Abstract
Background and Aims
A vast quantity of empirical evidence suggests that insufficient quantity or quality of pollen may lead to a reduction in fruit set, in particular for self-incompatible species. This study uses an integrative approach that combines field research with marker gene analysis to understand the factors affecting reproductive success in a widely distributed self-incompatible species, Prunus virginiana (Rosaceae).
Methods
Twelve patches of P. virginiana distributed within three populations that differed in degree of disturbance were examined. Two of the sites were small (7–35 km2) remnants of forest in an intensively used agricultural landscape, while the third was continuous (350 km2) and less disturbed. Field studies (natural and hand cross-pollinations) were combined with marker gene analyses (microsatellites and S-locus) in order to explore potential factors affecting pollen delivery and consequently reproductive success at landscape (between populations) and fine scales (within populations).
Key Results
Reductions in reproductive output were found in the two fragments compared with the continuous population, and suggest that pollen is an important factor limiting fruit production. Genetic analyses carried out in one of the fragments and in the continuous site suggest that even though S-allele diversity is high in both populations, the fragment exhibits an increase in biparental inbreeding and correlated paternity. The increase in biparental inbreeding in the fragment is potentially attributable to variation in the density of individuals and/or the spatial distribution of genotypes among populations, both of which could alter mating dynamics.
Conclusions
By using a novel integrative approach, this study shows that even though P. virginiana is a widespread species, fragmented populations can experience significant reductions in fruit set and pollen limitation in the field. Deatiled examination of one fragmented population suggests that these linitations may be explained by an increase in biparental inbreeding, correlated paternity and fine-scale genetic structure. The consistency of the field and fine-scale genetic analyses, and the consistency of the results within patches and across years, suggest that these are important processes driving pollen limitation in the fragment.
Keywords: Prunus virginiana, Rosaceae, pollen limitation, self-incompatibility, SI, S-locus, biparental inbreeding, spatial genetic structure, fragmentation
INTRODUCTION
Pollen limitation, caused by a reduction in the amount of pollen deposited (quantity of pollen) and/or a reduction in the effectiveness of available pollen in achieving fertilization (quality of pollen), has been advocated as one of the most important proximate causes of reduced reproductive success in plant populations (Aizen and Harder, 2007). A survey that included 306 plant species found that in 73 % of the studies, there was evidence of pollen limitation at some sites or during some years, suggesting that insufficient pollen receipt is a major cause of reduced fruit production (Knight et al., 2005).
Different evolutionary and ecological factors may affect the prevalence and magnitude of pollen limitation; including breeding systems (Larson and Barrett, 2000), ecological perturbations (Aguilar et al., 2006; Winfree et al., 2009) and/or fine-scale genetic processes (Aizen and Harder, 2007). These factors are not mutually exclusive and their combined effects shape many aspects of plant reproduction. At a local scale for example, fine genetic structure and density may affect the quality of pollen arriving at the stigmas if closely related individuals are arranged in dense patches throughout a population (Cunningham, 2000; Uchiyama et al., 2009). For species with self-incompatibility (SI) systems in particular, reductions in pollen quality can be particularly important (Larson and Barrett, 2000), because of the inherent restrictions that SI systems impose on mating.
Self-incompatibility is a genetically based mate recognition system, controlled by the gene products of the highly variable S-locus, in which only individuals that express different S-proteins can reproduce. In members of the Rosaceae and Solanaceae, SI is determined by allele-specific interactions between S-RNase proteins expressed in the pistil and F-box proteins expressed in the haploid pollen grain (so-called gametophytic self-incompatibility; GSI) (Ushijima et al., 2003; Sassa et al., 2010). In species with GSI, the probability of finding a compatible mate is dependent on the frequency, number and distribution of S-alleles in populations which, in turn, is dependent on population size and structure (Busch and Schoen, 2008). For instance, high levels of fine-scale genetic structure at the S-locus may increase pollen deposition from genetically related donor plants and result in biparental inbreeding and/or a reduction in reproductive success (Nason and Ellstrand, 1995; Hirao, 2010).
Here we use an integrative approach that combines field research with marker gene analysis to understand the factors affecting reproductive success in a widely distributed self-incompatible species, Prunus virginiana (Rosaceae). First, we performed a series of hand and open pollinations, to look for evidence of pollen limitation in patches of plants in fragmented vs. continuous landscapes. Then, to harness the greater sensitivity of the S-locus to changes in population size and structure, we compared levels of genetic variation in the forest fragment and continuous populations to look for evidence of genetic structure at both landscape (among populations) and fine (within populations) scales at both neutral loci (microsatellites) and the selected S-locus. To assess whether differences in levels of pollen limitation between populations were mirrored by changes in the mating system, we further analysed progeny arrays to determine if biparental inbreeding was greater and the estimated number of paternal donors per tree lower in the fragment population as expected. Overall, we find evidence of significantly greater pollen limitation in the forest fragments; moreover, this reduction in fruit set is associated with changes in mating system dynamics and the distribution of genotypes at a fine scale.
MATERIALS AND METHODS
Species description and sampling
Prunus virginiana is a common large shrub or small tree native to North America. It reproduces vegetatively via rhizomes which can form networks extending up to 15 m from the base of the tree (Geyer et al., 2008), and sexually using animal pollination by early-flying bees, especially in the genera Andrena and Bombus (Vicens and Bosch, 2000), but also by fly and ant species (pers. obs.). Each tree produces from one 140 inflorescences (mean 29·61, s.d. 36·16), and each inflorescence can bear from three to 47 flowers (mean 23·51, s.d. 7·43), each with a life span of 2–4 d. Since both flowers within an inflorescence and inflorescences within a tree bloom asynchronously, the entire bloom period may last 1–2 weeks. Multiple pieces of evidence collected in our lab indicate that the species harbours the SI system typical of other members of the Rosaceae (GSI). For example, we first identified pistillate proteins of the same size and pI as S-linked RNases in other members of the Rosaceae (32–33 kDa and highly basic pI; data not shown), and later amplified multiple S-alleles from genomic DNA using primers developed for other Prunus species (see methods and results below). Additionally, following self-pollination (assisted) and closed controls (unassisted) we found that the species is categorized as self-incompatible (Suarez-Gonzalez, 2011).
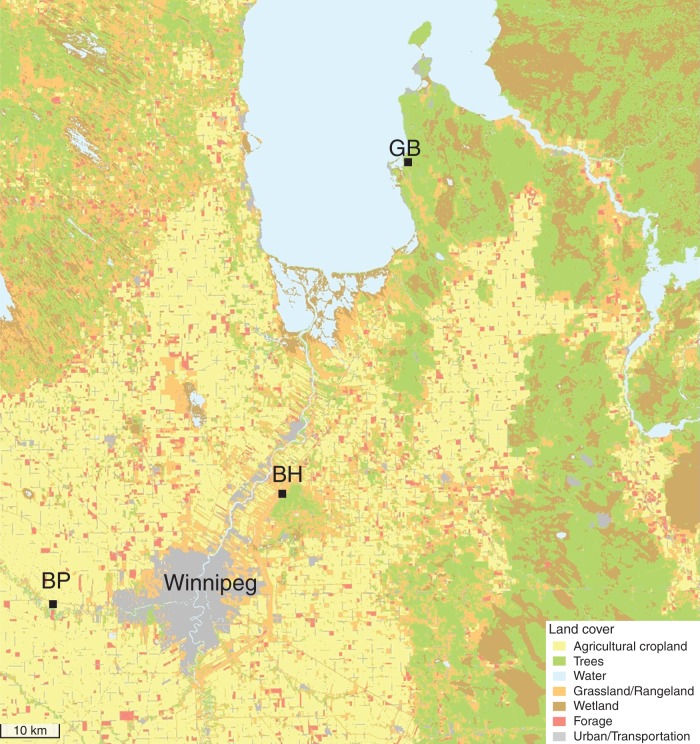
To assess variation in fruit set and pollen limitation, we sampled natural populations of P. virginiana at three sites that differed in degree of disturbance (Fig. 1). Two of the sites are small remnants of forest in an intensively used agricultural landscape (BP, 7 km2; and BH, 35 km2), while the third is continuous and less disturbed (GB, 350 km2). The short length of the flowering season (approx. 1 week) together with the vast extension of agriculture and urban areas in Southern Manitoba hampered our ability to include an additional continuous site for the analysis; however, our sampling design enabled us to assess ecological and genetic variation both within and among populations, and thereby explore the potential factors causing pollen limitation (see sampling design below).
Fig. 1.
Land cover map of the two fragments of forest (BP and BH) and the continuous population (GB) used to detect variation in fruit set and pollen limitation in P. virginiana. Adapted from AgriMap–Manitoba © 2001 Her Majesty the Queen in Right of Manitoba, as represented by the Minister of Conservation.
The fragments arose following modification of the landscape in Southern Manitoba, which occurred approx. 120 years ago, due to increasing urbanization in the Winnipeg area and intensive agricultural practices (Ramankutty and Foley, 1999). The three sites exhibit considerable variation in density, estimated as the number of flowering trees in ten patches of 50 m2. The mean density in BP and BH is higher than that in GB (Supplementary Data Fig. S1), but only significantly different in BH [tGB–BH(18) = 2·29, P < 0·05]. BP also holds patches with few or no P. virginiana trees due to regular flooding, although the mean density of trees was not significantly different from that of GB [tGB–BP(18) = 1·02, P = 0·17].
Ecological aspects
Reproductive success and pollen limitation
In 2009, 20 trees, each with at least ten inflorescences, were selected from four patches within each population (five trees per patch). By sampling patches randomly distributed at the edge and interior of each population, we were able to explore variation in reproductive success and pollen limitation within populations. The trees were located at least 20 m apart to avoid sampling of clones for the pollination experiments (see below). During two consecutive years (2009 and 2010), four inflorescences, each with at least 15 flowers (mean = 26·26, s.d. = 7·81), were chosen from separate branches of each tree to avoid competition for maternal resources (Vogler et al., 1998). Two of the inflorescences were used to estimate the production of fruits following open pollination, and the other two were used to look for pollen limitation by covering them with bridal veil bags prior to anthesis. Once the flowers inside the bags opened, fresh pollen from one donor located >20 m away (to avoid pollination within clones) was applied to all the stigmas in both inflorescences. By using a single pollen donor, we were able subsequently to assess the effect of relatedness of the paternal donor and recipient maternal tree (biparental inbreeding) on fruit production. These results have been reported elsewhere (Suarez-Gonzalez, 2011) and will be summarized in the Discussion. Although using a single pollen donor in supplemented pollinations may bias estimates of pollen limitation if control flowers receive higher pollen diversity (Ashman et al., 2004), we estimated that only a few pollen donors sired seeds per maternal tree following open pollinations (3–5; see the Results ‘Mating system analyis’). Furthermore, hand pollination treatments consistently produced higher fruit set, indicating that employing a single pollen donor in cross-pollinations provided a conservative estimate of pollen limitation in these P. virginiana populations.
To avoid seed predation, the inflorescences from both open and hand cross-pollination treatments were covered with bridal veil bags 3 weeks following flowering. Fruit set was estimated as the number of mature fruits divided by the number of flowers present at anthesis in each inflorescence, and, for subsequent analyses, the mean fruit set of both inflorescences in each treatment (open and cross-pollinations) was used. Pollen limitation was assessed per tree using the pollen limitation index (L) as 1 – Po/Pc, where Po is the fruit set from open controls, and Pc fruit set from hand cross-pollinations (Larson and Barrett, 2000).
Pollen production and receipt
Since fruit set may be limited by either the production or receipt of pollen, we additionally counted pollen grains from anthers and pistils in covered and uncovered controls, respectively, during 2011 in the same 60 trees that had been used for the pollen limitation analysis in 2009/2010. To quantify pollen production, three flowers per tree were collected in glass vials, and pollen grains were counted following the sonication protocol of Dafni et al. (2005). To estimate pollen receipt, 4–5 stigmas per tree were carefully extracted and stored in a 9:1 mixture of 70 % ethyl alcohol to glycerine. Because the amount of pollen that arrives on stigmas is affected by floral age, we collected all the flowers on the second day of bloom. To count the number of pollen grains on receptive stigmas, stigmas were stained with basic fuchsin gel, individually mounted on slides and visualized with a compound scope at ×100 magnification.
Data analysis
We used generalized linear mixed models (GLMMs) to test for variation among populations in open fruit set and pollen limitation (L) using year and population as fixed effects and patch nested in each population as random effects (to detect and account for variation among patches within each population). To asses variation in open fruit set, we used the binomial distribution and logit link function with a two-vector response variable that included the number of successes (number of fruits) and the number of failures (number of flowers that did not make it to fruits). To asses variation in pollen limitation, we used the Poisson distribution and log link function. The minimal adequate model for each response variable was then selected based on Akaike information criteria (AIC; Supplementary Data Table S1). Each model was simplified by removing year (fixed factor), since it did not show significant interactions with population (fixed factor) or a significant effect on the response variables (i.e. open fruit set and pollen limitation, respectively). To test for variation in pollen receipt and production, we implemented GLMM using population as the fixed effect and patch nested in each population as the random effect. We further used multiple comparisons of means for generalized linear models to perform post-hoc pairwise comparisons between populations. The GLMMs and multiple comparisons were performed in ‘lme4′ and ‘multcomp’ packages using ‘lmer’ and ‘glht’ functions, respectively, in R 2·13·2 (R Development Core Team, 2012). We also used one-tailed paired t-tests to see if the supplemental hand pollination treatments produced significantly more fruits than the open pollination treatments within each population.
Genetic aspects
Microsatellite genotyping
For the genetic analyses, only samples from BP (one of the fragments) and GB (continuous population) were analysed. BP was selected because it is geographically further from GB (Fig. 1), is embedded in agricultural fields and has an overall smaller population size and higher density than GB, making it a better contrast from the continuous population. Four trees located at least 20 m away from each other were additionally sampled in each population to increase the sample size from 20 to 24 maternal trees (parental generation) per site. To estimate mating system parameters, ten embryos (offspring generation) per mother were obtained from seeds by carefully removing the endocarp, testa and endosperm. DNA from a total of 150 embryos (from 15 maternal trees) was then extracted using ID labs extraction kits, while DNA from leaves (maternal trees) was extracted using a modified version of the cetyltrimethyl ammonium bromide (CTAB) method (Cheng et al., 1997). Eight microsatellite primers (Supplementary Data Table S2) designed for peach (Prunus persica), that were found to be polymorphic and transferable to other species of Prunus, were used to assess genetic variation within and between our populations of P. virginiana.
Microsatellite data and sample locations have been submitted to DRYAD (http://datadryad.org/).
S-allele genotyping
To explore genetic diversity at the putative S-locus within and among populations, eight trees (also located at least 20 m away from each other) were additionally sampled in BP and GB, for a total sample of 32 trees per population. S-alleles were identified based on the size of the PCR product by using consensus degenerate primers flanking the polymorphic second intron of the S-allele in the Rosaceae (Sutherland et al., 2004). After sequencing PCR products of equal sizes from different individuals multiple times (from two to 11 times), we confirmed that bands of equal size had the same nucleotide sequence, whereas products of different size corresponded to different haplotypes. A total of 22 putative S-RNase alleles (GenBank accessions JQ627789–JQ627810) were detected from the 82 sequences, and 55 additional alleles were genotyped based on the amplification patterns (Supplementary Data Table S3).
Genetic diversity and structure
To compare the total number of S-alleles in the parental generation of each population, Paxman's (1963) estimates of S-allele number were obtained via iteration and rounded to whole numbers. Since this estimator assumes that all alleles occur at the same frequency (i.e. isoplethy), we tested for isoplethy in each population using the Markov chain Fisher's exact test implemented in CHIFISH with 500 000 iterations (Ryman, 2006). A synthetic population of identical size, in which all S-alleles had equal frequencies, was compared with the observed distribution of S-alleles in the respective population (Hoebee et al., 2011).
To compare levels of genetic diversity and genetic differentiation in the parental generation at the simple sequence repeats (SSRs), allelic richness (AR) and the Weir and Cockerham estimator of FST were estimated using FSTAT 2·9.3 (Goudet, 2001). We also estimated the expected heterozygosity (He) and fixation index (FIS) using GENEALEX 6·2 (Peakall and Smouse, 2012). To avoid the effect of family structure on measurements of genetic diversity in the offspring generation, one offspring per maternal tree was randomly chosen 100 times to create 100 data sets consisting of 15 offspring each using a sampling algorithm written in R 2·13·2 (R Development Core Team, 2012). Differences in gene diversity parameters between populations or parental and offspring generations were considered significant if their 95 % confidence intervals (CIs) did not overlap.
Mating system analysis
The multilocus outcrossing rate (tm), single-locus outcrossing rate (ts), biparental inbreeding (tm – ts) and multilocus paternity correlation (rp) were estimated using MLTR V.3·2 (Ritland, 2002). Since there was no evidence of genetic differentiation based on the FST analysis, gene frequencies were assumed to be homogeneous between populations, and gene frequency estimates based on the pooled mean were employed in subsequent analyses to provide more statistical power. Standard errors (s.e.) were obtained using 1000 bootstrap replicates and family as the resampling unit, and then used to calculate 95 % CIs for (tm – ts) and rp. Significant differences between populations were declared if the 95 % CIs of the parameter estimates did not overlap. To determine whether the values were significantly lower than 1 (tm; ts) or greater than zero (tm – ts), the mean ± 1·96 × s.e. was used. The number of pollen donors contributing to a maternal tree (i.e. neighbourhood size) was then estimated as 1/rp.
The maximum-likelihood method of MLTR compares multilocus data within sibships to estimate the correlated paternity among siblings and the mean number of pollen donors (Ritland, 2002). To estimate correlated paternity based on the pattern of differentiation within sibships relative to that among sibships, we used the TwoGener model (Smouse et al., 2001). In TwoGener, the pairwise differentiation between pollen clouds of maternal trees (φFT) is estimated as a measure of pollen pool structure which itself is based on the correlated paternity between sibships. In the absence of inbreeding, it is expected that φFT = rp/2 (Hardy et al., 2004). To detect a significant difference in φFT between the fragment and continuous population, 95 % CIs around φFT were calculated based on the estimated standard deviation (sφ) assuming a normal distribution. The standard deviation was calculated from the variance, s2φ, following Smouse et al. (2001). The number of effective pollen donors (Nep) is estimated by the equation Nep = 1/(2φFT). The TwoGener-based analysis was performed in POLDISP 1·0c (http://poldisp.googlepages.com).
Spatial autocorrelation analysis
To look for spatial genetic structure (SGS) at both the neutral (SSR) and selected (S-locus) loci, the spatial genetic autocorrelation coefficient (r) was estimated and plotted at five distance classes. For this analysis, 999 permutations were used to estimate the two-tailed 95 % CI around the null hypothesis of no autocorrelation (r = 0). To take into account possible bias due to small sample size, we additionally used a bootstrap method to generate a CI around the observed estimate of r by drawing (with replacement) from pairwise comparisons within each distance class. The presence of a positive significant autocorrelation was declared within each distance class when the estimate of r was greater than the 95 % CI based on the permutation test indicating no autocorrelation, and when the bootstrap CI did not overlap with r = 0. This spatial autocorrelation analysis was performed using the software GENALEX 6·2.
RESULTS
Reproductive success and pollen limitation
Reproductive output and pollen limitation (L) were significantly different among populations as shown by the corresponding minimal adequate model (Table 1, Fig. 2A, B). This model indicates that mean fruit set was lowest in the most disturbed fragment (BP), intermediate in the other fragment (BH) and highest in the continuous population (GB) (Tukey's tests P < 0·001) and this effect was consistent across patches and in both years (see the Materials and Methods). Moreover, the mean level of pollen limitation was not significantly different among fragments but was significantly lower in the continuous population (GB) (Tukey's tests P < 0·05). Furthermore, when fruit set following cross-pollination was compared with that in the open controls, we found that crossed fruit set was significantly higher in the fragments (BP, t34 = 4·08, P < 0·001; BH, t38 = 2·36, P < 0·05) but not in the continuous population (GB, t38 = 0·54, P = 0·6) (Fig. 2A). Overall, these results show a consistent population effect on reproductive output and pollen limitation in P. virginiana.
Table 1.
Results of generalized mixed linear models (GLMMs) performed to assess the variation in open fruit set, pollen limitation, pollen production and pollen receipt among two fragments of forest (BP, BH) and one continuous population (GB) ofP. virginiana
| Response variable | Statistics | P-value | Random effect s.d. |
|---|---|---|---|
| Open fruit set* | F2,96 = 13·305 | < 0·001 | 2·72E-06 |
| Pollen limitation† | F2,87 = 6·8713 | < 0·01 | 0 |
| Pollen production† | F2,44 = 3·478 | 0·18 | 0·16267 |
| Pollen receipt† | F2,42 = 11·113 | < 0·01 | 0·14195 |
Only the minimal adequate models are shown, with population as the only fixed effect and patch nested in each population as random effect.
Significant values are given in bold.
*GLMM with binomial distribution function.
†GLMM with Poisson distribution.
Fig. 2.
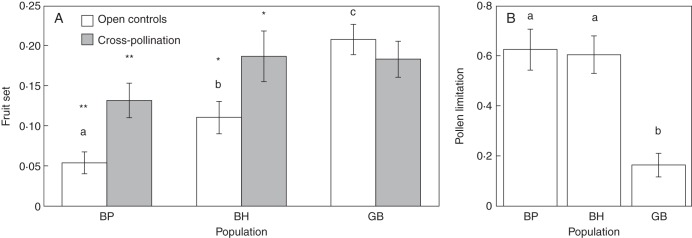
Histograms showing (A) the mean fruit set following open controls and cross-pollinations; and (B) the pollen limitation index as estimated during two consecutive years for three populations (BP, BH and GB) of P. virginiana trees. Shared letters above columns indicate that the average open controls are not statistically different among populations. Asterisks indicate when fruit set from cross-pollinations was significantly higher (**P < 0·01; *P < 0·05) than that in open controls after one-tailed paired t-tests between the two treatments within a population.
Pollen production and receipt
Pollen production did not differ among populations; however, the receipt of pollen was significantly different among populations in 2011, the only year it was tested (Fig. 3, Table 1). The total number of pollen grains in stigmas was significantly higher in GB compared with both fragments of forest (Tukey's tests P < 0·001), and there was no significant difference between the fragments (Tukey's test P = 0·186). Additionally, pollen receipt was found to be positively correlated with the mean reproductive success of trees in 2009 and 2010 (rs58 = 0·35, P < 0·05) and negatively correlated with the average pollen limitation in 2009 and 2010 (rs58 = –0·39, P < 0·05).
Fig. 3.
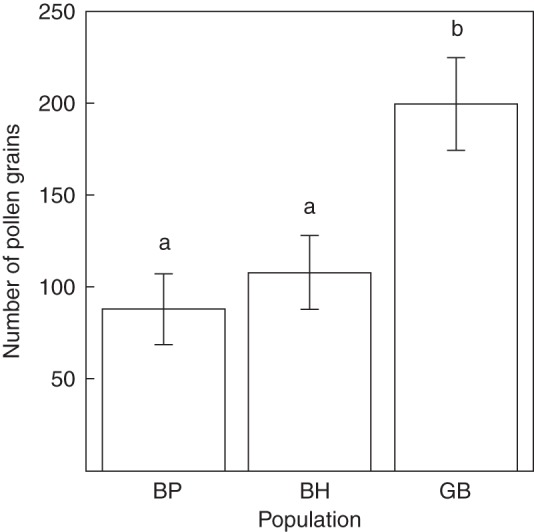
The number of pollen grains deposited on P. virginiana stigmas from two fragments of forest (BP and BH) and one continuous population, after 2 d of bloom. Shared letters above columns indicate that averages are not statistically different.
High genetic diversity and lack of genetic structure
SSR loci
All eight SSR loci were found to be polymorphic, collectively accounting for 82 alleles across loci and populations. In both the parental and offspring generations BP revealed a reduction in genetic diversity (AR and He) compared with GB; however, the differences were statistically significant only in the offspring generation (Table 2). In BP, comparison of genetic diversity parameters between generations revealed a reduction in diversity in the offspring compared with the adults, whereas in GB the trend was the reverse; however, none of the differences was significant. The fixation index was negative for both the offspring and adult generations in both populations, revealing no evidence in inbreeding, as expected. Finally, effectively no genetic differentiation was observed among populations at either the neutral loci (FST = 0·009, s.d. 0·002) or the S-locus (FST = 0·007).
Table 2.
Genetic diversity parameters, including expected heterozygosity (He), allelic richness (AR) and fixation index (FIS), and their 95 % CI at eight microsatellites for adult and offspring populations from one fragment of forest (BP) and one continuous population (GB) ofP. virginiana
| Adults |
Offspring |
|||
|---|---|---|---|---|
| BP | GB | BP | GB | |
| n | 24 | 24 | 150 | 150 |
| AR | 8·091 | 8·999 | 6·085 | 13·125 |
| (5·471–10·712) | (6·193–11·734) | (5·166–7·004) | (9·394–16·856) | |
| He | 0·726 | 0·752 | 0·706 | 0·735 |
| (0·686–0·766) | (0·708–0·796) | (0·704–0·709) | (0·732–0·738) | |
| FIS | –0·143 | –0·146 | –0·070 | –0·085 |
| (–0·236 to –0·050) | (–0·249 to –0·043) | (–0·08 to –0·061) | (–0·091 to –0·079) | |
S-locus
For the S-alleles, we found that PCR bands of equal size had identical nucleotide and amino acid sequences, whereas products of different size corresponded to different haplotypes. This supports the robustness of the method of using band size to identify unique S-alleles; however, substitutions occurring in some sequences of equal size may not have been detected, leading to a possible slight underestimation in the number of S-alleles (Busch et al., 2010). Although this and other commonly used techniques to detect S-allele polymorphism (e.g. SSP in Busch et al., 2010) share this problem, these approaches are convenient and considered robust when examining a large number of samples from natural populations.
The continuous population harboured 20 different S-alleles, of which two were unique. On the other hand, BP had 19 S-alleles, only one of which was unique, indicating a substantial overlap in S-alleles (18 out of a total of 21) between the two populations. The maximum likelihood estimation of the number of S-alleles in each population (after adjusting for incomplete sampling, Paxman, 1963) increased minimally, by one allele in each population, compared with the observed value (BP = 20, GB = 21), and the frequency of alleles within populations did not deviate from isoplethy (BP = 9·42, P = 0·97; GB = 17·38, P = 0·69).
Mating system analysis
Estimates of the mating system parameters from the fragment (BP) and continuous population (GB) are shown in Table 3. The multilocus outcrossing rates were not significantly lower than 1 (tm + 1·96 × s.e. ≥1) in either population, indicating an absence of self-fertilization. The single-locus outcrossing rate was significantly lower than 1 (ts + 1·96 × s.e. <1) and levels of biparental inbreeding were low but significantly greater than 0 (tm – ts – 1·96 × s.e. >0) in both populations. Biparental inbreeding and correlated paternity were significantly higher in BP (tm – ts = 0·313, rp 0·346) than in GB (tm – ts = 0·102, rp 0·180). Similar results for correlated paternity were obtained with the TwoGener method (Table 3); however, this analysis indicated a higher correlated paternity than that obtained with the MLTR model (rp = 2FT) in GB (2φFT = 0·260) but not in BP (2φFT = 0·326). Simulation studies have shown that MLTR's rp underestimates correlated paternity at the family level (Hardy et al., 2004), which may explain the higher correlated paternity found in GB based on TwoGener. In any case, using either method we observed fewer pollen donors in BP than in GB (Table 3), suggesting that more matings between related individuals are occurring in the forest fragment, even though the S-allele in the pollen was probably different from that in the maternal tree.
Table 3.
Results from the mixed mating system and pollen structure model analyses using MLTR and TwoGener, respectively, in one fragment (BP) and one continuous population (GB) ofPrunus virginiana
| Model | BP | GB | |
|---|---|---|---|
| Mixed mating (MLTR) | tm | 1·200 (0·002) | 0·994 (0·093) |
| ts | 0·887 (0·020) | 0·892 (0·025) | |
| tm – ts | 0·313 (0·079) | 0·102 (0·091) | |
| (0·303–0·323) | (0·056–0·148) | ||
| rp | 0·346 (0·053) | 0·180 (0·051) | |
| (0·319–0·373) | (0·154–0·206) | ||
| 1/rp | 2·9 | 5·5 | |
| Pollen pool structure | φFT | 0·163 | 0·130 |
| (TwoGener) | (0·150–0·180) | (0·129–0·145) | |
| Nep | 3·0 | 3·8 |
The parameter estimates included the multilocus outcrossing rate (tm), mean single-locus outcrossing rate (ts), biparental inbreeding rate (tm – ts), correlation of paternity (rp), paternity differentiation between families (φFT) and neighbourhood size as (1/rp) and Nep. Standard errors from the mixed mating model are shown in parentheses next to the estimates. 95 % confidence intervals are shown for tm – ts, rp and φFT.
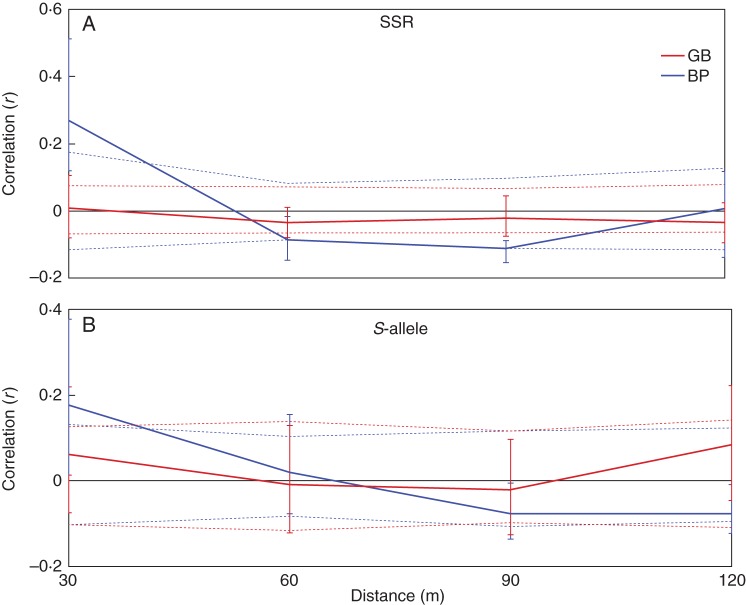
Spatial autocorrelation analysis
The results from the fine genetic structure analyses using the microsatellite loci showed that the values of r (autocorrelation coefficient) from BP at the first distance class (30 m) fell outside the confidence belt where no spatial structure is assumed (Fig. 4A). This and the fact that the r value was considerably higher in BP (0·268, 95 % CI = 0·119–0·512, P < 0·01) than in GB (0·009, 95 % CI = –0·08 to 0·106, P = 0·396) suggests that a pair of individuals between 0 and 30 m apart have a greater probability of being related if they are in BP compared with GB. We found similar structure at the S-locus; although the 95 % CIs of r at 30 m overlap (BP = 0·176, 95 % CI = 0·013 – 0·377; GB = 0·06. 95 % CI = –0·075 to 0·219), both statistical tests (i.e. bootstrap and permutation BP P < 0·05; GB P = 0·172) show evidence of genetic structure in BP, but not in GB (Fig. 4B). These results collectively suggest that the greater spatial genetic structure in the forest fragment compared with the continuous forest may be a factor limiting fruit production.
Fig. 4.
Correlograms showing the spatial genetic structure for BP and GB, using microsatellite loci (A) and the S-locus (B). The continuous line represents the autocorrelation coefficient (r). The dotted lines represent the 95 % CI around the null hypothesis of no autocorrelation (r = 0) based on the bootstrap method. The vertical bars represent the 95 % CI for the observed estimates of r based on the permutation test.
DISCUSSION
In this study, we explored ecological and genetic processes influencing reproductive success and pollen limitation in a species harbouring gametophytic SI, by analysing both breeding system data and genetic variation at neutral loci (SSRs) and the selected S-locus. Our results underscore how ecological and fine-scale genetic processes can influence pollen limitation and mating system dynamics, in particular for self-incompatible species. Widely distributed self-incompatible species, such as P. virginiana, have intrinsic characteristics that maintain homogeneity at a landscape scale; however, we found that small forest remnants had lower reproductive success and greater pollen limitation than a larger, continuous population. Furthermore these reductions in reproductive success were associated with lower pollen receipt, greater spatial genetic structure and increased biparental inbreeding and correlated paternity despite the finding of high levels of genetic diversity and little genetic differentiation between populations. This demonstrates that reductions in fruit set caused by pollen limitation were affected by fine-scale genetic processes (e.g. spatial structure of genotypes), and associated with ecological differences in habitat type (disturbed vs. continuous forest).
Ecological factors affecting reproductive success and pollen limitation
We found a striking reduction in reproductive success in two forest fragments compared with that in the continuous population during two consecutive years. Pollen limitation and limited maternal resources are among the most important factors associated with a decline in fruit set in natural populations (Stephenson, 1981). In a previous study, we evaluated the effect of maternal resources on fruit production using the first principal component of a principal component analysis (PC1) that included the size of the maternal tree and number of inflorescences, together with the incidence of disease (fungal infection) (Suarez-Gonzalez, 2011); however, neither PC1 nor fungus showed a direct effect on fruit set, indicating that maternal resources were not a primary factor limiting fruit production. The evidence presented here shows rather that pollen limitation is a primary driver of reduced fruit set as demonstrated in other studies examining the effect of fragmentation on plant reproduction (Aguilar et al., 2006). We also show that pollen limitation is exacerbated by fine-scale genetic events including increases in fine-scale genetic structure and biparental inbreeding in the fragmented sites.
In animal-pollinated species, many ecological factors can affect the behaviour and abundance of pollinators which may result in changes in the quantity and/or quality of pollination services (Aguilar et al., 2006; Winfree et al., 2009). Our results confirmed that flowers in the fragments are receiving significantly less pollen compared with those in the continuous population, and although we did not test for pollen production and receipt in 2009 and 2010, we found a significant correlation between the production of fruits/tree between years [rs(58) = 0·20, P < 0·001], and also a positive correlation between the amount of pollen received in 2011 and fruit production in 2009 and 2010. Thus, even though pollinator ecology is well known to vary between years (Aizen and Harder, 2007), we observed consistent reductions in reproductive success in the fragments across multiple years in either fruit set or pollen receipt.
Habitat alterations and spatial distribution of plants can influence pollination efficiency and consequently reduce fruit production by causing variation in the local assemblage of pollinators and/or changes in their foraging patterns (Aguilar et al., 2006). Some studies have found that higher plant density increases pollinator attraction and therefore visitation frequency (Ghazoul, 2005); however, this is not always the case (Bosch and Waser, 1999), as observed in P. virginiana where individuals in populations with higher tree density (i.e. fragments of forest) received less pollen than those in a less dense continuous population. Instead, the reduction in pollen receipt may indicate alterations in the local assemblage or abundance of pollinators caused by either habitat loss or an alteration in the type of matrix in and surrounding the forest remnants, both of which are embedded in agricultural fields (see Fig. 1). The majority of studies on pollinator diversity have found consistent decreases in pollinator richness and abundance in fragments compared with that in continuous populations (Aizen and Feinsinger, 2002; Winfree et al., 2009). Alterations in the composition of pollinator assemblages can affect pollen deposition following fragmentation when efficient pollinators are replaced by inefficient ones or by non-pollinating insects (Aguirrea and Dirzo, 2008), both of which may explain the reduced pollen receipt in both fragments of P. virginiana compared with the continuous site, despite the higher tree density in the former.
Effects of mating dynamics and fine-scale genetic structure on pollen limitation
Analysis of the mating system in P. virginiana showed that although outcrossing was absolute (tm = 1), some mating events occur among relatives (tm – ts >0). Biparental inbreeding was significantly higher in BP (31 %) compared with GB (10 %); a finding that agrees with several studies reporting higher levels of biparental inbreeding in fragmented compared with continuous populations (Fernandez-M. and Sork, 2005; Mimura et al., 2009). Results from the correlated paternity (rp) analogues using both the MLTR and TwoGener methods showed similar results and indicated that a higher proportion of progeny were produced by the same pollen donor in BP (tm × rp = 42 %) compared with GB (18 %).
If trees in BP are receiving related biparentally inbred pollen from neighbouring plants, the reduction in seed set could be attributed to early acting inbreeding depression, as has been found in other studies (Fenster, 1991; Byers, 1998; Robertson and Ulappa, 2004). The effect of biparental inbreeding on reproductive success was supported by a recent study that found that both fruit set and offspring fitness were reduced when the relatedness of breeding pairs increased (Hirao, 2010). On the other hand, reduced fruit set could be caused by reduced male fertilization success. In SI species, fertilization is prevented when the S-alleles expressed by the pollen donor are the same as those in the pistil (Ushijima et al., 2003; Sassa et al., 2010). We previously reported a greater reduction in fruit set following hand cross-pollination between full-siblings (Suarez-Gonzalez, 2011); however, we did not investigate whether this reduction was due to early acting inbreeding depression or differential success of non-inbred pollen. In either case, these data provide support for the hypothesis that the reduction in reproductive success in fragments is caused by a reduction in the ‘quality’ of pollen, a finding that has been observed in other studies of trees (Stacy, 2001; Lowe et al., 2005; Jones and Comita, 2008).
According to theoretical predictions and some empirical data, an increase in correlated paternity and biparental inbreeding may lead to higher levels of inbreeding in the fragments (Lienert, 2004; Coates et al., 2007). However, we found a negative fixation index in the offspring and adults of both populations, suggesting that biparentally inbred progeny do not persist to adulthood. This negative effect of inbreeding, commonly documented in plant populations (Husband and Schemske, 1996; Vogler and Stephenson, 2001), has been attributed to the high levels of genetic load maintained in SI species (Glemin et al., 2001; Porcher et al., 2005). Thus, inbreeding depression, in addition to SI, probably maintains high levels of outcrossing in P. virginiana.
Because P. virginiana harbours a gametophytic SI system, the arrival of inbred pollen at stigmas may also result in the reduction of fruit set if related parents share S-alleles. If fragmentation causes a reduction in genetic diversity, and an increase in genetic differentiation among populations, S-allele diversity can theoretically limit fruit set. However, these conditions do not easily arise, except in highly endangered species (Demauro, 1993), or in populations that have gone through long, protracted bottlenecks (Busch and Schoen, 2008; Young and Pickup, 2010). Although we observed an increase in both biparental inbreeding and fine-scale spatial genetic structure in the fragment compared with the continuous population (see below), there was no reduction in S-allele diversity and it is unlikely that mate limitation is occurring.
In fact, the analysis of variation at the S-locus revealed high allelic diversity, similar to that found in other trees in the Rosaceae (Hoebee et al., 2011), which is explained by negative frequency balancing selection operating on the S-locus (Wright, 1939). The strength of selection favouring new alleles strongly depends on the number of S-alleles in the population: if the number of S-alleles is near the maximum expected for a population of that size, new S-alleles have weaker selection coefficients (Schierup, 1998). In addition to maintaining many alleles in populations, frequency-dependent selection is expected to lead to equal S-allele frequencies, isoplethy, at equilibrium. Our finding that the S-alleles are in isoplethy suggests that there is equilibrium between mutation and selection in both the fragment and continuous population; however, we interpret this estimate with caution. The low statistical power of tests of equal allele frequencies (Castric and Vekemans, 2004) may obscure the occurrence of deviations from equilibrium. Overall, these results suggest that maternal trees are probably not limited by the number of compatible mates in the fragments, but rather by the type of pollen brought by pollinators, which appears to be biparentally inbred.
Analyses of SGS as inferred from microsatellite loci and the S-locus showed similar results, as expected when S-allele diversity is high and therefore negative frequency-dependent selection is weak (Leducq et al., 2011). Our results revealed a significant increase in the autocorrelation coefficient at small distances (30 m) in BP compared with GB, which may be explained by an increase in clonal propagation and limited pollen and/or seed movement in the former. If clonal propagation is increased in BP compared with GB, as suggested by the higher density in the former, the presence of greater clonal patch size in BP may augment spatial genetic structure and limit compatible pollen receipt and hence fruit production. This would be particularly true if the pollen dispersal distance is short, as shown by a previous analysis of the pollen dispersal curve in GB (Suarez-Gonzalez, 2011). We found a highly leptokurtic distribution of pollen dispersal distance indicating a large number of near-neighbour mating events. Although we could not analyse the pollen dispersal curve in BP, the consistent pattern of small population genetic effects in BP (greater SGS in both S-allele and SSR, and greater biparental inbreeding) suggests that biparentally inbred mating events are probably more frequent in the fragment and likely to result in S-allele sharing.
Lack of population structure
Theoretical analysis indicates that even low levels of migration prevent structure at the S-locus at a landscape scale (Schierup et al., 2000; Muirhead, 2001). Consistent with the expectation for a widely distributed self-incompatible species, we find overall high levels of genetic diversity and negligible population structure at both neutral and selected loci.
The high levels of gene flow and genetic diversity in this species explain the similarities in heterozygosity and gene diversity in fragmented and continuous populations. Given that the generation time of P. virginiana ranges from 30 to 40 years (Leigh, 1999), insufficient time has lapsed for a reduction in heterozygosity to arise since fragmentation probably occurred only 3–4 generations ago, when a massive amount of forest was converted to cropland in Southern Manitoba (approx. 120 years ago) (Ramankutty and Foley, 1999). However, the concomitant changes in mating system dynamics and SGS detected in the fragment suggest that population isolation, changes in the surrounding matrix and its corresponding effect on pollinators as well as genetic consequences at a local scale may collectively hinder reproductive success in this relatively widespread small tree.
Conclusions
By using a novel integrative approach, we showed that even though P. virginiana is a widespread species, fragmented populations can experience significant reductions in fruit set and pollen limitation in the field. These limitations were further explained in one of the fragments by an increase in biparental inbreeding, correlated paternity and fine-scale genetic structure. Even though we examined only a single continuous site and one of the fragments for the genetic analyses, the consistency of the field and fine-scale genetic analyses, and the consistency of the results within patches and across years, suggest that we have uncovered important processes driving pollen limitation in the fragment. However, this approach should be replicated in other species and/or populations to understand fully the interplay of genetic and ecological factors influencing species' response to forest fragmentation (Bacles and Jump, 2010).
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We are grateful to lab technician F. Martinez-Nunez for his precious help in the field and in the lab. We would also like to thank two anonymous reviewers for their comments on an earlier draft of the manuscript. This work was funded by the NSERC discovery grant to SVG, and a Manitoba Graduate Scholarship and University of Winnipeg Graduate Assistantship (UWGA) grant to AS-G.
LITERATURE CITED
- Aguilar R, Ashworth L, Galetto L, Aizen MA. Plant reproductive susceptibility to habitat fragmentation: review and synthesis through a meta-analysis. Ecology Letters. 2006;9:968–980. doi: 10.1111/j.1461-0248.2006.00927.x. [DOI] [PubMed] [Google Scholar]
- Aguirrea A, Dirzo R. Effects of fragmentation on pollinator abundance and fruit set of an abundant understory palm in a Mexican tropical forest. Biological Conservation. 2008;141:375–384. [Google Scholar]
- Aizen MA, Feinsinger P. Bees not to be? Responses of insect pollinator faunas and flower pollination to habitat fragmentation. In: Bradshaw GA, Marquet PA, editors. How landscapes change: human disturbance and ecosystem fragmentation in the Americas. Berlin: Springer-Verlag; 2002. pp. 111–129. [Google Scholar]
- Aizen MA, Harder LD. Expanding the limits of the pollen-limitation concept: effects of pollen quantity and quality. Ecology. 2007;88:271–281. doi: 10.1890/06-1017. [DOI] [PubMed] [Google Scholar]
- Ashman T, Knight TM, Steets JA, et al. Pollen limitation of plant reproduction: ecological and evolutionary causes and consequences. Ecology. 2004;85:2408–2421. [Google Scholar]
- Bacles CFE, Jump AS. Taking a tree's perspective on forest fragmentation genetics. Trends in Plant Science. 2010;16:13–18. doi: 10.1016/j.tplants.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Bosch M, Waser NM. Effects of local density on pollination and reproduction in Delphinium nuttallianum and Aconitum columbianum (Ranunculaceae) American Journal of Botany. 1999;86:871–879. [PubMed] [Google Scholar]
- Busch JW, Joly S, Schoen DJ. Does mate limitation in self-incompatible species promote the evolution of selfing? The case of Leavenworthia alabamica. Evolution. 2010;64:1657–1670. doi: 10.1111/j.1558-5646.2009.00925.x. [DOI] [PubMed] [Google Scholar]
- Busch JW, Schoen DJ. The evolution of self-incompatibility when mates are limiting. Trends in Plant Science. 2008;13:128–136. doi: 10.1016/j.tplants.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Byers D. Effect of cross proximity on progeny fitness in a rare and a common species of Eupatorium (Asteraceae) American Journal of Botany. 1998;85:644. [PubMed] [Google Scholar]
- Castric V, Vekemans X. Plant self-incompatibility in natural populations: a critical assessment of recent theoretical and empirical advances. Molecular Ecology. 2004;13:2873–2889. doi: 10.1111/j.1365-294X.2004.02267.x. [DOI] [PubMed] [Google Scholar]
- Cheng K, Chang H, Su C, Hsu F. Identification of dried rhizomes of Coptis species using random amplified polymorphic DNA. Botanical Bulletin of Academia Sinica. 1997;38:241–244. [Google Scholar]
- Coates DJ, Sampson JF, Yates CJ. Plant mating systems and assessing population persistence in fragmented landscapes. Australian Journal of Botany. 2007;55:239–249. [Google Scholar]
- Cunningham SA. Depressed pollination in habitat fragments causes low fruit set. Proceedings: Biological Sciences. 2000;267:1149–1152. doi: 10.1098/rspb.2000.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafni A, Kevan PG, Husband BC. Practical pollination biology. Cambridge, Ontario: Enviroquest; 2005. [Google Scholar]
- Demauro MM. Relationship of breeding system to rarity in the lakeside daisy (Hymenoxys acaulis var. glabra) Conservation Biology. 1993;7:542–550. [Google Scholar]
- Fenster CB. Gene flow in Chamaecrista fasciculata (Leguminosae) II. Gene establishment. Evolution. 1991;45:410–422. doi: 10.1111/j.1558-5646.1991.tb04414.x. [DOI] [PubMed] [Google Scholar]
- Fernandez-M JF, Sork VL. Mating patterns of a subdivided population of the Andean oak (Quercus humboldtii Bonpl., Fagaceae) Journal of Heredity. 2005;96:635–643. doi: 10.1093/jhered/esi104. [DOI] [PubMed] [Google Scholar]
- Geyer AW, Broyles PJ, Row JM. Plant fact sheet, chokecherry. Manhattan, KS: Manhattan Plant Materials Center; 2008. [Google Scholar]
- Ghazoul J. Pollen and seed dispersal among dispersed plants. Biological Reviews. 2005;80:413–443. doi: 10.1017/s1464793105006731. [DOI] [PubMed] [Google Scholar]
- Glemin S, Bataillon T, Ronfort J, Mignot A, Olivieri I. Inbreeding depression in small populations of self-incompatible plants. Genetics. 2001;159:1217–1229. doi: 10.1093/genetics/159.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2·9.3) 2001 [Google Scholar]
- Hardy OJ, Gonzalez-Martinez SC, Colas B, Freville H, Mignot A, Olivieri I. Fine-scale genetic structure and gene dispersal in Centaurea corymbosa (Asteraceae). II. Correlated paternity within and among sibships. Genetics. 2004;168:1601–1614. doi: 10.1534/genetics.104.027714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao AS. Kinship between parents reduces offspring fitness in a natural population of Rhododendron brachycarpum. Annals of Botany. 2010;105:637–646. doi: 10.1093/aob/mcq018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebee SE, Angelone S, Csencsics D, Maattanen K, Holderegger R. Diversity of S-alleles and mate availability in 3 populations of self-incompatible wild pear (Pyrus pyraster) Journal of Heredity. 2011;103:260–267. doi: 10.1093/jhered/esr126. [DOI] [PubMed] [Google Scholar]
- Husband BC, Schemske DW. Evolution of the magnitude and timing of inbreeding depression in plants. Evolution. 1996;50:54–70. doi: 10.1111/j.1558-5646.1996.tb04472.x. [DOI] [PubMed] [Google Scholar]
- Jones FA, Comita LS. Neighbourhood density and genetic relatedness interact to determine fruit set and abortion rates in a continuous tropical tree population. Proceedings of the Royal Society B: Biological Sciences. 2008;275:2759–2767. doi: 10.1098/rspb.2008.0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight T, Steets J, Vamosi J, et al. Pollen limitation of plant reproduction: pattern and process. Annual Review of Ecology, Evolution, and Systematics. 2005;36:467–497. [Google Scholar]
- Larson BMH, Barrett SCH. A comparative analysis of pollen limitation in flowering plants. Biological Journal of the Linnean Society. 2000;69:503–520. [Google Scholar]
- Leducq JB, Llaurens V, Castric V, Saumitou-Laprade P, Hardy OJ, Vekemans X. Effect of balancing selection on spatial genetic structure within populations: theoretical investigations on the self-incompatibility locus and empirical studies in Arabidopsis halleri. Heredity (Edinburgh) 2011;106:319–329. doi: 10.1038/hdy.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh M. Grow your own native landscape: a guide to identifying, propagating & landscaping with western washington native plants. Thurston County: Native Plant Salvage Project, Washington State University Cooperative Extension; 1999. [Google Scholar]
- Lienert J. Habitat fragmentation effects on fitness of plant populations – a review. Journal for Nature Conservation. 2004;12:53–72. [Google Scholar]
- Lowe AJ, Boshier D, Ward M, Bacles CFE, Navarro C. Genetic resource impacts of habitat loss and degradation; reconciling empirical evidence and predicted theory for neotropical trees. Heredity. 2005;95:255–273. doi: 10.1038/sj.hdy.6800725. [DOI] [PubMed] [Google Scholar]
- Mimura M, Barbour RC, Potts BM, Vaillancourt RE, Watanabe KN. Comparison of contemporary mating patterns in continuous and fragmented Eucalyptus globulus native forests. Molecular Ecology. 2009;18:4180–4192. doi: 10.1111/j.1365-294X.2009.04350.x. [DOI] [PubMed] [Google Scholar]
- Muirhead CA. Consequences of population structure on genes under balancing selection. Evolution. 2001;55:1532–1541. doi: 10.1111/j.0014-3820.2001.tb00673.x. [DOI] [PubMed] [Google Scholar]
- Nason JD, Ellstrand NC. Lifetime estimates of biparental inbreeding depression in the self- incompatible annual plant Raphanus sativus. Evolution. 1995;49:307–316. doi: 10.1111/j.1558-5646.1995.tb02243.x. [DOI] [PubMed] [Google Scholar]
- Paxman GJ. The maximum likelihood estimation of the number of self-sterility alleles in a population. Genetics. 1963;48:1029–1032. doi: 10.1093/genetics/48.8.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. GenAlEx 6·5: genetic analysis in Excel Population genetic software for teaching and research – an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcher E, Lande R, Fenster C. Loss of gametophytic self-incompatibility with evolution of inbreeding depression. Evolution. 2005;59:46–60. [PubMed] [Google Scholar]
- Ramankutty N, Foley J. Estimating historical changes in global land cover: croplands from 1700 to 1992. Global Biogeochemical Cycles. 1999;13:997–1028. [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2012. [Google Scholar]
- Ritland K. Extensions of models for the estimation of mating systems using n independent loci. Heredity. 2002;88:221–228. doi: 10.1038/sj.hdy.6800029. [DOI] [PubMed] [Google Scholar]
- Robertson IC, Ulappa AC. Distance between pollen donor and recipient influences fruiting success in slickspot peppergrass, Lepidium papilliferum. Canadian Journal of Botany. 2004;82:1705–1710. [Google Scholar]
- Ryman N. chifish: a computer program testing for genetic heterogeneity at multiple loci using chi-square and Fisher's exact test. Molecular Ecology Notes. 2006;6:285–287. [Google Scholar]
- Sassa H, Kakui H, Minamikawa M. Pollen-expressed F-box gene family and mechanism of S-RNase-based gametophytic self-incompatibility (GSI) in Rosaceae. Sexual Plant Reproduction. 2010;23:39–43. doi: 10.1007/s00497-009-0111-6. [DOI] [PubMed] [Google Scholar]
- Schierup MH. The number of self-incompatibility alleles in a finite, subdivided population. Genetics. 1998;149:1153–1162. doi: 10.1093/genetics/149.2.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup MH, Vekemans X, Charlesworth D. The effect of subdivision on variation at multi-allelic loci under balancing selection. Genetical Research. 2000;76:51–62. doi: 10.1017/s0016672300004535. [DOI] [PubMed] [Google Scholar]
- Smouse PE, Dyer RJ, Westfall RD, Sork VL. Two-generation analysis of pollen flow across a landscape. I Male gamete heterogeneity among females. Evolution. 2001;55:260–271. doi: 10.1111/j.0014-3820.2001.tb01291.x. [DOI] [PubMed] [Google Scholar]
- Stacy EA. Cross-fertility in two tropical tree species: evidence of inbreeding depression within populations and genetic divergence among populations. American Journal of Botany. 2001;88:1041–1051. [PubMed] [Google Scholar]
- Stephenson AG. Flower and fruit abortion: proximate causes and ultimate functions. Annual Review of Ecology and Systematics. 1981;12:253–279. [Google Scholar]
- Suarez-Gonzalez A. Reproductive and genetic consequences of fragmentation in chokecherry (Prunus virginiana, L.). Masters Thesis, University of Winnipeg; 2011. [Google Scholar]
- Sutherland BG, Robbins TP, Tobutt KR. Primers amplifying a range of Prunus S-alleles. Plant Breeding. 2004;123:582–584. [Google Scholar]
- Uchiyama K, Goto S, Ide Y. Effects of population density on male and female reproductive success in the wind-pollinated, wind-dispersed tree species Betula maximowicziana. Conservation Genetics. 2009;10:1265–1275. [Google Scholar]
- Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H. Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. The Plant Cell. 2003;15:771–781. doi: 10.1105/tpc.009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicens N, Bosch J. Weather-dependent pollinator activity in an apple orchard, with special reference to Osmia cornuta and Apis mellifera (Hymenoptera: Megachilidae and Apidae) Environmental Entomology. 2000;29:413–420. [Google Scholar]
- Vogler DW, Stephenson AG. The potential for mixed mating in a self-incompatible plant. International Journal of Plant Sciences. 2001;162:801–805. [Google Scholar]
- Vogler DW, Das C, Stephenson AG. Phenotypic plasticity in the expression of self-incompatibility in Campanula rapunculoides. Heredity. 1998;81:546–555. [Google Scholar]
- Winfree R, Aguilar R, Vazquez D, LeBuhn G, Aizen MA. A meta-analysis of bees' responses to anthropogenic disturbance. Ecology. 2009;90:2068–2076. doi: 10.1890/08-1245.1. [DOI] [PubMed] [Google Scholar]
- Wright S. The distribution of self-sterility alleles in populations. Genetics. 1939;24:538–552. doi: 10.1093/genetics/24.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AG, Pickup M. Low S-allele numbers limit mate availability, reduce seed set and skew fitness in small populations of a self-incompatible plant. Journal of Applied Ecology. 2010;47:541–548. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.