.
A three-step PCR approach was used to generate CTRP3, CTRP5 and CTRP10 that lack the entire collagen domain (referred to as globular g-CTRP3, g-CTRP5 and g-CTRP10). CTRP3ΔCys, CTRP5ΔCys and CTRP10ΔCys cloned into the pCRII TOPO vector were used as PCR templates. First, DNA sequences that correspond to the signal peptides and variable N-terminal regions of CTRP3 (residues 1–44), CTRP5 (residues 1–29) and CTRP10 (residues 1–75) were amplified out and the PCR products were purified. Secondly, DNA sequences that correspond to the entire globular domains of CTRP3 (residues 114–246), CTRP5 (residues 99–243) and CTRP10 (residues 120–287) were amplified out and the PCR products were purified. Thirdly, using PCR products (having overlap at the short variable region) from the first and second steps as a DNA template, we generated constructs encoding g-CTRP3, g-CTRP5 and g-CTRP10. Once these constructs were verified by DNA sequencing, g-CTRP3, g-CTRP5 and g-CTRP10 inserts were excised with EcoRI from the pCRII TOPO vector and cloned into the EcoRI site of the pCDNA3.1 vector.
Antibodies
A mouse monoclonal anti-FLAG M2 antibody was obtained from Sigma. A rat anti-HA antibody (clone 3F10) was obtained from Roche. Goat anti-adiponectin, goat anti-CTRP1/C1qTNF1, goat anti-CTRP3/C1qTNF3 and goat anti-CTRP5/C1qTNF5 antibodies were obtained from R&D Systems. Rabbit polyclonal antibodies specific for mouse CTRP2 were generated as described previously [14]. For the present study, we also generated rabbit polyclonal antibodies against full-length CTRP6 and CTRP10, and the globular domain of CTRP1. Recombinant mouse g-CTRP1, which corresponds to its C-terminal globular domain (amino acids 142–281), was produced by cloning the truncated form of the CTRP1 construct into the pTrcHis TOPO vector (Invitrogen) and maintained in Escherichia coli strain BL21(DE3)pLysE. The N-terminal His6-tagged fusion protein was produced in E. coli, isolated from the lysed bacterial pellet using a nickel-affinity column with Probond resin (Invitrogen), eluted with imidazole-containing buffer [50 mM NaPO4 (pH 7.0), 500 mM NaCl and 250 mM imidazole] and dialysed against PBS. Detoxi-Gel endotoxin-removing gel (Pierce) was used to remove potential endotoxin contaminants. Recombinant FLAG-tagged CTRP6 and CTRP10 were produced and purified from the supernatants of transfected HEK-293T cells. Purified proteins were dialysed against 20 mM Hepes buffer (pH 8.0) containing 135 mM NaCl in a 10 kDa cut-off Slide-A-Lyzer dialysis cassette (Pierce). Rabbit polyclonal antibodies directed at purified recombinant g-CTRP1, CTRP6 and CTRP10 were produced by immunizing NZW (New Zealand White) rabbits as previously described [14]. Sera were collected and tested for their ability to recognize HA-tagged CTRP1, CTRP6 and CTRP10 found in the supernatants of transfected HEK-293T cells.
Animals
C57BL/6, BALB/c, 129SvJ, FVB, DBA/2, AKR mice, ob/ob and their lean controls were all obtained from The Jackson Laboratory. Adiponectin-null mice [23] were kindly provided by Dr Iichiro Shimomura (Department of Metabolic Medicine, Graduate School of Medicine, Osaka University, Osaka, Japan). All animal protocols and experiments were approved by the Massachusetts Institute of Technology animal care and use committee.
Isolation of primary adipocytes and stromal cells from adipose tissue
Epididymal fat pads from ten 8-week-old C57BL/6 mice were removed, minced, pooled together and digested with collagenase (2 mg/g of tissue) at 37°C for 1 h. Digestions were stopped by adding DMEM (Dulbecco’s modified Eagle’s medium) containing 10% (v/v) FBS (foetal bovine serum) and filtered through a 100 μm hole size mesh cell strainer (BD Falcon) to remove undigested tissue. The collected cell suspension was incubated for 10 min at room temperature (25°C) or until adipocytes had floated to the top. The upper phase contained mature adipocytes, whereas the lower phase contained stromal/vascular cells. Primary adipocytes were collected from the floating cell layer, washed once with DMEM to remove collagenase and centrifuged at 180 g for 5–10 min. Total RNAs were extracted from purified primary adipocytes with tri-reagent (Molecular Research Center). The lower-phase layer that contained stromal/vascular cellswas centrifuged at 100–200 g for 5 min, resuspended in DMEM containing 10% (v/v) FBS, and filtered through a 25–70 μm pre-wet cell strainer (BD Falcon). The filtrates were washed once with DMEM and resuspended in erythrocyte lysis buffer (ammonium chloride solution; Stem Cell Technologies) at room temperature for 10 min. Cells were centrifuged at 100 g for 10 min and the supernatants were removed. Total RNA from stromal/vascular cells were extracted with tri-reagent. Potential DNA contaminants were removed by RNase-free DNase I (Ambion).
Quantitative PCR analysis of CTRP expression in mouse tissues
A quantitative real-time PCR approach was used to screen multiple tissue cDNA panels (Clontech), RNA isolated from adipose tissue of 8- and 12-week-old ob/ob mice and their lean controls, and RNA isolated from primary adipocytes and stromal cells for the presence of CTRP transcripts. ABI Prism Primer Express 2.0 software was used to design the following PCR primers: CTRP1 forward, 5′-TCCGAGCTCTGTTGACATGC-3′; CTRP1 reverse, 5′-AAAGATTGACCAGCCCCTGG-3′; CTRP2 forward, 5′-TCCTGGGTACTCTTGGCCTG-3′; CTRP2 reverse, 5′-AAGCATTGGGTCAGCAGCA-3′; CTRP3 forward, 5′-CATCTGGTGGCACCTGCTG-3′; CTRP3 reverse, 5′-TGACACAGGCAAAATGGGAG-3′; CTRP5 forward, 5′-TGGAGTCTGAGCCTCCG-3′; CTRP5 reverse, 5′-AGAAGGGCAAGAAGTGGCCT-3′; CTRP6 forward, 5′-ATCACAGACATGGGCCAAGG-3′; CTRP6 reverse, 5′-TCAACTCACAGACCCCGGAC-3′; CTRP7 forward, 5′-GACGAGTCTTGCCATCTGTGC-3′; CTRP7 reverse, 5′-TTAGCCTGATTGGCCCGAG-3′; CTRP10 forward, 5′-CGGCTTCATGACACTTCCTGA-3′; CTRP10 reverse, 5′-AGCAGGGATGTGTCTTTTCCA-3′; 18S RNA forward, 5′-GCAATTATTCCCCATGAACG-3′; 18S RNA reverse, 5′-GGCCTCACTAAACCATCCAA-3′; GAPDH (glyceraldehyde-3-phosphate dehydrogenase) forward, 5′-GGAGCGAGACCCCACTAACA-3′; and GAPDH reverse, 5′-ACATACTCAG CACCGGCCTC-3′. The default PCR protocol was used on an Applied Biosystems Prism 7000 Sequence Detection System. Epididymal fat pad cDNA from ob/ob mice or their lean controls (8- and 12-week-old), and primary adipocytes and stromal cells were synthesized from 2 μg of total RNA and 200 ng of random hexamers using the Superscript II RNase H-Reverse Transcriptase protocol (Invitrogen). For quantitative PCR, samples were analysed in triplicate 25 μl reaction volumes (10 ng of cDNA, 900 nmol of primer, 12.5 μl of Master Mix and water) according to the standard protocol provided in the SyBR® Green PCR Master Mix protocol (Applied Biosystems).
HEK-293T cell transfection
C-terminal FLAG- or HA-tagged adiponectin, CTRP1, CTRP2, CTRP3, CTRP5, CTRP6, CTRP7, CTRP9, CTRP10, adiponectinΔCys, CTRP1ΔCys, CTRP3ΔCys, CTRP5ΔCys, CTRP6ΔCys and CTRP10ΔCys constructs were used in the present transfection studies. HEK-293T cells were cultured in DMEM containing 10% (v/v) FBS supplemented with 2 mM l-glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin. Transient transfections were performed in HEK-293T cells using Lipofectamine™ 2000 reagent according to the manufacturer’s protocol (Invitrogen). At 24 h after transfection, cells were washed and then cultured in serum-free Opti-MEM I medium (Invitrogen) supplemented with vitamin C (0.1 mg/ml) for another 24–48 h before the conditioned media were collected for Western blot analysis using an anti-FLAG M2 or an anti-HA antibody. A sample of the conditioned media from FLAG-tagged adiponectin, CTRP1, CTRP2, CTRP3, CTRP5, CTRP6, CTRP7 and CTRP10 transfectants was also incubated with PNGase F (peptide N-glycosidase F; New England Biolabs) to determine the presence of N-linked glycans.
Detection of a carbohydrate moiety
Approx. 50 ng of purified recombinant FLAG-tagged CTRP3, CTRP5, CTRP10, g-CTRP3, g-CTRP5 and g-CTRP10 were each separated on duplicate SDS/PAGE gels and transferred on to nitrocellulose membranes. One blot was probed with an anti-FLAG antibody to show the presence of FLAG-tagged proteins. A duplicate blot was used in an ECL® (enhanced chemiluminescence) glycoprotein detection protocol according to the manufacturer’s protocol (Amersham Bioscience). Briefly, any carbohydrate moiety on recombinant proteins blotted on to the nitrocellulose membrane was oxidized with sodium metaperiodate and the oxidized sugar group was labelled with biotin using biotin-hydrazide. The presence of a carbohydrate moiety was then detected using streptavidin conjugated to HRP (horseradish peroxidase) and the chemiluminescence substrate (Millipore).
Gel-filtration chromatographic analysis
Serum from C57BL/6 mice (100 μl) or supernatants (500 μl) from transfected HEK-293T cells containing FLAG-tagged adiponectin, CTRP1, CTRP2, CTRP3, CTRP5, CTRP6, CTRP10, CTRP1ΔCys, CTRP3ΔCys, CTRP5ΔCys, CTRP6Δ-Cys, CTRP10ΔCys, adiponectin–FLAG/CTRP2–HA, CTRP2–FLAG/CTRP7–HA, CTRP1–FLAG/CTRP6–HA were sequentially loaded on to an ÄKTA FPLC and fractionated through a 10/30 Superdex 200 column (GE Healthcare) in PBS. The collected fractions were subjected to Western blot analysis using anti-FLAG, anti-CTRP2, anti-CTRP3, anti-CTRP5 or anti-CTRP6 antibodies.
Co-immunoprecipitation and Western blot analysis
C-terminal FLAG- or HA-tagged adiponectin, CTRP1, CTRP2, CTRP3, CTRP5, CTRP6, CTRP7, CTRP9 and CTRP10 were transfected individually or in combination into HEK-293T cells. Aliquots of the collected supernatants (250–350 μl) combined with 500 μl of IP buffer [150 mM Tris/HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA and 1% (v/v) Triton X-100] were subjected to immunoprecipitation using either 10 μl of EZview™ anti-FLAG affinity gel (Sigma) or anti-HA affinity matrix (Roche). Samples were rotated for 4 h at 4°C, washed four times with IP buffer, resuspended in 60 μl of NuPAGE sample buffer (Invitrogen) containing reducing agent [DTT (dithiothreitol)], heated at 90°C for 10 min, and separated on NuPAGE (10% gel) (Invitrogen). Proteins from gels were transferred on to 0.2 μm Protran BA83 nitrocellulose membranes (Whatman), blocked in 5% (w/v) non-fat dried skimmed milk for 1 h, and probed with a mouse anti-FLAG (1:2000) or rat anti-HA (1:2000) antibody in the presence of 5% (w/v) non-fat dried skimmed milk for 2 h or overnight. Immunoblots were washed three times (10 min each) in PBS containing 0.1% Tween 20 and incubated with HRP-conjugated sheep anti-mouse or goat anti-rat antibody (Amersham Biosciences) (1:2000–1:5000) for 2 h. Blots were washed three times (10 min each) in PBS containing 0.1% Tween 20, developed in chemiluminescence reagent for 2–5 min, and exposed to Blue XB-1 film (Kodak). We used the Molecular Analyst (Bio-Rad) or IMAGEJ software (http://rsb.info.nih.gov/ij) to quantify our Western blot results.
Mixing experiments
We performed mixing experiments to evaluate whether CTRP1/CTRP6, CTRP2/CTRP7 and adiponectin/CTRP2 interactions occurred during their biosyntheses or whether they associate as hetero-oligomers after proper folding and secretion. Conditioned media from transfected HEK-293T cells containing adiponectin–FLAG, CTRP2–HA, CTRP7–FLAG, CTRP1–FLAG and CTRP6–HA were harvested. Conditioned media from co-transfected HEK-293T cells containing co-expressed CTRP1–FLAG/CTRP6–HA, CTRP2–FLAG/CTRP7–HA and adiponectin–FLAG/CTRP2–HA were also harvested. Prior to immunoprecipitations, supernatants containing CTRP1–FLAG was mixed with CTRP6–HA, CTRP2–FLAG was mixed with CTRP7–HA, and adiponectin–FLAG was mixed with CTRP2–HA. The mixtures were allowed to rotate at 4°C for 4 h or overnight (16–20 h). Immunoprecipitations were performed on supernatants containing a single overexpressed protein, a mixture of two proteins expressed separately or medium containing co-expressed proteins using 10 μl of EZ view™ anti-FLAG affinity gel. Samples were washed four times with IP buffer, resuspended in 60 μl of NuPAGE sample buffer containing reducing agent (DTT), heated at 90°C for 10 min, and separated on NuPAGE (10%gel). Proteins from gels were transferred on to 0.2 μm Protran BA83 nitrocellulose membranes. Western blot analysis was carried out as described above using a rat anti-HA (1:2000) antibody.
Production and purification of recombinant CTRP1, CTRP3, CTRP5, CTRP10, g-CTRP3, g-CTRP5 and g-CTRP10
C-terminal FLAG-tagged CTRP1, CTRP3, CTRP5, CTRP10, g-CTRP3, g-CTRP5 and g-CTRP10 were produced and purified from the supernatants of transfected HEK-293T cells. Briefly, 24 h after transfection using Lipofectamine™ 2000 reagent, media were replaced with serum-free Opti-MEM I supplemented with vitamin C (0.1 mg/ml). Supernatants were collected three times, every 48 h, pooled and purified using an anti-FLAG affinity gel according to the manufacturer’s protocol (Sigma), and eluted with 150 μg/ml FLAG peptide (Sigma). Purified proteins were dialysed against 20 mM Hepes buffer (pH 8.0) containing 135 mM NaCl in a 10 kDa cut-off Slide-A-Lyzer dialysis cassette.
Generation of retrovirus vectors and infection of 3T3-L1 adipocytes
C-terminal FLAG-tagged CTRP1, CTRP2, CTRP3, CTRP5, CTRP6, CTRP7, CTRP9 and CTRP10 cDNA were subcloned into the MSCV retroviral vector (XZ201) [24] upstream of an IRES-GFP (internal ribosome entry site-green fluorescent protein) sequence. Expression of GFP is therefore proportional to that of the CTRPs [25]. Additionally, expression of GFP allows us to rapidly assess the infection rate. To generate retroviruses, MSCV–CTRP–FLAG vectors were co-transfected with the pCL-ECO vector (expressing ecotropic retroviral structural proteins) into HEK-293T cells and supernatants containing infectious virions were collected and frozen in 1 ml aliquots until use.
3T3-L1 preadipocytes were grown in 10 cm tissue culture plates and infected with retroviruses encoding GFP- or FLAG-tagged CTRP1, CTRP2, CTRP3, CTRP5, CTRP6, CTRP7, CTRP9 and CTRP10 when they were at ~70% confluence. At 1–2 days after the cells reached confluency, media were replaced with a differentiation cocktail [DMEM supplemented with 10% (v/v) FBS, 0.25 pM dexamethasone, 0.5 mM methyl-1-isobutylmethylxanthine and 160 pM insulin] and cells were allowed to differentiate for 2 days. On day 3, media were replaced with DMEM supplemented with 10% (v/v) FBS and 160 pM insulin. After this, media were replaced every 2 days with DMEM supplemented with 10% (v/v) FBS. On day 7, media were replaced with serum-free Opti-MEM and the supernatants were collected on day 8 for Western blot analysis, immunoprecipitation or gel-filtration chromatographic (FPLC) analysis.
Myotube stimulation
Mouse C2C12 myoblast cells (from A.T.C.C.) were differentiated into myotubes in DMEM containing 2% (v/v) horse serum. On day 6 or 7 post-differentiation, myotubes were starved for 1–2 h in serum-free DMEM and then stimulated with recombinant CTRP1 (2–4 μg/ml) or control buffer [20 mM Hepes (pH 8) and 135 mM NaCl] for 15 min before cells were lysed in RIPA buffer [50 mM Tris/HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA and 1 % Nonidet P40] containing phosphatase inhibitors (Sigma), sodium fluoride, sodium orthovanadate and a protease inhibitor cocktail (Roche). Western blot analysis was carried out on the cleared lysates using rabbit antibodies recognizing phospho-AMPK-α (Thr172), phospho-ACC (acetyl-CoA carboxylase; Ser79), phospho-p44/42 MAPK (Thr202 and Tyr204), phospho-Akt (Thr308), phospho-mTOR (mammalian target of rapamycin; Ser2448), phospho-IκB [inhibitor of NF-κB (nuclear factor κB); Ser32] (all from Cell Signaling Technology). Blots were stripped and reprobed with antibodies recognizing total AMPK, mTOR, Akt, p44/42-MAPK, IκB (all from Cell Signaling Technology) and ACC (Epitomics). An antibody conjugated to LI-COR IRDyeTM800 (Rockland Immunochemicals) was used as our secondary antibody for the purpose of quantification using the LI-COR Odyssey IR imaging system.
In vivo study
All mice (C57BL/6) aged 8–10-weeks-old were purchased from The Jackson Laboratory and were allowed to acclimatize to the animal facility for at least 1 week. Food was removed for 1–2 h in the morning prior to recombinant protein injection and only drinking water was supplied for the duration of the experiment. Approx. 2 μg/g of body weight of recombinant CTRP1, CTRP5 or the equivalent volume of control buffer [20 mM Hepes (pH 8) and 135 mM NaCl] was injected into mice (n=6–17) via the intraperitoneal route. Blood glucose levels were measured at zero time (prior to protein injection) and at 2, 4, 5 and 6 h post-injection using a glucometer (BD Pharmingen). Approx. 5 μl of blood was collected from the tip of the mouse tail for blood glucose measurements. In separate experiments, different batches of mice and recombinant CTRP1 were used.
RESULTS
Identification of CTRP10 cDNA
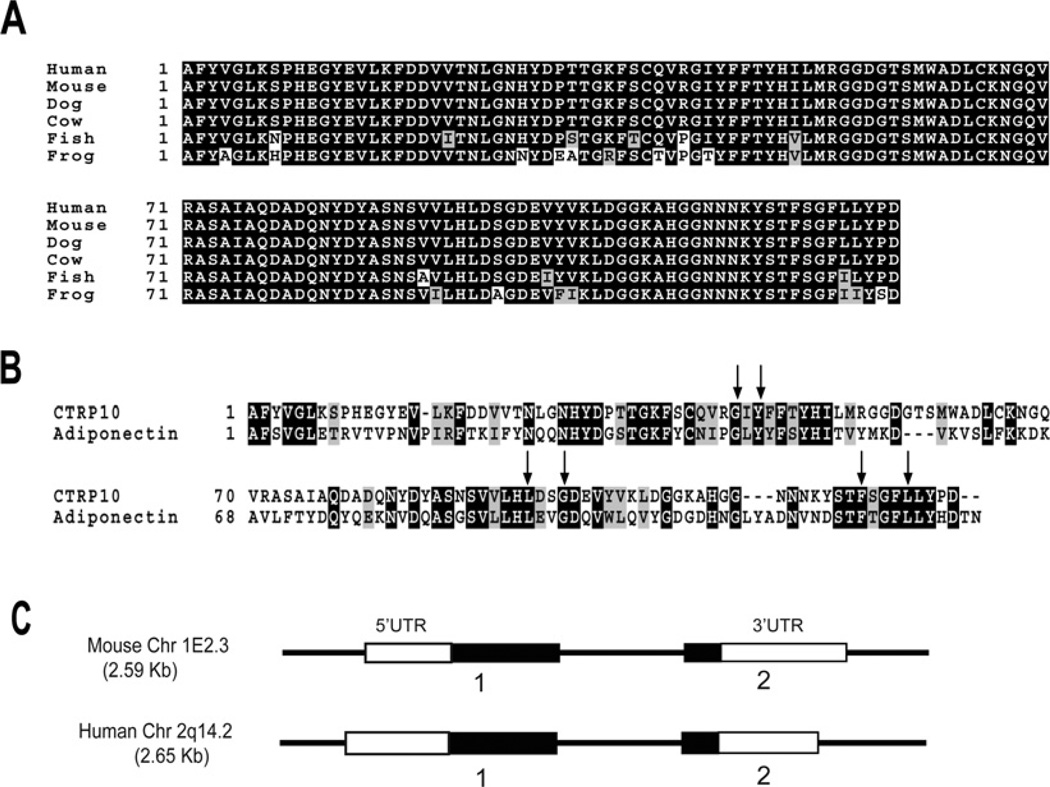
Using the globular C1q domain sequence of adiponectin to screen the NCBI GenBank® databases, we identified another novel and highly conserved paralogue of adiponectin distinct from the seven paralogues (designated as CTRP1–7) we previously identified [14]. We designated our novel cDNA and its encoded protein CTRP10 (Supplementary Figure S1 at http://www.BiochemJ.org/bj/416/bj4160161add.htm). Like adiponectin and other CTRPs, CTRP10 is composed of a signal peptide (residues 1–21), a short N-terminal variable region (residues 22–75), a collagen domain with 15 Gly-X-Y repeats (residues 76–120) and a C-terminal globular domain that is homologous with immune complement protein C1q (residues 121–287). Mouse CTRP10 contains four highly conserved cysteine residues: Cys29 and Cys33 are located at the N-terminal variable region, whereas Cys197 and Cys224 are located in the C-terminal globular domain. In contrast, the residue that corresponds to Cys33 in mouse CTRP10 is replaced by a tryptophan residue in human CTRP10. CTRP10 is highly conserved throughout evolution. Mouse CTRP10 and its corresponding human orthologue shared a striking 81%, 100% and 100% amino acid identity in their short N-terminal variable regions, collagen domains and C-terminal globular domains respectively (Figure 1A). Similarly, CTRP10 orthologues from the draft genome sequence of dog (Canis familiaris; XP_541006), cow (Bos taurus; XP_595145), puffer-fish (Tetraodon nigroviridis; CAG12827) and African clawed frog (Xenopus tropicalis; AAH88035) also showed remarkable conservations in their globular domains, with amino acid identity of 100%, 100%, 91% and 86% with mouse CTRP10 respectively. Four highly conserved residues (Gly159, Tyr161, Phe237 and Leu241 in adiponectin) important in the packing of the protomer’s hydrophobic core of C1q/TNF family members [15] are also conserved in CTRP10 (Figure 1B, arrows). Both human and mouse CTRP10 genes have similar exon/intron structures (Figure 1C). The mouse CTRP10 gene is 2.59 kb in size, consists of two exons and is located on chromosome 1E2.3. The human CTRP10 gene is 2.65 kb in size, consists of two exons and is located on chromosome 2q14.2.
Figure 1. Identification and cloning of CTRP10 cDNA.
(A) ClustalW alignments of the CTRP10 globular domain sequences of human (Homo sapiens, NP_872334), mouse (Mus musculus, AAY21933), dog (C. familiaris, XP_541006), cow (B. taurus, XP_595145), pufferfish (T. nigroviridis, CAG12827) and frog (X. tropicalis, AAH88035). Identical amino acids are shown as white text on a black background. (B) ClustalW alignment of the globular domain sequences between mouse CTRP10 and adiponectin. Identical amino acids are shown as white text on a black background and gaps are indicated by dashes. Arrows indicate amino acids that are conserved between the C1q and TNF family of proteins based on the crystal structure of adiponectin [15]. (C) Exon/intron organizations of the mouse and human CTRP10 gene. The size of the mouse CTRP10 exons 1 and 2 are 1220 and 779 bp respectively. The encoded mouse cDNA has a 535 bp 5′UTR (untranslated region) and a 599 bp 3′UTR. The size of the human CTRP10 exons 1 and 2 are 1310 and 738 bp respectively. The encoded human cDNA has a 625 bp 5′UTR and a 559 bp 3′UTR. Open boxes (white) refer to exons that encode the 5′ and 3′UTR, whereas the closed boxes (black) refer to exons that code for amino acids.
Expression profiles of CTRP1, CTRP2, CTRP3, CTRP5, CTRP6, CTRP7 and CTRP10 transcripts
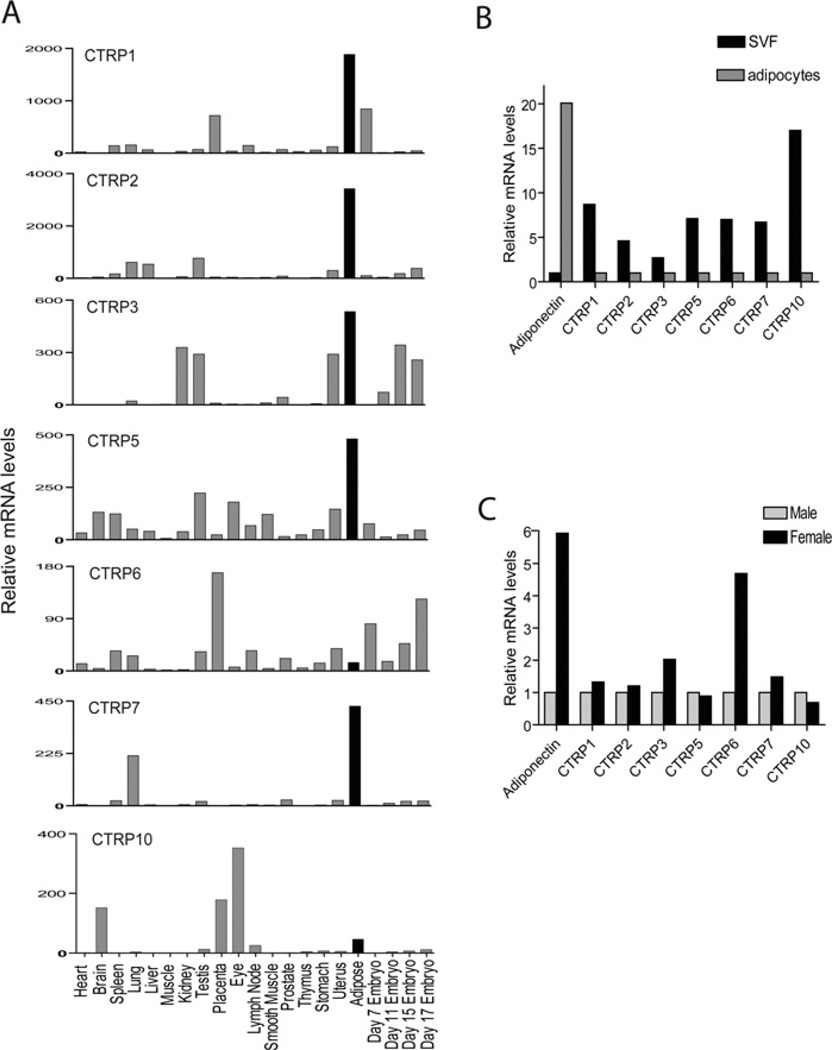
The expression of adiponectin transcripts is almost exclusively restricted to adipose tissue [3–5]. Although many tissues expressed low levels of CTRP transcripts, quantitative real-time PCR analysis revealed that adipose tissue was the predominant tissue that expressed CTRP1, CTRP2, CTRP3, CTRP5 and CTRP7 transcripts among the 17 different adult mouse tissues examined (Figure 2A). Expression levels of CTRP transcripts also varied across four developmental stages (day 7, 11, 15 and 17) of the mouse embryos, with some expressing early (CTRP1 and CTRP6), whereas others were late (CTRP3), during embryogenesis, suggesting that CTRP1, CTRP3 and CTRP6 may have embryonic and post-embryonic functions.
Figure 2. Analysis of CTRP1, CTRP2, CTRP3, CTRP5, CTRP6, CTRP7 and CTRP10 transcript levels in mouse tissues, primary adipocytes and stromal cells.
(A) Quantitative real-time PCR analyses of CTRP expression in mouse tissues. Expression levels of CTRP transcripts in different tissues were normalized to their corresponding GAPDH levels. The bar on the histograms that represents adipose tissue is highlighted in black. (B) Quantitative real-time PCR analyses of CTRP expressions in primary adipocytes and stromal cells [termed stromal vascular fraction (SVF)]. All PCR results were first normalized to their corresponding 18S RNA levels. The relative expression levels of CTRPs in primary adipocytes were normalized to that of SVF, which was set as 1.0. (C) Expression levels of CTRP transcripts in SVF between male and female mice were compared. For adiponectin, its expression levels in primary adipocytes were compared between male and female mice. All PCR results were first normalized to their corresponding 18S RNA levels. The relative expression levels of CTRPs in female were normalized to that of male mice, which was set as 1.0. All of these results were obtained from tissues or cells pooled from multiple mice and thus represent a single experiment.
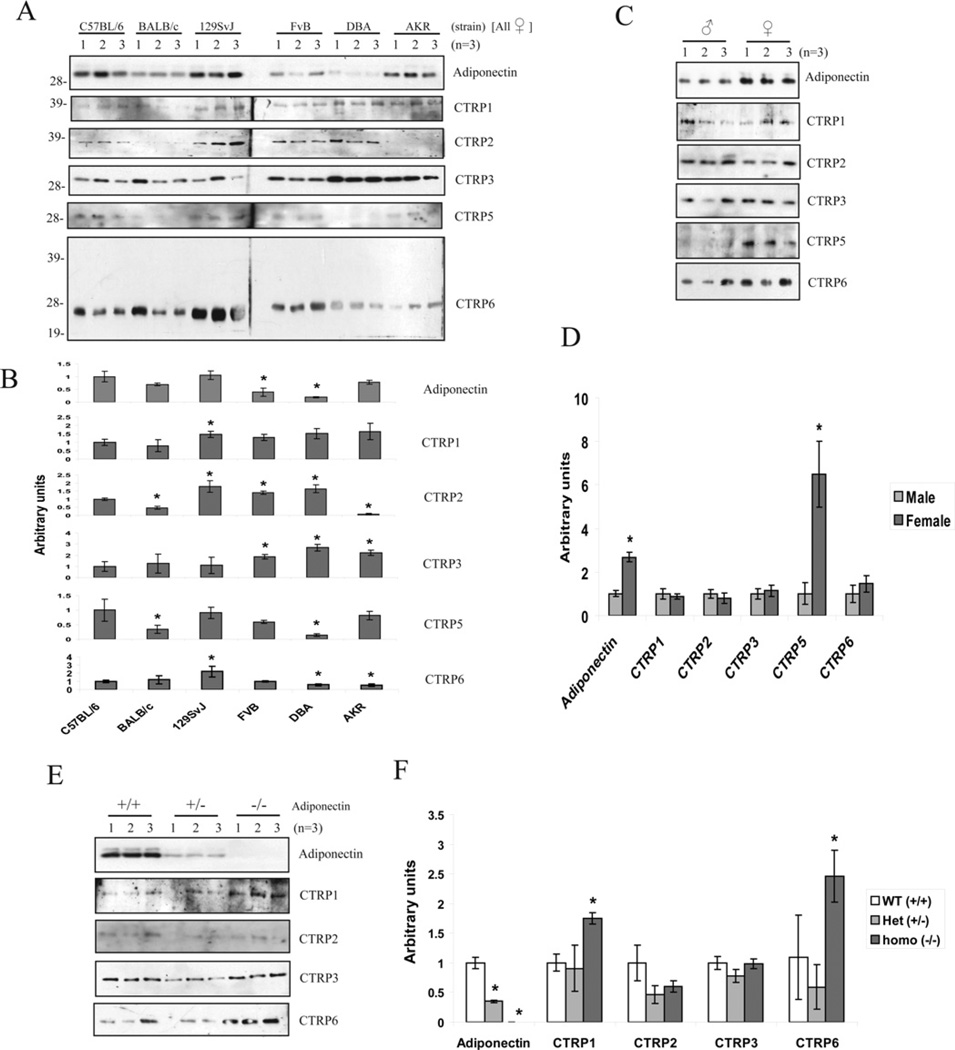
Since normal adipose tissue is composed of mature adipocytes and stromal cells that include preadipocytes, fibroblasts, mature endothelial cells and smooth muscle cells, we sought to determine the cellular source of CTRP1, CTRP2, CTRP3, CTRP5 and CTRP7 transcripts. Although primary adipocytes expressed 20-fold higher levels of adiponectin transcript relative to stromal cells, CTRP1, CTRP2, CTRP3, CTRP5 and CTRP7 transcripts were preferentially expressed by stromal cells (Figure 2B). Similar to adiponectin, CTRP3 and CTRP6 also displayed similar gender-biased expression profiles with females expressing 2- and 5-fold higher levels of the transcript in stromal cells compared with male mice respectively (Figure 2C).
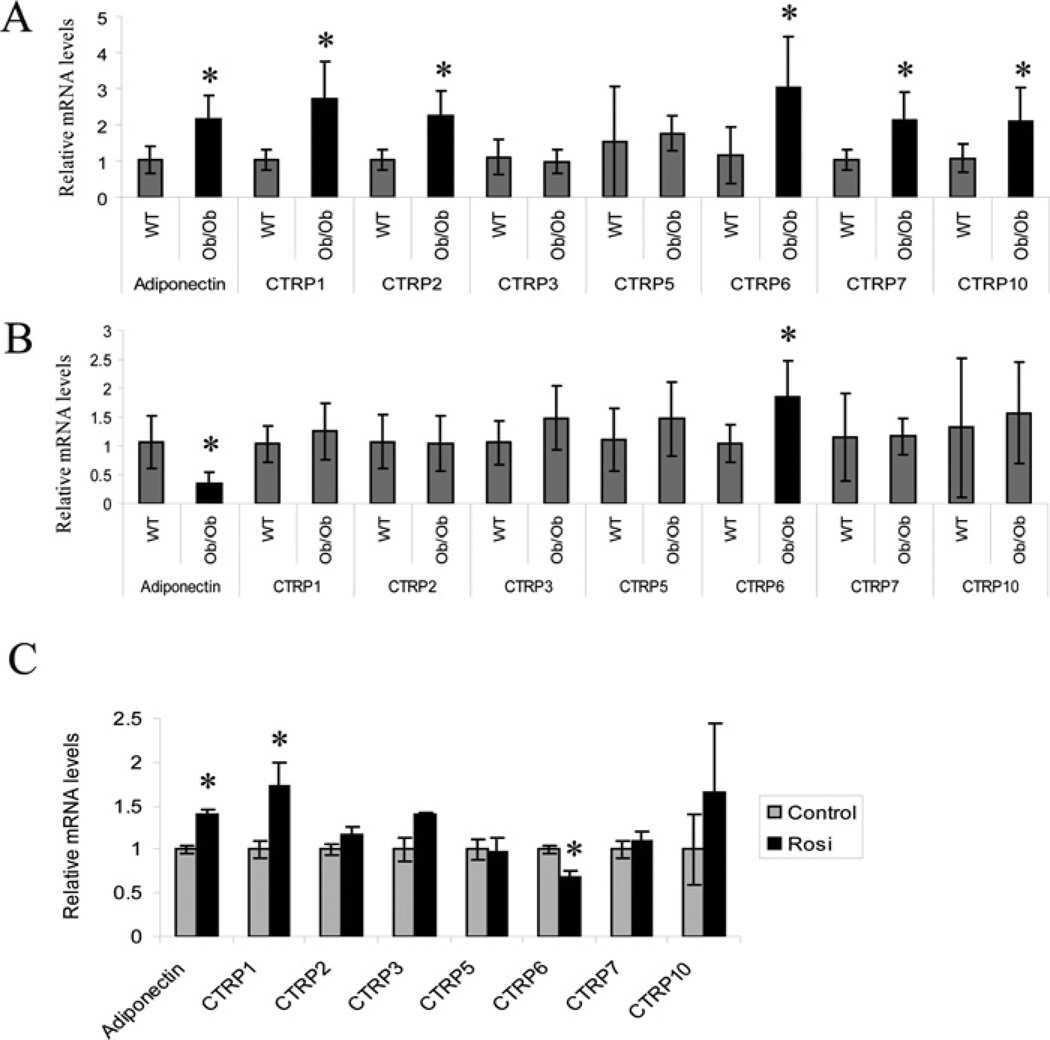
Expression of many adipokine genes are dysregulated in obesity and diabetes. Notably, adiponectin expression is significantly down-regulated in adipose tissue of ob/ob mice relative to lean, age-matched controls [4]. Consequently, we examined the expression levels of CTRP transcripts in the adipose tissues (epididymal fat pads) harvested from ob/ob mice and their lean controls. Expression levels of adiponectin, CTRP1, CTRP2, CTRP6, CTRP7 and CTRP10 transcripts were significantly upregulated in 8-week-old ob/ob mice relative to their age-matched controls (Figure 3A). In contrast, adiponectin transcript levels were significantly decreased in 12-week-old ob/ob mice relative to their lean controls, whereas CTRP6 transcript levels were increased relative to their lean controls (Figure 3B). No significant differences were seen in the expression levels of other CTRP transcripts in 12-week-old ob/ob mice relative to their lean controls.
Figure 3. Analysis of CTRP1, CTRP2, CTRP3, CTRP5, CTRP6, CTRP7 and CTRP10 transcript levels in adipose tissue of lean and ob/ob mice, and in mice treated with rosiglitazone.
(A) Quantitative real-time PCR analyses of adiponectin and CTRP expression in epididymal fat pads of 8-week-old (n = 10) and (B) 12-week-old (n = 8) ob/ob mice and their lean controls (WT; n = 8–10). Expression levels of CTRP transcripts were first normalized to their corresponding 18S RNA levels. The expression levels of CTRPs in ob/ob mice were normalized to that of lean mice. (C) Quantitative real-time PCR analyses of adiponectin and CTRP expressions in 9-week-old male ob/ob mice dosed with 15 mg of rosiglitazone (Rosi)/kg (n = 6) or vehicle control (n = 6) once per day for 3 weeks. Expression levels of CTRP transcripts were normalized to their corresponding 18S RNA. Values are means ± S.D. *P < 0.05 (measured using Student’s t test) for rosiglitazone-treated compared with control mice.
The TZD class of anti-diabetic drugs ameliorates insulin resistance in part through up-regulation of adiponectin gene expression [9]. We therefore examined whether the TZD compound rosiglitazone had any effect on the expression levels of CTRP transcripts in vivo. Administration of rosiglitazone daily for 3 weeks at a dose of 15 mg/kg significantly increased the expression of adiponectin and CTRP1, whereas it decreased the expression of CTRP6 transcript levels in adipose tissue (Figure 3C). At this dose and length of treatment, rosiglitazone had no appreciable effect on the transcript levels of CTRP2, CTRP3, CTRP5, CTRP7 and CTRP10.
CTRP1, CTRP2, CTRP3, CTRP5, CTRP6 and CTRP10 are secreted glycoproteins
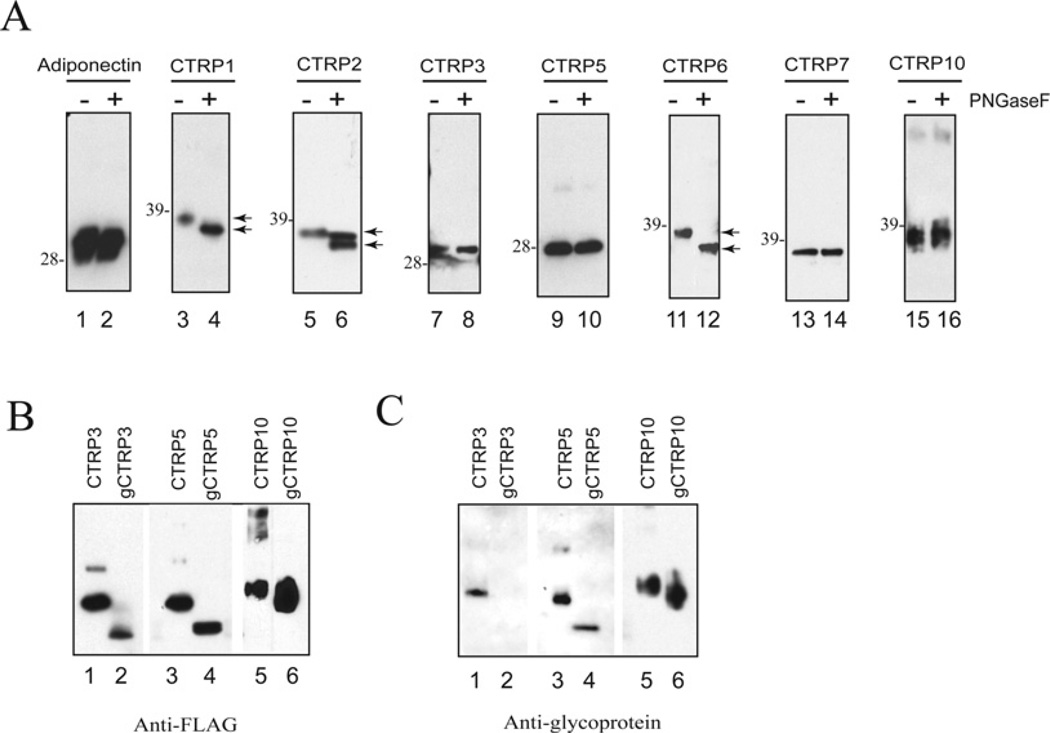
All CTRPs possess signal peptides at their N-termini and hence are predicted to be secreted proteins. When full-length constructs encoding C-terminal FLAG-tagged CTRP1, CTRP2, CTRP3, CTRP5, CTRP6, CTRP7 and CTRP10 were transfected into HEK-293T cells, the resultant proteins were readily detected in the supernatants of transfected cells, confirming that CTRP1, CTRP2, CTRP3, CTRP5, CTRP6, CTRP7 and CTRP10 are secreted proteins (Figure 4A). The deduced amino acid sequences of CTRP3, CTRP5 and CTRP10 do not contain any potential N-linked glycosylation sites that conform to the consensus motif N-X-(S/T). In contrast, CTRP1, CTRP2 and CTRP7 have one potential N-linked glycosylation site, whereas CTRP6 has two potential N-linked glycosylation sites. As expected, when treated with PNGase F, no shift in the apparent molecular mass of CTRP3, CTRP5, CTRP7 and CTRP10 was observed on SDS/PAGE, thus confirming the absence of N-linked glycans on these proteins (Figure 4A, lanes 7–10 and 13–16). In contrast, treatment with N-glycanase resulted in a shift in the apparent molecular masses of CTRP1, CTRP2 and CTRP6 proteins (Figure 4A, lanes 3–6 and 11–12), confirming that those N-linked glycosylation sites found on these proteins are functional and contain N-linked glycans. Although CTRP3, CTRP5 and CTRP10 proteins do not contain N-linked glycans, their apparent molecular masses (30, 28 and 37 kDa respectively) in SDS/PAGE are greater than their predicted molecular masses (24, 24 and 27 kDa respectively), suggesting the presence of carbohydrate moieties. Therefore we used a glycoprotein detection method to label all sugar groups on CTRP3, CTRP5, CTRP10, g-CTRP3, g-CTRP5 and g-CTRP10 and detected the presence of biotin-labelled sugar groups with streptavidin–HRP and chemiluminescence substrates. This allowed us to demonstrate that CTRP3, CTRP5 and CTRP10 are secreted glycoproteins (Figure 4C, lanes 1, 3 and 5). g-CTRP5 and g-CTRP10 that lack the entire collagen domain also contain sugar groups that can be specifically labelled with biotin and detected with streptavidin–HRP (Figure 4C, lanes 4 and 6), thus suggesting that some or all of the sugar groups on CTRP5 and CTRP10 are located on their N-terminal variable regions and/or C-terminal globular domains. In contrast, CTRP3, but not g-CTRP3, contained sugar groups that can be specifically labelled with biotin and detected with streptavidin–HRP (Figure 4C, lanes 1 and 2), suggesting that all the sugar groups on CTRP3 are localized to its collagen domain. The types of carbohydrate attached to these proteins remain to be determined.
Figure 4. CTRP1, CTRP2, CTRP3, CTRP5, CTRP6 and CTRP10 are secreted glycoproteins.
(A) Supernatants containing C-terminal FLAG-tagged CTRPs were incubated with (+) or without (−) PNGase F to determine the presence of N-linked glycans. Arrows indicate changes in protein size before and after PNGase F treatment. The molecular mass in kDa is indicated on the left-hand side of each gel. (B and C) Recombinant FLAG-tagged CTRP3, CTRP5, CTRP10 and their corresponding collagen domain-deleted globular forms (g-CTRP3, g-CTRP5 and g-CTRP10) were subjected to SDS/PAGE immunoblot analysis. The blot on the left-hand side (B) was probed with an anti-FLAG antibody. The blot on the right-hand side (C) was subjected to glycoprotein detection analysis (see the Materials and methods section) to reveal the presence of carbohydrates.
We next investigated whether CTRPs circulate in the mouse serum. This information would indicate whether CTRPs act in vivo in an autocrine, paracrine and/or endocrine manner. First, we showed that the antibodies we generated against CTRP1, CTRP2, CTRP6, CTRP9 and CTRP10 and the recently available, commercially derived antibodies against CTRP3 and CTRP5 were highly specific for each of the CTRPs (Supplementary Figure S2 at http://www.BiochemJ.org/bj/416/bj4160161add.htm). Using these CTRP-specific antibodies, we were able to detect the presence of CTRP1, CTRP2, CTRP3, CTRP5 and CTRP6 proteins in the serum (Figure 5A). The size of serum CTRP1, CTRP2, CTRP3 and CTRP5 on immunoblots is similar to the size of their respective full-length proteins secreted from transfected cells. In contrast, the size of serum CTRP6 on immunoblots corresponded to its globular head (~25 kDa) rather than the full-length protein (~38 kDa), suggesting that CTRP6 undergoes a proteolytic processing in vivo (Figure 5A, bottom panel and Supplementary Figure S2). Purified recombinant CTRP6 can compete out the binding of CTRP6 antibody to the 25 kDa serum protein on immunoblot, thus confirming the specificity of the CTRP6 antibody (results not shown).
Figure 5. Detection of CTRP1, CTRP2, CTRP3, CTRP5 and CTRP6 in mouse serum.
(A) Sera from six different strains of 8-week-old female mice (C57BL/6, BALB/c, 129SvJ, FVB, DBA and AKR) (n =3) were analysed by SDS/PAGE using antibodies specific to adiponectin or CTRPs. The molecular mass in kDa is indicated on the left-hand side of the gels. (B) Densitometric analysis of results presented in (A). Values are means±S.D. *P <0.05 (measured using Student’s t test) when compared with C57BL/6 mice. (C) Sera from 8-week-old male and female C57BL/6 mice (n = 3) were analysed by SDS/PAGE using antibodies specific to adiponectin or CTRPs. (D) Densitometric analysis of results presented in (C). Values are means±S.D. *P <0.05 (measured using Student’s t test) between male and female. (E) Sera from adiponectin wild-type (+/+), heterozygous (+/−) and homozygous null (−/−) male mice (on C57BL/6 background) were analysed by SDS/PAGE using antibodies specific to adiponectin, CTRP1, CTRP2, CTRP3 and CTRP6. (F) Densitometric analysis of results presented in (E). Values are means±S.D. *P <0.05 (measured using Student’s t test) between adiponectin-null (−/−) and wild-type (+/+; WT) mice.
Genetic background plays a prominent role in dictating the susceptibility of mice to diet-induced obesity and diabetes [26,27]. Strikingly, female mice of various genetic backgrounds have significantly different levels of circulating CTRPs (Figures 5A and 5B). Although both male and female mice expressed comparable levels of CTRP5 transcripts, the circulating serum levels of CTRP5 were significantly higher in female compared with male mice (Figure 5C, panel 5 and 5D), suggesting a potential posttranscriptional regulation of CTRP5 mRNA. We also evaluated the serum levels of various CTRPs in adiponectin-null mice and found that the serum levels of CTRP1 and CTRP6 were increased in the absence of adiponectin (Figures 5E and 5F).
CTRP1, CTRP2, CTRP3, CTRP5, CTRP6 and CTRP10 form higher-order oligomeric complexes
All C1q/TNF family proteins studied to date form trimers as their basic structural units and may be further assembled into higher-order oligomeric complexes via intermolecular disulfide bonding [16]. Adiponectin exists as trimers, hexamers and HMW (high-molecular-mass) oligomers of 18 units with distinct biological properties [28–30]. To address whether CTRP1, CTRP2, CTRP3, CTRP5, CTRP6 and CTRP10 are also assembled into higher-order structures, we performed gel-filtration chromatographic (FPLC) analysis on supernatants collected from transiently transfected HEK-293T cells. Because both adiponectin and CTRPs are reasonably similar in size and all have a globular head with a relatively rigid collagen triple helical stalk, we used the FPLC profile of adiponectin (Figure 6A, panel 1) as our benchmark to evaluate the oligomeric structures of CTRPs in our gel-filtration analysis. Although secreted CTRP1 and CTRP2 formed predominantly trimers (Figure 6A, panels 2 and 3), secreted CTRP3, CTRP5, CTRP6 and CTRP10 formed trimers and higher-order oligomers that presumably corresponded to hexamers and HMW oligomers (Figure 6A, panels 4–7). To investigate the higher-order structures of CTRPs in vivo, we performed gel-filtration analysis on mouse serum. The majority of the CTRP3 existed as HMW oligomers (Figure 6B, panel 2), whereas CTRP2 and CTRP5 existed mostly as trimers in serum (Figure 6B, panels 1 and 3). In contrast, the globular form of serum CTRP6 existed in two distinct oligomeric forms (Figure 6B, panel 4), but its precise oligomeric structure cannot be resolved in the present study.
Adiponectin and all CTRPs have a variable number of cysteine residues located in their N-termini preceding their globular domains (Figure 6C). These cysteine residues are presumed to participate in intermolecular disulfide bonding. Site-directed mutagenesis revealed that Cys39 is required for adiponectin to form hexamers and HMW oligomers [29,31] (Figure 6D, panel 1). Similarly, when all of the cysteine residues located in the N-termini of CTRP1, CTRP3, CTRP5, CTRP6 and CTRP10 were mutated to alanine, the secreted proteins formed only trimers, but not higher-order oligomeric forms, thus demonstrating that the N-terminal cysteine residues are required for their respective trimers to assemble into higher-order oligomeric complexes (Figures 6C and 6D).
Physical associations between different CTRPs
Members of the C1q/TNF family of proteins are known to form hetero-oligomers [32–34]. To systematically investigate possible hetero-oligomerization between different CTRPs, we constructed FLAG- and HA-tagged adiponectin, CTRP1, CTRP2, CTRP3, CTRP5, CTRP6, CTRP7, CTRP9 and CTRP10 constructs, co-transfected them in different combinations into HEK-293T cells, and performed co-immunoprecipitations on the secreted proteins found in the supernatants of transfected cells. This allowed us to demonstrate that: (i) all CTRPs form homooligomers; and (ii) CTRP1/CTRP6, CTRP2/CTRP7, adiponectin/ CTRP2 and adiponectin/CTRP9 can also be secreted as hetero-oligomers (Figures 7A–7D and Supplementary Figure S3 at http://www.BiochemJ.org/bj/416/bj4160161add.htm). These associations are specific because, out of the 63 combinations of co-expression, only four combinations resulted in robust and consistent formation of hetero-oligomers despite the fact that all of them were secreted into the conditioned medium of transfected cells. None of the CTRPs in the supernatants associated nonspecifically with the anti-FLAG or anti-HA resins (results not shown). We were also able to co-immunoprecipitate endogenous adiponectin with CTRP2 from the supernatant of differentiated 3T3-L1 adipocytes overexpressing FLAG-tagged CTRP2 (results not shown), thus confirming the adiponectin/CTRP2 association observed in transiently transfected HEK-293T cells. This association is specific because CTRP3, CTRP5, CTRP6, CTRP7 and CTRP10 did not co-immunoprecipitate with adiponectin although they were expressed and secreted into the supernatants of differentiated 3T3-L1 adipocytes (results not shown).
Requirements for CTRP hetero-oligomerization
Although adiponectin exists as trimers, hexamers and HMW oligomers, each of these oligomeric forms are stable and they do not interchange subunits nor interconvert [29,31]. These observations prompted us to address whether CTRP1/CTRP6, CTRP2/ CTRP7 and adiponectin/CTRP2 associations occurred during their biosyntheses or whether hetero-oligomerization occurred after proper folding and secretion. To do that, we performed experiments mixing supernatants containing individually expressed CTRPs. As shown in Figures 8(A)–8(C), CTRP1/CTRP6, CTRP2/CTRP7 and adiponectin/CTRP2 hetero-oligomeric complexes co-immunoprecipitated from the supernatants only if they were co-expressed in the same cells. Mixing supernatants containing CTRP1–FLAGwith CTRP6–HA, CTRP7–FLAG with CTRP2–HA or adiponectin–FLAG with CTRP2–HA, did not result in co-immunoprecipitation, consistent with the notion that CTRP1/CTRP6, CTRP2/CTRP7 and adiponectin/CTRP2 associate during their biosyntheses in the endoplasmic reticulum or during their transit through the Golgi compartment prior to secretion. Prolonged mixing of supernatants containing CTRP1–FLAG with CTRP6–HA, CTRP7–FLAG with CTRP2–HA and adiponectin–FLAG with CTRP2–HA overnight did not result in physical association, suggesting that once these proteins are properly folded and secreted, they are stable and do not interchange subunits (results not shown). One possibility is that CTRP1/CTRP6, CTRP2/CTRP7 and adiponectin/CTRP2 associate in a covalent manner involving their N-terminal cysteine residues. Despite the lack of N-terminal cysteine residues (mutated to alanine), adiponectin–FLAGΔCys/CTRP2–HAΔCys, CTRP2–FLAGΔCys/CTRP7–HAΔCys and CTRP1–FLAGΔCys/CTRP6–HAΔCys hetero-oligomeric complexes can be co-immunoprecipitated from the supernatants of co-transfected cells (Figures 8D–8F). Thus none of the N-terminal cysteine residues are needed for their physical associations.
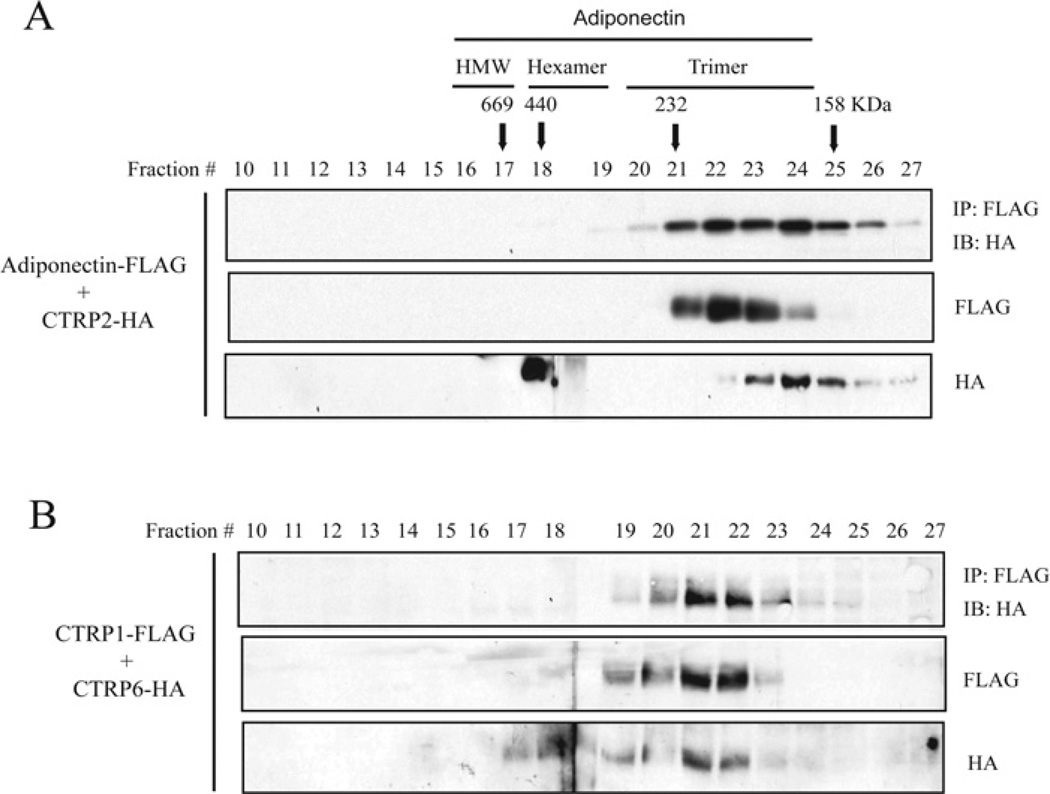
Adiponectin/CTRP2 and CTRP1/CTRP6 form heterotrimers
A trimer is the basic structural unit of all C1q/TNF family proteins. Since N-terminal cysteine residues are not required for adiponectin/CTRP2 and CTRP1/CTRP6 hetero-oligomerizations, we asked whether they could associate as heterotrimers. We performed gel-filtration analysis on supernatants collected from HEK-293T cells co-transfected with adiponectin–FLAG/CTRP2–HA or CTRP1–FLAG/CTRP6–HA constructs. Aliquots from FPLC fractions 10–27 were analysed by Western blotting. When co-transfected into cells, both secreted adiponectin and CTRP2, as well as CTRP1 and CTRP6 form predominantly trimers (Figures 9A and 9B, middle and bottom panels). We performed co-immunoprecipitations on FPLC fractions 10–27 to show that adiponectin/CTRP2 and CTRP1/CTRP6 co-immunoprecipitated from fractions 20–25 that contained the trimeric form of these proteins (Figures 9A and 9B, top panel), thus confirming that adiponectin/CTRP2 and CTRP1/CTRP6 can indeed form heterotrimers.
Figure 9. Adiponectin/CTRP2 and CTRP1/CTRP6 form heterotrimers.
Gel-filtration chromatographic analyses of co-expressed adiponectin–FLAG/CTRP2–HA (A) or CTRP1–FLAG/CTRP6–HA (B). Fractions 10–27 were analysed by immunoblot analysis using an anti-FLAG or an anti-HA antibody. Fractions 10–27 were subjected to co-immunoprecipitations using an anti-FLAG affinity gel and immunoblotted with an anti-HA antibody (top panel). FPLC fractions 16–17, 18–19 and 20–23 that correspond to adiponectin trimers, hexamers and HMW oligomers respectively are indicated across the top of the Figure. IP, immunoprecipitation; IB, immunoblot.
In vitro and in vivo activity of recombinant CTRP1
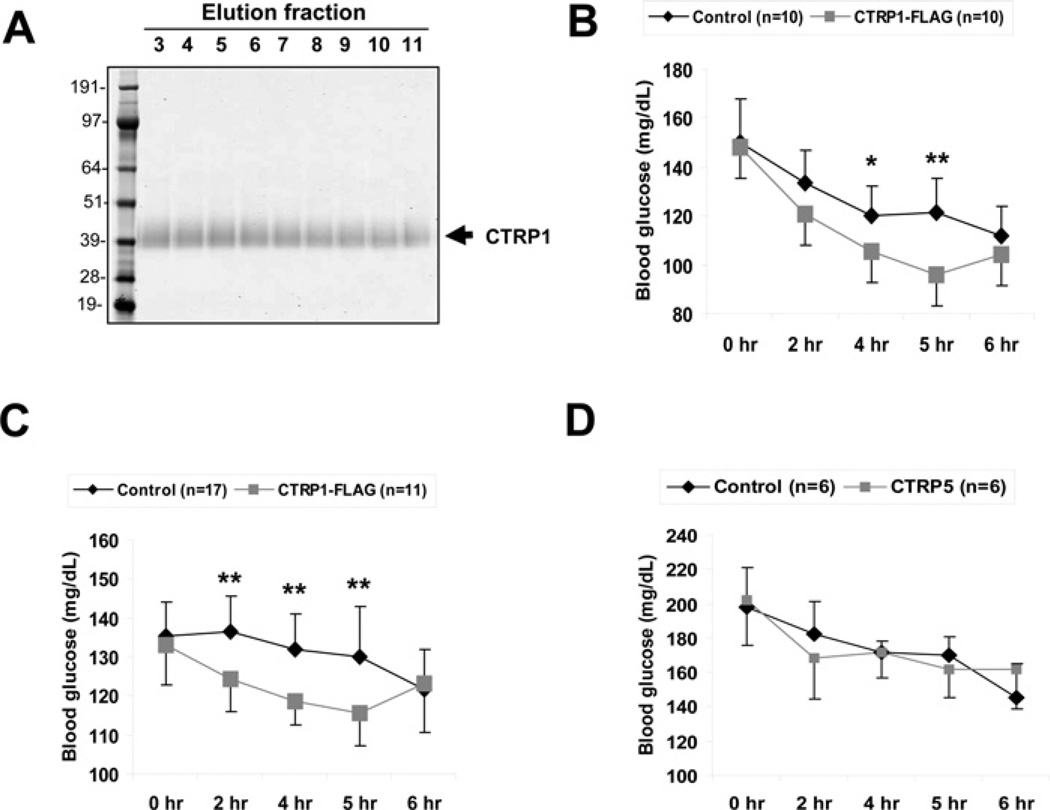
Because CTRP1 transcript levels are up-regulated in mice by the insulin-sensitizing drug rosiglitazone, and because CTRP1 protein levels are increased in the serum of adiponectin-null mice, we focused our initial functional characterization on CTRP1. In differentiated mouse C2C12 myotubes, stimulation with recombinant CTRP1 specifically activated Akt and p44/42-MAPK signalling pathways (Figure 10), but not the AMPK, ACC, mTOR or NF-κB pathways (Supplementary Figure S4 at http://www.BiochemJ.org/bj/416/bj4160161add.htm). To determine whether CTRP1 had any metabolic effect in vivo, we injected recombinant CTRP1 at 2 μg/g of body weight into C57BL/6 mice and demonstrated that it significantly lowered their blood glucose levels relative to control mice injected with control buffer (Figures 11A–11C). Different batches of mice and recombinant CTRP1 were used in multiple separate experiments to show the significant and reproducible glucose-lowering effect upon injection of the protein. The glucose-lowering effect induced by CTRP1 was specific because injection of another closely related protein, CTRP5, into mice had no effect on their glucose levels compared with controls (Figure 11D).
Figure 10. Recombinant CTRP1 activates Akt and p44/42 MAPK signaling pathways in C2C12 myotubes.
(A and B) Differentiated C2C12 myotubes were treated with control buffer or 4 μg/ml recombinant CTRP1 for 15 min. Western blot analysis were carried out on cleared cell lysates using antibodies specific to phospho-Akt (Thr308), total Akt, phospho-p44/42 MAPK (Thr202/Tyr204) and total p44/42 MAPK. The ratios of phospho to total proteins were quantified. Results shown are the fold difference between control buffer (set to a value of 1) compared with recombinant-CTRP1-treated myotubes. All experiments were performed in triplicate and similar results were obtained in two independent experiments. Values are means±S.D. *P <0.05 (measured using Student’s t test) between control buffer and CTRP1-treated myotubes.
Figure 11. Recombinant CTRP1 reduces blood glucose in mice.
(A) Representative Coomassie-Blue-stained gel of purified CTRP1–FLAG. (B and C) Different batches of recombinant CTRP1 were used in separate in vivo studies. Approx. 2 μg of recombinant CTRP1/g of body weight or the equivalent volume of control buffer [20 mM Hepes (pH 8) and 135 mM NaCl] were injected into 8–10-week-old C57BL/6 mice (n =9–17). Blood glucose levels were measured prior to protein injection (0 h) and at 2, 4, 5 and 6 h post-injection. Values are means±S.D. *P <0.05 and **P <0.005 (measured using Student’s t test) between mice injected with control buffer and mice injected with recombinant CTRP1.
DISCUSSION
In the present study, we made several new and important observations concerning the CTRP family of proteins. First, CTRP1, CTRP2, CTRP3, CTRP5 and CTRP7 transcripts are expressed predominantly by adipose tissue. Sex, age, genetic background of mice and PPAR-γ (peroxisome-proliferator-activated receptor-γ) agonist treatment all affected the relative levels of some, but not all, CTRP transcripts in adipose tissue. Secondly, all CTRPs are secreted glycoproteins and most of them are found circulating in the serum with levels varying according to the sex and genetic background of mice. Hence, they are potential endocrine hormones. Moreover, serum levels of CTRP1 and CTRP6 are increased in adiponectin-null mice. Thirdly, all CTRPs form trimers and some are further assembled into higher-order oligomeric complexes involving N-terminal cysteine residues. Different oligomeric forms of CTRPs may have distinct biological activities similar to those of adiponectin. Fourthly, we discovered combinatorial associations between different family members and showed that CTRP1/CTRP6, CTRP2/CTRP7 and adiponectin/CTRP2 can be secreted as heterotrimers, thus expanding the possible repertoire of functionally distinct ligands within the C1q/TNF family of proteins. Fifthly, we focused our functional characterization on CTRP1 and demonstrated that in cultured myotubes it specifically activates Akt and p44/42 MAPK signalling pathways. More importantly, when injected into mice, recombinant CTRP1 significantly lowered serum glucose levels compared with controls, thus revealing CTRP1 to be a novel adipokine.
Expression of CTRP transcripts in mice
Adipocytes synthesize and secrete adiponectin. In contrast, CTRP1, CTRP2, CTRP3, CTRP5 and CTRP7 are preferentially produced by adipose stromal cells. Although it includes preadipocytes, the stromal vascular fraction also contains fibroblasts, mature endothelial cells and smoothmuscle cells [35]. We believe preadipocytes may be the major source of these CTRP transcripts for two reasons. First, fibroblasts, mature endothelial cells and smooth muscle cells are found in many different types of tissues/organs besides adipose tissue, and yet expression of CTRP1, CTRP2 and CTRP7 is fairly restricted to adipose tissue. In contrast, preadipocytes are found almost exclusively in the adipose tissue. Secondly, using real-time PCR we could detect the presence of CTRP1, CTRP2, CTRP3, CTRP5 and CTRP7 transcripts in 3T3-L1 preadipocytes; their mRNA levels were unchanged, decreased or showed no sustained up-regulation over the course of adipocyte differentiation in culture (results not shown). It has been reported that CTRP3/CORS26 is induced in differentiated 3T3-L1 adipocytes, but not in preadipocytes, and its mRNA can be detected in the synovial adipocytes of the knee by in situ hybridization [36]. Since there are different types and anatomically distinct adipose depots [37], it is conceivable that expression of CTRP transcripts may differ in these depots. Therefore we also cannot rule out the possibility that other stromal cells beside preadipocytes may express CTRP transcripts.
PPAR-γ agonists of the TZDs class of compound have been used widely to treat patients with Type 2 diabetes [38]. TZDs enhance whole-body insulin sensitivity by increasing glucose disposal in muscles and suppressing hepatic glucose production [38]. Although the molecular mechanism of TZD action is not completely understood, its beneficial effect in ameliorating hyperglycaemia is due in part to its ability to up-regulate adiponectin gene expression and protein secretion by the adipose tissue [39]. In the absence of adiponectin, the ability of TZDs to improve glucose tolerance and insulin sensitivity is significantly blunted [40,41]. Depending on the dose of TZDs used, there seemed to be an adiponectin-dependent mechanism operating in the liver and adiponectin-independent mechanism operating in the muscle by which TZD exerts its metabolic effects [41]. In the present study, we showed that a 3 week treatment with rosiglitazone at a dose of 15 mg/kg significantly up-regulated both adiponectin and CTRP1, whereas it down-regulated CTRP6 transcripts levels in adipose tissue of ob/ob mice. It remains to be determined whether the adiponectin-independent mechanism of TZD actions reported by Kubota et al. [41] is due to its ability to up-regulate CTRP1 and/or down-regulate CTRP6. Metformin, another widely used anti-diabetic drug, exerts its glucose-lowering effects in vivo by reducing hepatic gluconeogenesis [42]. The molecular basis of metformin action in liver is not known. However, chronic treatment of diabetic (db/db) mice with metformin has no effect on serum levels of adiponectin or its mRNA levels in adipose tissue despite significant improvement in body weight, glucose and lipid profiles [43]. Therefore it remains to be determined whether metformin exerts its metabolic actions through any of the CTRPs independent of adiponectin.
Sex and genetic background of mice influence serum levels of CTRPs
Sex and genetic background confer variable degrees of susceptibility to developing obesity, insulin resistance and Type 2 diabetes in humans [44,45]. In various mouse models of obesity and diabetes, the genetic background plays a significant role in dictating the severity of the disease [26,27]. Presumably, genotypes influence the levels of various metabolic hormones and signalling pathways that in turn dictate the severity of metabolic dysfunction in conditions of obesity and diabetes. Interestingly, we found that the serum levels of adiponectin, CTRP1, CTRP2, CTRP3, CTRP5 and CTRP6 vary significantly in mice derived from six different genetic backgrounds; most of these strains were chosen for their varying degree of susceptibility or resistance to diet-induced obesity and diabetes [26]. For example, diabetes-prone DBA/2 mice have the lowest levels of adiponectin, whereas BALB/c and AKR mice have barely detectable CTRP2 in their serum. Interestingly, 129SvJ mice that are resistance to diet-induced obesity and diabetes relative to other strains of mice, also have correspondingly higher serum levels of adiponectin and some of the CTRPs.
Oligomeric structures of CTRPs
Adiponectin exists in three multimeric forms with distinct biological activities, representing a unique way to control signalling pathways in cells and tissue [28]. Although trimers and hexamers activate AMPK in muscle to increase glucose uptake, glycogen deposition and fatty acid oxidation, HMW oligomers preferentially exert its effects in liver to reduce glucose output [46,47]. In contrast with the three oligomeric forms of adiponectin, CTRP1 and CTRP2 are secreted primarily as trimers in transfected HEK-293T cells. Consistent with this, CTRP2 in mouse serum also exists predominantly as trimers. Although CTRP3 is secreted as trimers and presumably hexamers and HMW oligomers in transfected cells, it exists predominantly in an HMW oligomeric form in mouse serum, suggesting that its assembly in vivo is regulated differently or that it binds to other serum proteins. Similarly, although CTRP5 is secreted in three multimeric forms in transfected HEK-293T cells, it exists predominantly as trimers in the mouse serum, again suggesting that its assembly is tightly regulated in vivo. Factors such as exercise and TZD treatment that affect the metabolic states of obese and diabetic humans result in changes in the proportion of oligomeric forms of circulating serum adiponectin [47]. The ratio of HMW oligomers to trimers has been previously proposed to serve as an index of insulin sensitivity [47]. It remains to be determined whether different metabolic or disease states affect the distributions of CTRP oligomeric forms in serum and whether different oligomeric forms of these proteins have distinct biological activities.
Combinatorial associations between different CTRPs
Hetero-oligomer formations have been documented for several members of the C1q/TNF family of proteins. These include complement C1q (consisting of A-, B- and C-chains forming a hexamer of trimers) [32], chipmunk hibernation proteins (consisting of HP20, HP25 and HP27 heterotrimers) [34,48], and cerebellins (consisting of homo-oligomers or Cbln-1/Cbln-3 or Cbln-3/Cbln-4 hetero-oligomers) [33]. In the present study, we observed robust and consistent interactions between secreted CTRP1/CTRP6, CTRP2/CTRP7, CTRP2/adiponectin and CTRP9/adiponectin. These associations are not random. Sequence and phylogenetic analysis suggest that CTRP1 is most closely related to CTRP6, CTRP2 is most closely related to CTRP7, and CTRP2 and CTRP9 are most closely related to adiponectin (results not shown). Because these proteins form heterotrimers, amino acids that lie at the interface between three protomer subunits must be able to interact appropriately to allow trimerization. Consistent with our results, C1q/TNF family proteins that are more distantly related to each other in protein sequences were not able to form heterotrimers. The combinatorial associations we uncovered between closely related CTRPs may represent a mechanism to generate functionally distinct ligands that can potentially interact with different cell-surface receptors to activate distinct signalling pathways in different cell types.
Although CTRP2/adiponectin and CTRP1/CTRP6 can form heterotrimers, the stoichiometry of adiponectin to CTRP2 and CTRP1 to CTRP6 in the heterotrimeric complexes remains to be determined. All of the proteins with Gly-X-Y repeats form characteristic triple helical structures [49]. Both CTRP1 and CTRP6 have 14 Gly-X-Y repeats, whereas CTRP2 and CTRP7 have 34 Gly-X-Y repeats in their collagen domains. Thus the equal numbers of Gly-X-Y repeats may facilitate the formation of CTRP1/CTRP6 and CTRP2/CTRP7 heterotrimers. In contrast, adiponectin has 22 Gly-X-Y repeats, whereas CTRP2 has 34 Gly-X-Y repeats in their collagen domain respectively. Proteins with unequal numbers of Gly-X-Y repeats and/or ‘interrupting amino acids’ within the Gly-X-Y repeats have been shown to form heterotrimers in vivo. Examples include mouse complement C1q (heterotrimer composed of A-, B- and C-chains) [32], chipmunk hibernating proteins (heterotrimers composed of HP20, HP25 and HP27) [34], and the ubiquitous type VI collagen in connective tissues (heterotrimers composed of α1, α2 and α3 subunits) [50,51]. Thus CTRP2/adiponectin is another example of a heterotrimer composed of proteins with an unequal length of collagen domain.
Metabolic function of CTRP1
Several adiponectin-null mouse lines have recently been generated to address and confirm the in vivo metabolic function of adiponectin [21–23]. In all cases, the adiponectin-null mice show mild to no metabolic abnormalities when maintained on a normal chow diet. This raises the interesting possibility that other secreted protein(s) may partially be able to compensate for the loss of serum adiponectin in mice under this condition. Like adiponectin, CTRP1 transcript levels are up-regulated in mice treated with a PPAR-γ agonist, rosiglitazone. Additionally, serum levels of CTRP1 are increased in adiponectin-null mice. Moreover, injection of recombinant CTRP1 into mice significantly lowered their serum glucose levels compared with control mice. Taken together, these findings suggest that CTRP1 may represent one of the serum factors produced by adipose tissue that share overlapping functions with adiponectin. It remains to be determined whether liver, muscle or other organs are the natural target tissues where CTRP1 exerts its action. In differentiated mouse C2C12 myotubes, recombinant CTRP1 specifically activates Akt and p44/42 MAPK signalling pathways. Activation of Akt has been shown by many groups to enhance glucose uptake in muscle due to increased trafficking of GLUT4 (glucose transporter 4) to the plasma membrane [52]. This result suggests that CTRP1 may act on muscle among other target tissues to exert its metabolic effects.
In summary, we have provided a detailed molecular, biochemical and initial functional analysis of the CTRP family of proteins. This information will lay the groundwork to further explore their physiological functions and mechanisms of action in both normal and disease states using molecular, cellular and in vivo approaches. In the light of the diagnostic utility of adiponectin as a serum marker for a wide variety of diseases, the CTRP family of serum glycoproteins may also have diagnostic values beyond their normal physiological functions.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Prakash Rao, Dr Joseph Marszalek, Dr Christopher Hug and Dr Kelly Wong for helpful comments during the course of this work. We thank Ferenc Reinhardt for animal technical support.
FUNDING
This work was supported in part by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [grant number R37 DK47618-19] (to H. F. L.); the National Institutes of Health KO8 grant [grant number HL077499-01] (to C.H.); and a National Institutes of Health National Research Service Award postdoctoral fellowship [grant number F32DK067835] (to G.W.W.).
Abbreviations used
- ACC
acetyl-CoA carboxylase
- AMPK
AMP-activated protein kinase
- ASFV
African Swine Fever virus
- CTRP
C1q/TNF-related protein
- DMEM
Dulbecco’smodified Eagle’smedium
- DTT
dithiothreitol
- EST
expressed sequence tag
- FBS
foetal bovine serum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GFP
green fluorescent protein
- HA
haemagglutinin
- HEK
human embryonic kidney
- HMW
high-molecular-mass
- HRP
horseradish peroxidase
- IκB
inhibitor of nuclear factor κB
- MAPK
mitogen-activated protein kinase
- mTOR
mammalian target of rapamycin
- NF-κB
nuclear factor κB
- PNGase F
peptide N-glycosidase F
- PPAR-γ
peroxisome-proliferator-activated receptor-γ
- TNFα
tumour necrosis factor α
- TZD
thiazolidinedione.
REFERENCES
- 1.Trujillo ME, Scherer PE. Adipose tissue-derived factors: impact on health and disease. Endocrin. Rev. 2006;27:762–778. doi: 10.1210/er.2006-0033. [DOI] [PubMed] [Google Scholar]
- 2.Ahima RS, Qi Y, Singhal NS, Jackson MB, Scherer PE. Brain adipocytokine action and metabolic regulation. Diabetes. 2006;55(Suppl. 2):S145–S154. doi: 10.2337/db06-s018. [DOI] [PubMed] [Google Scholar]
- 3.Scherer PE, Williams S, Fogliano M, Baldini G, Lodish HF. A novel serum protein similar to C1q, produced exclusively in adipocytes. J. Biol. Chem. 1995;270:26746–26749. doi: 10.1074/jbc.270.45.26746. [DOI] [PubMed] [Google Scholar]
- 4.Hu E, Liang P, Spiegelman BM. AdipoQ is a novel adipose-specific gene dysregulated in obesity. J. Biol. Chem. 1996;271:10697–10703. doi: 10.1074/jbc.271.18.10697. [DOI] [PubMed] [Google Scholar]
- 5.Maeda K, Okubo K, Shimomura I, Funahashi T, Matsuzawa Y, Matsubara K. cDNA cloning and expression of a novel adipose specific collagen-like factor, apM1 (AdiPose Most abundant Gene transcript 1) . Biochem. Biophys. Res. Commun. 1996;221:286–289. doi: 10.1006/bbrc.1996.0587. [DOI] [PubMed] [Google Scholar]
- 6.Arita Y, Kihara S, Ouchi N, Takahashi M, Maeda K, Miyagawa J, Hotta K, Shimomura I, Nakamura T, Miyaoka K, et al. Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem. Biophys. Res. Commun. 1999;257:79–83. doi: 10.1006/bbrc.1999.0255. [DOI] [PubMed] [Google Scholar]
- 7.Ruan H, Hacohen N, Golub TR, Van Parijs L, Lodish HF. Tumor necrosis factor-α suppresses adipocyte-specific genes and activates expression of preadipocyte genes in 3T3-L1 adipocytes: nuclear factor-κ B activation by TNF-α is obligatory. Diabetes. 2002;51:1319–1336. doi: 10.2337/diabetes.51.5.1319. [DOI] [PubMed] [Google Scholar]
- 8.Vasseur F, Helbecque N, Lobbens S, Vasseur-Delannoy V, Dina C, Clement K, Boutin P, Kadowaki T, Scherer PE, Froguel P. Hypoadiponectinaemia and high risk of type 2 diabetes are associated with adiponectin-encoding (ACDC) gene promoter variants in morbid obesity: evidence for a role of ACDC in diabesity. Diabetologia. 2005;48:892–899. doi: 10.1007/s00125-005-1729-z. [DOI] [PubMed] [Google Scholar]
- 9.Maeda N, Takahashi M, Funahashi T, Kihara S, Nishizawa H, Kishida K, Nagaretani H, Matsuda M, Komuro R, Ouchi N, et al. PPARγ ligands increase expression and plasma concentrations of adiponectin, an adipose-derived protein. Diabetes. 2001;50:2094–2099. doi: 10.2337/diabetes.50.9.2094. [DOI] [PubMed] [Google Scholar]
- 10.Esposito K, Pontillo A, Di Palo C, Giugliano G, Masella M, Marfella R, Giugliano D. Effect of weight loss and lifestyle changes on vascular inflammatory markers in obese women: a randomized trial. JAMA. J. Am. Med. Assoc. 2003;289:1799–1804. doi: 10.1001/jama.289.14.1799. [DOI] [PubMed] [Google Scholar]
- 11.Fruebis J, Tsao TS, Javorschi S, Ebbets-Reed D, Erickson MR, Yen FT, Bihain BE, Lodish HF. Proteolytic cleavage product of 30-kDa adipocyte complement-related protein increases fatty acid oxidation in muscle and causes weight loss in mice. Proc. Natl. Acad. Sci. U.S.A. 2001;98:2005–2010. doi: 10.1073/pnas.041591798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamauchi T, Kamon J, Waki H, Terauchi Y, Kubota N, Hara K, Mori Y, Ide T, Murakami K, Tsuboyama-Kasaoka N, et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat. Med. 2001;7:941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- 13.Berg AH, Combs TP, Du X, Brownlee M, Scherer PE. The adipocyte-secreted protein Acrp30 enhances hepatic insulin action. Nat. Med. 2001;7:947–953. doi: 10.1038/90992. [DOI] [PubMed] [Google Scholar]
- 14.Wong GW, Wang J, Hug C, Tsao TS, Lodish HF. A family of Acrp30/adiponectin structural and functional paralogs. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10302–10307. doi: 10.1073/pnas.0403760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shapiro L, Scherer PE. The crystal structure of a complement-1q family protein suggests an evolutionary link to tumor necrosis factor. Curr. Biol. 1998;8:335–338. doi: 10.1016/s0960-9822(98)70133-2. [DOI] [PubMed] [Google Scholar]
- 16.Kishore U, Gaboriaud C, Waters P, Shrive AK, Greenhough TJ, Reid KB, Sim RB, Arlaud GJ. C1q and tumor necrosis factor superfamily: modularity and versatility. Trends Immunol. 2004;25:551–561. doi: 10.1016/j.it.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Lasser G, Guchhait P, Ellsworth JL, Sheppard P, Lewis K, Bishop P, Cruz MA, Lopez JA, Fruebis J. C1qTNF-related protein-1 (CTRP-1): a vascular wall protein that inhibits collagen-induced platelet aggregation by blocking VWF binding to collagen. Blood. 2006;107:423–430. doi: 10.1182/blood-2005-04-1425. [DOI] [PubMed] [Google Scholar]
- 18.Akiyama H, Furukawa S, Wakisaka S, Maeda T. Cartducin stimulates mesenchymal chondroprogenitor cell proliferation through both extracellular signal-regulated kinase and phosphatidylinositol 3-kinase/Akt pathways. FEBS. J. 2006;273:2257–2263. doi: 10.1111/j.1742-4658.2006.05240.x. [DOI] [PubMed] [Google Scholar]
- 19.Hayward C, Shu X, Cideciyan AV, Lennon A, Barran P, Zareparsi S, Sawyer L, Hendry G, Dhillon B, Milam AH, et al. Mutation in a short-chain collagen gene, CTRP5, results in extracellular deposit formation in late-onset retinal degeneration: a genetic model for age-related macular degeneration. Hum. Mol. Genet. 2003;12:2657–2667. doi: 10.1093/hmg/ddg289. [DOI] [PubMed] [Google Scholar]
- 20.Chang AC, Zsak L, Feng Y, Mosseri R, Lu Q, Kowalski P, Zsak A, Burrage TG, Neilan JG, Kutish GF, et al. Phenotype-based identification of host genes required for replication of African swine fever virus. J. Virol. 2006;80:8705–8717. doi: 10.1128/JVI.00475-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma K, Cabrero A, Saha PK, Kojima H, Li L, Chang BH, Paul A, Chan L. Increased β-oxidation but no insulin resistance or glucose intolerance in mice lacking adiponectin. J. Biol. Chem. 2002;277:34658–34661. doi: 10.1074/jbc.C200362200. [DOI] [PubMed] [Google Scholar]
- 22.Kubota N, Terauchi Y, Yamauchi T, Kubota T, Moroi M, Matsui J, Eto K, Yamashita T, Kamon J, Satoh H, et al. Disruption of adiponectin causes insulin resistance and neointimal formation. J. Biol. Chem. 2002;277:25863–25866. doi: 10.1074/jbc.C200251200. [DOI] [PubMed] [Google Scholar]
- 23.Maeda N, Shimomura I, Kishida K, Nishizawa H, Matsuda M, Nagaretani H, Furuyama N, Kondo H, Takahashi M, Arita Y, et al. Diet-induced insulin resistance in mice lacking adiponectin/ACRP30. Nat. Med. 2002;8:731–737. doi: 10.1038/nm724. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Ren R. Bcr-Abl efficiently induces a myeloproliferative disease and production of excess interleukin-3 and granulocyte-macrophage colony-stimulating factor in mice: a novel model for chronic myelogenous leukemia. Blood. 1998;92:3829–3840. [PubMed] [Google Scholar]
- 25.Liu X, Constantinescu SN, Sun Y, Bogan JS, Hirsch D, Weinberg RA, Lodish HF. Generation of mammalian cells stably expressing multiple genes at predetermined levels. Anal. Biochem. 2000;280:20–28. doi: 10.1006/abio.2000.4478. [DOI] [PubMed] [Google Scholar]
- 26.West DB, Boozer CN, Moody DL, Atkinson RL. Dietary obesity in nine inbred mouse strains. Am. J. Physiol. 1992;262:R1025–R1032. doi: 10.1152/ajpregu.1992.262.6.R1025. [DOI] [PubMed] [Google Scholar]
- 27.Haluzik M, Colombo C, Gavrilova O, Chua S, Wolf N, Chen M, Stannard B, Dietz KR, Le Roith D, Reitman ML. Genetic background (C57BL/6J versus FVB/N) strongly influences the severity of diabetes and insulin resistance in ob/ob mice. Endocrinology. 2004;145:3258–3264. doi: 10.1210/en.2004-0219. [DOI] [PubMed] [Google Scholar]
- 28.Tsao TS, Murrey HE, Hug C, Lee DH, Lodish HF. Oligomerization state-dependent activation of NF-κB signaling pathway by adipocyte complement-related protein of 30 kDa (Acrp30) J. Biol. Chem. 2002;277:29359–29362. doi: 10.1074/jbc.C200312200. [DOI] [PubMed] [Google Scholar]
- 29.Pajvani UB, Du X, Combs TP, Berg AH, Rajala MW, Schulthess T, Engel J, Brownlee M, Scherer PE. Structure-function studies of the adipocyte-secreted hormone Acrp30/adiponectin. Implications for metabolic regulation and bioactivity. J. Biol. Chem. 2003;278:9073–9085. doi: 10.1074/jbc.M207198200. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki S, Wilson-Kubalek EM, Wert D, Tsao TS, Lee DH. The oligomeric structure of high molecular weight adiponectin. FEBS Lett. 2007;581:809–814. doi: 10.1016/j.febslet.2007.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsao TS, Tomas E, Murrey HE, Hug C, Lee DH, Ruderman NB, Heuser JE, Lodish HF. Role of disulfide bonds in Acrp30/adiponectin structure and signaling specificity. Different oligomers activate different signal transduction pathways. J. Biol. Chem. 2003;278:50810–50817. doi: 10.1074/jbc.M309469200. [DOI] [PubMed] [Google Scholar]
- 32.Kishore U, Reid KB. C1q: structure, function, and receptors. Immunopharmacology. 2000;49:159–170. doi: 10.1016/s0162-3109(00)80301-x. [DOI] [PubMed] [Google Scholar]
- 33.Bao D, Pang Z, Morgan MA, Parris J, Rong Y, Li L, Morgan JI. Cbln1 is essential for interaction-dependent secretion of Cbln3. Mol. Cell. Biol. 2006;26:9327–9337. doi: 10.1128/MCB.01161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kondo N, Kondo J. Identification of novel blood proteins specific for mammalian hibernation. J. Biol. Chem. 1992;267:473–478. [PubMed] [Google Scholar]
- 35.Mitchell JB, McIntosh K, Zvonic S, Garrett S, Floyd ZE, Kloster A, Di Halvorsen Y, Storms RW, Goh B, Kilroy G, Wu X, Gimble JM. Immunophenotype of human adipose-derived cells: temporal changes in stromal-associated and stem cell-associated markers. Stem Cells. 2006;24:376–385. doi: 10.1634/stemcells.2005-0234. [DOI] [PubMed] [Google Scholar]
- 36.Schaffler A, Ehling A, Neumann E, Herfarth H, Tarner I, Gay S, Scholmerich J, Muller-Ladner U. Genomic organization, chromosomal localization and adipocytic expression of the murine gene for CORS-26 (collagenous repeat-containing sequence of 26 kDa protein) . Biochim. Biophys. Acta. 2003;1628:64–70. doi: 10.1016/s0167-4781(03)00114-3. [DOI] [PubMed] [Google Scholar]
- 37.Cinti S. Anatomy of the adipose organ. Eat. Weight Disord. 2000;5:132–142. doi: 10.1007/BF03354443. [DOI] [PubMed] [Google Scholar]
- 38.Rangwala SM, Lazar MA. Peroxisome proliferator-activated receptor γ in diabetes and metabolism. Trends Pharmacol. Sci. 2004;25:331–336. doi: 10.1016/j.tips.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 39.Yu JG, Javorschi S, Hevener AL, Kruszynska YT, Norman RA, Sinha M, Olefsky JM. The effect of thiazolidinediones on plasma adiponectin levels in normal, obese, and type 2 diabetic subjects. Diabetes. 2002;51:2968–2974. doi: 10.2337/diabetes.51.10.2968. [DOI] [PubMed] [Google Scholar]
- 40.Nawrocki AR, Rajala MW, Tomas E, Pajvani UB, Saha AK, Trumbauer ME, Pang Z, Chen AS, Ruderman NB, Chen H, et al. Mice lacking adiponectin show decreased hepatic insulin sensitivity and reduced responsiveness to peroxisome proliferator-activated receptor γ agonists. J. Biol. Chem. 2006;281:2654–2660. doi: 10.1074/jbc.M505311200. [DOI] [PubMed] [Google Scholar]
- 41.Kubota N, Yamauchi T, Tobe K, Kadowaki T. Adiponectin-dependent and -independent pathways in insulin-sensitizing and antidiabetic actions of thiazolidinediones. Diabetes. 2006;55(Suppl. 2):S32–S38. [Google Scholar]
- 42.Hundal RS, Krssak M, Dufour S, Laurent D, Lebon V, Chandramouli V, Inzucchi SE, Schumann WC, Petersen KF, Landau BR, Shulman GI. Mechanism by which metformin reduces glucose production in type 2 diabetes. Diabetes. 2000;49:2063–2069. doi: 10.2337/diabetes.49.12.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujita H, Fujishima H, Koshimura J, Hosoba M, Yoshioka N, Shimotomai T, Morii T, Narita T, Kakei M, Ito S. Effects of antidiabetic treatment with metformin and insulin on serum and adipose tissue adiponectin levels in db/db mice. Endocr. J. 2005;52:427–433. doi: 10.1507/endocrj.52.427. [DOI] [PubMed] [Google Scholar]
- 44.Gale EA, Gillespie KM. Diabetes and gender. Diabetologia. 2001;44:3–15. doi: 10.1007/s001250051573. [DOI] [PubMed] [Google Scholar]
- 45.O’Rahilly S, Barroso I, Wareham NJ. Genetic factors in type 2 diabetes: the end of the beginning? Science. 2005;307:370–373. doi: 10.1126/science.1104346. [DOI] [PubMed] [Google Scholar]
- 46.Tomas E, Tsao TS, Saha AK, Murrey HE, Zhang CcC, Itani SI, Lodish HF, Ruderman NB. Enhanced muscle fat oxidation and glucose transport by ACRP30 globular domain: acetyl-CoA carboxylase inhibition and AMP-activated protein kinase activation. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16309–16313. doi: 10.1073/pnas.222657499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pajvani UB, Hawkins M, Combs TP, Rajala MW, Doebber T, Berger JP, Wagner JA, Wu M, Knopps A, Xiang AH, et al. Complex distribution, not absolute amount of adiponectin, correlates with thiazolidinedione-mediated improvement in insulin sensitivity. J. Biol. Chem. 2004;279:12152–12162. doi: 10.1074/jbc.M311113200. [DOI] [PubMed] [Google Scholar]
- 48.Takamatsu N, Ohba K, Kondo J, Kondo N, Shiba T. Hibernationassociated gene regulation of plasma proteins with a collagen-like domain in mammalian hibernators. Mol. Cell. Biol. 1993;13:1516–1521. doi: 10.1128/mcb.13.3.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vitagliano L, Berisio R, Mazzarella L, Zagari A. Structural bases of collagen stabilization induced by proline hydroxylation. Biopolymers. 2001;58:459–464. doi: 10.1002/1097-0282(20010415)58:5<459::AID-BIP1021>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 50.Chu ML, Pan TC, Conway D, Kuo HJ, Glanville RW, Timpl R, Mann K, Deutzmann R. Sequence analysis of α1(VI) and α2(VI) chains of human type VI collagen reveals internal triplication of globular domains similar to the A domains of von Willebrand factor and two α 2(VI) chain variants that differ in the carboxy terminus. EMBO. J. 1989;8:1939–1946. doi: 10.1002/j.1460-2075.1989.tb03598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stokes DG, Saitta B, Timpl R, Chu ML. Human α 3(VI) collagen gene. Characterization of exons coding for the amino-terminal globular domain and alternative splicing in normal and tumor cells. J. Biol. Chem. 1991;266:8626–8633. [PubMed] [Google Scholar]
- 52.Huang S, Czech MP. The GLUT4 glucose transporter. Cell Metab. 2007;5:237–252. doi: 10.1016/j.cmet.2007.03.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.