Abstract
Pulp engineering with dental mesenchymal stem cells is a promising therapy for injured teeth. An important point is to determine the fate of implanted cells in the pulp over time and particularly during the early phase following implantation. Indeed, the potential engraftment of the implanted cells in other organs has to be assessed, in particular, to evaluate the risk of inducing ectopic mineralization. In this study, our aim was to follow by nuclear imaging the radiolabeled pulp cells after implantation in the rat emptied pulp chamber. For that purpose, indium-111-oxine (111In-oxine)-labeled rat pulp cells were added to polymerizing type I collagen hydrogel to obtain a pulp equivalent. This scaffold was implanted in the emptied pulp chamber space in the upper first rat molar. Labeled cells were then tracked during 3 weeks by helical single-photon emission computed tomography (SPECT)/computed tomography performed on a dual modality dedicated small animal camera. Negative controls were performed using lysed radiolabeled cells obtained in a hypotonic solution. In vitro data indicated that 111In-oxine labeling did not affect cell viability and proliferation. In vivo experiments allowed a noninvasive longitudinal follow-up of implanted living cells for at least 3 weeks and indicated that SPECT signal intensity was related to implanted cell integrity. Notably, there was no detectable systemic release of implanted cells from the tooth. In addition, histological analysis of the samples showed mitotically active fibroblastic cells as well as neoangiogenesis and nervous fibers in pulp equivalents seeded with entire cells, whereas pulp equivalents prepared from lysed cells were devoid of cell colonization. In conclusion, our study demonstrates that efficient labeling of pulp cells can be achieved and, for the first time, that these cells can be followed up after implantation in the tooth by nuclear imaging. Furthermore, it appears that grafted cells retained the label and are viable to follow the repair process. This technique is expected to be of major interest for monitoring implanted cells in innovative therapies for injured teeth.
Introduction
Teeth are particularly prone to severe injury because of their location and function. They can be affected by dental caries, a progressive degradation of dental tissues by the by-products of cariogenic bacteria contained in the oral biofilm. If not treated promptly, this process results in pulp infection rapidly spreading in the periapical area. Healthy teeth can also be severely damaged by fracture as a result of trauma. The most common treatment of both conditions, endodontic treatment, consists of pulp removal, mechanical enlargement of the root canal, and placement of a bioinert material sealed with root canal cement in the cleaned pulp cavity. However, endodontically treated teeth are also prone to both fracture due to dryness and absence of sensitivity, and periapical infections resulting from inadvertently left pulp remnants, which are good substrata for bacterial growth. Such infections may further disseminate and induce systemic complications, such as infective endocarditis, nephropathies, or rheumatic fever.1,2
The evidence of cells endowed with stem cell properties in adult pulp, the dental pulp stem cells (DPSCs), promoted the research on the development of alternative therapeutic approaches.3–5 Among them, several teams have proposed a therapy of the pulpal lesions through tissue engineering using these stem cells.6–10 Indeed, DPSCs are capable of differentiating into dentin-forming cells after ectopic implantation,11 in tooth slice models12–15 and in response to pulp aggression in a canine pulp injury model.10
In situ partial pulp regeneration by using tissue engineering is based on the observation that pulpal infection and inflammation are compartmentalized until the entire pulp tissue undergoes necrosis.4,9,16 Current evidence suggests that the remaining healthy portion of the pulp could be recoverable before the final stages and may have the potential to regenerate the lost portion under certain conditions.9 To enhance this regeneration, engineered pulp constructs by growing pulp cells onto collagen or synthetic scaffolds may be inserted into the pulp space to facilitate the total recovery of pulp tissue.9
Although the proof of concept of pulp therapy with stem cells is now obtained, transfer to human teeth requires deciphering cellular and molecular mechanisms underlying tissue repair. An important point is to determine the fate of implanted cells over time and, particularly, during the early period following implantation. Indeed, the success of tissue engineering is known to be dependent on an initial homing of implanted cells and subsequent engraftment to the target tissue. To this regard, in vivo imaging has been reported as a tool used to track the initial homing.17,18 In addition, the potential engraftment of the implanted cells in other organs has to be assessed to evaluate the risk of inducing ectopic mineralization or tumorigenicity.19,20
In the specific perspective of pulp engineering, the follow-up of implanted cells is hampered by the fact that pulp is embedded in highly calcified tissues, especially the enamel that prevents the use of classical cell labeling techniques, such as fluorescence21 or bioluminescence.22 Nuclear imaging (either single-photon emission computed tomography [SPECT] or positron emission tomography) is an alternative approach that is likely to overcome such a constraint. Indeed, this highly sensitive imaging method is particularly suited for cell tracking since it is based on the detection of photons in the gamma range, which are only faintly attenuated by mineralized tissues.23 Cell labeling with indium-111-oxine (111In-oxine) is a well-established method validated in preclinical and clinical studies24–26 and routinely used in humans in the setting of infection scintigraphy since the 1970s.27,28 Indium forms a neutral and lipid-soluble complex with three molecules of 8-hydroxyquinoline (oxine), which enables it to penetrate through the phospholipid bilayer cell membrane. Within the cell, the indium becomes firmly attached to cytoplasmic components (such as lactoferrin). Once labeled, cells may be tracked by monophotonic scintigraphy (SPECT) during the days or weeks after implantation due to physical half-life (T=2.83 days) of 111In.
In the context of developing a cell therapy for damaged dental tissues, we have specifically focused in the present study, on the fate of radiolabeled pulp cells after implantation in a rat emptied pulp chamber. We were able to follow-up the implanted 111In-oxine-labeled pulp cells during 3 weeks postimplantation. In parallel, we assessed the repair process occurring during this period by microcomputed tomography (micro-CT) and histology.
Materials and Methods
Animals
All experiments in this study were designed according to the ARRIVE guidelines and were performed under a protocol approved by the Animal Care Committee of the University Paris Descartes. No. CEEA34.CC.010.11.
Cell isolation and culture
Multicolony-derived rat pulp cells were obtained using a protocol adapted from Gronthos et al.11 from the molars of 5-day Lewis rats. Under sterile conditions, rat molars were extracted using a scalpel blade and artery forceps, and the dental pulp tissue was then removed with a sterile barbed broach. The tissue was evenly sliced into 1–2-mm sections, and then incubated with phosphate-buffered saline (PBS) containing 3 mg/mL collagenase type I (Worthington Biochem) and 4 mg/mL dispase (Boehringer) in a shaking water bath (at 37°C) for 1 h.
The isolated cells were then centrifuged (100 g) at 4°C for 5 min, washed in PBS, resuspended, and plated in the DMEM (Gibco) supplemented with 20% fetal bovine serum (FBS) and 100 mg/mL streptomycin at 37°C under 5% CO2 atmosphere.
The cultures were visually monitored regularly using the light microscopy. The required cell number (106/experiment) for the in vivo experiments was reached after 3–4 passages.
Radiolabeling of rat pulp cells with 111In-oxine
Dental pulp cells were detached under mild conditions in 0.05% trypsin, 0.02% EDTA (Sigma-Aldrich) for 5 min at 37°C, and then centrifuged at 100 g for 5 min. The cell pellet was resuspended in the serum- and glucose-free DMEM, and the cell concentration was measured by microscopic examination.
Labeling was carried out by mixing 25×106 pulp cells with 50 MBq 111In-oxine and a 1 mL TRIS buffer (Covidien Pharmaceuticals). After incubation at 37°C for 30 min, the labeled cells were separated from 111In-oxine solution excess by centrifugation. Subsequently, cells were washed twice with the serum-free DMEM. To determine the radiolabeling efficiency, radioactivity of labeled cell pellets and supernatants was measured in an activity meter (Medi 404; Medisystem). 111In-oxine-labeling efficiency, expressed as a ratio of cell suspension activity to total activity (supernatants and pellet activities), was ∼30%. In vitro stability of radiolabeling was evaluated 2 h after labeling; only 5% of the total activity was eluted from labeled cells. To assess the role of cell membrane integrity in retention of 111In-oxine in pulp equivalents, lysed cells were also prepared. Cell lysis was obtained by a 5-min contact between radiolabeled cells and the hypotonic solution (distilled water).
Cell proliferation assay and cell viability testing
Radiolabeled cells and unradiolabeled cells were cultured in the DMEM (Invitrogen) supplemented with 20% FBS (Invitrogen) and 1% penicillin/streptomycin (Invitrogen) at 37°C under 5% CO2 atmosphere. Labeled and unlabeled pulp cell proliferation and viability were performed at 0, 1, and 5 days after labeling, respectively, using the hemocytometer and Trypan blue test.
Pulp equivalent preparation
Type I collagen was extracted from rat-tail tendons in acetic acid solutions to constitute a stock solution at 2 mg/mL and stored at 4°C, according to.29 Briefly, to prepare four pulp equivalents, 0.04 mL of a concentrated Eagle's minimum essential medium, 0.2 mL of a type I collagen solution, 0.04 mL 0.1 N NaOH, and 0.119 mL of either the entire or lysed radiolabeled cells corresponding to 2.4×106 cells (6×105 cells/matrix) were added and mixed at 0°C. The final concentration of collagen was 1 mg/mL. This mixture was poured into a culture plate for bacteriological culture to prevent cell adhesion at the bottom of the well, with DMEM, 1% penicillin-streptomycin serum free and incubated 1 h at 37°C under 5% CO2 atmosphere for complete collagen polymerization. The pulp equivalent preparation can be kept up to 4 h at 37°C under 5% CO2 before implantation.
Rat pulpotomy model
Twenty 2-month-old male inbred Lewis rats weighing between 160 and 220 g were used for all experiments. Rats were housed in pairs and fed standard rat chow and water ad libitum (Supplementary Video S1; Supplementary Data are available online at www.liebertpub.com/tec).
Animals were anesthetized by intraperitoneal injection of a combination of ketamine 10% (Imalgène 500; Merial) and xylazine 2% (Rompun; Bayer). On the maxillary first molars, an occlusal cavity was drilled to the pulp chamber with high-speed rotary 0.6-mm-diameter round bur (Maillefer Dentsply) under an endodontic microscope (Carl Zeiss). The cameral pulp parenchyma was eliminated with a rotary instrument. Hemostasis was obtained by sterile cotton pellet compression. One pulp equivalent (100 μL) was placed within each empty pulp chamber space. Each animal received both treatments (entire cells and lysed cells) on the left and right first maxillary molars. The occlusal cavity was sealed with a calcium silicate-based restorative cement (Biodentine™; Septodont) and covered with a light-cured composite (FloRestore; Denmat). Animals were then allowed to recover.
For histological analysis, animals were sacrificed by intracardiac perfusion with a 4% paraformaldehyde/glutaraldehyde solution buffered with sodium cacodylate 0.1 M at pH 7.2–7.4, at days 14 and 28 after the surgery.
SPECT/CT imaging procedure
A first scan was performed immediately after implantation of pulp equivalent, and then every 7 days until the signal was no longer detectable (equal to background signal). Each scan included the first acquisition centered on the craniofacial area, designed to quantify the activity related to implanted pulp equivalent in teeth, as well as potential signals in lymph nodes secondary to clearance of radiolabeled cell debris. A second whole-body acquisition was intended to detect a potential signal in the reticuloendothelial system secondary to systemic diffusion of cells or cell debris.
Acquisition and reconstruction parameters
Helical SPECT/CT was performed on a dual modality dedicated small animal camera (NanoSPECT/CT plus; Bioscan, Inc.) under intraperitoneal pentobabital anesthesia (40 mg/kg b.w.; Ceva Santé Animale). SPECT used a multiplexed multipinhole technology (four detectors equipped with nine 0.9-mm-diameter pinhole tungsten collimators each, allowing a spatial resolution of 0.8 mm). Acquisitions centered on the craniofacial area were performed using the fine mode (helical scan with 28 projections/rotation plus circular scans at the beginning and at the end of the scan range, 50 s/projection). Whole-body acquisitions were performed using a standard mode (helical scan with 28 projections/rotation, 30 s/projection). All datasets were reconstructed using dedicated HiSPECT software (Bioscan, Inc.) with an iterative algorithm (three subsets and three iterations), pixel size: 0.5 mm.
Flat-panel detector CT was performed with the following parameters: helical acquisition with 240 projections/rotation, zoom: 1.3, pitch: 1, time/step: 1000 ms, tube voltage: 55 kV, tube current: 145 mAs, and reconstructed using filtered backprojection algorithm with a Ram-Lak (ramp) filter, in-plane voxel size of 147×147 μm, and slice thickness of 147 μm.
Data analysis
Reconstructed SPECT and CT scans were coregistered using InVivo Scope software (Bioscan, Inc.), allowing to localize areas of increased activity. The presence of signals within the pulp chamber on visual analysis was quoted, as well as the presence of uptake in ectopic foci if present. To assess the amount of cells implanted within the pulp chamber, as well as its evolution over time, we quantified the 111In-oxine activity by drawing three-dimensional regions of interest (ROI) on CT, corresponding with treated teeth. ROI were then automatically applied on SPECT images and provided activities (total count value), which were then corrected from radioactive decay. This correction is due to physical half-life of the radionuclide and allows for comparison of sequential measurements by normalization of activities. For decay correction, the following formula was applied: A0=At. (eλt), where A0 represents the total count value at the time of pulp equivalent implantation (t=0); At represents the total count value at the time of measurement (t>0); e is the exponential function; λ is the radioactive constant (ln 2/physical half-life of the radionuclide expressed in days), which is approximated to (0.693/2.83) for 111In; and t is the time elapsed since pulp implantation expressed in days.
Micro-X-ray CT analysis
For exploration of the repair process at 2 weeks, 1 month postsurgery, maxillary blocks were fixed in 4% paraformaldehyde and stored in 70% ethanol at 4°C. Half maxillas were subjected to a desktop micro-CT (Skyscan 1172; Skyscan). Scanning time was ∼20 min. Samples were reconstructed using NRecon software (Skyscan) and visualized using OsiriX imaging software (3.7.1 version).
Histology and immunohistochemistry
Blocks, including the maxillary molar, were decalcified in 4.13% EDTA for ∼3 months. After tissue embedding in Paraplast, serial sections (7 μm) were stained for hematoxylin and eosin. For immunohistochemical analysis, the sections were incubated with primary antibodies against proliferating cell nuclear antigens (Calbiochem) diluted at 1/100, against von Willebrand factor (ABCAM ab-6994) diluted at 1/200, and against the anti-calcitonin gene related peptide (CGRP) (Sigma-Aldrich C819) at 1/2000. Tissue sections were then incubated with anti-mouse immunoglobin (Dakocytomation) diluted at 1/100. Color reaction was developed by using 3,3′-diaminodenzidine tetrahydrochloride. A negative control was included by using an equal concentration of rabbit preimmune serum as the primary antibody.
Statistical analysis
Continuous variables were expressed as mean±SEM. Comparisons between two groups were performed using the Mann–Whitney U test and between more than two groups using the Kruskal–Wallis test. The level of significance was set at p<0.05.
Results
To evaluate pulp therapy with mesenchymal stem cells for injured teeth, we used a rat pulpotomy model consisting of removing the cameral pulp parenchyma. A pulp equivalent with rat pulp cells seeded in a 3D collagen matrix is then placed in the emptied pulp space (Fig. 1).
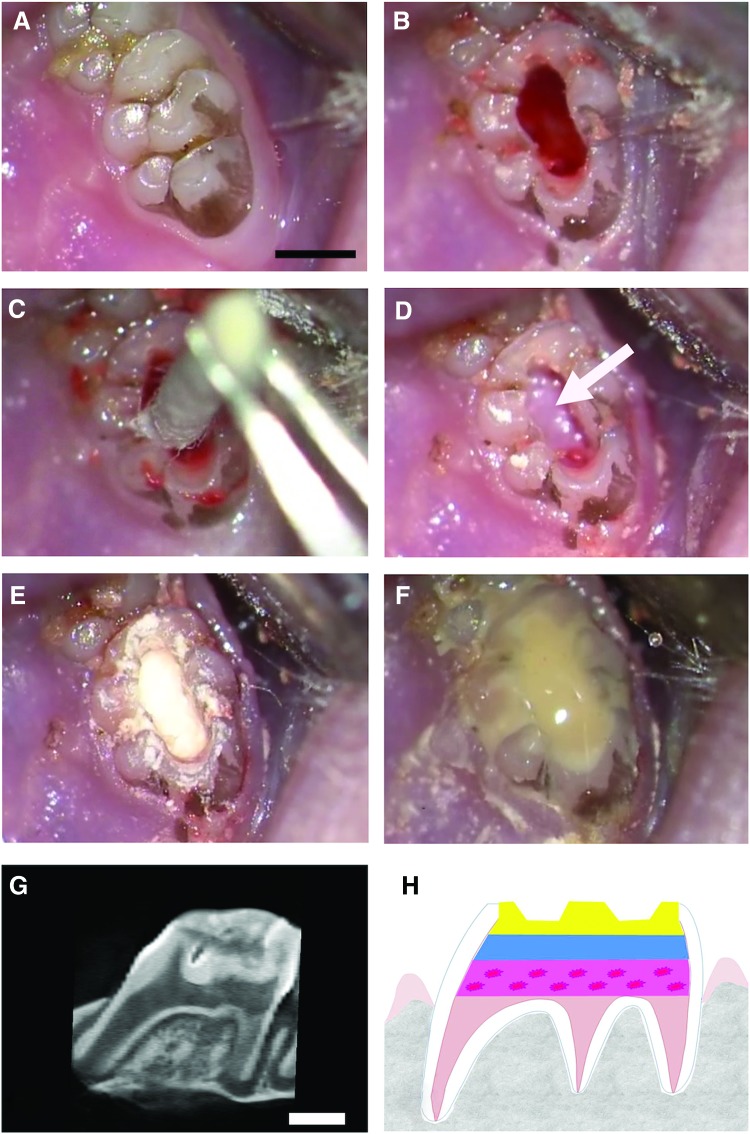
FIG. 1.
Description of the rat pulpotomy model. The pulpotomy model based on the implantation of pulp equivalent (pulp cells seeded in a 3D collagen hydrogel) in the rat pulp chamber (scale bar, 1 mm). After anesthesia, drilling of an occlusal cavity of the first upper molar under an endodontic microscope (A). Elimination of the cameral pulp parenchyma with a rotary instrument (B). After hemostasis by compression (C), placement of radioactive pulp equivalent (with either entire cells or lysed cells) in the pulp chamber space (white arrow shows the pulp equivalent) (D). Sealing of the cavity with a calcium silicate-based cement (Biodentine™; Septodont) (E). Covering with light-cured flow composite (FloRestore; Denmat) (F). Microcomputed tomography (micro-CT) imaging of the tooth immediately after implantation showing the different layers (G), which are illustrated on a schema (dotted-pink layer: pulp equivalent seeded with cells; blue layer: calcium silicate-based cement; yellow layer: composite restoration) (H). Color images available online at www.liebertpub.com/tec
Analysis of repair process
We analyzed the repair process at 1 month following pulp cell implantation (Fig. 2). Micro-CT imaging showed no pulp or root canal obliteration by mineral apposition in both the treatment groups. Histological analysis showed numerous fibroblasts into the collagen matrix implanted with living cells. Among these cells, several were immunolabeled for PCNA, an antibody used to identify mitotically active cells (Fig. 2F). In contrast, no cell was observed in the matrix implanted with lysed cells, either by histology or by immunolabeling for PCNA (Fig. 2C, E).
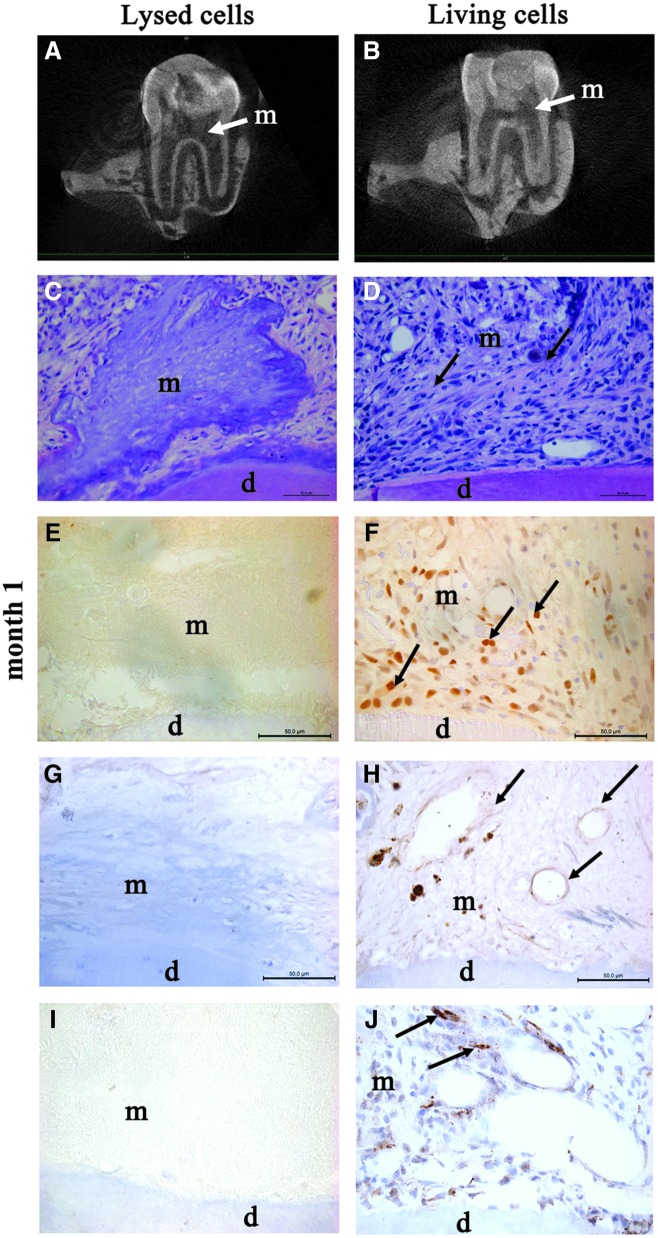
FIG. 2.
Repair process at 1-month postpulp cell implantation. Micro-CT imaging showed no pulp or root canal obliteration 4 weeks after implantation of lysed cells (A) or living cells (B) (white arrows show the implanted matrix). At month 1, hematoxylin and eosin staining showed the absence of cells in the collagen matrix implanted with lysed cells (C). In contrast, numerous cells were observed in the matrix (arrows) implanted with living cells (D). Immunohistochemistry for proliferating cell nuclear antigen showed proliferating cells (arrows) in the matrix seeded with living cells (F), but not in controls (E). Vascularization (arrows) was observed by immunostaining for von Willebrand factor in the matrix seeded with living cells (H), whereas no vessel was observed in controls (G). Innervation was evidenced in the matrix seeded with living cells by immunostaining for the calcitonin gene related peptide, which labels sensory nervous fibers (arrows) (J). In contrast, no labeling was observed in the control matrix (I). d, dentin; m, implanted matrix.
Vascularization structures were observed in contact with the implanted matrix seeded with living cells. As expected, these structures were immunolabeled for von Willebrand factor (Fig. 2H) and alpha smooth muscle actin (data not shown). In addition, we investigated for a potential neurogenesis in the implanted matrix and clearly observed the structures immunolabeled for CGRP, which attests the presence of sensory nerve fibers in the matrix seeded with living cells (Fig. 2J). In contrast, neither vessel nor nervous fiber was observed in the implanted matrix seeded with lysed cells (Fig. 2G, I).
Viability of radiolabeled cells
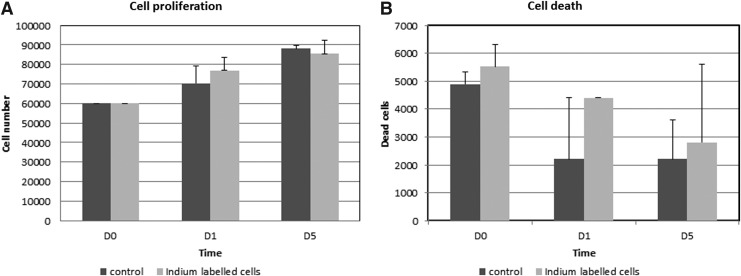
To assess a possible effect of the 111In-oxine radiolabeling process on rat pulp cells, we compared proliferation and viability of labeled and unlabeled pulp cells. Radiolysis phenomena are likely to affect cell viability. As shown in Figure 3A, the proliferation rate of labeled cells was comparable to that of unlabeled cells at day 1 and 5. Furthermore, at the same time points, both labeled and unlabeled pulp cells exhibited a similar viability rate, which was >90% (Fig. 3B).
FIG. 3.
Effect of indium-111-oxine (111In-oxine) labeling on pulp cell proliferation and viability. The absence of detrimental effect of 111In-oxine incorporation on the proliferation rate of labeled cells at early time points (0, 1, and 5 days). Mean results of three individual experiments are presented with 95% confidence intervals for cultures of 111In-oxine-labeled cells and controls. There was no significant difference between the groups (A). The absence of detrimental effect of 111In-oxine incorporation on the viability of labeled cells at same time points (no significant difference) (B).
SPECT/CT imaging of 111In-oxine-labeled DPSCs in pulpotomy models in vivo
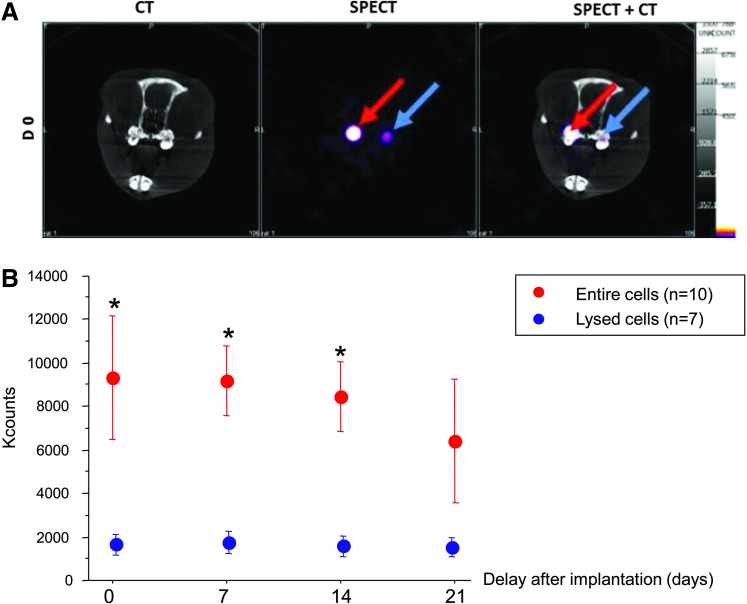
The in vivo follow-up of 111In-oxine-labeled cells was performed using SPECT and CT scans, allowing a comparison between living and lysed cells (Fig. 4A). A signal was detectable in teeth implanted with pulp equivalent for at least 3 weeks after implantation, without a significant decrease (corrected from radioactive decay), according to time whatever pulp cells embedded were entire or lysed. However, total counts were 5-fold higher in pulp equivalents prepared with entire cells compared with lysed cells, although pulp equivalents embedded the same 111In-oxine activity at baseline before implantation. This result indicates a correlation between the cellular integrity and the strength of the signal. This difference of detected signal intensity remained steady from the day of implantation (9334±2867 vs. 1764±528 counts, p<0.004) to the end of the experiment (Fig. 4B).
FIG. 4.
In vivo follow-up of implanted pulp equivalents by single-photon emission computed tomography (SPECT)—comparison of entire and lysed cells. 111In-oxine-labeled cells, entire (red arrow) and lysed (blue arrow), were followed up after implantation with SPECT/CT. The signal displays a greater intensity in tooth implanted with living cells when compared to samples implanted with lysed cells (A). Quantification of 111In-oxine activity was performed during 3 weeks. Data are presented as mean±SEM after radioactive decay correction. Although a similar 111In activity was embedded in both pulp equivalents, detected counts are 5-fold lower in the preparation with lysed cells in all time points, including immediately postimplantation. This suggests a release of cell fragments from the collagen network during implantation. There is no significant count decrease according to time in both pulp equivalents (Kruskal–Wallis p=0.6 for entire cells and p=1 for lysed cells) (B). *p<0.004 (Mann–Whitney) between entire and lysed cells.
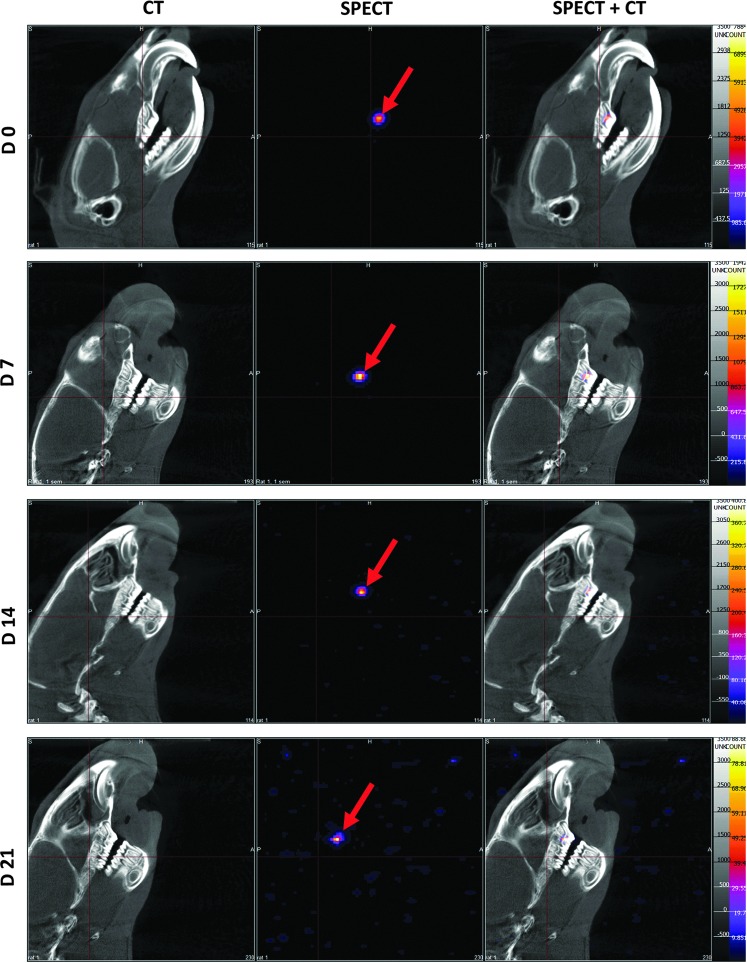
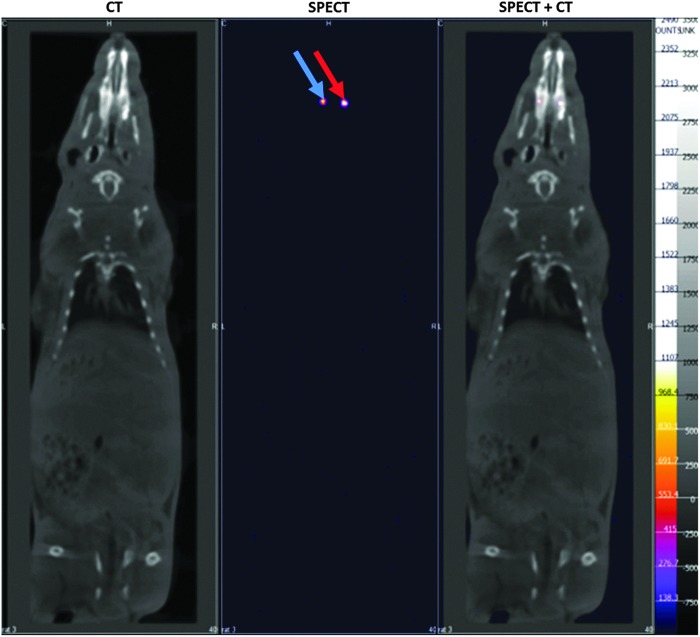
To determine whether implanted cells remained localized in the pulp chamber or underwent dissemination, we analyzed both the craniofacial area (lymph nodes) (Fig. 5) and whole-body acquisitions (reticuloendothelial system) (Fig. 6) by SPECT/CT. No signal was detected outside the tooth, whatever the delay after implantation, suggesting that radiolabeled cells remained located in the pulp chamber.
FIG. 5.
Craniofacial imaging. SPECT/CT imaging of 111In-oxine-labeled cell implantation. Implanted living cells were successfully tracked during 3 weeks (red arrows). Localization of implanted cells was possible with merging SPECT and CT images (right column). No signal was detectable out of the teeth and, in particular, no cell spreading was observed in the craniofacial area.
FIG. 6.
Whole-body imaging. Whole-body imaging at D14 did not show spreading of labeled cells, either in the ectopic sites or in the reticuloendothelial system. red arrow: living cells; blue arrow: lysed cells.
Discussion
The fate of implanted cells is crucial in tissue engineering and, to the best of our knowledge, this issue has never been investigated in the dental pulp context. In this study, we tracked 111In-oxine-labeled rat pulp cells implanted in the rat upper first molar by SPECT. Our data showed that (1) 1 month after implantation, living and mitotically active fibroblasts as well as new vessels and nervous fibers were present in the pulp equivalents seeded with entire cells, whereas pulp equivalents prepared from lysed cells were devoid of cell colonization; (2) in vivo SPECT signal intensity was related to implanted cell integrity and allowed a noninvasive longitudinal follow-up for at least 3 weeks; and (3) there was no detectable systemic release of implanted cells from the pulp chamber.
Cell labeling with radioisotope such as 111In followed by SPECT detection is a well-established method for monitoring implanted cells in several models, including acute brain trauma and cerebral or myocardial ischemia.25,30 For the specific engraftment in the dental pulp, nuclear imaging and MRI31 were the two possible noninvasive imaging methods. However, these techniques had to be tailored for the exploration of highly calcified dental tissues, especially the enamel (96% of mineral). In particular, for MRI, specific radio frequency fields have to be developed.
In this study, we used nuclear imaging, which offers a high sensitivity together with the possibility of signal quantification, both determinants for cell tracking during the first weeks after implantation (for review, see Ref.19). In addition, nuclear imaging provides information on whole-body biodistribution of labeled cells. This permits to check that there was no accumulation of cell debris in organs involved in blood clearance: the kidneys, liver, and spleen, as well as lungs in case of venous diffusion. Furthermore, whole-body imaging allows the detection of a possible dissemination of implanted cells, which, in the case of DPSCs, could lead to ectopic mineralization, an important safety issue in tissue engineering. Therefore, our study provides reliable results for the first 3 weeks following implantation, which is a decisive period in terms of biological events (cell engraftment, proliferation, potential migration, angiogenesis, etc.). In the future, the use of MRI, a technique allowing a longer follow-up of implanted cells, may provide complementary information regarding the fate of implanted cells in the dental pulp.
The present study showed that the radiolabeling process did not alter both the viability and proliferation ability of pulp cells in vitro. Additionally, histological analysis evidenced the presence of cells in the collagen matrix seeded with entire cells 1 month after implantation, conversely to those seeded with lysed cells. Finally, implanted pulp equivalents displayed signal intensity much intense when seeded with entire cells. Taken together, those results suggest that the in vivo SPECT signal was related to the viability of implanted cells and thus could be a useful surrogate marker to monitor pulp equivalent engraftment. Indeed, evidences of cell colonization, neovascularization, and neurogenesis of the collagen matrix seeded with entire cells, conversely to those seeded with lysed cells, support the critical role of cell viability in promoting engraftment of pulp equivalent. The ability offered by SPECT to assess viability of the implant in living animals is expected to be of major interest in a challenging environment (alterations of physicochemical conditions), which is clinically relevant. In the present study, cells were implanted in healthy teeth, whereas in dental clinic, cells will be implanted in damaged and inflamed teeth. This underlies the limit of our animal model and requires further investigations under less favorable conditions. However, for the first time, our study shows that it is possible to track cells in the dental tooth and paves the way for future investigations.
SPECT/CT results revealed complete retention of radioactivity in the implanted teeth over the observation time period, and no evidence of redistribution to other organs. Conversely, pulp equivalents prepared from lysed cells were associated with a drop in signals originating from the time of implantation, suggesting a lack of cell debris retention by the collagen matrix. Indeed, even if the signal is not directly related to cell viability, a decrease in intensity is likely to account for the elimination of cell residues; in the event of cell death, the radiotracer would be eliminated via the kidneys and the bile duct.32,33 In this respect, the marked difference in signal intensity between pulp equivalents containing entire cells and lysed cells is observed at the time of implantation. Although labeling is equivalent in both cases, membrane rupture leads to the loss of 111In-oxine retention, and the radioactivity was released in the culture plate and cotton pellets used.
Cell proliferation in the matrix was detected at 1 month following implantation of intact cells in contrast with the controls. This suggests that our implanted cells remained viable in the pulp equivalent. However, we cannot exclude the possibility that these cells are recruited host-derived cells since both are from the same species. Further studies using human cells in immunodeficient rats or FISH analysis with male/female donors/receivers may allow to distinguish implanted cells from host cells.
The pulpotomy model consists of eliminating cameral pulp parenchyma, yet maintaining vascular pedicle to allow neoangiogenesis (colonization of pulp substitute by neovessels) from dental roots. Our analysis showed that indeed neoangiogenesis is occurring, but only in implanted tissues seeded with living cells. Angiogenesis plays a key role in tissue production since without the existence of a functional vascular network, cells have to rely on diffusion for nutrient supply and waste removal.9 A rapid vascularization of the implanted tissue is therefore a major issue in the success of the pulp therapy. Noteworthy, we were able to detect sensory nervous fibers in the implanted matrix, which attest the functionality of our reconstructed tissue.34,35 Interestingly, we did not detect a mineralization of the pulp chamber that might be expected through the implantation of cells that have been shown to easily undergo an odonto/osteogenic differentiation process.36 This confirms that the use of a low-osteogenic carrier, such as the loose type 1 collagen matrix, is adapted to engineer the dental pulp.37 The formation of tertiary dentin at the boundary of the scaffold with the dentin walls remains to be investigated at longer time points.
In conclusion, our study demonstrates that pulp cell labeling using 111In-oxine is feasible and allows for their noninvasive follow-up during the first weeks after implantation. Signal intensity detected by SPECT is related to cell integrity and further seems to be related to pulp equivalent engraftment. This technique is expected to be of major interest in monitoring DPSCs used in cell therapy for the damaged tooth.
Supplementary Material
Acknowledgments
This work was supported by the University Paris Descartes and the University Paris Diderot, by Assistance Publique - Hopitaux de Paris and by grants from Fondation de l'Avenir (ET1-610) for Sibylle Opsahl Vital (EA2496) and François Rouzet (Inserm U698) and Fondation les Gueules cassées for EA2496. The authors thank Mrs Annie Llorens for her assistance with immunohistochemistry for nervous fibers and Mr Cyril Willig (present address: Université Pierre et Marie Curie, Plateforme d'histologie, Bâtiment Jussieu, Paris, France) for his participation to the surgeries and Dr. Anne Gruaz-Guyon (U773, UFR de Médecine, site Bichat) for her assistance equipment.
Disclosure Statement
The authors have no conflicts of interest.
References
- 1.Parahitiyawa N.B., Jin L.J., Leung W.K., Yam W.C., and Samaranayake L.P.Microbiology of odontogenic bacteremia: beyond endocarditis. Clin Microbiol Rev 22, 46, Table of Contents, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Somma F., Castagnola R., Bollino D., and Marigo L.Oral inflammatory process and general health. Part 2: how does the periapical inflammatory process compromise general health? Eur Rev Med Pharmacol Sci 15,35, 2011 [PubMed] [Google Scholar]
- 3.Sun H.H., Jin T., Yu Q., and Chen F.M.Biological approaches toward dental pulp regeneration by tissue engineering. J Tissue Eng Regen Med 5,e1, 2011 [DOI] [PubMed] [Google Scholar]
- 4.Huang A.H., Chen Y.K., Chan A.W., Shieh T.Y., and Lin L.M.Isolation and characterization of human dental pulp stem/stromal cells from nonextracted crown-fractured teeth requiring root canal therapy. J Endod 35,673, 2009 [DOI] [PubMed] [Google Scholar]
- 5.Rosa V., Della Bona A., Cavalcanti B.N., and Nor J.E.Tissue engineering: from research to dental clinics. Dent Mater 28,341, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iohara K., Nakashima M., Ito M., Ishikawa M., Nakasima A., and Akamine A.Dentin regeneration by dental pulp stem cell therapy with recombinant human bone morphogenetic protein 2. J Dent Res 83,590, 2004 [DOI] [PubMed] [Google Scholar]
- 7.Cordeiro M.M., Dong Z., Kaneko T., Zhang Z., Miyazawa M., Shi S., et al. Dental pulp tissue engineering with stem cells from exfoliated deciduous teeth. J Endod 34,962, 2008 [DOI] [PubMed] [Google Scholar]
- 8.Prescott R.S., Alsanea R., Fayad M.I., Johnson B.R., Wenckus C.S., Hao J., et al. In vivo generation of dental pulp-like tissue by using dental pulp stem cells, a collagen scaffold, and dentin matrix protein 1 after subcutaneous transplantation in mice. J Endod 34,421, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang G.T.Pulp and dentin tissue engineering and regeneration: current progress. Regen Med 4,697, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iohara K., Zheng L., Ito M., Ishizaka R., Nakamura H., Into T., et al. Regeneration of dental pulp after pulpotomy by transplantation of CD31(-)/CD146(-) side population cells from a canine tooth. Regen Med 4,377, 2009 [DOI] [PubMed] [Google Scholar]
- 11.Gronthos S., Mankani M., Brahim J., Robey P.G., and Shi S.Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci U S A 97,13625, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavalcanti B.N., Zeitlin B.D., and Nor J.E.A hydrogel scaffold that maintains viability and supports differentiation of dental pulp stem cells. Dent Mater 29,97, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki T., Lee C.H., Chen M., Zhao W., Fu S.Y., Qi J.J., et al. Induced migration of dental pulp stem cells for in vivo pulp regeneration. J Dent Res 90,1013, 2011 [DOI] [PubMed] [Google Scholar]
- 14.Galler K.M., Cavender A.C., Koeklue U., Suggs L.J., Schmalz G., and D'Souza R.N.Bioengineering of dental stem cells in a PEGylated fibrin gel. Regen Med 6,191, 2011 [DOI] [PubMed] [Google Scholar]
- 15.Sakai V.T., Zhang Z., Dong Z., Neiva K.G., Machado M.A., Shi S., et al. SHED differentiate into functional odontoblasts and endothelium. J Dent Res 89,791, 2010 [DOI] [PubMed] [Google Scholar]
- 16.Huang A.H., Snyder B.R., Cheng P.H., and Chan A.W.Putative dental pulp-derived stem/stromal cells promote proliferation and differentiation of endogenous neural cells in the hippocampus of mice. Stem Cells 26,2654, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berman S.C., Galpoththawela C., Gilad A.A., Bulte J.W., and Walczak P.Long-term MR cell tracking of neural stem cells grafted in immunocompetent versus immunodeficient mice reveals distinct differences in contrast between live and dead cells. Magn Reson Med 65,564, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manley N.C., and Steinberg G.K.Tracking stem cells for cellular therapy in stroke. Curr Pharm Des 18,3685, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu E., Chen W.Y., Gu J., Burridge P., and Wu J.C.Molecular imaging of stem cells: tracking survival, biodistribution, tumorigenicity, and immunogenicity. Theranostics 2,335, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindvall O., and Kokaia Z.Stem cells in human neurodegenerative disorders—time for clinical translation? J Clin Invest 120,29, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chudakov D.M., Matz M.V., Lukyanov S., and Lukyanov K.A.Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev 90,1103, 2010 [DOI] [PubMed] [Google Scholar]
- 22.de Almeida P.E., van Rappard J.R., and Wu J.C.In vivo bioluminescence for tracking cell fate and function. Am J Physiol Heart Circ Physiol 301,H663, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sinusas A.J., Bengel F., Nahrendorf M., Epstein F.H., Wu J.C., Villanueva F.S., et al. Multimodality cardiovascular molecular imaging, part I. Circ Cardiovasc Imaging 1,244, 2008 [DOI] [PubMed] [Google Scholar]
- 24.Aicher A., Brenner W., Zuhayra M., Badorff C., Massoudi S., Assmus B., et al. Assessment of the tissue distribution of transplanted human endothelial progenitor cells by radioactive labeling. Circulation 107,2134, 2003 [DOI] [PubMed] [Google Scholar]
- 25.Schachinger V., Aicher A., Dobert N., Rover R., Diener J., Fichtlscherer S., et al. Pilot trial on determinants of progenitor cell recruitment to the infarcted human myocardium. Circulation 118,1425, 2008 [DOI] [PubMed] [Google Scholar]
- 26.Gildehaus F.J., Haasters F., Drosse I., Wagner E., Zach C., Mutschler W., et al. Impact of indium-111 oxine labelling on viability of human mesenchymal stem cells in vitro, and 3D cell-tracking using SPECT/CT in vivo. Mol Imaging Biol 13,1204, 2011 [DOI] [PubMed] [Google Scholar]
- 27.Thakur M.L., Coleman R.E., and Welch M.J.Indium-111-labeled leukocytes for the localization of abscesses: preparation, analysis, tissue distribution, and comparison with gallium-67 citrate in dogs. J Lab Clin Med 89,217, 1977 [PubMed] [Google Scholar]
- 28.Roca M., de Vries E.F., Jamar F., Israel O., and Signore A.Guidelines for the labelling of leucocytes with (111)In-oxine. Inflammation/Infection Taskgroup of the European Association of Nuclear Medicine. Eur J Nucl Med Mol Imaging 37,835, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaussain Miller C., Septier D., Bonnefoix M., Lecolle S., Lebreton-Decoster C., Coulomb B., et al. Human dermal and gingival fibroblasts in a three-dimensional culture: a comparative study on matrix remodeling. Clin Oral Investig 6,39, 2002 [DOI] [PubMed] [Google Scholar]
- 30.Yoon J.K., Park B.N., Shim W.Y., Shin J.Y., Lee G., and Ahn Y.H.In vivo tracking of 111In-labeled bone marrow mesenchymal stem cells in acute brain trauma model. Nucl Med Biol 37,381, 2010 [DOI] [PubMed] [Google Scholar]
- 31.Struys T., Ketkar-Atre A., Gervois P., Leten C., Hilkens P., Martens W., et al.Magnetic resonance imaging of human dental pulp stem cells in vitro and in vivo. Cell Transplant 2012. [Epub ahead of print]; http://dx.doi.org/10.3727/096368912X657774 [DOI] [PubMed] [Google Scholar]
- 32.Brenner W., Aicher A., Eckey T., Massoudi S., Zuhayra M., Koehl U., et al. 111In-labeled CD34+ hematopoietic progenitor cells in a rat myocardial infarction model. J Nucl Med 45,512, 2004 [PubMed] [Google Scholar]
- 33.Kraitchman D.L., Tatsumi M., Gilson W.D., Ishimori T., Kedziorek D., Walczak P., et al. Dynamic imaging of allogeneic mesenchymal stem cells trafficking to myocardial infarction. Circulation 112,1451, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishizaka R., Hayashi Y., Iohara K., Sugiyama M., Murakami M., Yamamoto T., et al. Stimulation of angiogenesis, neurogenesis and regeneration by side population cells from dental pulp. Biomaterials 34,1888, 2013 [DOI] [PubMed] [Google Scholar]
- 35.Nakashima M., Iohara K., and Sugiyama M.Human dental pulp stem cells with highly angiogenic and neurogenic potential for possible use in pulp regeneration. Cytokine Growth Factor Rev 20,435, 2009 [DOI] [PubMed] [Google Scholar]
- 36.Yu J., Wang Y., Deng Z., Tang L., Li Y., Shi J., et al. Odontogenic capability: bone marrow stromal stem cells versus dental pulp stem cells. Biol Cell 99,465, 2007 [DOI] [PubMed] [Google Scholar]
- 37.Zhang W., Walboomers X.F., van Kuppevelt T.H., Daamen W.F., Bian Z., and Jansen J.A.The performance of human dental pulp stem cells on different three-dimensional scaffold materials. Biomaterials 27,5658, 2006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.