Abstract
Apolipoprotein F (ApoF) is a sialoglycoprotein that is a component of the HDL and LDL fractions of human serum. We sought to test the hypothesis that ApoF plays an important role in atherosclerosis in mice by modulating lipoprotein function. Atherosclerosis was assessed in male low density lipoprotein receptor knockout (LDLR KO) and ApoF/LDLR double knockout (DKO) mice fed a Western diet for 16 weeks. ApoF/LDLR DKO mice showed a 39% reduction in lesional area by en face analysis of aortas (p<0.05), despite no significant differences in plasma lipid parameters. ApoF KO mice had reduced expression of Interferon alpha (IFNα) responsive genes in liver and spleen, as well as impaired macrophage activation. Interferon alpha induced gene 27 like 2a (Ifi27l2a), Oligoadenylate synthetases 2 and 3 (Oas2 and Oas3) were significantly reduced in the ApoF KO mice relative to wild type controls. These effects were attributable to hypomorphic expression of Stat2 in the ApoF KO mice, a critical gene in the Type I IFN pathway that is situated just 425 base pairs downstream of ApoF. These studies implicate STAT2 as a potentially important player in atherosclerosis, and support the growing evidence that the Type I IFN pathway may contribute to this complex disease.
Keywords: ApoF, STAT2, Atherosclerosis, Type I IFN
Introduction
Apolipoprotein F (ApoF) is a component of the HDL and LDL fractions of human serum 1 that occurs at concentrations of about 80 μg / mL in human plasma 2. Approximately 75% of the circulating ApoF is associated with small dense HDL3, while the remaining 25% resides on electronegative LDL 3, 4. We previously showed that Apo F overexpression reduces HDL cholesterol levels in mice, and results in HDL particles that are more efficient acceptors of macrophage cholesterol 4. Surprisingly, ApoF deficiency does not substantially alter plasma lipid or lipoprotein levels in mice 5. In the current study, we considered the possibility that Apo F may reside on lipoproteins for a function other than simple lipid transport and may play an important role in atherosclerosis.
Signal Transducer and Activator of Transcription 2 (Stat2) is a transcription factor that plays a critical role in the Type I Interferon signaling. Type I interferons (IFN) consisting of IFN-α and IFN-β are secreted by hematopoietic and non-hematopoietic cell types in response to viral infection 6. Binding of Type I IFN to the IFN α Receptor (IFNAR) induces cross-phosphorylation of tyrosine residues on Tyk2 and Jak1, which are physically associated with the cytoplasmic tails of the receptor 7. This results in phosphorylation of Stat2, which then recruits and phosphorylates Stat1. The active pStat2/pStat1 complex then translocates to the nucleus and associates with (Interferon response factor) IRF-9 to form a trimolecular transcription factor complex also known as ISGF3 8. ISGF3 binds to Interferon Stimulated Response Elements (ISRE) in the promoters of target genes to activate transcription. This activates an orchestrated program of genes that work together to mount the innate antiviral immune response by coordinately shutting down transcription, translation and replication of the virus. For example, 2′–5′ Oligoadenylate Synthetases (OAS) are direct Stat2 target genes, and a key part of the antiviral response. OAS proteins catalyze the formation of 2′–5′ oligoadenylate from ATP, which binds to and activates RNase L. Activated RNase L then degrades viral RNAs to prevent replication and dissemination of the virus9. Activation of the Type I IFN pathway is critical for the innate immune response to viral infection, and is also required for effective priming of the adaptive immune response 10. While Stat1 readily partners with other Stat family members and participates in IFN gamma signaling, Stat2 is more specific to the Type I IFN pathway 11. As such, it offers a unique leverage point to investigate the role of Type I IFN signaling in atherosclerosis.
We report that ApoF-deficient mice are protected from western diet induced atherosclerosis on an LDLR KO background. This was accompanied by substantially reduced expression of Type I IFN responsive genes and impaired macrophage activation. These effects were attributable to reduced expression of the neighboring Stat2 gene; and support an important role for the Type I interferon pathway in atherogenesis.
Methods
Animals
ApoF KO mice generated on a C57BL6 background 5 were crossed with Ldlr KO mice to generate ApoF/Ldlr DKO mice. Mice were housed in an AAALAC accredited animal facility in polycarbonate cages with filter tops and maintained on a 6 am to 6 pm light cycle, with free access to food and water. Mice were fed a chow diet (Purina LabDiet 50100) from weaning until 10–12 weeks of age, and then switched to a western (0.21 % cholesterol 1 % corn oil (w/w), Research Diets D12079B) for 16 weeks until sacrifice. Aortas were harvested by dissection, fixed in formalin and stained with Oil Red O 12. Total lesional area was measured under 0.4X magnification using Image Pro software. Fasting plasma lipids were analyzed on a Roche Cobas-Mira autoanalyzer using commercially available reagents (Sigma). Intraperitoneal (IP) administration of poly(I:C) (Sigma-Aldrich) was performed by injecting wild type and ApoF KO mice with a 200 μl of 0.9% saline or 100 μg of poly(I:C) in saline. Plasma was taken at 6 hours post injection for IFNα measurements, and animals were sacrificed at 24 hours to examine innate immune cell activation status and gene expression in the liver and spleen. Adeno-associated viral (AAV) vectors containing an empty expression cassette (null) or encoding the mouse Apo F protein were generated by the Penn Vector Core and have been described 4. Mice were injected with AAV vectors at a dose of 5 x 1011 genome copies (GC) via IP injection and examined 14 days later. Animal procedures were performed with the approval of the University of Pennsylvania Institutional Animal Care and Use Committee (IACUC).
Microarray
Total liver RNA was used for microarray analysis on the Agilent 4 X 44 Whole Mouse Genome Array. Microarray and data analysis were performed by the Penn Functional Genomics Core. Differential expression was determined using a False Discovery Rate of 10% and a minimum fold change of 1.5. All expression data will be deposited into GEO upon publication of this manuscript.
Real time RT-PCR
Approximately 20–50 mg of tissue were rapidly flash frozen in liquid nitrogen. Total RNA was isolated using the Trizol method according to the manufacturer’s instructions (Invitrogen). One microgram of total RNA was reverse transcribed using random hexameric primers with the Superscript III RT (Invitrogen) according to the manufacturer’s instructions. Real time RT-PCR was performed on five microliters of a 1:50 dilution of the original cDNA sample in a final volume of 12 μL in an ABI 7900 384-well PCR machine. TaqMan primer/probe sets from Applied Biosystems were used to detect mouse ApoF and Stat2, while the remaining genes were detected using SYBR green detection (list of primers in supplemental table 1). The relative quantity of each mRNA was calculated using the delta delta CT method with β-Actin as the housekeeping gene.
Flow cytometry
Spleens were minced with a razor blade and digested with collagenase (0.5mg/ml) and DNase (20ug/ml) for 30 mins at 37°C before being passed through a 100 micron cell strainer to obtain a single cell suspension. Red blood cells were then lysed using ACK lysing buffer (Gibco). Cells were stained with Live/dead fixable stain in PBS (Invitrogen) before surface antibodies were added. To examine antigen presenting cells (APCs), a dump gate of anti-CD3, anti-CD5 and anti-CD19 were used to gate out T and B cells. Macrophages were identified by the high expression of F4/80 and CD11b. Cell surface expression of CD80 was used as a marker of macrophage activation. Samples were collected using the LSR II flow cytometer (BD) and analyzed using Flowjo.
Immunohistochemistry
Paraffin embedded formalin fixed heart sections were prepared by the Penn Cardiovascular Institute Histology Core. Sections of the aortic sinus were subjected to staining with hematoxylin and eosin, Masson’s Trichrome, or Rat anti-Mac 3 IgG (BD Pharmingen 553322), Goat anti Sm22 alpha IgG (Abcam ab10135), and Mouse anti-Stat2 IgG (Millipore 07-140) antibodies. Mac3 and Sm22 were visualized using a horseradish peroxidase conjugated secondary antibody (ImPRESS TSA system), and Stat2 was imaged by immunofluorescence. Analysis of positively stained areas was performed using ImageJ software.
Statistics
All data are reported as the mean +/− standard deviation. Comparisons between groups were performed using a two-tailed student’s t-test. One way ANOVA was used for experiments involving 3 or more groups, and Tukey’s posttest was then used to test for significant differences between individual groups. In all cases, significance is assigned at p < 0.05.
Results
Targeted deletion of ApoF results in hypomorphic expression of Stat2 and ApoN
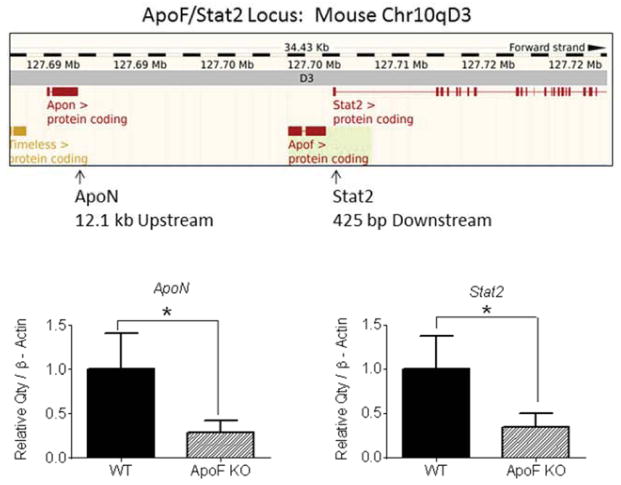
We previously generated mice with a targeted deletion of the ApoF gene5 in which the ApoF coding sequence was replaced with a Beta galactosidase reporter cassette. Surprisingly, deletion of the ApoF gene had no discernible effect on circulating lipids or lipoproteins5. The murine ApoF gene is situated 12.1 kb downstream of ApoN (an uncharacterized apolipoprotein), and only 425 upstream of Stat2, a critical mediator of Type I IFN signaling. During the course of these studies we found that the ApoF KO mice also exhibited significantly reduced expression of Stat2 and ApoN in the liver: Stat2: −66%, p<0.05; ApoN: −71%, p<0.05 (Figure 1, Supplemental Figure 1). The ApoF KO mice were generated isogenically using C57BL6 derived ES cells, meaning that these expression differences could not be due to passenger gene effects from the more commonly used 129 derived ES cells. Rather, it seemed more likely that the manipulation of the ApoF gene itself directly resulted in silencing of the neighboring ApoN and Stat2 genes, possibly through deletion of important cis-acting regulatory elements (Supplemental Figure 2).
Figure 1. Decreased expression of flanking genes at the mouse ApoF locus.
The positions of ApoN and Stat2 are shown relative to ApoF using a display from the Ensembl genome browser. Message levels of ApoN and Stat2 in mouse liver were measured in wild type and ApoF KO mice by real time RT-PCR with β-Actin as the housekeeping gene.
Microarray analysis reveals alterations in the Type I IFN signaling pathway
Apo F is expressed exclusively by the liver1, 5 where it is secreted into the circulation and then attaches to lipoproteins. Since there was no discernible lipid phenotype in the ApoF KO mice, we performed a microarray analysis on livers from these animals to gain insight into the function of Apo F. Interestingly, only a very short list of transcripts showed statistically significant differences in regards to ApoF genotype based on a False Discovery Rate (FDR) threshold of 5% (Table 1). Surprisingly, the list was enriched in interferon alpha responsive genes (blue), including Interferon Induced transcript 27 like 2a (Ifi27l2a), and a number of 2′–5′ oligoadenylate synthetases (Oas3, Oas2, Oasl2, and Oas1a) which were significantly lower in the ApoF KO mice.
Table 1. Microarray results.
Genes are listed in order of lowest false discovery rate (FDR) and then greatest fold-change from top to bottom. Genes with a false discovery rate of less than 5% were considered statistically significant (above the bold line). Highlighted in blue are known IFNα target genes or genes with activities involved in innate immunity.
| Gene Name | Description | FC | FDR | Adj P. |
|---|---|---|---|---|
|
| ||||
| Apof | Apolipoprotein F | - | 0 | 0.00000 |
| Apon | Apolipoprotein N | −7.41 | 0 | 0.00740 |
| Ifi27l2a | IFNα-induced protein 27-like 2a | −5.46 | 0 | 0.01062 |
| Gm4955 | −4.53 | 0 | 0.01920 | |
| ENSMUST37976 | −4.37 | 0 | 0.01482 | |
| Oas3 | 2′–5′ oligoadenylate synthetase 3 | −3.70 | 0 | 0.01062 |
| Oas2 | 2′–5′ oligoadenylate synthetase 2 | −3.25 | 0 | 0.06874 |
| Oasl2 | 2′–5′ oligoadenylate synthetase-like 2 | −2.92 | 0 | 0.00740 |
| Oas1a | 2′–5′ oligoadenylate synthetase-like 2a | −2.85 | 0 | 0.00740 |
| Oas1f | 2′–5′ oligoadenylate synthetase 1f | −2.51 | 0 | 0.03218 |
| D030028A08Rik | −2.48 | 0 | 0.03218 | |
| Oas1a | 2′–5′ oligoadenylate synthetase-like 1a | −2.47 | 0 | 0.01987 |
| Gm7998 | −2.10 | 0 | 0.03218 | |
| Trim79 | Tripartite Motif Protein 79 | −1.92 | 0 | 0.05003 |
|
| ||||
| EG545385 | −4.07 | 5.44 | 0.16180 | |
| LOC100044227 | −3.18 | 5.44 | 0.19062 | |
| Erdr1 | Erythroid differentiation regulator 1 | −3.14 | 5.44 | 0.19062 |
| Gm5431 | −2.89 | 5.44 | 0.20147 | |
| Cd200r4 | Cluster of Differentiation 200 receptor 4 | −2.39 | 5.44 | 0.11829 |
| Ifit3 | IFNα-induced protein with tetratricopeptide repeats 3 | −2.02 | 5.44 | 0.13465 |
| Tex14 | Testes expressed gene 14 | −2.02 | 5.44 | 0.15613 |
| Xaf1 | XIAP-associated factor 1 | −2.01 | 5.44 | 0.12067 |
| Ly6a | lymphocyte antigen 6 complex, locus A | −1.94 | 5.44 | 0.16180 |
| Slfn8 | Schlafen 8 | −1.91 | 5.44 | 0.16789 |
| Ccl3 | Chemokine (C-C motif) Cigand 3 | −1.83 | 5.44 | 0.11810 |
| Trim30 | Tripartite Motif Protein 30 | −1.78 | 5.44 | 0.12797 |
| Csprs | Component of Sp100_rs | −1.71 | 5.44 | 0.12797 |
| LOC100041903 | −1.67 | 5.44 | 0.15613 | |
| Phf11 | PHD Finger Protein 11 | −1.67 | 5.44 | 0.13465 |
| Mx1 | Interferon-Induced GTP-binding Protein Mx1 | −2.79 | 8.65 | 0.21366 |
| Siglec1 | Sialoadhesin (CD169) | −2.50 | 8.65 | 0.23638 |
| LOC631406 | −2.30 | 8.65 | 0.20973 | |
| Elovl3 | Elongation of very long chain fatty acids like 3 | −2.03 | 8.65 | 0.19062 |
| AA414992 | −1.95 | 8.65 | 0.19557 | |
| Oasl1 | 2′–5′ oligoadenylate synthetase-like 1 | −1.93 | 8.65 | 0.20973 |
| Gm9629 | −1.86 | 8.65 | 0.19557 | |
| Gm684 | −1.80 | 8.65 | 0.19062 | |
| ENSMUST100417 | −1.75 | 8.65 | 0.20973 | |
Impaired Type I IFN response and macrophage activation in the ApoF KO mice
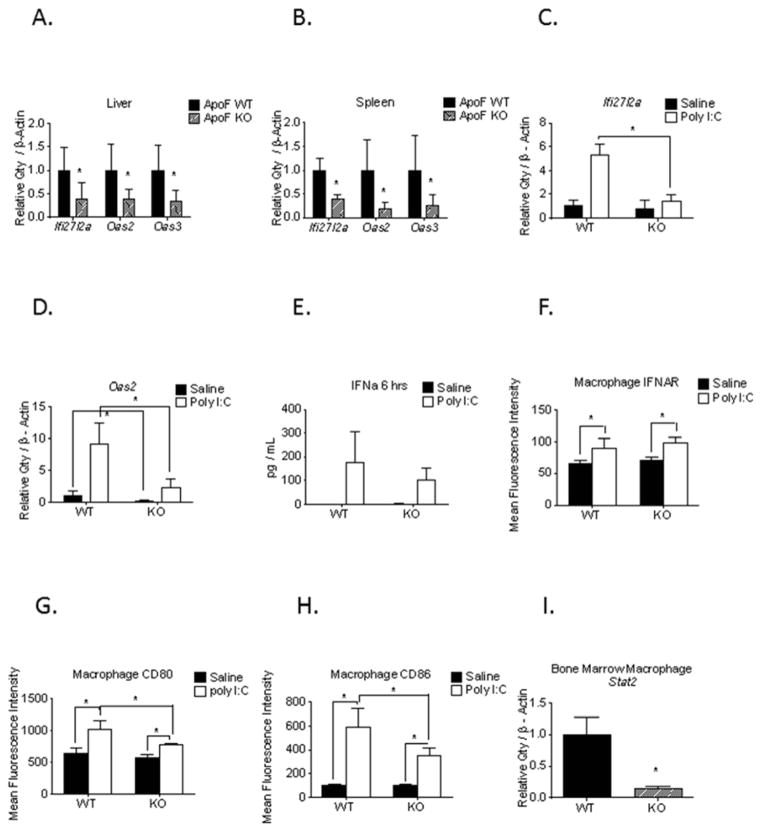
Real time RT-PCR confirmed significantly lower mRNA expression of Ifi27l3a, Oas2, and Oas3 in both the livers and the spleens of the ApoF KO mice (Figure 2 A,B). We next challenged these animals with polyinosinic:polycytidilic acid (poly I:C) to assess whether the Type I interferon (IFN) pathway is functionally intact in the ApoF KO mice. Poly I:C is a Toll Like Receptor 3 (TLR3) agonist that mimics the double stranded RNA ligands present during a viral infection, resulting in secretion of IFNα/β. Poly(I:C) challenge produced a robust upregulation of Ifi27l2a and Oas2 mRNA levels in the spleen at 24 hours post-injection which was substantially blunted in the ApoF KO mice (Figure 2 C,D). Interestingly, plasma levels of IFN alpha and cell surface expression of Interferon alpha receptor (IFNAR) both increased after Poly I:C treatment, but did not differ between the WT and ApoF KO mice (Figure 2 E,F). Poly I:C treatment resulted in a robust increase in macrophage CD80 and CD86 expression, early activation co-stimulatory molecules, which was dramatically blunted in the ApoF KO mice (Figure 2 G,H). This led us to suspect that these differences may actually be due to a cell intrinsic effect in immune cells. In line with this, we observed an 85% decrease in Stat2 mRNA expression in isolated bone marrow macrophages (BMM) (Figure 2I). This is consistent with the fact that Ifi27l2a, Oas, and Mx1 are well established direct transcriptional targets of Stat2. We therefore surmised that the decrease in expression of these genes and impaired immune cell activation could be due to reduced Stat2 expression rather than a signaling event caused by loss of Apo F.
Figure 2. Impaired activation of the Type I IFN pathway in ApoF KO mice.
Total RNA was isolated from the liver and spleen of wild type and ApoF KO mice for validation by real time RT-PCR. Message levels of Ifi27l2a, Oas2, and Oas3 in A) liver and B) spleen normalized to β-Actin. Spleen RNA was isolated from wild type and ApoF KO mice that received an intraperitoneal injection of either saline or 100 μg of poly(I:C). Message levels of C) Ifi27l2a and D) Oas2 were measured by real time RT-PCR. E) Plasma levels of IFN α at 6 hours after poly(I:C) injection were measured by ELISA. Cell surface expression of F) Interferon alpha receptor, G) CD80, and H) CD86 on splenic macrophages that were harvested twenty four hours after poly(I:C) administration. I) Message levels of Stat2 in primary bone marrow derived macrophages from wild type and ApoF KO mice.
ApoF Deficient mice are resistant to Western Diet induced atherosclerosis
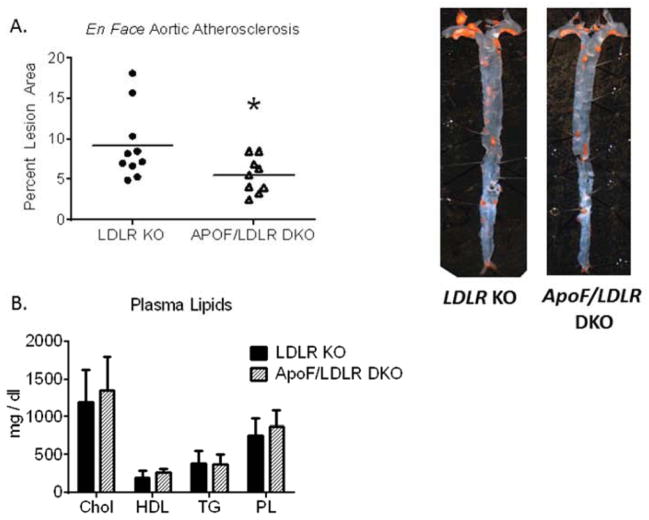
We sought to test the hypothesis that Apo F may affect lipoprotein function and hence susceptibility to atherosclerosis. To accomplish this, male Ldlr KO and ApoF/Ldlr DKO mice were fed a Western Diet for 16 weeks for assessment of atherosclerosis by en face analysis and oil red O staining. We observed a significant 39% reduction in the total lesional area in the ApoF/Ldlr DKO mice relative to controls, p < 0.05 (Figure 3 A). Aortic roots contained complex lesions composed of necrotic core, cholesterol crystals, macrophage foam cells, and smooth muscle cells (Supplemental Figure 3). Smooth muscle cell (Sm22) and collagen (blue Trichrome) content did not significantly differ by genotype (Supplemental Figures 4,5). We did however observe a 38% reduction in macrophage positive area in the Apof/Ldlr DKO mice (p< 0.05) (Supplemental Figure 6,7). Immunostaining for Stat2 produced robust signal on the aortic valves, with less organized positive staining dispersed throughout the lesions (Supplemental Figure 8). Perhaps most intriguing, the ApoF/Ldlr DKO mice did not exhibit any changes in plasma lipid parameters that could explain the decreased lesion burden observed by en face analysis (Figure 3 B).
Figure 3. Decreased Atherosclerosis in ApoF Deficient Mice.
Male LDLR KO (n= 10) or ApoF/LDLR DKO (n = 9) mice were fed a Western diet for 16 weeks for assessment of atherosclerosis development. A) En face lesional area in Oil Red O stained aortas from LDLR KO and ApoF/LDLR DKO mice (* p<0.05). B) Fasted plasma lipid parameters after 12 weeks on Western Diet.
Reconstitution of Apo F does not restore expression of Stat2 and Type I IFN responsive genes
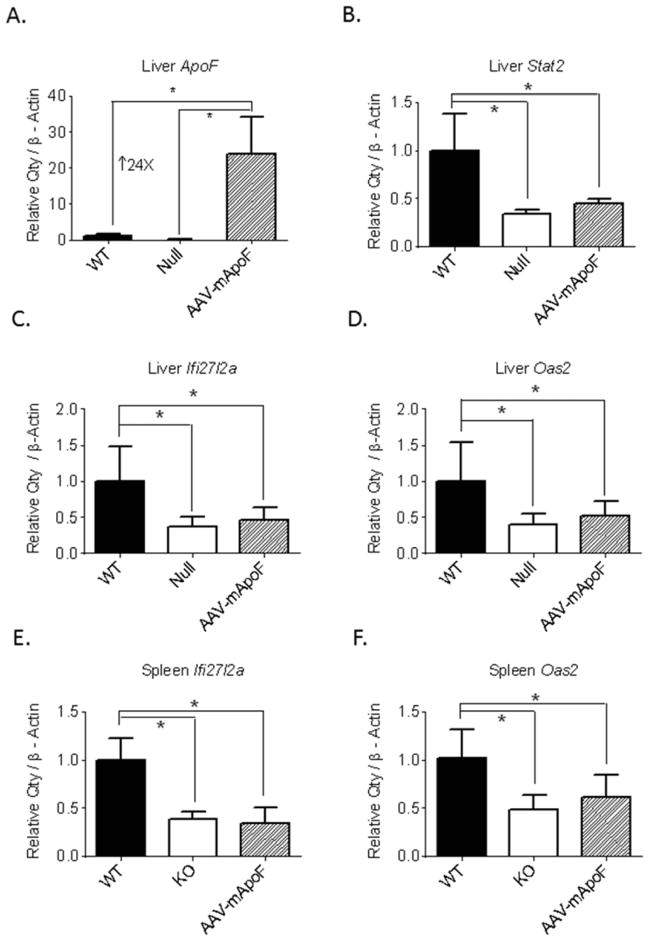
We performed an experiment to test whether the loss of Apo F could be responsible for the immune phenotype and altered Type I IFN signaling in these animals. We used an AAV8 based vector driven by the liver specific thyroxine binding globulin (TBG) promoter to overexpress Apo F only in the livers of the ApoF knockout mice. In parallel, a control group of ApoF KO mice was given an empty vector (Null). AAV-mediated delivery of Apo F resulted in a 24-fold increase in liver ApoF mRNA levels relative to wild type mice (Figure 4A). In contrast, liver Stat2 mRNA levels in these animals were unaffected by ApoF overexpression (Figure 4B). Apo F overexpression did not rescue the lower expression of IFNα-responsive genes in the liver (Figure 4 C,D) or in the spleen (Figure 4 E,F), indicating that these gene expression differences are Apo F-independent.
Figure 4. AAV-mediated overexpression of ApoF in the ApoF knockout mice.
ApoF KO mice were injected with 5 x 1011 genome copies of either an empty AAV vector (Null), or one encoding mouse ApoF (mApoF) under the control of a liver specific promoter. A) Expression of ApoF and Stat2 in the livers of these mice. Expression of C) Ifi27l2a and D) Oas2 in the livers of these animals. Expression of E) Ifi27l2a and F) Oas2 in the spleen (*indicates significant differences at p<0.05 by one way ANOVA followed by Tukey’s post-test).
Discussion
The overall goal of this study was to determine whether physiological levels of Apo F expression have an impact on atherosclerosis. We found that the ApoF KO mice are protected from Western Diet induced atherosclerosis on an Ldlr deficient background- an effect that could not be explained by changes in plasma lipids or lipoproteins. We discovered that the ApoF KO mice have expression of IFNα responsive genes, a diminished response to poly(I:C) challenge, and impaired macrophage activation. These effects are most likely due to hypomorphic expression of the neighboring Stat2 gene.
The reduced expression of Ifi27l2a, Oas2, Oas3 and other genes involved in the innate antiviral response agrees with studies in other Stat2 deficient mouse models. A Stat2 hypomorphic mouse designated “P117” harbors a splice site mutation resulting in only trace amounts of full length Stat2 protein expression 13. Primary mouse embryonic fibroblasts (PMEFs) from these animals are completely incapable of activating Oas and Pkr expression in response to IFNα. This is accompanied by impaired dendritic cell activation13. Mice with a targeted deletion of Stat2 have been reported and studied extensively for their response to viral infection 10, 14, 15. PMEFs from the Stat2 deficient mice do not activate the Oas and Mx genes in response to IFNα stimulation. Importantly, Oas and Mx expression are induced less than 50% in the Stat2 heterozygous mice 10. These defects in IFNα signaling render the P117 mice as well as the Stat2 KO mice particularly susceptible to viral infections. Taken together, the 70–85% reduction in Stat2 mRNA in the ApoF KO mice is adequate to explain the reduced expression of IFNα-responsive genes and the defective macrophage activation.
The lower levels of ApoN and Stat2 are most likely a direct result of the targeting of the ApoF gene. It is worth noting that Stat2 resides only 425 bp downstream of the ApoF gene. Despite our best efforts to generate these mice on a pure C57BL6 background to avoid strain differences and passenger genes, unintended regulatory effects of the insertion may have been unavoidable. ChiP Sequencing data on the human genome indicates the presence of acetylated histones within the second exon of ApoF that extend through the 3′UTR and well into the 425 bp gap between APOF and STAT2 16, suggesting that the APOF gene is contiguous with the STAT2 promoter (Supplemental Figure 2). It is plausible that important cis-acting regulatory elements contained within the ApoF coding sequence, intron or 3′UTR were removed by our targeting vector. It is also conceivable that the silencing of ApoN and Stat2 is due to negative effects of the beta galactosidase reporter cassette or the neomycin resistance (Neo) gene. There are other examples of local silencing of neighboring genes at a targeted locus by insertion of foreign DNA sequences such as Neo 17, 18.
Recent data from other mouse models support a causal relationship between Type I IFN signaling and atherosclerosis. Ldlr KO mice fed a western diet have increased atherosclerosis with low dose IFN α treatment. In this study, treatment with IFN α increases atherosclerotic plaque area (↑64%, p<0.008) relative to saline injected control mice 19. Likewise, IFN β administration promoted atherosclerosis in both a collar-induced model in ApoE KO mice, as well as in western diet fed Ldlr KO mice20. In contrast to the report with IFN α stimulation, treatment with IFN β did not alter plasma cholesterol levels. Bone marrow transplant experiments showed that Ldlr KO mice with a myeloid-specific deletion of the Ifnar gene were in fact protected from atherosclerosis in the absence of exogenously supplied IFN β. Interestingly, the control Ldlr KO mice in this study expressed the Stat2 target genes Oas1, Oas2, Mx1 and Irf9 in aortic arches, supporting a role for constitutive activation of this pathway- even in the absence of viral infection. Plasmacytoid dendritic cells (pDCs) are the most potent producers of Interferon alpha in the body, producing as much as 1000-fold more than other immune cell populations21. Recent work has demonstrated that pDCs exist within atherosclerotic lesions, and that antibody mediated depletion of these cells offers protection from atherosclerosis22. Accordingly, these authors also found that specific stimulation of pDCs with CpG oligonucleotides led to an increase in plaque burden in ApoE KO mice fed a high fat diet22. It is tempting to speculate that pDCs present within the lesion may be the source of the endogenous IFN alpha, secreted in response to either viral infection or necrotic cell death during lesion formation. Collectively, these data support a proatherogenic role for IFN α/β in atherogenesis, both of which require obligate signaling through Stat2.
The reduced macrophage content in the lesions of the ApoF/Stat2 DKO mice (Supplemental Figures 4 and 7), combined with the 85% reduction in macrophage Stat2 expression (Figure 2I), are consistent with an important role for monocyte/macrophage Type I Interferon signaling in atherogenesis. The impaired macrophage activation in these animals raises the possibility that Stat2 may act at the level of monocyte infiltration or foam cell formation. The lack of a difference in Stat2 immunostaining by genotype may be due to the advanced nature of the lesions (Supplemental Figure 8), which is not ideally suited for examining early processes such as monocyte infiltration and foam cell formation. However, it is also worth noting that other cell types of the vessel wall may also play a role (such as endothelial and smooth muscle cells), and this merits further investigation in appropriate model systems.
Genetic evidence linking STAT2 to CAD or MI risk in humans has not yet been reported. Interestingly, patients with systemic lupus erythematosus (SLE) have markedly elevated Type I IFN levels and downstream signalling 23–27, which may contribute to their increased CAD risk 28. Likewise, a growing body of literature also supports a relationship between viral infections (particularly cytomegalovirus) and atherosclerotic disease risk and severity 29. It is tempting to speculate that elevated Type I IFN signaling through STAT2 could be an underlying mechanistic link between autoimmune diseases, viral infection, and atherosclerosis. Future studies using the Stat2 knockout mice will allow us to cleanly and definitively assess the role of this protein in atherosclerosis and the molecular mechanisms involved.
Supplementary Material
Highlights.
ApoF KO mice are protected from western diet induced atherosclerosis on an LDLR KO background, via a lipid independent mechanism.
Reduced atherosclerosis is accompanied by decreased expression of IFN alpha responsive genes and impaired immune cell activation.
Deletion of the ApoF gene results in hypomorphic expression of Stat2, supporting a role for the Type I IFN response in atherogenesis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Day JR, Albers JJ, Gilbert TL, et al. Purification and molecular cloning of human apolipoprotein F. Biochem Biophys Res Commun. 1994;203:1146–1151. doi: 10.1006/bbrc.1994.2302. [DOI] [PubMed] [Google Scholar]
- 2.Morton RE, Gnizak HM, Greene DJ, et al. Lipid transfer inhibitor protein (apolipoprotein F) concentration in normolipidemic and hyperlipidemic subjects. J Lipid Res. 2008;49:127–135. doi: 10.1194/jlr.M700258-JLR200. [DOI] [PubMed] [Google Scholar]
- 3.Bancells C, Canals F, Benitez S, et al. Proteomic analysis of electronegative low-density lipoprotein. J Lipid Res. 2010;51:3508–3515. doi: 10.1194/jlr.M009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagor WR, Brown RJ, Toh SA, et al. Overexpression of apolipoprotein F reduces HDL cholesterol levels in vivo. Arterioscler Thromb Vasc Biol. 2009;29:40–46. doi: 10.1161/ATVBAHA.108.177105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lagor WR, Fields DW, Khetarpal SA, et al. The effects of apolipoprotein F deficiency on high density lipoprotein cholesterol metabolism in mice. PloS one. 2012;7:e31616. doi: 10.1371/journal.pone.0031616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 7.Yeh TC, Pellegrini S. The Janus kinase family of protein tyrosine kinases and their role in signaling. Cell Mol Life Sci. 1999;55:1523–1534. doi: 10.1007/s000180050392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell IL. Cytokine-mediated inflammation, tumorigenesis, and disease-associated JAK/STAT/SOCS signaling circuits in the CNS. Brain Res Brain Res Rev. 2005;48:166–177. doi: 10.1016/j.brainresrev.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Silverman RH. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. Journal of virology. 2007;81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park C, Li S, Cha E, et al. Immune response in Stat2 knockout mice. Immunity. 2000;13:795–804. doi: 10.1016/s1074-7613(00)00077-7. [DOI] [PubMed] [Google Scholar]
- 11.Schindler C, Plumlee C. Inteferons pen the JAK-STAT pathway. Semin Cell Dev Biol. 2008;19:311–318. doi: 10.1016/j.semcdb.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kassim SH, Li H, Vandenberghe LH, et al. Gene therapy in a humanized mouse model of familial hypercholesterolemia leads to marked regression of atherosclerosis. PLoS One. 2010;5:e13424. doi: 10.1371/journal.pone.0013424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen LS, Wei PC, Liu T, et al. STAT2 hypomorphic mutant mice display impaired dendritic cell development and antiviral response. J Biomed Sci. 2009;16:22. doi: 10.1186/1423-0127-16-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perry ST, Buck MD, Lada SM, et al. STAT2 mediates innate immunity to Dengue virus in the absence of STAT1 via the type I interferon receptor. PLoS Pathog. 2011;7:e1001297. doi: 10.1371/journal.ppat.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashour J, Morrison J, Laurent-Rolle M, et al. Mouse STAT2 restricts early dengue virus replication. Cell Host Microbe. 2010;8:410–421. doi: 10.1016/j.chom.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olson EN, Arnold HH, Rigby PW, et al. Know your neighbors: three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell. 1996;85:1–4. doi: 10.1016/s0092-8674(00)81073-9. [DOI] [PubMed] [Google Scholar]
- 18.Fiering S, Epner E, Robinson K, et al. Targeted deletion of 5′HS2 of the murine beta-globin LCR reveals that it is not essential for proper regulation of the beta-globin locus. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 19.Levy Z, Rachmani R, Trestman S, et al. Low-dose interferon-alpha accelerates atherosclerosis in an LDL receptor-deficient mouse model. Eur J Intern Med. 2003;14:479–483. doi: 10.1016/j.ejim.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 20.Goossens P, Gijbels MJ, Zernecke A, et al. Myeloid type I interferon signaling promotes atherosclerosis by stimulating macrophage recruitment to lesions. Cell Metab. 2010;12:142–153. doi: 10.1016/j.cmet.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Siegal FP, Kadowaki N, Shodell M, et al. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- 22.Doring Y, Manthey HD, Drechsler M, et al. Auto-antigenic protein-DNA complexes stimulate plasmacytoid dendritic cells to promote atherosclerosis. Circulation. 2012;125:1673–1683. doi: 10.1161/CIRCULATIONAHA.111.046755. [DOI] [PubMed] [Google Scholar]
- 23.Hooks JJ, Moutsopoulos HM, Geis SA, et al. Immune interferon in the circulation of patients with autoimmune disease. N Engl J Med. 1979;301:5–8. doi: 10.1056/NEJM197907053010102. [DOI] [PubMed] [Google Scholar]
- 24.Hooks JJ, Jordan GW, Cupps T, et al. Multiple interferons in the circulation of patients with systemic lupus erythematosus and vasculitis. Arthritis Rheum. 1982;25:396–400. doi: 10.1002/art.1780250406. [DOI] [PubMed] [Google Scholar]
- 25.Dall’era MC, Cardarelli PM, Preston BT, et al. Type I interferon correlates with serological and clinical manifestations of SLE. Ann Rheum Dis. 2005;64:1692–1697. doi: 10.1136/ard.2004.033753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santer DM, Yoshio T, Minota S, et al. Potent induction of IFN-alpha and chemokines by autoantibodies in the cerebrospinal fluid of patients with neuropsychiatric lupus. J Immunol. 2009;182:1192–1201. doi: 10.4049/jimmunol.182.2.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett L, Palucka AK, Arce E, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manzi S, Meilahn EN, Rairie JE, et al. Age-specific incidence rates of myocardial infarction and angina in women with systemic lupus erythematosus: comparison with the Framingham Study. Am J Epidemiol. 1997;145:408–415. doi: 10.1093/oxfordjournals.aje.a009122. [DOI] [PubMed] [Google Scholar]
- 29.Ji YN, An L, Zhan P, et al. Cytomegalovirus infection and coronary heart disease risk: a meta-analysis. Mol Biol Rep. 2012;39:6537–6546. doi: 10.1007/s11033-012-1482-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.