Abstract
Aims: Distinctive states of redox-dependent cysteine (Cys) modifications are known to regulate signaling homeostasis under various pathophysiological conditions, including myocardial injury or protection in response to ischemic stress. Recent evidence further implicates a dynamic interplay among these modified forms following changes in cellular redox environment. However, a precise delineation of multiplexed Cys modifications in a cellular context remains technically challenging. To this end, we have now developed a mass spectrometry (MS)-based quantitative approach using a set of novel iodoacetyl-based Cys-reactive isobaric tags (irreversible isobaric iodoacetyl Cys-reactive tandem mass tag [iodoTMT]) endowed with unique irreversible Cys-reactivities. Results: We have established a sequential iodoTMT-switch procedure coupled with efficient immunoenrichment and advanced shotgun liquid chromatography-MS/MS analysis. This workflow allows us to differentially quantify the multiple redox-modified forms of a Cys site in the original cellular context. In one single analysis, we have identified over 260 Cys sites showing quantitative differences in multiplexed redox modifications from the total lysates of H9c2 cardiomyocytes experiencing hypoxia in the absence and presence of S-nitrosoglutathione (GSNO), indicative of a distinct pattern of individual susceptibility to S-nitrosylation or S-glutathionylation. Among those most significantly affected are proteins functionally implicated in hypoxic damage from which we showed that GSNO would protect. Innovation: We demonstrate for the first time how quantitative analysis of various Cys-redox modifications occurring in biological samples can be performed precisely and simultaneously at proteomic levels. Conclusion: We have not only developed a new approach to map global Cys-redoxomic regulation in vivo, but also provided new evidences implicating Cys-redox modifications of key molecules in NO-mediated ischemic cardioprotection. Antioxid. Redox Signal. 20, 1365–1381.
Introduction
Reactive cysteine (Cys) residues in proteins are often specifically and preferentially modified during oxidative stresses because of the unique redox chemistry of this amino acid (33, 58). Such redox-mediated Cys modifications, that is, oxidation of the Cys residues to sulfenic acid (S-OH), S-nitrosylated (S-nitrosylation [SNO]), S-glutathionylated (S-glutathionylation [S-SG]), or other mixed disulfide (S-SR) forms, are increasingly being identified as key post-translational modifications that regulate myriad protein functions in a wide range of health and disease related cellular processes including cancer, inflammatory, neurodegenerative, and cardiovascular disorders [as reviewed in (15, 20, 49, 55)]. SNO, in particular, plays a critical role in protecting functional catalytic Cys from irreversible oxidation, thus preserving its ability to revert back to free thiol (SH) status befitting physiological normality upon stress relief (8, 51). It is now well appreciated that both the SNO and denitrosylation processes may additionally involve an equally reversible S-thiolation, mostly in the form of S-SG, as mediated by the abundant cellular glutathione or other mixed thiol intermediates (48). Moreover, S-SG was reported to uncouple endothelial nitric oxide synthase (eNOS), switching it from NO to O2•− generation, thus leading to impaired endothelium-dependent vasodilation (7). This observation not only revealed the unique role of S-SG in vascular function, but also emphasized the interplay between Cys modifications. Since each Cys modification may convey unique effects on protein functions (1, 35), signaling convergence or divergence orchestrated through such distinct but related modifications can only be sensibly delineated by first defining the stoichiometry of conversions at high sensitivity and precision.
Innovation.
Our understanding of redox-directed biological processes is significantly delayed by lack of method for global visualization of multiplexed cysteine (Cys) modifications. The recent development of irreversible iodoacetyl-based Cys-reactive isobaric tags has now allowed us to establish a novel mass spectrometry-based workflow for multiplexed quantification of reversibly modified Cys sites at proteomic level. This method enabled us to identify a network of nitric oxide-regulated Cys-redoxome comprising S-nitrosylation and S-glutathionylation that act in concert to protect cardiomyocytes against hypoxic injury. For the first time, the differential in vivo susceptibilities of Cys sites to redox-mediated modifications in the presence and absence of S-nitrosoglutathione were quantified.
Despite progressive analytical methodologies development over the last decade, identification of physiologically relevant reversible Cys modifications on important classes of proteins remains technically challenging. By far, the most commonly used strategies were variations derived from the biotin switch technique (BST) developed by Jaffrey et al. (30, 31), which involves the initial blocking of all SH, followed by the use of ascorbate to selectively reduce SNO back to free SH, and its subsequent tagging with a sulfhydryl-specific biotinylated probe that would allow avidin-based affinity capture from treated cell lysates. Since the transient nature of in vivo Cys modifications defies direct analysis, “switching” of the redox modifications to more stable Cys reactive tags through specific reduction, followed by selective enrichment of the Cys-labeled peptides and mass spectrometry (MS)-based proteomic analysis, has essentially become the standard approach for redox proteomic studies (17, 43, 60, 66), with impressive list of SNO-modified sites being catalogued over the last few years. However, none of the analytical methods reported to date including the use of various isotope-coded Cys reactive tags (42, 65, 67) allows simultaneous multiplexed monitoring and quantification of the various forms of coexisting site-specific Cys modifications.
More recently, an isobaric Cys-reactive tandem mass tag (reversible isobaric Cys-reactive tandem mass tag [cysTMT]) has become commercially available and is used for quantitative analysis of S-nitrosoproteome (52), and it is being adopted to profile the thiol redox sensitivity (54) and occupancy of SNO (37). Unlike the use of other isotope-coded Cys-reactive tags, which are largely limited to pair-wise comparison (26), the isobaric cysTMT offers the advantage of multiplexing with up to six channels of reporter ions (126–131 Da). Moreover, enrichment of the cysTMT-tagged peptides is facilitated by the accompanying anti-TMT resin, which proves to be more specific than a biotin-avidin-based capture that might be interfered by endogenous biotinylation. A major shortcoming, however, is that the disulfide linkage between the tag and protein thiol group cannot survive the strong reducing conditions normally used to promote efficient enzymatic proteolysis for subsequent shotgun proteomic analysis. It immediately became apparent that if the chemistry can be substituted instead by irreversible Cys tagging, a more versatile enabling tool for MS-based Cys-redox proteomics will be in hand to address a wide range of redox-mediated biological processes involving multiple forms of reversible Cys modifications.
We now report the development and applications of an irreversible Cys-reactive TMT reagent (irreversible isobaric iodoacetyl Cys-reactive tandem mass tag [iodoTMT]) containing a sulfhydryl-reactive iodoacetyl reactive group, a mass normalizing spacer arm and a mass reporter (Fig. 1). As with other related isobaric tandem mass tags (23), MS-based quantification using iodoTMT tags is achieved by inspection of the reporter ion region in MS/MS spectra. The isobaric sixplex iodoTMT reagents have the same nominal mass containing five variable isotope-coded positions, in which 12C or 14N located in the regions of normalizer and/or reporter are replaced by 13C or 15N. Under collision-induced dissociation (CID)-MS/MS, the mass reporters readily dissociate from the parent molecules, thus generating reporter ions with unique m/z values (126–131), the relative intensities of which represent the relative quantity of the labeled peptides from up to six designated samples. As reported here, this newly available reagent enabled us to develop a quantitative Cys-redox proteomic workflow based on a novel sequential iodoTMT switch procedure coupled with efficient enrichment of the mass-tagged peptides and advanced shotgun liquid chromatography (LC)-MS/MS analysis. Instead of single switch as in conventional BST, redox-modified Cys are sequentially reduced and differentially labeled with distinctive isobaric iodoTMT tags depending on their initial Cys modification status.
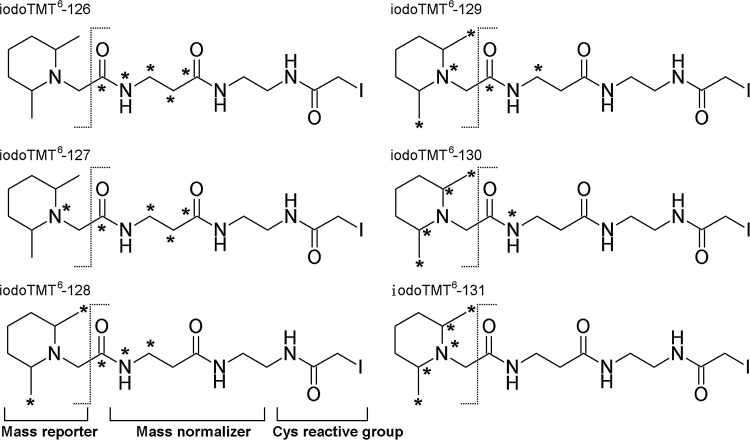
FIG. 1.
Chemical structures of isobaric iodoacetyl tandem mass tag sixplex reagents (iodoTMT6). Each of the isobaric iodoTMT6 reagents has the same nominal mass and comprises a sulfhydryl-reactive iodoacetyl group, a mass normalizing spacer arm and a mass reporter. Asterisks (*) indicate the positions in each of the mass tags where 12C and 14N are replaced by 13C and 15N. Under MS/MS conditions, the isobaric mass tags are readily cleaved at the bond indicated by dash line, thus generating reporter ions with unique m/z of 126–131. iodoTMT, irreversible isobaric iodoacetyl Cys-reactive tandem mass tag; MS, mass spectrometry.
Applied to whole cell lysates treated with S-nitrosylating and glutathionylating agents, we established the feasibility, precision, and sensitivity of this optimized workflow in defining the respective ratios of SNO versus other reversible modifications on a proteomic scale. We further showed an NO-dependent protective function by S-nitrosoglutathione (GSNO) against hypoxic damage in cultured cardiomyocytes and, under such conditions, the global changes in Cys-redox proteomic pattern as revealed by the extent of distinctive reversible Cys modifications relative to “unprotected” control. In total, 266 redox modified Cys residues from 169 proteins were identified and quantified. Among them, 13 Cys sites from 12 proteins showed significant changes of SNO levels, with another 14 sites from 14 proteins underwent higher levels of other reversible modifications. The most affected cellular functions represented by these proteins include cell death, free radical scavenging, and cellular assembly and organization, indicative of NO-mediated protection of hypoxic cardiomyocytes from apoptosis and loss of intercellular adhesion through disruption of F-actin cytoskeleton.
Results
The key performance indices of any MS-based quantitative proteomics are the sensitivity, accuracy, and precision afforded (Fig. 2). In the case of an isobaric mass tag, additional optimization of the MS/MS parameters is needed to produce the best combination of fragment ions for both peptide identification and multiplexed quantification (10, 73). Since only a very restricted subset of the digested peptide pool, namely those carrying a reversibly modified Cys residue are to be targeted for MS/MS analysis, an efficient enrichment of the mass-tagged peptides is also a requisite for attaining high sensitivity. We therefore systematically investigated each of these factors (Supplementary Figs. S1–S3; Supplementary Data are available online at www.liebertpub.com/ars) before applying the established workflow (Fig. 3) to quantify the differential Cys modifications at the proteomic level.
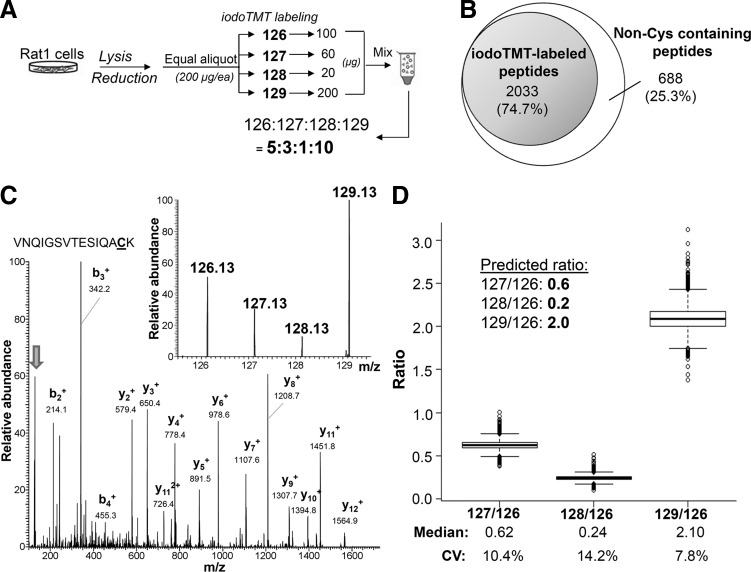
FIG. 2.
The accuracy and precision of iodoTMT-based multiplexed quantification. (A) Schematics of the sample preparation strategy used to evaluate the performance of iodoTMT-based quantification as mentioned in text. After tryptic digestion, the anti-TMT affinity enriched peptides were subjected to liquid chromatography (LC)-MS/MS analysis, which resulted in identification of over 2000 iodoTMT-labeled peptides representing an enrichment specificity of ∼75% (B). A representative MS/MS spectrum of an iodoTMT-labeled peptide is shown in (C), with its major sequence-informative b/y fragment ions fully annotated and the narrow mass region containing the TMT reporter ions (marked by an arrow) further expanded in the inset. Deviation of the experimentally derived reporter ion ratio values from the predicted ratios, namely 0.6, 0.2, and 2.0, for m/z 127, 128, and 129 to 126, respectively, for all 1669 quantifiable iodoTMT-labeled peptides as shown by Boxplot in (D), was taken as a measure of the quantification accuracy. CV, coefficient of variation.
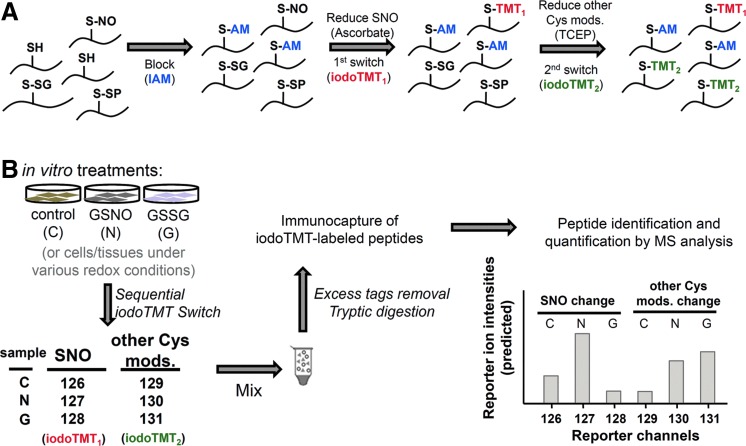
FIG. 3.
Experimental workflow of the sequential iodoTMT switch strategy for simultaneous monitoring of multiple Cys-modifications and quantitative determination of their respective ratios. (A) The principle requires that samples to be assayed be first treated with IAM to block all SH. Subsequently, S-nitrosylated Cys residues (SNO) are specifically reduced by ascorbate followed by iodoTMT1 labeling. Both S-SG and S-SP are then reduced by TCEP prior to iodoTMT2 alkylation. Note that the second switch will also convert any other reversible modifications that may be present to iodoTMT2. Since up to sixplexed quantification can be effected and the sequential switch strategy utilizes two channels per sample, up to three cell states can be monitored simultaneously. In the example illustrated in (B), the same cells can either be treated with GSNO (N) or GSSG (G), or non-treated (control, C). Following the sequential iodoTMT switch with various channels of iodoTMT tags as indicated, all samples were mixed, digested, and enriched for iodoTMT-labeled peptides prior to LC-MS/MS analysis. Relative abundance of the reporter ions detected for each positively identified peptide can then be used to calculate fold changes of SNO versus other reversible Cys modifications upon treatments with either GSNO or GSSG, relative to control. Cys, cysteine; GSNO, S-nitrosoglutathione; GSSG, glutathione disulfide; IAM, iodoacetamide; SH, free thiol; SNO, S-nitrosylation; S-SG, S-glutathionylation; S-SP, protein disulfides; TCEP, tris(2-carboxyethyl) phosphine. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Enrichment efficiency using anti-TMT resin
Apart from irreversible labeling of SH, a significant advantage afforded by the iodoTMT tag over otherwise comparable iTRAQ is the availability of an anti-TMT resin immobilized with monoclonal antibody recognizing the common structural motif in the mass reporter region of iodoTMT tags. This allows for enrichment of TMT-labeled peptides, the efficiency of which was first evaluated against tryptic peptides derived from bovine serum albumin (Supplementary Fig. S1). To further ascertain whether high enrichment efficiency could be attained at proteomic level, and to optimize for the iodoTMT-based quantification workflow, four equal amounts of a tris(2-carboxyethyl) phosphine (TCEP)-reduced total Rat-1 cell lysates were respectively labeled with four different iodoTMT tags and mixed in defined ratios, before removal of excess tags and tryptic digestion (Fig. 2A). Peptides from anti-TMT-immunocaptured eluates were then subjected to LC-MS/MS analysis under “HCD10” mode (see “Materials and Methods” and Supplementary Data) and the resulting dataset searched against database to identify all TMT-labeled and non-labeled peptides at high confidence (false discovery rate [FDR]<1%). In total, up to 75% of all identified peptides (2033 from a total of 2721 tryptic peptides) in the resulting enriched fraction were found to be TMT-labeled (Fig. 2B). For the following experiments of labeling reversible redox Cys modifications, depending on the stoichiometry of the modifications and complexity of the peptide mixtures, enrichment efficiency ranging from 40% to 85% was routinely achieved.
Reliability in the identification and quantification of iodoTMT-labeled peptides
In general, a higher normalized collision energy (NCE) setting would produce reporter ions with higher intensity, leading to better quantitative precision, but at the expense of sequence informative ions (Supplementary Fig. S2). We found that adjusting the NCE from 40% to 50% led to 1.5 and 1.8-fold increase of identifiable and quantifiable iodoTMT-labeled peptides, respectively, whereas further increase of NCE to 55% resulted in about 12% less peptide counts for both identification and quantification. Another notable consequence of iodoTMT-labeling is that it significantly increases the average charge state of labeled peptides ionized by electrospray (Supplementary Fig. S3A) (72), which adversely affects the CID or higher-energy C-trap dissociation (HCD) MS/MS pattern and ensuing identification efficiency (Supplementary Fig. S3B) (21, 27, 70). Moreover, several fragment ions derived from cleavage of the mass tag (marked with # in Supplementary Fig. S2A) would reduce the overall scoring by commonly used search engine such as Mascot (56). We showed that these deficiencies can be somewhat rectified by applying the extra Mascot Percolator function (5, 34, 68), through which the overall number of confidently identified peptides was improved by more than threefolds at a given FDR (Supplementary Fig. S3C, D).
A representative CID MS/MS spectrum produced under the optimized NCE, with intense reporter ions and clear b- and y-series of assigned fragment ions, is shown in Figure 2C, which could be confidently identified as corresponding to the peptide “VNQIGSVTESIQACK” from enolase. Importantly, the relative intensity of the reporter ions exactly matched the anticipated protein ratios (5:3:1:10) dictated by the experimental design (Fig. 2A). Overall, 1669 unique peptides were identified and quantified, and their measured reporter ion ratios summarized in Figure 2D. The median of all measured reporter ions 127 to 126 ratio was 0.62, which is close to the theoretical ratio of 0.6, with a 10.4% coefficient of variation (CV). Measured 128 to 126 and 129 to 126 ratios also closely matched their theoretical ratios with reasonable CV of 14.2% and 7.8%, respectively. These observations collectively established the high accuracy and precision in quantification afforded by the newly developed iodoTMT reagents.
Sequential iodoTMT switch
Having settled on the MS data acquisition and analysis parameters, we then proceeded to incorporate a sequential iodoTMT switch strategy into the overall workflow (Fig. 3A). Samples to be assayed, which can be any tissues or cells, are lysed in the presence of iodoacetamide (IAM) to block all SH to prevent their further conversion in subsequent steps. Next, S-nitrosylated Cys were specifically reduced to free SH by ascorbate treatment and labeled by iodoTMT1. Finally, all other remaining reversible Cys-modifications were reduced by TCEP and the generated thiols were labeled by iodoTMT2. Under this scheme, each sample requires two different iodoTMT tags for sequential labeling and therefore up to three different samples and/or treatment conditions can be simultaneously analyzed using a maximum of the sixplex iodoTMT reagents. As a feasibility study, we first applied the workflow to H9c2 cardiomyocyte lysates treated in vitro with either GSNO or glutathione disulfide (GSSG) and the resulting alterations in both SNO and other reversible modifications were quantified against non-treated control (Fig. 3B). The three samples were mixed after sequential labeling, and the iodoTMT-labeled peptides immunocaptured for LC-MS/MS analysis.
By the experimental design, relative intensities of the reporter ions 126–128 (from iodoTMT1) in the MS/MS spectrum of each identified peptides represent the change of SNO level of the implicated Cys site, whereas reporter ions 129–131 (from iodoTMT2) indicate the level of other reversible modifications, which correspond in this case mainly to protein disulfides and S-SG induced by either GSNO or GSSG. Protein structural disulfides could additionally be distinguished from S-SG or other oxidative stress-induced S-thiolations since the Cys involved would be insensitive to GSNO/GSSG treatment and ascorbate reduction. Consequently, these sites would not be tagged by iodoTMT1 in the first switching step, and almost equally tagged by iodoTMT2 in the subsequent step after TCEP reduction (Fig. 4A, reporter channel profile exemplified by the tryptic peptide of cathepsin B, which Cys is known to be involved in disulfide linkage). A total of 1977 unique peptides with about 2500 Cys residues were unambiguously identified with 1% FDR (Supplementary Table S2), ∼42% of which may be attributed to protein disulfides (cluster 1, Fig. 4B) based on the aforementioned reporter ion profiles. More importantly, several patterns attesting to the usefulness of our quantitative approach can be noted as following section from this global profiling.
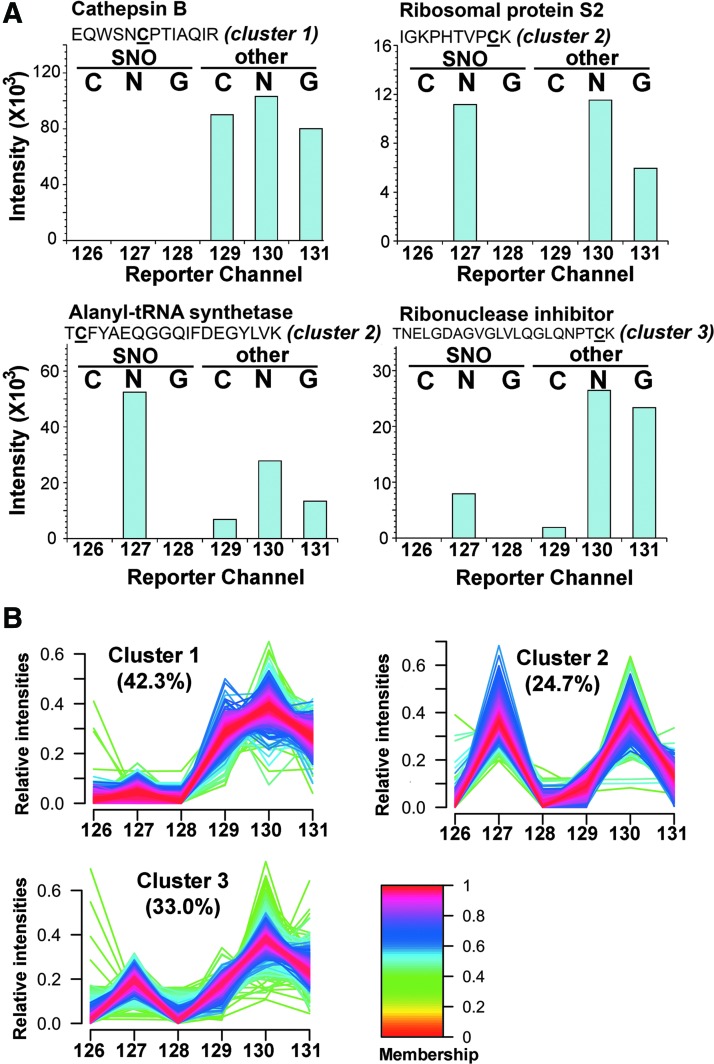
FIG. 4.
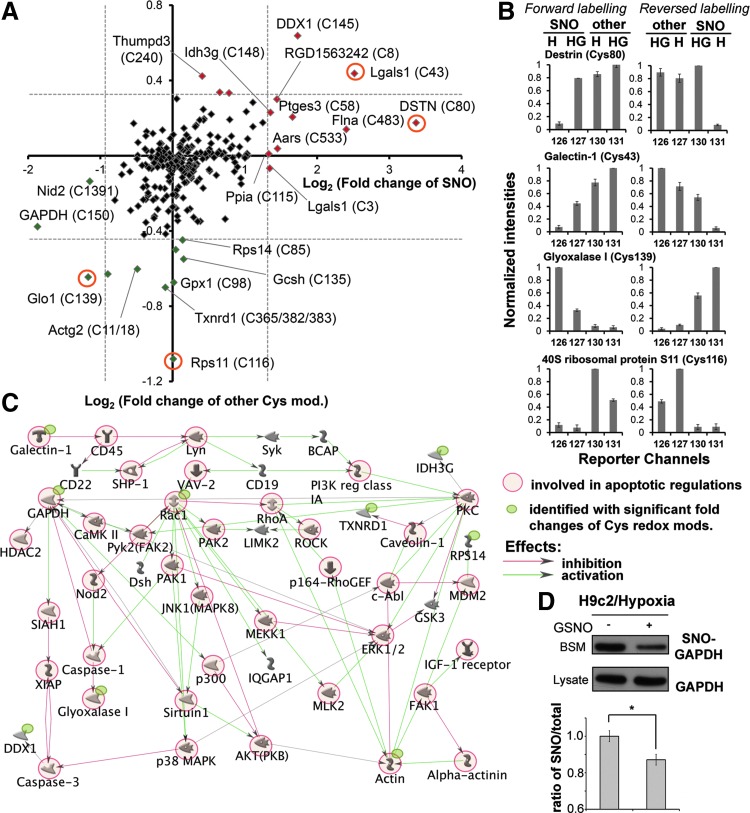
Clustering of the identified Cys residues based on the relative intensities of the reporter ions. Four representative intensity patterns of the reporter ion channels were shown in (A). Notably, the Cys in cathepsin B is known to be involved in disulfide bridge and therefore only “other” form and no SNO was detected. Good specificity of ascorbate reduction was shown by detecting SNO only in channel 127. The varying amount of SNO and other Cys-modification induced by GSNO on each specific Cys site is represented by the relative intensity ratio of reporter ions 127 to 130. The other three examples shown represent sites where approximately equal amounts (ribosomal protein S2), more SNO (alanyl-tRNA synthetase) and less SNO (ribonuclease inhibitor) than other Cys modifications were induced by added GSNO. All identified Cys residues were classified by the patterns of the relative intensities of reporter ions into three clusters using GproX (B). Percentage of the involved Cys residues in each cluster was shown as indicated with the color-coded intensities of membership shown at right.
Induced Cys modification by GSNO versus GSSG
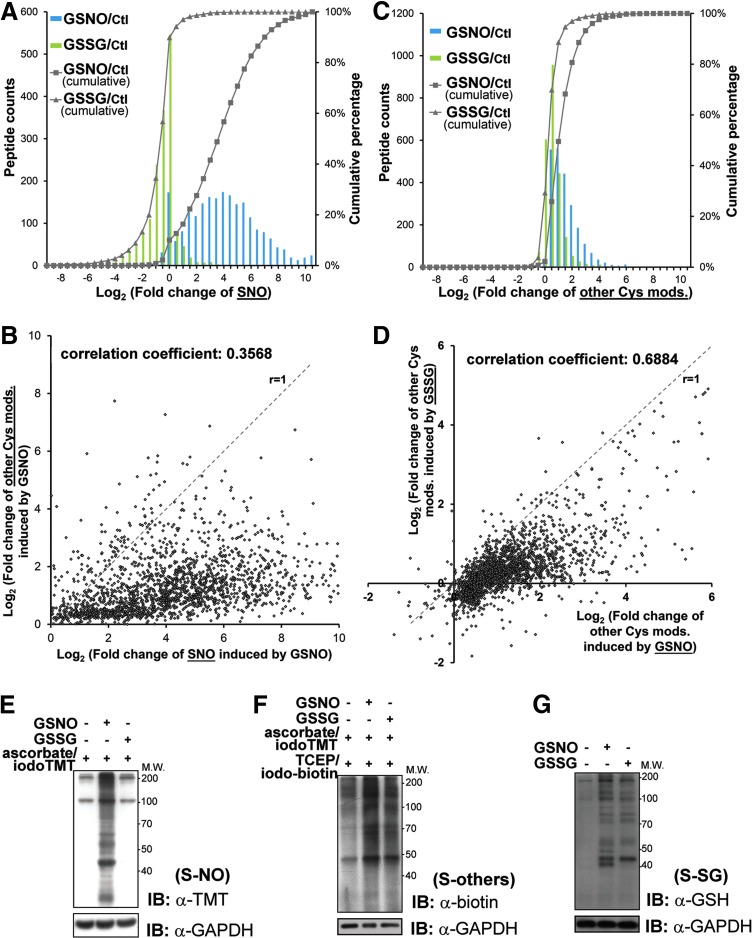
First of all, it is clear that SNO is rather specifically induced by addition of GSNO and not by GSSG, with a negligible basal level shown by control, which are measured by reporter ion 127, 128, and 126, respectively. Addition of GSSG was expected to perturb the reduced glutathione (GSH)/GSSG ratio of the cell lysates and may thus induce additional S-SG but not SNO. GSNO, on the other hand, is both a transnitrosylating and glutathionylating agent (19, 32, 36, 39, 71). Our global profiling revealed that about 25% of all Cys sites (cluster 2, Fig. 4B) were either equally susceptible (e.g., Cys139 of 40S ribosomal protein S2) or more prone to SNO than S-SG (reporter ions 127>130, e.g., Cys533 of alanyl-tRNA synthetase) by GSNO (Fig. 4A). In addition, ∼33% of all Cys sites (cluster 3, Fig. 4B) were more readily glutathionylated by GSNO, or initially preoccupied by other TCEP-reversible modifications to render them relatively resistant to SNO (e.g., Cys80 of ribonuclease inhibitor).
Focusing on the quantitative aspects of SNO, GSNO induced up to three orders of increase in SNO relative to non-treated control, with more than 80% showed twofold or higher increment while about 10% not affected (Fig. 5A). This distribution suggests that without the constrains of cellular compartmentalization and controlled intracellular redox potential, different Cys sites exhibit a wide range of inherent susceptibility to transnitrosylation by GSNO. Moreover, although GSNO is capable of effecting S-SG, a scatter plot of all identified Cys sites shows that a majority (>90%) were preferentially S-nitrosylated rather than glutathionylated, while the reverse is true for a distinctive minority (Fig. 5B). Consistent with a previous report (52), GSSG largely effected no change or a slight to moderate decrease of SNO level on more than 90% of all identified Cys sites, with about 50% showing less than twofold changes (Fig. 5A). In the case of other reversible Cys modifications (Fig. 5C), about 50% were not affected by GSSG treatment while another 40% showed less than twofold increase. This compares with about 50% showed two or more fold increment by GSNO treatment, with the remaining mostly showing a slight increase. It therefore appears that, overall, GSNO is a better agent in causing S-SG despite the competing SNO event. This is also reflected by the scatter plot (Fig. 5D), which shows that the majority of spots were distributed under the r=1 slope, namely higher level of S-SG were induced by GSNO than by GSSG. The global quantitative results of SNO and other reversible Cys modifications were confirmed by immunoblotting (Fig. 5E, F) after applying similar sequential switch approach to the treated samples and untreated control (see “Materials and Methods”). We also confirmed the induction of S-SG under the in vitro experiments by immunoblotting with anti-glutathione antibody (Fig. 5G).
FIG. 5.
Relative amount of induced SNO and other Cys-modification upon in vitro treatment of H9c2 cell lysates with GSNO or GSSG. Global analysis by the sequential iodoTMT switch method revealed that GSNO but not GSSG treatment would induce significant SNO formation as shown by the histograms of fold changes of SNO induced by different treatments (A). The solid lines with closed square or triangle labels represent the cumulative percentage of induced SNO by GSNO or GSSG treatments, respectively. This is further supported by anti-TMT immunoblotting in parallel experiments involving only the first switching step (inset, gel pattern). On average, GSNO was shown to induce more SNO formation than other Cys-modifications on most Cys sites, which led to the observed scatter plot pattern (B). Dash line in the scatter plot (slope=1) represented equal fold change of SNO and S-SG of particular Cys residues under GSNO treatment. Correlation coefficient of induced fold changes of SNO versus other Cys modifications was shown as indicated. Moreover, GSNO induced an overall higher amount of other Cys modification than GSSG did (C), as revealed by the corresponding scatter plot pattern (D). All measured fold changes were relative to untreated control. The changes of SNO and other Cys modifications caused by in vitro treatments were also validated by immunoblotting using anti-TMT and anti-biotin, respectively, after sequential reduction/alkylation with IAM, ascorbate/iodoTMT, and TCEP/Polyethyleneoxide-iodoacetyl-biotin as described in “Materials and Methods” (E, F). The formation of S-SG was confirmed by immunoblotting of treated samples using anti-glutathione antibody without any labeling (G). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Quantitative redox proteomics of hypoxic cardiomyocyte with GSNO protection
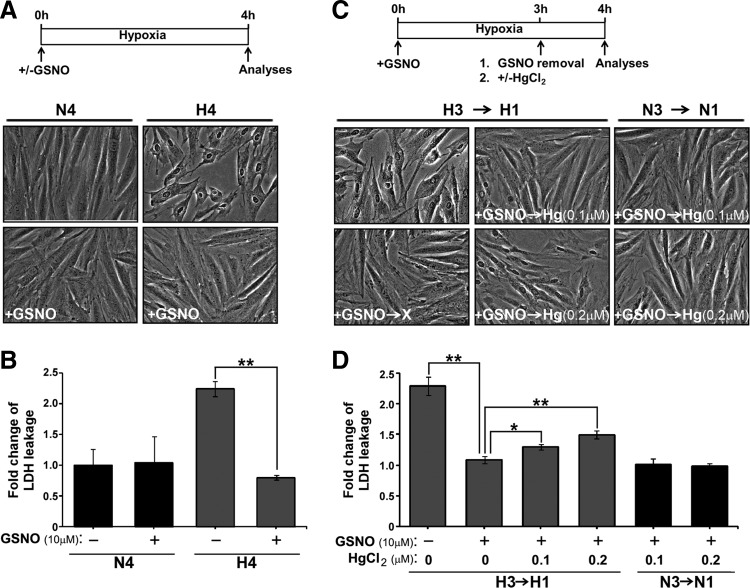
The inherent susceptibility of Cys sites to SNO and/or S-SG by GSNO and the global pattern thus generated provided a comparative basis for us to next examine the global effect of adding GSNO to H9c2 cardiomyocyte undergoing hypoxia. It has been shown that an increased level of protein SNO contributes to myocardial ischemic preconditioning-mediated protection against reperfusion injury (38, 69). We reconstituted SNO-dependent cytoprotection by treating hypoxic H9c2 cells (0.5% O2, 37°C for 4 h) with GSNO and monitored changes of morphology by microscope, and the index of cell injury by detection of cellular enzyme lactate dehydrogenase (LDH) in the conditioned medium. As highlighted in Figure 6A, a retraction of cell shape and a loss of cell–cell contact shown by hypoxic cells were alleviated by the GSNO treatment. In addition, hypoxia-induced leakage of LDH was suppressed by incubation of cells with GSNO (Fig. 6B), suggesting that the plasma membrane integrity in hypoxic H9c2 cells was restored in the presence of GSNO. We further examined whether GSNO-mediated cytoprotection was SNO-dependent. For this, mercury dichloride (HgCl2), which is a cell-permeable agent of denitrosylation that selectively removes NO moiety from S-nitrosothiols thus forming a stable Cys-Hg bond (22, 46), was added to H9c2 cells already pre-exposed to GSNO under the hypoxic condition. Removal of excess GSNO at the time of HgCl2 addition (Fig. 6C) allowed Hg-mediated denitrosylation to occur specifically on thiols of endogenous proteins in H9c2 cells. Employing this workflow, we demonstrated that while supplement of GSNO prevented H9c2 cells from hypoxic injuries, such protective effects on morphological changes (Fig. 6C) and leakage of LDH (Fig. 6D) were significantly attenuated by the treatment of HgCl2 following a dose dependent manner. Importantly, HgCl2 treatment did not cause any detectable effect in normoxic H9c2 cells supplied with GSNO (Fig. 6C, D). Together, these results demonstrated that SNO-dependent cytoprotection was specific to cardiomyocytes suffering hypoxic stress.
FIG. 6.
GSNO treatment protects H9c2 cells against hypoxic injuries via protein SNO. (A, B) Rat cardiomyocyte H9c2 cells were exposed to normoxia (N) or hypoxia (H) for 4 h in the absence or presence of GSNO (10 μM). (A) Cell morphology was assessed by the phase contrast microscopy. (B) Aliquots of conditioned medium were subjected to LDH activity assay. (C, D) Hypoxic H9c2 cells supplemented with GSNO (10 μM) for 3 h were exposed to HgCl2 (0.1 or 0.2 μM) for additional 1 h. Normoxic H9c2 cells with the same exposure to GSNO and HgCl2 were included for comparison. At the end of 4 h of treatment, morphological changes (C) and level of LDH released to conditioned medium (D) of H9c2 cells were analyzed. Data shown in (B) and (D) are presented as mean±SD [n=6 in (B) and n=3 in (D); *p<0.05, **p<0.001]. HgCl2, mercury dichloride; LDH, lactate dehydrogenase; SD, standard deviation.
Under the same experimental conditions, the cell lysates were subjected to Cys-redox proteomic analysis after being taken through our established sequential iodoTMT switch workflow. In parallel, a reverse labeling was also performed to cross validate the reproducibility of the results. In total, 266 Cys sites from 169 proteins were determined as redox modified (Supplementary Table S3). Ingenuity Pathway Analysis (IPA) was used to explore the potential roles on NO-dependent protective effects of the identified proteins in hypoxic cardiomyocyte. Our data revealed that the proteins with redox modifications are involved in a variety of cellular processes (Supplementary Table S4). The determined change of both SNO and other reversible modifications on each identified Cys residues was summarized in Figure 7. Among them, 12 and 14 proteins were shown to have significantly changed (Z score >2) SNO or other reversible modifications, respectively (Table 1). Interestingly, if only those proteins with significant change of redox modifications are considered in IPA, the most related cellular functions include cell death (p-value: 4.13E-05), free radical scavenging (p-value: 5.86E-5), and cellular assembly and organization (p-value: 8.13E-5) (Supplementary Table S5). Potential signaling networks proposed through the MetaCore server using the same dataset pinpointed several critical “nodes,” such as Rac1, RhoA, protein kinase C, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), in the redox signaling, where still focused on regulating apoptosis and cytoskeleton organization (Fig. 7C). As a potential key regulator in the redox signaling, we thus further confirmed the decrease of SNO level on GAPDH by conventional BST in GSNO-treated cardiomyocyte under hypoxic insult (Fig. 7D). In summary, our results suggest that regulation of redox-related modifications play a role in NO-mediated protection of hypoxic cardiomyocytes from apoptosis and loss of intercellular adhesion, consistent with the biological assays (Fig. 6).
FIG. 7.
Effect of GSNO treatment on in vivo Cys-SNO and other reversible Cys modifications in H9c2 cells exposed to hypoxia. Global analysis by the sequential iodoTMT switch method revealed different degree of in vivo SNO and other Cys modification in cardiomyocytes subjected to hypoxia conditions for 4 h, with and without addition of GSNO. Four-plexed quantification with the 126/127 and 130/131 channels reporting SNO and other Cys modifications respectively for conditions with and without GSNO addition, was cross validated by similar four-plexed reverse labeling in which the SNO and other Cys modification were reported instead by the 130/131 and 126/127 channels, respectively. The overall global pattern derived from forward labeling was displayed on a scatter plot (A) and those with significant changes in either SNO or other Cys modifications, or both, are annotated by their gene names and the modified Cys residues. Dash lines represent the significance threshold of the reported changes in Cys modifications (Z score >2). Red and green color-coded data points represent increase and decrease in both Cys modifications, respectively. The normalized intensity patterns of the reporter ion channels corresponding to four circled data points in (A) are shown in (B), along with the reverse labeling pattern. Cys43 of Gal-1 represents a site where increased formation of SNO and other reversible forms were observed with added GSNO, whereas Cys139 of Glo1 represents another where significant decrease in SNO was registered. The Cys80 of DSTN and Cys116 of 40S ribosomal protein S11 showed instead only significant increase in SNO and decrease in other Cys modification, respectively. In general, fold change in other Cys modification was less distinct than that of SNO, indicating that SNO is the prime effect of adding GSNO to hypoxic cardiomyocytes. Potential molecular networks proposed based on the proteins with significantly changed redox Cys modifications using MetaCore algorithm was shown in (C). Decreased SNO level of GAPDH in NO-protected hypoxic H9c2 was also confirmed by conventional BST and the relatively quantitative results were determined from the image intensities with mean±SD of three independent experiments (*p<0.01) (D). BST, biotin switch technique; DSTN, destrin; Gal-1, galectin 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Glo1, glyoxalase I.
Table 1.
List of Cysteine Sites with Significantly Changed Redox-Cysteine Modifications in NO-Mediated Protection of Hypoxic Cardiomyocytes
| Log2 ratio | Z score | ||||||
|---|---|---|---|---|---|---|---|
| Protein name | Gene name | Peptide sequence | Cys site positions | SNO | other | SNO | other |
| SNO increased | |||||||
| Destrin | DSTN | HFVGMLPEKDCR | 80 | 3.37 | 0.18 | 5.5 | 1.2 |
| Galectin-1 | LGALS1 | SFVLNLGKDSNNLCLHFNPR | 43 | 2.52 | 0.44 | 4.1 | 2.5 |
| Filamin A, alpha | FLNA | SPYTVTVGQACamNPTACR | 483 | 2.40 | 0.14 | 3.9 | 1.0 |
| DEAD box helicase 1 | DDX1 | HYYEVSCamHDQGLCR | 145 | 1.73 | 0.64 | 2.7 | 3.5 |
| Prostaglandin E synthase 3 | Ptges3 | HLNEIDLFHCIDPNDSK | 58 | 1.66 | 0.21 | 2.6 | 1.3 |
| Alanyl-tRNA synthetase | AARS | TCFYAEQGGQIFDEGYLVK | 533 | 1.45 | 0.04 | 2.2 | 0.5 |
| Riboflavin kinase | RFK | SLPFFCR | 8 | 1.44 | 0.30 | 2.2 | 1.8 |
| Isocitrate dehydrogenase 3 gamma | IDH3G | TSLDLYANVIHCK | 148 | 1.35 | 0.23 | 2.0 | 1.5 |
| Galectin-1 | LGALS1 | ACGLVASNLNLKPGECamLK | 3 | 1.34 | −0.07 | 2.0 | 0.0 |
| Peptidylprolyl isomerase A | PPIA | HTGPGILSMANAGPNTNGSQFFICTAK | 115 | 1.33 | 0.01 | 2.0 | 0.3 |
| SNO decreased | |||||||
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | IVSNASCTTNCamLAPLAK | 150 | −1.87 | −0.38 | −3.6 | −1.6 |
| Glyoxalase I | GLO1 | GFGHIGIAVPDVYEACK | 139 | −1.17 | −0.65 | −2.4 | −3.0 |
| Nidogen 2 | NID2 | SHLYGITAVYPYCPTGR | 1391 | −1.15 | −0.14 | −2.3 | −0.4 |
| Other redox modifications increased | |||||||
| DEAD box helicase 1 | DDX1 | HYYEVSCamHDQGLCR | 145 | 1.73 | 0.64 | 2.7 | 3.5 |
| Galectin-1 | LGALS1 | SFVLNLGKDSNNLCLHFNPR | 43 | 2.52 | 0.44 | 4.1 | 2.5 |
| THUMP domain containing 3 | THUMPD3 | HCFTSNEAAR | 240 | 0.41 | 0.42 | 0.4 | 2.5 |
| Transmembrane 9 superfamily member 2 | TM9SF2 | HTHIDKPDCSGPAMDISNK | 250 | 0.65 | 0.34 | 0.8 | 2.0 |
| Ras-related C3 botulinum toxin substrate 1 | RAC1 | HHCPNTPIILVGTK | 105 | 0.78 | 0.33 | 1.0 | 2.0 |
| Other redox modifications decreased | |||||||
| Ribosomal protein S11 | RPS11 | HKNMSVHLSPCFR | 116 | 0.01 | −1.08 | −0.3 | −5.2 |
| Thioredoxin reductase 1 | TXNRD1 | CDYDNVPTTVFTPLEYGCCGLSEEK | 365/382/383 | −0.10 | −0.70 | −0.5 | −3.3 |
| Glutathione peroxidase 1 | GPX1 | YIIWSPVCR | 98 | 0.02 | −0.67 | −0.3 | −3.1 |
| Glyoxalase I | GLO1 | GFGHIGIAVPDVYEACK | 139 | −1.17 | −0.65 | −2.4 | −3.0 |
| Actin, alpha 2 | ACTA2 | CamEEEDSTALVCDNGSGLCK | 12/19 | −0.90 | −0.63 | −1.9 | −2.9 |
| Actin, gamma 2 | ACTG2 | CamEEETTALVCDNGSGLCK | 11/18 | −0.49 | −0.60 | −1.2 | −2.8 |
| Glycine cleavage system protein H | GCSH | SCYEDGWLIK | 135 | 0.15 | −0.55 | −0.1 | −2.5 |
| Ribosomal protein S14 | RPS14 | CKELGITALHIK | 85 | 0.04 | −0.50 | −0.3 | −2.2 |
| Serine/arginine-rich splicing factor 7 | Srsf7 | GHYAYDCHR | 119 | 0.14 | −0.45 | −0.1 | −2.0 |
SNO, S-nitrosylation.
C, Cys residue identified with increased or decreased level of SNO.
Discussion
It is now well accepted that redox status of specific Cys sites can impact intricate cellular pathways, and that dysregulation of cellular redox homeostasis has severe physiological consequences. Although several reversible Cys modifications on the same and/or different sites are understood to cross-talk and collectively fine-tune the protein functions, it remains extremely difficult to identify and quantify all the relevant forms simultaneously due to the lack of suitable methodology. Here, taking advantage of the newly available multiplex iodoTMT reagents, we have developed a sequential iodoTMT switch procedure, which enables site-specific identification and quantification of multiple redox-related Cys-modifications at a proteomic level. For the first time, we provided a quantitative assessment of the global in vivo thiol status of proteins in cardiomyocytes undergoing hypoxia and how these were altered by applied GSNO, which elicited a protective consequence against hypoxic damage.
There remains a few technical shortcomings in our approach particularly in further dissecting the so-called other reversible modifications, which are resistant to ascorbate but susceptible to TCEP reduction. However, we have shown that Cys involved in structural disulfides can be readily distinguished by unique reporter ion profiles, whereas incorporation of a few additional reagents within the overall sequential iodoTMT switch workflow could potentially further resolve the pattern. For example, prior to or instead of TCEP-reduction, arsenite may be used to specifically reduce S-OH (57, 60), and S-SG can also be more specifically reduced by glutaredoxin-based reaction (45), which can discriminate against other thiolation and oxidation. In this context, the less well-defined specificities of these reducing reagents (2, 18, 43) other than the ascorbate are in fact the main limiting factors and not the iodoTMT switching itself since the multiplex nature of the isobaric tags is more than capable of accommodating the extra sequential steps of reduction and tagging cycle. As an alternative approach, contribution of the sulfenic form can be subtracted from the “other reversible modifications” by arsenite treatment during the initial cell lysates, or by irreversible trapping using a reagent such as dimedone (4, 64). The former approach would lead to it being tagged by IAM in the initial alkylating steps of all available SH, whereas the latter would convert it to forms no longer reducible by TCEP. In parallel analysis, we have, in fact, added dimedone and sodium arsenite in IAM blocking buffer after GSNO treatment (Supplementary Fig. S4A) and observed no significant changes in the relative ratio of other modifications compared to that without dimedone or sodium arsenite addition (Supplementary Fig. S4B). This implies very little contribution of sulfenic forms to what we identified as “other modifications”, which therefore predominantly comprises S-SG if the disulfides are also discounted.
By applying the sequential iodoTMT switch, we have initially identified and quantified about 2500 modified Cys sites that can be variably S-nitrosylated or S-glutathionylated in vitro by GSNO. In this case, we were examining the intrinsic susceptibility of each of the identified Cys sites under circumstances not further constrained by intracellular compartmentalization or redox potential. Although it is widely accepted that GSNO can induce the formation of both SNO and S-SG in a competitive manner, the mechanism underlying the tendencies of each reactive Cys to undergo either modifications remains poorly understood (13, 17, 19, 32, 48), due to technical difficulties. Giustarini et al. (19) have previously shown a diverse response, in terms of the amounts of SNO or S-SG formation, exhibited by select few proteins exposed to GSNO, and suggested that the individual Cys reactivity defined by its surrounding microenvironment was the key factor for the differential preferences. We now extended this observation to include many more sites on a proteomic scale and likewise found quite a diverse predisposition to form either SNO or S-SG (Fig. 5B, correlation coefficient=0.3568). A much stronger positive correlation was shown instead for the formation of S-SG induced by GSNO or GSSG (Fig. 5D, correlation coefficient=0.6884). It was suggested that Cys thiols with higher nucleophilicity, such as histidine-activated thiolate, can attack the SNO bond of GSNO, leading to the preferential formation of GSH-mixed protein disulfides (11, 19, 50). Our results showed that those Cys more prone to form S-SG under GSNO treatment were also more likely to form S-SG when exposed to GSSG (Fig. 5D), which indicated that either these Cys contained thiols are better nucleophiles regardless of the glutathionyl donor, or the formed S-SG on these Cys are less readily removed by either the enzymatic system (e.g., glutaredoxins) or GSH.
We further classified the modified Cys into three clusters based on the relative amount of induced SNO and other reversible Cys modifications (Fig. 4B). The potential sequence motifs of the three clusters were examined using Motif-X (9, 62). Interestingly, the three highest scored motifs (p<0.000001) as determined from cluster 1 all contained two or more nearby Cys, which may form disulfide bonds (Supplementary Fig. S5A). We further evaluated the Cys residues classified in cluster 1, 38% of them are already annotated as disulfide-linked sites in UniProt Protein knowledgebase, whereas another 47% of them are not annotated in database but containing at least one nearby Cys in their primary sequences, which increases the possibility for these Cys residues to form disulfide linkages during the in vitro experiments. Indeed, 51% of those non-annotated Cys sites in cluster 1 are predicted as disulfide-linked residues by applying the DisLocate algorithm (61). Remarkably, the only potential motif determined for cluster 2 showed an exclusive aspartate residue in position −3 while the only potential motif in cluster 3 exhibited a lysine located at position +1. (Supplementary Fig. S5B, C). Negatively charged amino acids adjacent to Cys has previously been suggested as the key feature of S-nitrosylated Cys in vivo (11), which is consistent with current finding that Cys sites in cluster 2 were preferentially S-nitrosylated. It should, however, be cautioned that the motif scores for clusters 2 and 3 are relatively low and may not represent the whole population. In general, the lack of the strong consensus motif for redox-modified Cys as found here and by others (52) further support the notion that susceptibility of a particular Cys to redox modifications is not solely determined by its flanking primary sequence but rather dependent on higher ordered structure and other superimposed intracellular constrains such as proximity or localization of reactive nitrosative species. Each different cell types also maintains different relative amounts of redox agents such as the GSH and GSSG pools, different activities of redox-related enzyme systems including the thioredoxins, glutaredoxins, various reductases, and the eNOS systems, which collectively define a cell- and pathophysiology-specific redox potentials acting on the susceptible Cys. We have, for example, found from our quantitative analysis that overall a higher degree of reversible Cys modifications could be induced by GSNO on the endothelial Rat-1 lysates than the cardiomyocytes H9c2 (Supplementary Fig. S6).
Quantitative measurement of multiplexed redox Cys modifications in hypoxic cardiomycotyes provides a paramount view on how NO may exert its protective function under myocardial ischemia. It has been well documented that ischemic stress caused by coronary occlusion led to myocardial injuries, including disruption of F-actin and microtubule cytoskeleton (25, 29). In response to reperfusion of the heart, myocardium suffers severe damage at least in part due to production of reactive oxygen species (ROS) that may oxidize various cellular proteins, thus inducing proteolysis or cardiac cell death (3, 53). Obviously, myocardial protection is essential for reducing injuries of the heart in both phases of ischemia and reperfusion. It has been previously shown that reversible SNO prevents proteins from ROS-induced irreversible Cys oxidation (8), suggesting that NO may act as a cytoprotector against reperfusion-induced myocardial injury (12). In the present study, we provide evidence demonstrating that NO could also protect cardiomyocytes under the hypoxic stress, consistent with the cytoprotective role of SNO in myocardium of the ischemic heart. Therefore, our identification of multiplexed Cys modifications, including SNO and other reversible modifications in hypoxic cardiomyocytes exposed to NO, is informative. This dataset illustrates the fact that NO action is not confined to SNO, but likely promotes a complicated shift among various forms of reversible Cys modifications. We propose that such a global change of Cys-redoxome is indeed the fundamental basis for the prominent cytoprotective role of NO in myocardium under the ischemic stress.
While the full scale of evaluation of our dataset requires additional investigations, some of the candidates identified by this analysis are potentially important for NO-mediated protection in hypoxic cardiomyocyte. Among them, Galectin-1 is involved in a wide range of cellular functions, including cell proliferation, apoptosis, inflammation, and cell adhesion (6). It executes distinct functions depending on its redox status and only the reduced form possesses β-galactoside-binding activity, which is crucial for functional recovery after brain ischemia (28, 44, 74). Previous reports also indicated that galectin-1 is the target of SNO and S-SG (16, 41). Together, these evidences suggested that galectin-1 is sensitive to environmental redox change and its functions are modulated through complicated redox-related Cys modifications. The present results showed that the two Cys residues (Cys3 and Cys43) of galectin-1 were significantly S-nitrosylated in NO-protected cardiomyocyte under hypoxia. In addition, the level of other reversible modifications on Cys43 was increased under the same condition. We propose that such robust Cys modifications in response to GSNO treatment might preclude galectin-1 from induction of apoptosis in hypoxic cardiomyocyte. In contrast, a decreased level of SNO was observed on Cys150 of GAPDH in GSNO-treated hypoxic H9c2 cells, which was also supported by the results from conventional BST. Interestingly, it was previously shown that SNO of Cys150 not only abolished the catalytic activity of GADPH but also directed nuclear translocalization of GAPDH for promoting apoptosis through binding to Siah1 (24). Our findings thus suggest that NO might protect cardiomyocytes against hypoxia-induced apoptosis through a yet to be identified mechanism for maintaining denitrosylation and cytosolic localization of GAPDH. It is worth to note that the potential molecular networks proposed by MetaCore from our dataset also support the possible routes that (i) galectin-1 functions as a redox-sensing probe and passes signals into cell through affecting receptors on plasma membrane; and (ii) GAPDH transmits the redox signals to different cellular compartments and affects the regulation of cell death through converting redox status on particular Cys residues (Fig. 7C). In addition, by acting as a transnitrosylase, GAPDH might also augment the level of acetylation through inhibiting sirtuin-1 and activating p300-CBP (40, 63), as revealed on the proposed networks. On the other hand, the proposed networks indicate that Rac1, which we found significantly decreased other reversible modifications on Cys105 in NO-protected cardiomyocyte, plays a central role in response to hypoxia to regulate either apoptotic processes through P38 mitogen-activated protein kinases or cellular barrier function through myosin light chain kinase. Interestingly, two protein tyrosine phosphatases, PTPRC (CD45) and SHP-1, were also involved in the proposed redox networks suggesting that tyrosine phosphorylation is also the key mechanism for hypoxic responses. In fact, we did observe dramatic decrease of overall protein tyrosine phosphorylation in hypoxic cardiomyocyte accompanied with nearly complete recover in NO-mediated protection (article in preparation). Overall, it suggests that although more studies are required to clarify the exact mechanisms, redox-sensitive reversible Cys modifications might be the initiator to regulate acetylation and phosphorylation levels in different cellular compartments thus achieving NO-mediated myocardial protection.
On the basis of our data (comparison between Supplementary Tables S2 and S3), it is important to note that the Cys sites with significant changes of SNO or other modifications in GSNO-protected hypoxic H9c2 cells may not necessarily be the most redox susceptible sites in total lysates exposed to NO in vitro. For example, Cys61 of Galectin-1 was not identified to be redox-modified in vivo, but was highly susceptible to SNO (∼3-fold higher than Cys3 and Cys43) when the protein was treated with GSNO in vitro. These findings suggest that, in contrast to the intrinsic reactivity of any particular thiol being merely determined by its chemical properties, site-specific regulation of Cys modifications inside cells is precisely controlled by endogenous enzyme systems and redox microenvironment of subcellular compartments, consistent with a concept proposed previously (12, 47).
In conclusion, with the newly developed iodoTMT tags available for quantitative measurement of multiplexed Cys-redox modifications by efficient immunoenrichment and advanced shotgun LC-MS/MS analysis, our study provides the first evidence that NO-mediated ischemic protection may be achieved by a process far more complicated than mere increase of SNO level in myocardium. In addition to SNO for shielding Cys residues against oxidation suggested by recent studies (37, 38), other forms of reversible redox modifications are involved in NO-dependent cardioprotection. A comprehensive delineation of Cys-redoxome by the experimental workflow introduced in the present study will significantly contribute to our understanding of how NO maintains physiological homeostasis and thus protecting heart from ischemia-reperfusion injury.
Materials and Methods
Cell culture, hypoxic exposure, detection of LDH leakage, and treatment with GSNO and HgCl2
All cell lines were purchased from American Type Culture Collection. H9c2 rat cardiac myoblast was cultivated in high glucose Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). Rat-1 cells were grown in the same medium plus 10 mM of glutamine. To simulate experimental hypoxia, the serum- and glucose-free Hank's Balanced Salt Solution (HBSS) was first pre-equilibrated in an air-tight glove box (Don Whitley Scientific) under hypoxic condition (0.5% O2–95% N2–5% CO2) for 48 h. The H9c2 cells were then incubated in the pre-equilibrated hypoxic HBSS in the glove box at 37°C for 4 h after two times of wash. To examine the NO-mediated protection effects on plasma membrane integrity, 10 μM of GSNO was included in the HBSS during the incubation and the degree of cell injury was assessed by the changes of morphology using microscope and by the level of LDH leakage through measuring the LDH activity in both conditioned HBSS and lysate using a commercial colorimetric kit (Promega). LDH leakage was expressed as percentage of the LDH activity in the conditioned HBSS versus the total LDH activity (conditioned HBSS+lysate) and then normalized against the control sample, that is, H9c2 cell under normoxia without GSNO treatment. To test the role of SNO-dependent cytoprotection, the conditioned HBSS supplemented with GSNO was removed from the cells after 3 h of hypoxic exposure, followed by immediate addition of the HBSS containing HgCl2 (0.1 or 0.2 μM) to the cells for one more hour of hypoxic exposure. The conditioned HBSS was collected for the measurement of LDH released during the entire course of hypoxic exposure.
In vitro SNO and S-SG
Lysis buffer (20 mM HEPES pH 7.4, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid [EDTA], and 1% (v/v) NP-40) was first purged with N2 for 1 h at room temperature to remove dissolved oxygen. H9c2 cells (total ∼6×106) were then harvested and lysed in 0.5 ml of the purged lysis buffer containing freshly added protease inhibitor cocktail (Roche). Cell debris was removed by centrifugation and protein concentration of the resulting supernatant was determined by Bradford protein assay (Bio-Rad). Equal aliquots (0.3 ml with protein concentration of ∼1 mg/ml) were taken and treated with 100 μM of GSNO, GSSG, or mock-treated for 20 min at 37°C. The small molecules were removed by acetone precipitation as described (31). Protein pellets were resuspended by HENS buffer (250 mM HEPES pH 8.0, 1 mM EDTA, and 0.1 mM neocuproine containing 2.5% sodium dodecyl sulfate) for iodoTMT labeling. All steps were performed under the protection from light to avoid unwanted changes of redox Cys modifications.
Sequential iodoTMT switch
For each experiment, about 300 μg of proteins were used. All the alkylation steps were performed in dark. First, all SH in the sample were blocked by IAM to preclude further change of Cys modifications during subsequent labeling procedures. For in vitro GSNO/GSSG treatments, 200 mM of IAM was added in the 300 μl of HENS buffer used for resuspending protein pellets after the completion of treatments. The alkylation was conducted through incubation in end-to-end rotator at 37°C for 1 h. For examining the NO-mediated protective effects on hypoxic cardiomyocyte, 200 mM of IAM was included in lysis buffer during cell lysis. After removing the cell debris, 300 μg of proteins were taken from each sample individually and IAM blocking was continued by adding three volumes of HENS buffer containing 100 mM of IAM and incubated at 37°C for 1 h. Acetone precipitation was used here and followed by steps to remove excess reagents. S-nitrosylated thiols were specifically reduced and labeled by resuspending protein pellets in HENS containing 20 mM of ascorbate and 1 mM of iodoTMT1 reagents carrying the desirable reporter ions and incubated at 37°C for 2 h (14, 31, 52). After removing the excess reagents, all other remaining reversible modified thiols were reduced and labeled by reacting with 5 mM of TCEP and 1 mM of iodoTMT2 carrying a distinct set of reporter ions different from iodoTMT1, at 37°C for 1 h. Upon completion of labeling, all samples to be analyzed were mixed. Acetone precipitation was performed again and the resulting pellet was dissolved in 25 mM of ammonium bicarbonate, pH 8.0, containing 0.5% of RapiGest SF (Waters), followed by adding sequential grade trypsin (Promega) at an enzyme to protein ratio of 1:50 (w/w) and incubated at 37°C overnight.
For additional cysTMT switch, all steps were performed as described above for iodoTMT switch, except for the substitution of cysTMT-X for iodoTMT-X to label S-nitrosylated thiols. In addition, due to the reversibility of disulfide linkage between cysTMT and the Cys-thiol group, no further reducing reaction was performed. An aliquot of labeled proteins (5 μg) from each treatment were subjected to immunoblotting with anti-TMT antibody (Thermo Fisher Scientific), which recognizes all TMT-related tags.
To determine the levels of SNO and other remaining reversible modifications in treated samples through immunoblotting, all NO moieties on S-nitrosylated thiols were first “switched” to iodoTMT tag as aforementioned. The other reversible modifications were then reduced as in the sequential iodoTMT switch, but 5 mM of polyethyleneoxide-iodoacetyl-biotin was used to label the generated SH. Aliquots of sequentially labeled samples (5 μg) were subjected to immunoblotting with anti-TMT antibody for detecting SNO, or anti-biotin antibody (Cell Signaling Technology) for other reversible modifications. To further confirm the formation of SSG under in vitro GSNO and GSSG treatments, immunoblotting with anti-glutathione antibody (Virogen) was also performed.
Enrichment of iodoTMT- or cysTMT-labeled peptides
The iodoTMT- or cysTMT-labeled peptides were immunocaptured following the manufacturer's instruction, with some modifications. Briefly, 250 μl of combined sample with peptide concentration of 1 mg/ml was diluted with three volumes of Tris-buffered saline (TBS, 25 mM Tris, 150 mM NaCl, and 2 mM KCl, pH 7.4) and incubated overnight at 4°C with pre-equilibrated immobilized anti-TMT resin. In general, 250 μl of the immobilized anti-TMT resin slurry was used for the indicated amount of peptide mixture after three times of washes with three volumes of TBS by repeated centrifugation and supernatant removal. Unlabeled peptides were removed by sequential washes with TBS and TBS containing 4 M urea, followed by incubation at room temperature for 30 min in the presence of urea. Beads were then washed three times with TBS and five times with water. Labeled peptides were eluted with 40% (v/v) acetonitrile (ACN) containing 0.5% (v/v) trifluoroacetic acid and dried in SpeedVac.
MS analysis
All samples to be analyzed were pre-cleaned by C18 Micro TipColumn (Hoefer) according to the manufacture's protocol. Peptides (0.5 μg) were injected into a trapping column (180 μm×2 cm) with 0.1% formic acid for 3 min and then separated by a 75 μm×250 mm C18 capillary column (1.7 μm; BEH130) using a nanoACQUITY UPLC system (Waters) controlled by MassLynx 4.1 with a linear gradient of 5–50% ACN (in 0.1% formic acid) in 150 min at a constant flow rate of 300 nl/min. Peptides were detected in an LTQ-Orbitrap Velos hybrid mass spectrometer (Thermo Fisher Scientific) by a data-dependent “HCD10” acquisition method. For each cycle, full-scan MS spectra (m/z 350–1600) were acquired in the Orbitrap at a resolution of 60,000 and the 10 most intense ions with charge states ≥2 were sequentially isolated (1.2 Da of isolation window) to a target value of 50,000 ions and fragmented by HCD in the multipole collision cell with NCE as described in the text. Fragment ions were then detected in the Orbitrap at a resolution of 7500.
Data analysis
All MS and MS/MS raw data were processed with Proteome Discoverer version 1.3 (Thermo Fisher Scientific), and the peptides were identified from the MS/MS spectra searched against the UniProtKB/Swiss-Prot (20110531/529056 sequences entries) database using the Mascot 2.3.02 (Matrix Science) with the following constraints: only tryptic peptides with up to two missed cleavage sites; taxonomy: Rattus; and mass accuracy of 10 ppm for the parent ion and 0.05 Da for the fragment ions. iodoTMT or cysTMT-labeled (C), carbamidomethyl (C), deamidation (NQ), oxidation (M), and acetyl (protein N-term) were specified as dynamic modifications. FDRs were calculated by target-decoy strategy with or without using Mascot Percolator. All peptide hits were filtered with a 1% FDR cutoff. All reporter ion areas were calculated by Proteome Discoverer with the integration tolerance of 20 ppm. Missing values were replaced with minimal intensities. The annotation of protein disulfide bonds formation was manually reviewed using the UniProt Protein knowledgebase (http://uniprot.org/). For those not well annotated proteins from the taxonomy of rat, homologues from mouse or human were used as the basis for reviewing. DisLocate algorithm was applied to predict protein disulfide linkage based on their primary sequences (61). Protein annotations were obtained from the IPA program (Ingenuity® Systems, www.ingenuity.com). Potential redox networking of NO-mediated protection on hypoxic cardiomyocyte was determined by MetaCore from Thomson Reuters with several manual adjustments. Data clustering was determined by GProX (59) based on relative intensities of all reporter ions of each identified peptides bearing iodoTMT tags. Overrepresented primary sequence motif patterns in each cluster were extracted by Motif-X algorithm with default parameters (p<0.000001) (9, 62). Potential networks and involved subcellular pathways of identified proteins were analyzed by IPA.
Supplementary Material
Abbreviations Used
- ACN
acetonitrile
- BSA
bovine serum albumin
- BST
biotin switch technique
- CID
collision-induced dissociation
- CV
coefficient of variation
- Cys
cysteine
- CysTMT
reversible isobaric Cys-reactive tandem mass tag
- DMEM
Dulbecco's modified Eagle's medium
- DSTN
destrin
- EDTA
ethylenediaminetetraacetic acid
- eNOS
endothelial nitric oxide synthase
- FDR
false discovery rate
- Gal-1
galectin 1
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- Glo1
glyoxalase I
- GSH
reduced glutathione
- GSNO
S-nitrosoglutathione
- GSSG
glutathione disulfide
- HBSS
Hank's Balanced Salt Solution
- HCD
higher-energy C-trap dissociation
- HgCl2
mercury dichloride
- IAM
iodoacetamide
- iodoTMT
irreversible isobaric iodoacetyl Cys-reactive tandem mass tag
- IPA
Ingenuity Pathway Analysis
- LC
liquid chromatography
- LDH
lactate dehydrogenase
- MS
mass spectrometry
- NCE
normalized collision energy
- PEO
polyethylene oxide
- PEP
posterior error probability
- PSM
peptide spectrum match
- ROS
reactive oxygen species
- SD
standard deviation
- SH
free thiol
- SNO
S-nitrosylation
- S-OH
sulfenic acid
- S-SG
S-glutathionylation
- S-SP
protein disulfides
- TBS
tris-buffered saline
- TCEP
tris(2-carboxyethyl) phosphine
Acknowledgments
This work was supported by Academia Sinica Program grant (to K.-H.K. and T.-C.M.). The MS data were acquired at the Core Facilities for Protein Structural Analysis at Academia Sinica, supported under the Taiwan National Core Facility Program for Biotechnology, NSC100-2325-B-001-029 and NSC101-2319-B-001-003.
Author Disclosure Statement
The authors have no conflicts of interest to disclose.
References
- 1.Aracena P, Tang W, Hamilton SL, and Hidalgo C. Effects of S-glutathionylation and S-nitrosylation on calmodulin binding to triads and FKBP12 binding to type 1 calcium release channels. Antioxid Redox Signal 7: 870–881, 2005 [DOI] [PubMed] [Google Scholar]
- 2.Aracena-Parks P, Goonasekera SA, Gilman CP, Dirksen RT, Hidalgo C, and Hamilton SL. Identification of cysteines involved in S-nitrosylation, S-glutathionylation, and oxidation to disulfides in ryanodine receptor type 1. J Biol Chem 281: 40354–40368, 2006 [DOI] [PubMed] [Google Scholar]
- 3.Becker LB. New concepts in reactive oxygen species and cardiovascular reperfusion physiology. Cardiovasc Res 61: 461–470, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Brennan JP, Miller JI, Fuller W, Wait R, Begum S, Dunn MJ, and Eaton P. The utility of N,N-biotinyl glutathione disulfide in the study of protein S-glutathiolation. Mol Cell Proteomics 5: 215–225, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Brosch M, Yu L, Hubbard T, and Choudhary J. Accurate and sensitive peptide identification with Mascot Percolator. J Proteome Res 8: 3176–3181, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camby I, Le Mercier M, Lefranc F, and Kiss R. Galectin-1: a small protein with major functions. Glycobiology 16: 137R–157R, 2006 [DOI] [PubMed] [Google Scholar]
- 7.Chen CA, Wang TY, Varadharaj S, Reyes LA, Hemann C, Talukder MA, Chen YR, Druhan LJ, and Zweier JL. S-glutathionylation uncouples eNOS and regulates its cellular and vascular function. Nature 468: 1115–1118, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen YY, Chu HM, Pan KT, Teng CH, Wang DL, Wang AH, Khoo KH, and Meng TC. Cysteine S-nitrosylation protects protein-tyrosine phosphatase 1B against oxidation-induced permanent inactivation. J Biol Chem 283: 35265–35272, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou MF. and Schwartz D. Biological sequence motif discovery using motif-x. In: Curr Protoc Bioinformatics Chapter 13, Unit 13: 15–24, 2011 [DOI] [PubMed] [Google Scholar]
- 10.Dayon L, Pasquarello C, Hoogland C, Sanchez JC, and Scherl A. Combining low- and high-energy tandem mass spectra for optimized peptide quantification with isobaric tags. J Proteomics 73: 769–777, 2010 [DOI] [PubMed] [Google Scholar]
- 11.Doulias PT, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, and Ischiropoulos H. Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation. Proc Natl Acad Sci U S A 107: 16958–16963, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evangelista AM, Kohr MJ, and Murphy E. S-nitrosylation: specificity, occupancy, and interaction with other post-translational modifications. Antioxid Redox Signal 19: 1209–1219, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filipovska A. and Murphy MP. Overview of protein glutathionylation. In: Current Protocols in Toxicology, edited by Costa LG, et al., John Wiley & Sons, Inc., Chapter 6: Unit 6.10, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Forrester MT, Foster MW, Benhar M, and Stamler JS. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radic Biol Med 46: 119–126, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster MW, Hess DT, and Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med 15: 391–404, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fratelli M, Demol H, Puype M, Casagrande S, Eberini I, Salmona M, Bonetto V, Mengozzi M, Duffieux F, Miclet E, Bachi A, Vandekerckhove J, Gianazza E, and Ghezzi P. Identification by redox proteomics of glutathionylated proteins in oxidatively stressed human T lymphocytes. Proc Natl Acad Sci U S A 99: 3505–3510, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fratelli M, Gianazza E, and Ghezzi P. Redox proteomics: identification and functional role of glutathionylated proteins. Expert Rev Proteomics 1: 365–376, 2004 [DOI] [PubMed] [Google Scholar]
- 18.Gao XH, Bedhomme M, Veyel D, Zaffagnini M, and Lemaire SD. Methods for analysis of protein glutathionylation and their application to photosynthetic organisms. Mol Plant 2: 218–235, 2009 [DOI] [PubMed] [Google Scholar]
- 19.Giustarini D, Milzani A, Aldini G, Carini M, Rossi R, and Dalle-Donne I. S-nitrosation versus S-glutathionylation of protein sulfhydryl groups by S-nitrosoglutathione. Antioxid Redox Signal 7: 930–939, 2005 [DOI] [PubMed] [Google Scholar]
- 20.Giustarini D, Rossi R, Milzani A, Colombo R, and Dalle-Donne I. S-glutathionylation: from redox regulation of protein functions to human diseases. J Cell Mol Med 8: 201–212, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Good DM, Wirtala M, McAlister GC, and Coon JJ. Performance characteristics of electron transfer dissociation mass spectrometry. Mol Cell Proteomics 6: 1942–1951, 2007 [DOI] [PubMed] [Google Scholar]
- 22.Gow AJ, Chen Q, Hess DT, Day BJ, Ischiropoulos H, and Stamler JS. Basal and stimulated protein S-nitrosylation in multiple cell types and tissues. J Biol Chem 277: 9637–9640, 2002 [DOI] [PubMed] [Google Scholar]
- 23.Grimsrud PA, Swaney DL, Wenger CD, Beauchene NA, and Coon JJ. Phosphoproteomics for the masses. ACS Chem Biol 5: 105–119, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, Ferris CD, Hayward SD, Snyder SH, and Sawa A. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol 7: 665–674, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Hein S, Scheffold T, and Schaper J. Ischemia induces early changes to cytoskeletal and contractile proteins in diseased human myocardium. J Thorac Cardiovasc Surg 110: 89–98, 1995 [DOI] [PubMed] [Google Scholar]
- 26.Held JM. and Gibson BW. Regulatory control or oxidative damage? Proteomic approaches to interrogate the role of cysteine oxidation status in biological processes. Mol Cell Proteomics 11: R111 013037, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Y, Triscari JM, Tseng GC, Pasa-Tolic L, Lipton MS, Smith RD, and Wysocki VH. Statistical characterization of the charge state and residue dependence of low-energy CID peptide dissociation patterns. Anal Chem 77: 5800–5813, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishibashi S, Kuroiwa T, Sakaguchi M, Sun L, Kadoya T, Okano H, and Mizusawa H. Galectin-1 regulates neurogenesis in the subventricular zone and promotes functional recovery after stroke. Exp Neurol 207: 302–313, 2007 [DOI] [PubMed] [Google Scholar]
- 29.Iwai K, Hori M, Kitabatake A, Kurihara H, Uchida K, Inoue M, and Kamada T. Disruption of microtubules as an early sign of irreversible ischemic injury. Immunohistochemical study of in situ canine hearts. Circ Res 67: 694–706, 1990 [DOI] [PubMed] [Google Scholar]
- 30.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, and Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol 3: 193–197, 2001 [DOI] [PubMed] [Google Scholar]
- 31.Jaffrey SR. and Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE 2001: pl1, 2001 [DOI] [PubMed] [Google Scholar]
- 32.Ji Y, Akerboom TP, Sies H, and Thomas JA. S-nitrosylation and S-glutathiolation of protein sulfhydryls by S-nitroso glutathione. Arch Biochem Biophys 362: 67–78, 1999 [DOI] [PubMed] [Google Scholar]
- 33.Jones DP. and Go YM. Mapping the cysteine proteome: analysis of redox-sensing thiols. Curr Opin Chem Biol 15: 103–112, 2011 [DOI] [PubMed] [Google Scholar]
- 34.Kall L, Canterbury JD, Weston J, Noble WS, and MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods 4: 923–925, 2007 [DOI] [PubMed] [Google Scholar]
- 35.Kim SO, Merchant K, Nudelman R, Beyer WF, Jr., Keng T, DeAngelo J, Hausladen A, and Stamler JS. OxyR: a molecular code for redox-related signaling. Cell 109: 383–396, 2002 [DOI] [PubMed] [Google Scholar]
- 36.Klatt P. and Lamas S. Regulation of protein function by S-glutathiolation in response to oxidative and nitrosative stress. Eur J Biochem 267: 4928–4944, 2000 [DOI] [PubMed] [Google Scholar]
- 37.Kohr MJ, Aponte A, Sun J, Gucek M, Steenbergen C, and Murphy E. Measurement of S-nitrosylation occupancy in the myocardium with cysteine-reactive tandem mass tags: short communication. Circ Res 111: 1308–1312, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohr MJ, Sun J, Aponte A, Wang G, Gucek M, Murphy E, and Steenbergen C. Simultaneous measurement of protein oxidation and S-nitrosylation during preconditioning and ischemia/reperfusion injury with resin-assisted capture. Circ Res 108: 418–426, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konorev EA, Kalyanaraman B, and Hogg N. Modification of creatine kinase by S-nitrosothiols: S-nitrosation vs. S-thiolation. Free Radic Biol Med 28: 1671–1678, 2000 [DOI] [PubMed] [Google Scholar]
- 40.Kornberg MD, Sen N, Hara MR, Juluri KR, Nguyen JV, Snowman AM, Law L, Hester LD, and Snyder SH. GAPDH mediates nitrosylation of nuclear proteins. Nat Cell Biol 12: 1094–1100, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lam YW, Yuan Y, Isaac J, Babu CV, Meller J, and Ho SM. Comprehensive identification and modified-site mapping of S-nitrosylated targets in prostate epithelial cells. PloS One 5: e9075, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leichert LI, Gehrke F, Gudiseva HV, Blackwell T, Ilbert M, Walker AK, Strahler JR, Andrews PC, and Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci U S A 105: 8197–8202, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leonard SE. and Carroll KS. Chemical ‘omics’ approaches for understanding protein cysteine oxidation in biology. Curr Opin Chem Biol 15: 88–102, 2011 [DOI] [PubMed] [Google Scholar]
- 44.Lim SY, Raftery MJ, and Geczy CL. Oxidative modifications of DAMPs suppress inflammation: the case for S100A8 and S100A9. Antioxid Redox Signal 15: 2235–2248, 2011 [DOI] [PubMed] [Google Scholar]
- 45.Lind C, Gerdes R, Hamnell Y, Schuppe-Koistinen I, von Lowenhielm HB, Holmgren A, and Cotgreave IA. Identification of S-glutathionylated cellular proteins during oxidative stress and constitutive metabolism by affinity purification and proteomic analysis. Arch Biochem Biophys 406: 229–240, 2002 [DOI] [PubMed] [Google Scholar]
- 46.Maejima Y, Adachi S, Morikawa K, Ito H, and Isobe M. Nitric oxide inhibits myocardial apoptosis by preventing caspase-3 activity via S-nitrosylation. J Mol Cell Cardiol 38: 163–174, 2005 [DOI] [PubMed] [Google Scholar]
- 47.Martinez-Ruiz A, Araujo IM, Izquierdo-Alvarez A, Hernansanz-Agustin P, Lamas S, and Serrador JM. Specificity in S-nitrosylation: a short-range mechanism for NO signaling? Antioxid Redox Signal 19: 1220–1235, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez-Ruiz A. and Lamas S. Signalling by NO-induced protein S-nitrosylation and S-glutathionylation: convergences and divergences. Cardiovasc Res 75: 220–228, 2007 [DOI] [PubMed] [Google Scholar]
- 49.Mieyal JJ. and Chock PB. Posttranslational modification of cysteine in redox signaling and oxidative stress: focus on S-glutathionylation. Antioxid Redox Signal 16: 471–475, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohr S, Hallak H, de Boitte A, Lapetina EG, and Brune B. Nitric oxide-induced S-glutathionylation and inactivation of glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem 274: 9427–9430, 1999 [DOI] [PubMed] [Google Scholar]
- 51.Murphy E, Kohr M, Sun J, Nguyen T, and Steenbergen C. S-nitrosylation: a radical way to protect the heart. J Mol Cell Cardiol 52: 568–577, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murray CI, Uhrigshardt H, O'Meally RN, Cole RN, and Van Eyk JE. Identification and quantification of S-nitrosylation by cysteine reactive tandem mass tag switch assay. Mol Cell Proteomics 11: M111 013441, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pagliaro P, Moro F, Tullio F, Perrelli MG, and Penna C. Cardioprotective pathways during reperfusion: focus on redox signaling and other modalities of cell signaling. Antioxid Redox Signal 14: 833–850, 2011 [DOI] [PubMed] [Google Scholar]
- 54.Parker J, Zhu N, Zhu M, and Chen S. Profiling thiol redox proteome using isotope tagging mass spectrometry. J Vis Exp 61: e3766, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paulsen CE. and Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem Biol 5: 47–62, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pichler P, Kocher T, Holzmann J, Mazanek M, Taus T, Ammerer G, and Mechtler K. Peptide labeling with isobaric tags yields higher identification rates using iTRAQ 4-plex compared to TMT 6-plex and iTRAQ 8-plex on LTQ Orbitrap. Anal Chem 82: 6549–6558, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Radi R, Bush KM, Cosgrove TP, and Freeman BA. Reaction of xanthine oxidase-derived oxidants with lipid and protein of human plasma. Arch Biochem Biophys 286: 117–125, 1991 [DOI] [PubMed] [Google Scholar]
- 58.Reddie KG. and Carroll KS. Expanding the functional diversity of proteins through cysteine oxidation. Curr Opin Chem Biol 12: 746–754, 2008 [DOI] [PubMed] [Google Scholar]
- 59.Rigbolt KT, Vanselow JT, and Blagoev B. GProX, a user-friendly platform for bioinformatics analysis and visualization of quantitative proteomics data. Mol Cell Proteomics 10: O110 007450, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saurin AT, Neubert H, Brennan JP, and Eaton P. Widespread sulfenic acid formation in tissues in response to hydrogen peroxide. Proc Natl Acad Sci U S A 101: 17982–17987, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Savojardo C, Fariselli P, Alhamdoosh M, Martelli PL, Pierleoni A, and Casadio R. Improving the prediction of disulfide bonds in eukaryotes with machine learning methods and protein subcellular localization. Bioinformatics 27: 2224–2230, 2011 [DOI] [PubMed] [Google Scholar]
- 62.Schwartz D. and Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol 23: 1391–1398, 2005 [DOI] [PubMed] [Google Scholar]
- 63.Sen N, Hara MR, Kornberg MD, Cascio MB, Bae BI, Shahani N, Thomas B, Dawson TM, Dawson VL, Snyder SH, and Sawa A. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat Cell Biol 10: 866–873, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seo YH. and Carroll KS. Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proc Natl Acad Sci U S A 106: 16163–16168, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seo YH. and Carroll KS. Quantification of protein sulfenic acid modifications using isotope-coded dimedone and iododimedone. Angew Chem Int Ed Engl 50: 1342–1345, 2011 [DOI] [PubMed] [Google Scholar]
- 66.Seth D. and Stamler JS. The SNO-proteome: causation and classifications. Curr Opin Chem Biol 15: 129–136, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sethuraman M, McComb ME, Heibeck T, Costello CE, and Cohen RA. Isotope-coded affinity tag approach to identify and quantify oxidant-sensitive protein thiols. Mol Cell Proteomics 3: 273–278, 2004 [DOI] [PubMed] [Google Scholar]
- 68.Spivak M, Weston J, Bottou L, Kall L, and Noble WS. Improvements to the percolator algorithm for peptide identification from shotgun proteomics data sets. J Proteome Res 8: 3737–3745, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun J, Morgan M, Shen RF, Steenbergen C, and Murphy E. Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ Res 101: 1155–1163, 2007 [DOI] [PubMed] [Google Scholar]
- 70.Swaney DL, McAlister GC, and Coon JJ. Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat Methods 5: 959–964, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tao L. and English AM. Protein S-glutathiolation triggered by decomposed S-nitrosoglutathione. Biochemistry 43: 4028–4038, 2004 [DOI] [PubMed] [Google Scholar]
- 72.Thingholm TE, Palmisano G, Kjeldsen F, and Larsen MR. Undesirable charge-enhancement of isobaric tagged phosphopeptides leads to reduced identification efficiency. J Proteome Res 9: 4045–4052, 2010 [DOI] [PubMed] [Google Scholar]
- 73.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, and Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem 75: 1895–1904, 2003 [DOI] [PubMed] [Google Scholar]
- 74.Whitney PL, Powell JT, and Sanford GL. Oxidation and chemical modification of lung beta-galactoside-specific lectin. Biochem J 238: 683–689, 1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.