Abstract
Recent advances reveal mRNA 3′end processing as a highly regulated process that fine-tunes posttranscriptional gene expression. This process can affect the site and/or the efficiency of 3′end processing, controlling the quality and the quantity of substrate mRNAs. The regulation of 3′end processing plays a central role in fundamental physiology such as blood coagulation and innate immunity. In addition, errors in mRNA 3′end processing have been associated with a broad spectrum of human diseases, including cancer. We summarize and discuss the paradigmatic shift in the understanding of 3′end processing as a mechanism of posttranscriptional gene regulation that has reached clinical medicine.
Keywords: clinical importance of 3′ end mRNA processing, control of polyadenylation, post-transcriptional gene regulation, regulation of 3′ end mRNA processing
Introduction
Until recently, mRNA 3′end processing was viewed as a constitutive step of RNA biogenesis. Current advances have shown that the mechanism is regulated via a network of cis-acting RNA sequence elements and trans-acting proteins, contributing to the qualitative and quantitative adjustment of gene expression (Millevoi & Vagner, 2010). Reports about the impact of regulated mRNA polyadenylation and cleavage on physiological as well as pathological pathways increase steadily. These days, the underlying molecular mechanisms are the subject of intensive research. In this review, we provide the background on the recent progress in the field, leading to a better understanding of the complex process of mRNA 3′end processing; more detailed reviews on the mechanism of constitutive 3′end processing and alternative polyadenylation site (PAS) choice have recently been published (Elkon et al, 2013; Tian & Manley, 2013). We will especially highlight quantitative regulation of 3′end processing and its role in human pathophysiology.
Molecular mechanisms of mRNA 3′end processing
Constitutive 3′end processing
As the first step in the maturation of eucaryotic mRNAs, transcripts are synthesized by RNA polymerase II (PolII), capped, spliced and polyadenylated. Cleavage and polyadenylation of cellular pre-mRNAs at the 3′end is accomplished by a fine-tuned mechanism involving a number of RNA-binding proteins (RBPs) and regulatory cis-acting RNA sequence elements (Fig 1). In addition to the core machinery required for mRNA 3′end processing, auxiliary regulatory protein factors, miRNAs and supporting RNA sequence elements contained in the 3′-untranslated region (3′UTR) ensure efficient processing of a target mRNA (Matoulkova et al, 2012; Millevoi & Vagner, 2010). The processing steps of tRNA and replication-dependent histone mRNAs differ from the mechanism described here and are extensively covered elsewhere (Maraia & Lamichhane, 2011; Yang et al, 2013).
Figure 1.
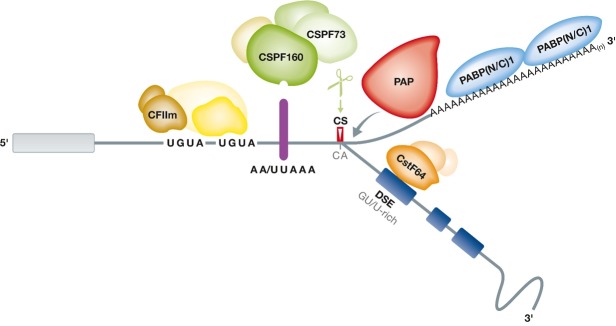
Endonucleolytic cleavage of pre-mRNAs and their subsequent polyadenylation requires the four multisubunit complexes CPSF, CstF, cleavage factor I (CFIm) and cleavage factor II (CFIIm) as well as the single-subunit polyadenylation-polymerase PAP. At the PAS CPSF and CstF bind to the central hexameric sequence (AA/UUAAA) and to the GU/U-rich DSE, respectively, as the first step in mRNA 3′end processing. The other protein complexes assemble at specific RNA sequence elements both up- and down-stream of the PAS, including several UGUA-repeats, thereby ensuring efficient cleavage and polyadenylation of an mRNA. The pre-mRNA is cleaved at the CS by the endonuclease CSPF73 before the nuclear poly(A) polymerase adds ∼200 As to the 3′end. This poly(A) tail stabilizes the processed mRNA for nuclear export upon binding of the nuclear poly(A) binding protein (PABPN1) which is subsequently exchanged for its cytoplasmic counterpart PABPC which promotes translation and RNA stability.
cis-acting RNA elements required for 3′end processing
In an mRNA's 3′UTR, the arrangement of cis-acting sequence elements determine the efficiency of a poly(A) (polyadenylation) site.
The most prominent among these cis-acting elements is a hexameric sequence motif that was first described by Proudfoot and Brownlee (1976). In ∼70% of human RNAs, the hexamer consists of the nucleotide sequence AAUAAA or AUUAAA. The remaining ∼30% of RNAs contain other sequences, such as UAUAAA, AACAAA, or ACUAAA (MacDonald & Redondo, 2002). The “strength” of a specific PAS is determined by surrounding auxiliary RNA elements occurring both down- and upstream of the hexamer, which serve as binding platforms for core and auxiliary 3′end processing factors (Millevoi & Vagner, 2010). The PAS not only affects mRNA 3′end processing but also plays an important role in other steps of the gene expression pathway, such as coupling 3′end processing to transcription termination (Proudfoot et al, 2002).
A less well-conserved auxiliary cis-acting element is the U/GU-rich downstream sequence element (DSE), which is located 30 to 45 nt 3′ of the hexameric sequence motif (Gil & Proudfoot, 1987). As a first step in mRNA 3′end processing, the cleavage and polyadenylation specificity factor (CPSF) and the cleavage stimulation factor (CstF) bind to the hexamer and the DSE, respectively, and stimulate polyadenylation (Keller et al, 1991; MacDonald et al, 1994; Proudfoot, 2011). The relative positions of the hexamer and the DSE define the site of cleavage of a pre-mRNA, which typically occurs within a 13-nucleotide window between these two elements (Chen et al, 1995). Most pre-mRNAs are cleaved 3′ of an adenosine residue and CA is defined as the optimal cleavage site (CS), although the nucleotide sequence of the exact CS is not highly conserved (Sheets et al, 1990).
Additional auxiliary cis-acting elements located both downstream and upstream of the central hexamer have been reported to be involved in mRNA 3′end processing, including U-, GU- and AU-rich sequences (Tian & Graber, 2012).
Protein factors involved in constitutive 3′end processing
Efficient mRNA 3′end formation is ensured by a network of interacting proteins. Although the three-dimensional structure of this machinery is still unknown, its ∼20 core proteins have been identified by biochemical analyses. This protein complex not only ensures efficient mRNA cleavage and polyadenylation, but also links 3′end formation to other steps of mRNA biosynthesis and processing (Shi et al, 2009b). Endonucleolytic cleavage of pre-mRNAs and their subsequent polyadenylation requires the four multisubunit complexes CPSF, CstF, CFIm and CFIIm, referred to as the core 3′end processing factors, and the single-subunit nuclear poly(A)-polymerase PAP plus the nuclear poly(A)-binding protein (PABPN) (Millevoi & Vagner, 2010). Additional factors stimulating the polyadenylation of a nascent mRNA comprise RBBP6 and the serine/threonine phosphatase PP1α/β (Shi et al, 2009a).
Mechanisms of regulated 3′end processing
Regulated 3′end processing has attracted attention because gene expression can be adjusted in a quantitative manner by tuning the amount of polyadenylated mRNA and protein produced (Millevoi et al, 2009; Wahle & Ruegsegger, 1999). In addition, ∼70% of human pre-mRNAs and ∼66% of long noncoding RNAs contain more than one functional polyadenylation signal and are candidates for alternative polyadenylation (APA) (Tian & Manley, 2013) (Fig 2). Depending on the location of the alternative PAS relative to the exon/intron structure of the pre-mRNA, APA either triggers the production of mRNA isoforms differing in the length of their 3′UTRs or varying in their C-terminal coding region. The former frequently entails a regulation of gene expression via the exclusion or inclusion of regulatory elements such as miRNA- and RBPs-binding sites, whereas the latter contributes to the diversity of the proteome. Current research focuses on the mechanisms controlling polyadenylation efficiency and alternative PAS usage.
Figure 2.
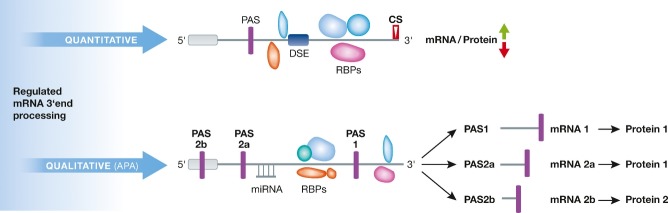
In a quantitative manner, the regulation of 3′end processing stimulates or inhibits the gene expression via the formation of specific mRNPs. Qualitatively, mRNAs containing more than one polyadenylation signal (PAS) can be subjected to APA. In case both PAS are present in the 3′UTR (PAS1 and PAS 2a), APA results in the expression of mRNA isoforms that encode the same protein (protein 1) but differ in the length of their 3′UTR (mRNA 1, mRNA 2a), including or excluding regulatory elements such as miRNA or RBP binding sites. If one poly(A) signal is contained within the coding region (PAS 2b), APA produces mRNA isoforms with distinct C-terminal coding regions (mRNA 2a and mRNA 2b), which leads to the expression of different protein isoforms (protein 1 and protein 2).
Proteins regulating 3′end processing
The regulation of mRNA 3′end processing is mainly achieved by RBPs and other interacting proteins assembling at cis-acting regulatory RNA elements within a transcript (Tian & Manley, 2013). Some of these proteins, such as TFIID (transcription factor II D) (Dantonel et al, 1997), have previously been shown to also be involved in other steps of RNA biogenesis, like transcription, splicing or constitutive 3′end processing. Others, including the tumour suppressor p53 (Nazeer et al, 2011) and the heat shock factor protein HSF1 (Xing et al, 2004), have previously been documented to contribute to completely unrelated annotated cellular functions, such as protein stability, stress response or RNA degradation (supplementary Table S1).
It has been shown that the cellular levels of core 3′end processing factors play a role in regulated 3′end processing. Single knockdown experiments of both the subunits of human cleavage factor CFIm (CFIm 25 and CFIm 68) resulted in a transcriptome-wide shift towards the use of proximal PASs, revealing a role of CFIm in APA (Gruber et al, 2012; Kubo et al, 2006; Martin et al, 2012; Sandberg et al, 2008). Further, isoforms of CstF-64 are associated with APA in different cell types, such as T-effector cells and male germ cells (Chennathukuzhi et al, 2001; Chuvpilo et al, 1999; Dass et al, 2007; Hockert et al, 2011; Monarez et al, 2007; Shell et al, 2005; Yao et al, 2012). Specifically, CstF-64 levels influence the class switch of immunoglobulin heavy chain synthesis (Takagaki et al, 1996). Finally, PAS usage has been recently shown to be affected by nuclear poly(A)-binding protein 1 (PAPBN1) levels. Biochemical studies demonstrated that PAPBN1 binds to poly(A) sites and that the depletion of PAPBN1 increases the usage of more upstream poly(A) sites (de Klerk et al, 2012; Jenal et al, 2012). Recent work suggests that PAPBN1 may influence PAS selection by directly binding to and thus inhibiting promoter-proximal PAS (Jenal et al, 2012). However, other findings propose that PAPBN1 preferentially binds to canonical PAS and stimulates 3′end processing (de Klerk et al, 2012). The exact underlying molecular mechanism(s) by which PAPBN1 affects poly(A) site usage remains to be elucidated.
3′End processing of pre-mRNAs is strongly interconnected with other steps of RNA biogenesis. Splicing factors, e.g. hnRNPs, and transcription-associated proteins, including the transcription factor E2F, have been found to regulate mRNA 3′end formation (supplementary Table S1). This is exemplified by hnRNPI/PTB1, which is a well-known global splicing repressor when bound to polypyrimidine tracts. hnRNPI/PTB1 can act as a regulator of 3′end processing efficiency upon binding to cis-acting RNA sequence elements playing a role in polyadenylation, such as the upstream sequence element (USE) and the DSE (Blechingberg et al, 2007b; Castelo-Branco et al, 2004; Danckwardt et al, 2011; Hall-Pogar et al, 2007; Millevoi et al, 2009). Likewise, the basal splicing factor U2AF65 can influence the functionality of the 3′end processing machinery by assembling at different polyadenylation-associated RNA elements, including the USE. U2AF65 can directly interact with core 3′end processing factors, including PAP, and can regulate the efficiency of 3′end processing by competing with other RBPs that assemble at such elements (Danckwardt et al, 2011; Hall-Pogar et al, 2007; Ko & Gunderson, 2002; Millevoi et al, 2006). Furthermore, the transcription factors TFIID and E2F can directly or indirectly affect mRNA polyadenylation either by binding to core 3′end processing factors or by stimulating their expression (Dantonel et al, 1997; Elkon et al, 2012; Martincic et al, 2009; Nagaike et al, 2011).
Regulated 3′end processing and its impact on pathophysiology
During the last decade, biomedical research has revealed how regulated 3′end processing influences important physiological pathways and how misregulated mRNA 3′end processing can cause human disease (Danckwardt et al, 2008; Elkon et al, 2013). Below, we describe the pathophysiological impact of misregulated mRNA 3′end processing in detail.
Physiological consequences of regulated 3′end processing
Quantitative regulation of gene and protein expression
By controlling the efficiency of the 3′end processing reaction, mRNA and protein levels can be post-transcriptionally adjusted according to cellular needs in response to cellular signalling or environmental conditions. As described above, this is achieved via auxiliary cis-acting RNA elements and regulatory trans-acting proteins, which can either stimulate or inhibit the utilization of PASs in response to exogenous stimuli (Fig 3). As a consequence, the amount of mature mRNA and protein is altered appropriately, thus modulating physiological processes. First described for viral RNA, the USE has subsequently been identified to stimulate the efficiency of 3′end processing in mammalian mRNAs (Gilmartin et al, 1995; Graveley et al, 1996). The prothrombin RNA represents the prototype for the function of the USE (Danckwardt et al, 2007). Prothrombin is the zymogen of the serine protease thrombin and is one of the key proteins of blood coagulation, complement activation, angiogenesis and tumour invasion. The expression of prothrombin is tightly regulated in a USE-dependent fashion in response to inflammation and stress (Danckwardt et al, 2011), as already a small inappropriate increase by less than twofold gives rise to thrombophilia, a predisposition to develop thrombosis (Gehring et al, 2001). Normally, the USE is occupied by the inhibitory proteins FBP2 and FBP3. Following inflammation or other stress stimuli, p38MAPK is activated leading to the phosphorylation of these two proteins and their concomitant release from the USE. The USE then becomes available for the binding of the 3′end processing activator proteins U2AF35, U2AF65 and hnRNPI/PTB1. This results in elevated prothrombin mRNA and protein synthesis and the stimulation of prothrombin downstream pathways (Danckwardt et al, 2011). A similar USE-regulated mechanism of gene expression has been found in several other mRNAs, namely those encoding the complement factor C2, the collagen genes COL1A1, COL1A2 and COL2A1, lamin B2 and the cyclooxygenase COX-2 (Brackenridge & Proudfoot, 2000; Hall-Pogar et al, 2007; Moreira et al, 1998; Natalizio et al, 2002). The USE system thus exemplifies how regulated mRNA 3′end processing can post-transcriptionally contribute to the fine-tuning of physiological processes in response to external stimuli.
Figure 3.
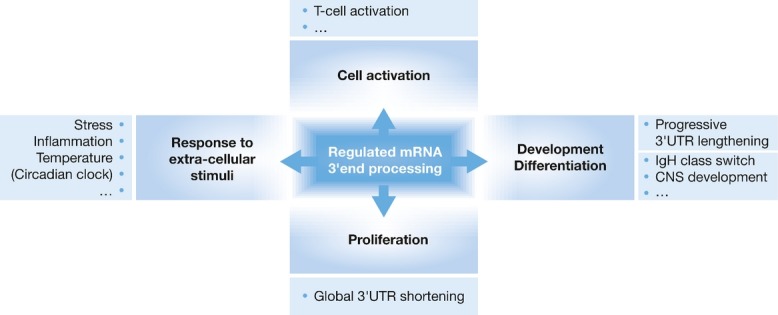
The control of 3′end processing post-transcriptionally adjusts mRNA and protein levels in response to environmental conditions, such as stress or inflammation. Further, it plays a role in development and differentiation, as in the case of the immunoglobulin heavy chain class switch and in the development of the central nervous system (CNS). In addition, alternative 3′end processing functions as a regulatory mechanism upon cell activation, e.g. T-cell activation, and proliferation. While a progressive lengthening of 3′UTRs via APA is typically observed during development and differentiation, higher proliferation states are associated with global 3′UTR shortening.
Although not yet understood in its mechanistic details, the expression Hsp70.3 is another example of the importance of regulated 3′end processing. Thermal stress leads to alternatively polyadenylated transcripts with shortened 3′UTRs, potentially lacking miRNA binding, which are more stable and associated with an increased biosynthesis of the Heat shock protein 70 (Hsp70) (Tranter et al, 2011). Moreover, the polyadenylation of a set of different mRNAs is catalysed by an inducible non-canonical poly(A) polymerase, termed STAR-PAP (nuclear speckle targeted PIPKIα regulated-poly(A) polymerase). STAR-PAP activity can be stimulated by stress-induced signalling pathways. Interestingly, this non-canonical poly(A) polymerase controls the expression of mRNAs encoding proteins associated with cellular stress response and disease, including cancer (Gonzales et al, 2008; Laishram et al, 2011; Li et al, 2013; Mellman et al, 2008).
Apart from the quantitative regulation of the expression of specific (sets of) genes via stimulated 3′end processing, the transcriptome-wide modulation of polyadenylation efficiency has been reported in response to various stress conditions, including thermal stress and DNA damage (Nazeer et al, 2011; Xing et al, 2004). Interestingly, thermal stress or DNA damage have been demonstrated to globally inhibit polyadenylation efficiency, whereas specific stress response genes are not affected by this repression (Di Giammartino et al, 2013; Nazeer et al, 2011). Taken together, regulated 3′end processing contributes to the maintenance of important physiological pathways and specifically appears to play a role in protective responses of cells to exogenous stimuli, such as stress.
Qualitative regulation of gene and protein expression via alternative polyadenylation
Physiological processes can be further influenced by APA that triggers the expression of different transcript or protein isoforms (Fig 3).
Development
The importance of APA in cellular differentiation was first demonstrated when Brown and Morrison reported that the immunoglobulins heavy chain (IgH) class shift is triggered by APA (Brown & Morrison, 1989). The production of the secreted or the membrane bound form of immunoglobulins, respectively, is achieved via a splicing-dependent poly(A) site switch in IgH genes which is based on a competition between splicing and 3′end processing (Peterson, 2007). While developmentally “younger” B cells express immunoglobulins in a membrane-bound form on their cell surface, the more mature plasma cells produce immunoglobulins as secreted proteins, which is mediated by the usage of a PAS located further 5′ (Peterson, 2011).
The role of regulated 3′end processing during development and differentiation was extended by the analysis of the time-dependent protein expression program of GFAP (glial fibrillary acidic protein) in the central nervous system (CNS). GFAP forms the intermediate cytoskeleton in mature astrocytes. APA results in the dynamic expression of two GFAP protein isoforms (GFAPɛ and GFAPκ) (Blechingberg et al, 2007b). The GFAPκ/ɛ-ratio has been found to decrease during cortex development suggesting that APA of this pre-mRNA plays a pivotal role in CNS development (Blechingberg et al, 2007a). Biochemical analyses suggest a role for splicing factors, such as hnRNPI/PTB1 and SR-proteins, in triggering the APA-dependent GFAP isoform switch but the exact underlying molecular mechanism remains to be elucidated (Blechingberg et al, 2007b).
Development-associated APA can also trigger the expression of mRNAs that differ in 3′UTR-length, leading to the exclusion or inclusion of miRNA binding sites and a change in the regulatory state of a specific transcript. This is exemplified by PAX3, a key regulator of myogenesis during development. PAX3 promotes proliferation, inhibits differentiation and is transiently expressed during the activation of adult muscle stem cells. In quiescent stem cell populations, its expression is usually suppressed via miR-206 that has been found to be highly abundant in these cells. In specific muscles, such as in the diaphragm muscle, PAX3 is able to escape the miRNA-driven regulation in quiescent muscle stem cells. This is due to 3′UTR shortening via APA at a more 5′PAS (Boutet et al, 2012). These observations predict a role of regulated 3′end processing in controlling stem cell function and implicate APA as a powerful mechanism to modulate miRNA-dependent regulation of gene expression during development.
More generally, APA functions as a regulatory mechanism on a transcriptome-wide scale in response to environmental demands by changing the regulatory state of mRNAs. Highly parallel RNA sequencing methods have recently helped to gain insights into the global modulation of mRNA 3′end processing. In the course of development and differentiation, regulated 3′end processing can function in a tissue-specific fashion, such as triggering APA during spermatogenesis or in the brain (Dass et al, 2007; Hockert et al, 2011; Li et al, 2012; Liu et al, 2007; Miura et al, 2013). The utilization of promoter-distal PASs generally increases with the developmental and differentiation state, and mRNAs with longer 3′UTRs generally contain a higher number of miRNA and protein binding sites (Hoque et al, 2013; Shepard et al, 2011). During later stages of development, mRNAs thus tend to be subject to more complex mechanisms of post-transcriptional control.
Proliferation and cell activation
In contrast to the progressive lengthening of 3′UTRs observed during development and differentiation, the usage of promoter-proximal PASs and therefore diminished regulation via miRNAs or RBPs is linked to proliferation (Elkon et al, 2012; Sandberg et al, 2008). Not only do proliferating cells exhibit significantly shortened 3′UTRs when compared to cells in a resting state, but are also subject to enhanced cleavage at intronic PAS. Members of the E2F family which influence cell cycle progression have been found to stimulate the expression of 3′end processing factors and proximal poly(A) site usage in proliferating cells (Elkon et al, 2012).
Further, APA has been shown to play a role in cell activation. This is illustrated by the inducible expression of NF-ATc (nuclear factor of activated T cells) upon antigen exposure. The stimulation of the T-cell antigen receptor triggers a switch from long protein isoforms in naïve T cells to a short isoform in mature T cells. Whereas in naïve T cells, the “weak” proximal PAS is not efficiently used, mature T cells show proximal poly(A) site usage and the short NF-ATc isoform is produced. The short NF-ATc isoform stimulates the expression of T-cell-specific genes, including lymphokine and cytokine genes (Chuvpilo et al, 1999; Serfling et al, 2000). Moreover, when comparing transcript expression in different immune cells (T lymphocytes, B cells, monocytes) before and after stimulation, a decrease of the expression of longer isoforms, which are processed at more distal PASs, has been reported to be associated with higher proliferation states (Sandberg et al, 2008). A transcriptome-wide shift towards promoter-proximal PAS usage has also been found in activated neurons (Flavell et al, 2008). Recent studies suggest a role of U1snRNP in APA. Low U1snRNP levels in activated neuronal cells have been shown to lead to widespread shortening of mRNAs due to a shift to proximal PASs, and that U1snRNP overexpression can inhibit this effect (Berg et al, 2012). Nonetheless, the molecular mechanism underlying U1snRNP-dependent APA is still unknown.
Cold shock and circadian gene expression
mRNA polyadenylation can be qualitatively regulated in response to cold shock and to the circadian clock. Two cold-induced RBPs, CIRBP and RBM3, have been found to repress promoter-proximal poly(A) site usage in a set of genes under cold shock conditions, leading to elongated 3′UTRs (Liu et al, 2013). Interestingly, further investigations revealed that many of the affected genes show 3′UTR-lengthening or -shortening via APA with strong circadian oscillations depending on the ambient temperature. These findings suggest that APA contributes to the regulation of the temperature-dependent circadian clock.
Human pathology associated with altered 3′end processing
Errors in mRNA processing leading to human disease can be caused by mutations in RNA sequence elements with a critical role in mRNA 3′end processing, defects in the biological function of trans-acting protein factors, or differential usage of PASs within otherwise normal transcripts (Fig 4).
Figure 4.
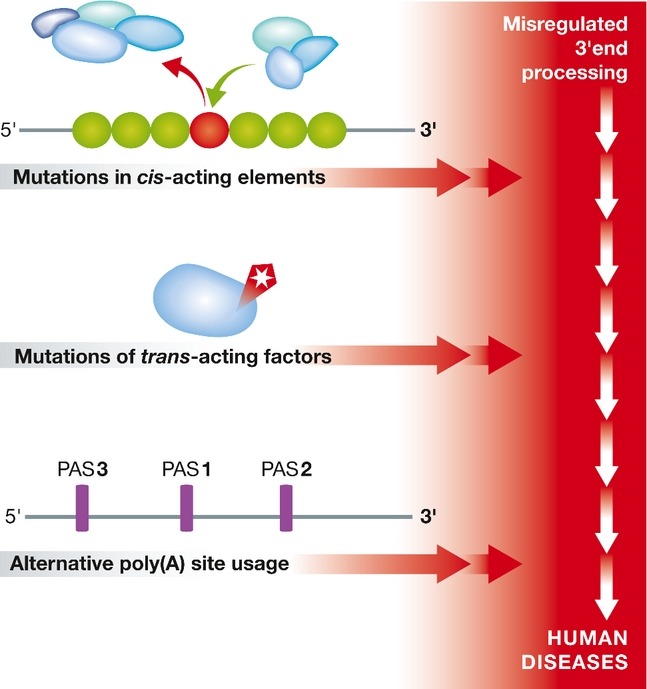
Misregulation of mRNA 3′end processing can be caused by constitutive errors incis-acting RNA elements, mutations of trans-acting proteins or changes in poly(A) site usage of otherwise normal transcripts. Mutations in RNA sequence elements or trans-acting proteins both potentially interfere with the interaction of RBPs and the RNA, leading to gain or loss of function. APA triggers the production of mRNAs varying in the length of their 3′UTR or in their C-terminal coding region, which leads to the expression of different protein isoforms. Certain diseases, such as cancer, are characterized by a polyadenylation pattern significantly different from the one in a healthy state.
Mutations in cis-acting elements
Haematologic diseases
The importance of 3′end processing for normal gene function and human health was first illustrated by an inactivating mutation in the hexamer of the poly(A) signal (AATAAA → AATAAG) of the α 2-globin gene causing α-thalassaemia (Higgs et al, 1983). Subsequently, other mutations affecting the α-globin and the β-globin polyadenylation signal were found in patients with α- and β-thalassaemia (Harteveld & Higgs, 2010). Interestingly, human disease cannot only result from the inactivation of efficient 3′end processing but also from pathological gain-of-function-mutations. This has first been demonstrated by a commonly occurring mutation of the CS of the prothrombin pre-mRNA, with a prevalence of heterozygotes of 1.7–3% in the Caucasian population (Gehring et al, 2001; Rosendaal et al, 1998). Physiologically, cleavage occurs at a suboptimal CG dinucleotide. A CG → CA mutation (G20210A) at this position transforms the physiological, inefficient site into a more active site thus increasing prothrombin synthesis. Because prothrombin is one of the key pro-coagulatory proteins of the network controlling blood coagulation, this mutation results in thrombophilia, i.e. a predisposition to develop thrombosis.
Cancer
Back in the 1860s, Armand Trousseau was the first to connect a prothrombotic state to cancer (Trousseau, 1865). This early finding foreshadowed an association of regulated 3′end processing with more complex diseases by some 150 years. Recently, mutations in cis-acting RNA elements of ubiquitously expressed genes leading to defective 3′end processing have indeed been linked to cancer. The tumour suppressor gene p53, a crucial regulator of the cell cycle and apoptosis, is mutated in approximately 60% of human cancers (Muller & Vousden, 2013). A germline mutation in the p53 polyadenylation signal (AAUAAA → AAUACA), which occurs in approximately 0.5–2% of different European populations, has been reported to predispose to basal cell carcinoma, prostate cancer, glioma and colorectal cancer (Enciso-Mora et al, 2013; Stacey et al, 2011).
Other disorders resulting from PAS mutations
Mutations of mRNA 3′end processing elements are further known to cause complex syndromes. An example is a polyadenylation signal mutation (AAUAAA → AAUGAA) in the FOXP3 (forkhead box P3) gene causing the IPEX syndrome, which is characterized by immune dysfunction, polyendocrinopathy and enteropathy (Bennett & Ochs, 2001). The mutation leads to a very long 3′UTR of the FOXP3 mRNA, which becomes relatively unstable, thus resulting in decreased FOXP3 protein levels. FOXP3 functions as a transcription factor in regulatory T cells, explaining the immune dysfunction of affected patients.
In addition, panic disorders and altered fear extinction memory have been associated with a common mutation in the distal polyadenylation signal of the serotonin (5-hydroxytryptamine) transporter gene (SERT or 5-HTT) (Gyawali et al, 2010; Hartley et al, 2012). This mutation leads to the usage of the more proximal PAS and to the expression of a transcript with a shortened 3′UTR and a potentially higher stability. These findings associate the PAS polymorphism occurring in the serotonin transporter gene with mood and anxiety disorders in humans.
Mutations of trans-acting factors
Defects in trans-acting proteins with a role in 3′end processing appear to be rare. An example of such an error is a triplet-repeat expansion of PABPN1 (trePABPN1) that causes autosomal-dominant oculopharyngeal muscle dystrophy (OPMD). trePABPN1 interacts with WT PABPN1 in a dominant negative fashion and interferes with its function as a suppressor of proximal poly(A) site usage. Mouse models of OPMD as well as human cells expressing trePABPN1 show a global shift towards proximal, usually “weaker” PAS (de Klerk et al, 2012; Jenal et al, 2012). How exactly this change in poly(A) site usage causes the phenotype of OPMD remains to be investigated.
Diseases caused by alternative polyadenylation
Differential poly(A) site usage in otherwise normal transcripts is associated with malignancies. A number of diseases have been linked to either APA of single transcripts or to global changes in polyadenylation. The symptoms of such diseases include growth retardation, cancer predisposition syndromes and neurodegeneration.
Cancer predisposition syndromes
Lynch syndrome is a hereditary predisposition to several types of cancer, including colorectal and endometrial cancer, which is caused by a loss of function of one of the four DNA mismatch repair genes MSH2, MLH1, MSH6, PMS2. Some patients with this syndrome carry germline mutations in the coding region of one of those genes. Recently, a 20 nucleotide duplication in close vicinity to the poly(A) signal of MSH6 mRNA has been detected in two patients, associated with decreased MSH6 mRNA levels (Decorsiere et al, 2012). Although the underlying molecular mechanism is still unknown, the duplication has been found to negatively affect polyadenylation efficiency (Decorsiere et al, 2012). These findings link decreased MSH6 mRNA expression caused by impaired mRNA 3′end processing to Lynch syndrome.
Neurodegenerative diseases
The development of the central nervous system is a complex process, involving tight regulation of gene expression. The fragile X syndrome is the most common form of inherited intellectual disability. This disease is caused by the methylation and the expansion (>200 repeats) of a CGG element within the promoter region of the fragile X mental retardation 1 (FMR1) gene, with the premutation (55–200 CGG repeats) already showing a clinically relevant phenotype (McLennan et al, 2011). Several isoforms of the FMR1 mRNA are generated via alternative splicing, as well as via APA (Tassone et al, 2011; Verkerk et al, 1993). The analysis of brain specimen of CGG knock-in mice and of post-mortem brain tissues from a carrier of the premutation revealed differential usage of poly(A) sites within the FMR1 3′UTR when compared to control samples (Tassone et al, 2011). Together with observed variations in the 5′UTR, this poly(A) site choice likely contributes to the pathology of the fragile X syndrome.
Cancer
In recent years, alterations in cleavage and polyadenylation of single genes have been linked to the development and progression of numerous different forms of cancer. Specifically, tumour cell lines and primary human cancer material from patients with mantle cell lymphoma (MCL) exhibited different polyadenylation patterns than control cells or tissues. MCL is related to the APA of cyclin D1. In proliferating MCL tumours, truncated cyclin D1 mRNA isoforms originating from processing at a proximal PAS are over-represented (Mayr & Bartel, 2009; Wiestner et al, 2007). Although the underlying molecular mechanism triggering the APA of cyclin D1 mRNA is unknown, the shorter transcript variants have been found to be more stable than their full-length counterparts, which may be attributable to a loss of miRNA-driven degradation. The increased mRNA stability leads to elevated Cyclin D1 protein expression and correlates with increased overall survival of patients (Rosenwald et al, 2003). Strikingly, cancer has been also associated with APA-dependent shortening of mRNAs observed on a transcriptome-wide scale. High-throughput sequencing methods revealed a global shift towards promoter-proximal poly(A) site usage in tumour cell lines as well as in tumour samples derived from different organs, such as breast, colon, kidney, liver and lung, which might contribute to the stabilization and/or translational activation of mRNAs encoding oncogenes and other cancer-relevant proteins (Lin et al, 2012; Mayr & Bartel, 2009).
3′End processing on its way to the clinic
Based on the current knowledge about regulated 3′end processing in human pathophysiology, it seems reasonable to consider its potential role in diagnosis and as a therapeutic target in the future. High-throughput methods used to quantitatively and qualitatively profile RNA PASs on a global scale revealed tissue-specific polyadenylation patterns suggesting coordinated regulation of these processes (Wang et al, 2008). Furthermore, there are population genetic variations of mRNA 3′end processing in general and in poly(A) site usage in particular (Wang et al, 2008; Yoon et al, 2012; Zhang et al, 2005). Inter-individual differences in 3′end architecture and, therefore, in the regulation of 3′end processing, have been found to cause the observed changes in mRNA polyadenylation and cleavage between individuals, contributing to “personal” gene expression profiles. These observations suggest that such variations may modulate medically relevant phenotypes. As described above, samples derived from numerous forms of cancer exhibit polyadenylation profiles that significantly differ from samples derived from healthy tissue (Fu et al, 2011; Lin et al, 2012; Mayr & Bartel, 2009; Morris et al, 2012). Genes undergoing cancer-associated 3′UTR shortening or, in rarer cases, elongation, can be clustered according to functional groups and categories, foreshadowing their potential role as significant biomarkers, which may help to distinguish tumour subtypes with distinct molecular signatures, thus signifying biological heterogeneity which potentially impacts on treatment response and prognosis (Morris et al, 2012).
Apart from using 3′end processing profiles as diagnostic tools, the polyadenylation step might serve as a target of future medical therapies. Since mRNA 3′end processing is regulated via trans-acting proteins and cis-acting RNA sequence elements, successful therapies must aim at this interplay. Inhibitors as well as activators of the proteins involved in mRNA polyadenylation have been shown to have the potential to treat 3′end processing-associated diseases in model systems. This is exemplified by the anti-inflammatory effect of cordycepin (3′ deoxyadenosine, dATP), a compound that inhibits polyadenylation by arresting the cleavage complex (Kondrashov et al, 2012). Interestingly, dATP treatment was reported to inhibit the 3′end processing of inflammatory genes without affecting the polyadenylation of control transcripts, indicating drug specificity for gene-specific differences in the 3′end processing step. Importantly, the gene-specific differences of the polyadenylation process provide the perspective to develop therapies targeting the 3′end processing machinery of distinct genes and the associated pathologies. Specifically, the interactions of trans-acting proteins with their cognate RNA binding motifs such as those that have been reported between the USE and stimulatory and inhibitory proteins of 3′end processing (Danckwardt et al, 2011) could, in principle, be envisaged to be targeted by small molecules thus modulating 3′end processing efficiency of specific classes of genes.
In addition, directly modulating the function of RNA molecules in mammalian cells via the introduction of antisense oligonucleotides has gained much attention as a potential therapeutic strategy (Bennett & Swayze, 2010). Antisense oligonucleotides that inhibit PASs have been successfully used to modulate poly(A) site selection in cultured cells (Vickers et al, 2001). Potentially, targeting cis-acting elements playing a role in the cleavage and polyadenylation reaction might help to treat associated diseases in the future.
Conclusions and future perspectives
The regulation of mRNA 3′end processing has gained much attention in recent years as a mechanism that can fine-tune gene expression in a quantitative and qualitative manner. Its role in pathophysiology and the underlying molecular mechanisms are intensively investigated at present. Certainly, the number of physiological and pathological cellular pathways controlled via regulated mRNA polyadenylation and cleavage will progressively increase. Prospectively, regulated mRNA 3′end processing could instruct new diagnostic approaches and serve as a therapeutic target, aiding in treating diseases, including cancer.
Conflict of interest
The authors declare that they have no conflict of interest.
Pending issues.
Develop strategies to therapeutically target misregulated mRNA 3′end processing.
Unravel the molecular mechanisms underlying misregulated mRNA 3′end processing to increase the level of understanding of its impact on pathological and physiological pathways.
Explore the differences in mRNA 3′end processing as a diagnostic tool.
Glossary
- mRNA 3′end processing
Cleavage of pre-mRNA and its subsequent polyadenylation, ensured by a highly regulated network of proteins in the nucleus.
- 3′UTR
Untranslated region at the 3′end of an mRNA molecule that contains regulatory RNA sequence elements, including the polyadenylation signal, miRNA- and RBP-binding sites.
- Trans-acting
Acting from a different molecule (e.g. protein regulates RNA).
- Cis-acting
Acting from within the same molecule (e.g. RNA sequence element controls the RNA it is contained in).
- Germline mutation
Any variation in cells that differentiate into gametes and somatic cells.
- Thalassaemia
A group of diseases caused by mutations of the α- or β-globin genes leading to decreased production of globin chains and severe anemia.
- Oculopharyngeal muscle dystrophy (OPMD)
Autosomal dominant neuromuscular disease that is caused by a mutation in the PABPN1 gene. It is characterized by ptosis (droopy eyelids), muscle weakness and difficulty in swallowing (dysphagia).
- Antisense oligonucleotides
Oligonucleotides that can bind to RNA through Watson-Crick base pairing and can modulate the function of the target RNA
References
- Bennett CL, Ochs HD. IPEX is a unique X-linked syndrome characterized by immune dysfunction, polyendocrinopathy, enteropathy, and a variety of autoimmune phenomena. Curr Opin Pediatr. 2001;13:533–538. doi: 10.1097/00008480-200112000-00007. [DOI] [PubMed] [Google Scholar]
- Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang Z, Cho S, Sherrill-Mix S, Wan L, et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blechingberg J, Holm IE, Nielsen KB, Jensen TH, Jorgensen AL, Nielsen AL. Identification and characterization of GFAPkappa, a novel glial fibrillary acidic protein isoform. Glia. 2007a;55:497–507. doi: 10.1002/glia.20475. [DOI] [PubMed] [Google Scholar]
- Blechingberg J, Lykke-Andersen S, Jensen TH, Jorgensen AL, Nielsen AL. Regulatory mechanisms for 3′-end alternative splicing and polyadenylation of the Glial fibrillary acidic protein, GFAP, transcript. Nucleic Acids Res. 2007b;35:7636–7650. doi: 10.1093/nar/gkm931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet SC, Cheung TH, Quach NL, Liu L, Prescott SL, Edalati A, Iori K, Rando TA. Alternative polyadenylation mediates microRNA regulation of muscle stem cell function. Cell Stem Cell. 2012;10:327–336. doi: 10.1016/j.stem.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackenridge S, Proudfoot NJ. Recruitment of a basal polyadenylation factor by the upstream sequence element of the human lamin B2 polyadenylation signal. Mol Cell Biol. 2000;20:2660–2669. doi: 10.1128/mcb.20.8.2660-2669.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SL, Morrison SL. Developmental regulation of membrane and secretory Ig gamma 2b mRNA. J Immunol. 1989;142:2072–2080. [PubMed] [Google Scholar]
- Castelo-Branco P, Furger A, Wollerton M, Smith C, Moreira A, Proudfoot N. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol Cell Biol. 2004;24:4174–4183. doi: 10.1128/MCB.24.10.4174-4183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, MacDonald CC, Wilusz J. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 1995;23:2614–2620. doi: 10.1093/nar/23.14.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chennathukuzhi VM, Lefrancois S, Morales CR, Syed V, Hecht NB. Elevated levels of the polyadenylation factor CstF 64 enhance formation of the 1kB Testis brain RNA-binding protein (TB-RBP) mRNA in male germ cells. Mol Reprod Dev. 2001;58:460–469. doi: 10.1002/1098-2795(20010401)58:4<460::AID-MRD15>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Chuvpilo S, Zimmer M, Kerstan A, Glockner J, Avots A, Escher C, Fischer C, Inashkina I, Jankevics E, Berberich-Siebelt F, et al. Alternative polyadenylation events contribute to the induction of NF-ATc in effector T cells. Immunity. 1999;10:261–269. doi: 10.1016/s1074-7613(00)80026-6. [DOI] [PubMed] [Google Scholar]
- Danckwardt S, Kaufmann I, Gentzel M, Foerstner KU, Gantzert AS, Gehring NH, Neu-Yilik G, Bork P, Keller W, Wilm M, et al. Splicing factors stimulate polyadenylation via USEs at non-canonical 3′ end formation signals. EMBO J. 2007;26:2658–2669. doi: 10.1038/sj.emboj.7601699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Hentze MW, Kulozik AE. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2008;27:482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Gantzert AS, Macher-Goeppinger S, Probst HC, Gentzel M, Wilm M, Grone HJ, Schirmacher P, Hentze MW, Kulozik AE. p38 MAPK controls prothrombin expression by regulated RNA 3′end processing. Mol Cell. 2011;41:298–310. doi: 10.1016/j.molcel.2010.12.032. [DOI] [PubMed] [Google Scholar]
- Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- Dass B, Tardif S, Park JY, Tian B, Weitlauf HM, Hess RA, Carnes K, Griswold MD, Small CL, Macdonald CC. Loss of polyadenylation protein tauCstF-64 causes spermatogenic defects and male infertility. Proc Natl Acad Sci USA. 2007;104:20374–20379. doi: 10.1073/pnas.0707589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klerk de E, Venema A, Anvar SY, Goeman JJ, Hu O, Trollet C, Dickson G, Dunnen den JT, Maarel van der SM, Raz V, et al. Poly(A) binding protein nuclear 1 levels affect alternative polyadenylation. Nucleic Acids Res. 2012;40:9089–9101. doi: 10.1093/nar/gks655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decorsiere A, Toulas C, Fouque F, Tilkin-Mariame AF, Selves J, Guimbaud R, Chipoulet E, Delmas C, Rey JM, Pujol P, et al. Decreased efficiency of MSH6 mRNA polyadenylation linked to a 20-base-pair duplication in Lynch syndrome families. Cell Cycle. 2012;11:2578–2580. doi: 10.4161/cc.20625. [DOI] [PubMed] [Google Scholar]
- Giammartino Di DC, Shi Y, Manley JL. PARP1 represses PAP and inhibits polyadenylation during heat shock. Mol Cell. 2013;49:7–17. doi: 10.1016/j.molcel.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Drost J, Haaften van G, Jenal M, Schrier M, Vrielink JA, Agami R. E2F mediates enhanced alternative polyadenylation in proliferation. Genome Biol. 2012;13:R59. doi: 10.1186/gb-2012-13-7-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14:496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- Enciso-Mora V, Hosking FJ, Stefano Di AL, Zelenika D, Shete S, Broderick P, Idbaih A, Delattre JY, Hoang-Xuan K, Marie Y, et al. Low penetrance susceptibility to glioma is caused by the TP53 variant rs78378222. Br J Cancer. 2013;108:2178–2185. doi: 10.1038/bjc.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, Xu A. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 2011;21:741–747. doi: 10.1101/gr.115295.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Frede U, Neu-Yilik G, Hundsdoerfer P, Vetter B, Hentze MW, Kulozik AE. Increased efficiency of mRNA 3′ end formation: a new genetic mechanism contributing to hereditary thrombophilia. Nat Genet. 2001;28:389–392. doi: 10.1038/ng578. [DOI] [PubMed] [Google Scholar]
- Gil A, Proudfoot NJ. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA 3′ end formation. Cell. 1987;49:399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- Gilmartin GM, Fleming ES, Oetjen J, Graveley BR. CPSF recognition of an HIV-1 mRNA 3′-processing enhancer: multiple sequence contacts involved in poly(A) site definition. Genes Dev. 1995;9:72–83. doi: 10.1101/gad.9.1.72. [DOI] [PubMed] [Google Scholar]
- Gonzales ML, Mellman DL, Anderson RA. CKIalpha is associated with and phosphorylates star-PAP and is also required for expression of select star-PAP target messenger RNAs. J Biol Chem. 2008;283:12665–12673. doi: 10.1074/jbc.M800656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Fleming ES, Gilmartin GM. RNA structure is a critical determinant of poly(A) site recognition by cleavage and polyadenylation specificity factor. Mol Cell Biol. 1996;16:4942–4951. doi: 10.1128/mcb.16.9.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Martin G, Keller W, Zavolan M. Cleavage factor Im is a key regulator of 3′ UTR length. RNA Biol. 2012;9:1405–1412. doi: 10.4161/rna.22570. [DOI] [PubMed] [Google Scholar]
- Gyawali S, Subaran R, Weissman MM, Hershkowitz D, McKenna MC, Talati A, Fyer AJ, Wickramaratne P, Adams PB, Hodge SE, et al. Association of a polyadenylation polymorphism in the serotonin transporter and panic disorder. Biol Psychiatry. 2010;67:331–338. doi: 10.1016/j.biopsych.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Pogar T, Liang S, Hague LK, Lutz CS. Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3′-UTR. RNA. 2007;13:1103–1115. doi: 10.1261/rna.577707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harteveld CL, Higgs DR. Alpha-thalassaemia. Orphanet J Rare Dis. 2010;5:13. doi: 10.1186/1750-1172-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley CA, McKenna MC, Salman R, Holmes A, Casey BJ, Phelps EA, Glatt CE. Serotonin transporter polyadenylation polymorphism modulates the retention of fear extinction memory. Proc Natl Acad Sci USA. 2012;109:5493–5498. doi: 10.1073/pnas.1202044109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs DR, Goodbourn SE, Lamb J, Clegg JB, Weatherall DJ, Proudfoot NJ. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature. 1983;306:398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- Hockert KJ, Martincic K, Mendis-Handagama SM, Borghesi LA, Milcarek C, Dass B, MacDonald CC. Spermatogenetic but not immunological defects in mice lacking the tauCstF-64 polyadenylation protein. J Reprod Immunol. 2011;89:26–37. doi: 10.1016/j.jri.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods. 2013;10:133–139. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal M, Elkon R, Loayza-Puch F, Haaften van G, Kuhn U, Menzies FM, Oude Vrielink JA, Bos AJ, Drost J, Rooijers K, et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012;149:538–553. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- Keller W, Bienroth S, Lang KM, Christofori G. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J. 1991;10:4241–4249. doi: 10.1002/j.1460-2075.1991.tb05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko B, Gunderson SI. Identification of new poly(A) polymerase-inhibitory proteins capable of regulating pre-mRNA polyadenylation. J Mol Biol. 2002;318:1189–1206. doi: 10.1016/s0022-2836(02)00240-1. [DOI] [PubMed] [Google Scholar]
- Kondrashov A, Meijer HA, Barthet-Barateig A, Parker HN, Khurshid A, Tessier S, Sicard M, Knox AJ, Pang L, Moor De CH. Inhibition of polyadenylation reduces inflammatory gene induction. RNA. 2012;18:2236–2250. doi: 10.1261/rna.032391.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Wada T, Yamaguchi Y, Shimizu A, Handa H. Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3′-UTRs. Nucleic Acids Res. 2006;34:6264–6271. doi: 10.1093/nar/gkl794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laishram RS, Barlow CA, Anderson RA. CKI isoforms alpha and epsilon regulate Star-PAP target messages by controlling Star-PAP poly(A) polymerase activity and phosphoinositide stimulation. Nucleic Acids Res. 2011;39:7961–7973. doi: 10.1093/nar/gkr549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Yeh HJ, Shankarling GS, Ji Z, Tian B, MacDonald CC. The tauCstF-64 polyadenylation protein controls genome expression in testis. PloS ONE. 2012;7:e48373. doi: 10.1371/journal.pone.0048373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Laishram RS, Anderson RA. The novel poly(A) polymerase Star-PAP is a signal-regulated switch at the 3′-end of mRNAs. Adv Biol Regul. 2013;53:64–76. doi: 10.1016/j.jbior.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Li Z, Ozsolak F, Kim SW, Arango-Argoty G, Liu TT, Tenenbaum SA, Bailey T, Monaghan AP, Milos PM, et al. An in-depth map of polyadenylation sites in cancer. Nucleic Acids Res. 2012;40:8460–8471. doi: 10.1093/nar/gks637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Brockman JM, Dass B, Hutchins LN, Singh P, McCarrey JR, MacDonald CC, Graber JH. Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis. Nucleic Acids Res. 2007;35:234–246. doi: 10.1093/nar/gkl919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Hu W, Murakawa Y, Yin J, Wang G, Landthaler M, Yan J. Cold-induced RNA-binding proteins regulate circadian gene expression by controlling alternative polyadenylation. Sci Rep. 2013;3:2054. doi: 10.1038/srep02054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald CC, Redondo JL. Reexamining the polyadenylation signal: were we wrong about AAUAAA? Mol Cell Endocrinol. 2002;190:1–8. doi: 10.1016/s0303-7207(02)00044-8. [DOI] [PubMed] [Google Scholar]
- MacDonald CC, Wilusz J, Shenk T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol Cell Biol. 1994;14:6647–6654. doi: 10.1128/mcb.14.10.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia RJ, Lamichhane TN. 3′ processing of eukaryotic precursor tRNAs. Wiley Interdiscip Rev RNA. 2011;2:362–375. doi: 10.1002/wrna.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Gruber AR, Keller W, Zavolan M. Genome-wide analysis of pre-mRNA 3′end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Rep. 2012;1:753–763. doi: 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Martincic K, Alkan SA, Cheatle A, Borghesi L, Milcarek C. Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat Immunol. 2009;10:1102–1109. doi: 10.1038/ni.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoulkova E, Michalova E, Vojtesek B, Hrstka R. The role of the 3′ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 2012;9:563–576. doi: 10.4161/rna.20231. [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan Y, Polussa J, Tassone F, Hagerman R. Fragile X syndrome. Curr Genomics. 2011;12:216–224. doi: 10.2174/138920211795677886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, Anderson RA. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–1017. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S. An interaction between U2AF 65 and CF I(m) links the splicing and 3′end processing machineries. EMBO J. 2006;25:4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Decorsiere A, Loulergue C, Iacovoni J, Bernat S, Antoniou M, Vagner S. A physical and functional link between splicing factors promotes pre-mRNA 3′end processing. Nucleic Acids Res. 2009;37:4672–4683. doi: 10.1093/nar/gkp470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura P, Shenker S, Andreu-Agullo C, Westholm JO, Lai EC. Widespread and extensive lengthening of 3′ UTRs in the mammalian brain. Genome Res. 2013;23:812–825. doi: 10.1101/gr.146886.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monarez RR, MacDonald CC, Dass B. Polyadenylation proteins CstF-64 and tauCstF-64 exhibit differential binding affinities for RNA polymers. Biochem J. 2007;401:651–658. doi: 10.1042/BJ20061097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley JL, Proudfoot NJ. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AR, Bos A, Diosdado B, Rooijers K, Elkon R, Bolijn AS, Carvalho B, Meijer GA, Agami R. Alternative cleavage and polyadenylation during colorectal cancer development. Clin Cancer Res. 2012;18:5256–5266. doi: 10.1158/1078-0432.CCR-12-0543. [DOI] [PubMed] [Google Scholar]
- Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- Nagaike T, Logan C, Hotta I, Rozenblatt-Rosen O, Meyerson M, Manley JL. Transcriptional activators enhance polyadenylation of mRNA precursors. Mol Cell. 2011;41:409–418. doi: 10.1016/j.molcel.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natalizio BJ, Muniz LC, Arhin GK, Wilusz J, Lutz CS. Upstream elements present in the 3′-untranslated region of collagen genes influence the processing efficiency of overlapping polyadenylation signals. J Biol Chem. 2002;277:42733–42740. doi: 10.1074/jbc.M208070200. [DOI] [PubMed] [Google Scholar]
- Nazeer FI, Devany E, Mohammed S, Fonseca D, Akukwe B, Taveras C, Kleiman FE. p53 inhibits mRNA 3′ processing through its interaction with the CstF/BARD1 complex. Oncogene. 2011;30:3073–3083. doi: 10.1038/onc.2011.29. [DOI] [PubMed] [Google Scholar]
- Peterson ML. Mechanisms controlling production of membrane and secreted immunoglobulin during B cell development. Immunol Res. 2007;37:33–46. doi: 10.1007/BF02686094. [DOI] [PubMed] [Google Scholar]
- Peterson ML. Immunoglobulin heavy chain gene regulation through polyadenylation and splicing competition. Wiley Interdiscip Rev RNA. 2011;2:92–105. doi: 10.1002/wrna.36. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ, Brownlee GG. 3′ non-coding region sequences in eukaryotic messenger RNA. Nature. 1976;263:211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Rosendaal FR, Doggen CJ, Zivelin A, Arruda VR, Aiach M, Siscovick DS, Hillarp A, Watzke HH, Bernardi F, Cumming AM, et al. Geographic distribution of the 20210 G to A prothrombin variant. Thromb Haemost. 1998;79:706–708. [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, Gascoyne RD, Grogan TM, Muller-Hermelink HK, Smeland EB, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3:185–197. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serfling E, Berberich-Siebelt F, Chuvpilo S, Jankevics E, Klein-Hessling S, Twardzik T, Avots A. The role of NF-AT transcription factors in T cell activation and differentiation. Biochim Biophys Acta. 2000;1498:1–18. doi: 10.1016/s0167-4889(00)00082-3. [DOI] [PubMed] [Google Scholar]
- Sheets MD, Ogg SC, Wickens MP. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990;18:5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shell SA, Hesse C, Morris SM, Jr, Milcarek C. Elevated levels of the 64-kDa cleavage stimulatory factor (CstF-64) in lipopolysaccharide-stimulated macrophages influence gene expression and induce alternative poly(A) site selection. J Biol Chem. 2005;280:39950–39961. doi: 10.1074/jbc.M508848200. [DOI] [PubMed] [Google Scholar]
- Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Chan S, Martinez-Santibanez G. An up-close look at the pre-mRNA 3′-end processing complex. RNA Biol. 2009a;6:522–525. doi: 10.4161/rna.6.5.9554. [DOI] [PubMed] [Google Scholar]
- Shi Y, Giammartino Di DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, III, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009b;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey SN, Sulem P, Jonasdottir A, Masson G, Gudmundsson J, Gudbjartsson DF, Magnusson OT, Gudjonsson SA, Sigurgeirsson B, Thorisdottir K, et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet. 2011;43:1098–1103. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- Tassone F, Rubeis De S, Carosi C, Fata La G, Serpa G, Raske C, Willemsen R, Hagerman PJ, Bagni C. Differential usage of transcriptional start sites and polyadenylation sites in FMR1 premutation alleles. Nucleic Acids Res. 2011;39:6172–6185. doi: 10.1093/nar/gkr100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Graber JH. Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip Rev RNA. 2012;3:385–396. doi: 10.1002/wrna.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Manley JL. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci. 2013;38:312–320. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranter M, Helsley RN, Paulding WR, McGuinness M, Brokamp C, Haar L, Liu Y, Ren X, Jones WK. Coordinated post-transcriptional regulation of Hsp70.3 gene expression by microRNA and alternative polyadenylation. J Biol Chem. 2011;286:29828–29837. doi: 10.1074/jbc.M111.221796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trousseau A. Plegmasia alba dolens. Lectures on Clinical Medicine, delivered at the Hotel-Dieu. 1865;5:281–332. [Google Scholar]
- Verkerk AJ, Graaff de E, Boulle De K, Eichler EE, Konecki DS, Reyniers E, Manca A, Poustka A, Willems PJ, Nelson DL, et al. Alternative splicing in the fragile X gene FMR1. Hum Mol Genet. 1993;2:399–404. doi: 10.1093/hmg/2.4.399. [DOI] [PubMed] [Google Scholar]
- Vickers TA, Wyatt JR, Burckin T, Bennett CF, Freier SM. Fully modified 2′ MOE oligonucleotides redirect polyadenylation. Nucleic Acids Res. 2001;29:1293–1299. doi: 10.1093/nar/29.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E, Ruegsegger U. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiestner A, Tehrani M, Chiorazzi M, Wright G, Gibellini F, Nakayama K, Liu H, Rosenwald A, Muller-Hermelink HK, Ott G, et al. Point mutations and genomic deletions in CCND1 create stable truncated cyclin D1 mRNAs that are associated with increased proliferation rate and shorter survival. Blood. 2007;109:4599–4606. doi: 10.1182/blood-2006-08-039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Mayhew CN, Cullen KE, Park-Sarge OK, Sarge KD. HSF1 modulation of Hsp70 mRNA polyadenylation via interaction with symplekin. J Biol Chem. 2004;279:10551–10555. doi: 10.1074/jbc.M311719200. [DOI] [PubMed] [Google Scholar]
- Yang XC, Sabath I, Debski J, Kaus-Drobek M, Dadlez M, Marzluff WF, Dominski Z. A complex containing the CPSF73 endonuclease and other polyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3′-end processing. Mol Cell Biol. 2013;33:28–37. doi: 10.1128/MCB.00653-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C, Biesinger J, Wan J, Weng L, Xing Y, Xie X, Shi Y. Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation. Proc Natl Acad Sci USA. 2012;109:18773–18778. doi: 10.1073/pnas.1211101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon OK, Hsu TY, Im JH, Brem RB. Genetics and regulatory impact of alternative polyadenylation in human B-lymphoblastoid cells. PLoS Genet. 2012;8:e1002882. doi: 10.1371/journal.pgen.1002882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lee JY, Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


