Abstract
Purpose
Active surveillance (AS) is a viable patient option for prostate cancer where a clinical determination of low-risk and presumably organ-confined disease can be made. In an effort to standardize risk stratification schemes, the National Comprehensive Cancer Network (NCCN) has provided guidelines for the AS option. Our purpose was to determine the effectiveness of expressed prostatic secretion (EPS) biomarkers in detecting occult risk factors in NCCN AS candidates.
Materials and Methods
EPS specimens were obtained prior to Robot-Assisted Radical Prostatectomy (RARP). Secretion capacity biomarkers: total RNA and EPS specimen volume were measured by standard techniques. RNA expression biomarkers: TXNRD1-mRNA, PSA-mRNA, TMPRSS2:ERG fusion mRNA and PCA3-mRNAs were measured by quantitative reverse-transcription PCR.
Results
Of the 528 patients from whom EPS was collected, 216 were eligible for AS under NCCN guidelines. Variable Selection in logistic regression identified two models, one featuring Type III and Type VI TMPRSS2:ERG variants, and one featuring two secretion capacity biomarkers. Of the two high performing models, the secretion capacity model was the most effective in detecting patients within this group that were upstaged or both upstaged and upgraded. It reduced the risk of upstaging in patients with a negative test by nearly 8 fold, and reduced the risk of being both upstaged and upgraded by about 5 fold, while doubling the prevalence upstaging in the positive test group.
Conclusions
Non-invasive EPS testing may improve patient acceptance of AS by dramatically reducing the presence of occult risk factors among patients eligible for AS under NCCN guidelines.
Keywords: NCCN Guidelines, Active Surveillance, Expressed Prostatic Secretions, Secretion Capacity, TMPSS2:ERG Fusions
Introduction
Screening for PCa appears to have produced considerable over-diagnosis1–3. Moreover, the reported 21% decrease in overall mortality from PCa4 may be due in part to the increased detection of low risk cancers5 resulting in over-treatment3, 5 of patients whose cancers would not have been life threatening, often involving significant morbidity6. However, over-treatment is likely to continue as a patient driven phenomenon unless better non-invasive tests for the detection and stratification of PCa are developed.
AS has been shown to be a viable patient alternative to over-treatment where a clinical determination of low-risk and presumably organ-confined disease can be made. Several classification schemes for AS eligibility7–11 have proven successful in multiple studies with significant numbers of patients. Although there is strong evidence that implementation of these stratification schemes in AS programs would do no harm in low risk patients, clinical practice appears to be guided by a personal opinion approach that often favors curative intervention as a patient choice12, effectively rendering AS an underutilized approach.
The availability of the AS option varies from about 15% for the most stringent criteria to-about 50% of the diagnosed population for the more inclusive criteria7–11. In an effort to standardize these classification schemes the NCCN has recently provided guidelines for AS. Unfortunately, the NCCN eligibility requirements are met by significant numbers of patients with non-organ confined and higher-risk disease that appears to have been undetected13.
Non-invasive testing in NCCN eligible patients could potentially augment current eligibility requirements and improve the acceptance of AS. Most tests for PCa detection and stratification exploit the anatomical alterations in the gland itself. For example, the DRE depends on the palpable nature of larger tumor masses, while the serum PSA test in its many incarnations14–16 exploits anatomical damage to the glandular epithelium and the basement membrane that permits luminal PSA to enter the circulation. However, neither DRE nor PSA testing have been sucessful in statifying patients in AS17.
Pressure applied to the prostate is known to release PSA into the circulation even in normal prostates lacking anatomical damage18, and this phenomenon has been considered for risk assessment. For example, nucleic acids released during prostatic massage in exosomes, stromal or epithelial cells or cell fragments have been used successfully in PCa screening19–22. Currently, two types of specimen are commonly considered. The most widely employed specimen of this type is urine collected after attentive DRE (i.e. post-DRE urine). It has been used primarily in PCa detection, based on nucleic acid amplification to detect expression of tumor biomarkers like PCA3 and TMPRSS2:ERG19, 21, 22, as well as hypermethylation of selected gene promoters23. EPS can also be collected directly by milking the urethra after prostatic massage. This specimen is not diluted in urine and generally has a higher content of these same markers. Although it has received less attention in the PCa literature, it has been sucessfully employed in PCa detection using telomerase RNA, TMPRSS2:ERG fusions, PCA3 and DNA methylation analysis20, 24. A key disadvantage of post-DRE urine is the requirement for normalization of variables like PCA3 level19, 21, 22. This problem arises because urine catch is generally incomplete, so a dilution factor for prostatic secretions in urine cannot be obtained. With EPS, the collected volume is not diluted and can be measured directly. Consequently, variables like collected volume, total nucleic acid, or the total amount of a metabolite can be accurately estimated only by using EPS specimens. Variables of this type are of interest in light of the potential for an extensive tumor to impair the secretory function of the prostate gland.
In this report, we prospectively collected data on EPS specimens obtained from 528 patients who subsequently underwent RARP. Of these 216 were eligible for AS based on the NCCN guidelines. Our results identify two models that are effective in reducing the risk of finding an upstaged tumor or a tumor that has been both upstaged and upgraded in the NCCN eligible group.
Materials and Methods
Patients
Between 2008 and 2012, men were identified with the inclusion criteria of age 18 or older, and a diagnosis of PCa, followed by RARP as primary treatment. 536 men who met these criteria were consented under an Institutional Review Board-approved protocol for the collection and evaluation of biomarkers in EPS prior to PARP. Of the 536 consented subjects, 528 had presurgical clinical staging and evaluable ECE status. The characteristics of the study population and the NCCN criteria for AS eligibility are given in Table 1.
Table 1.
Patient Demographics: Full cohort (N = 528), NCCN AS-Ineligible (N = 312), and NCCN AS Eligible (N = 216).
| Full cohort (N = 528) | Non-NCCN (N = 312) | * NCCN pts (N = 216) | ||||
|---|---|---|---|---|---|---|
| Factor | Mean ± SE | Mean ± SE | Mean ± SE | |||
| Age | 63y ± 0.3y | 63y ± 0.4y | 62y ± 0.6y | |||
| Pre-Bx Serum PSA | 7.0 ± 0.4 | 8.2 ± 0.7 | 5.3 ± 0.2 | |||
| Factor | N | (%) | N | (%) | N | (%) |
| Serum PSA | ||||||
| <10 | 456 | (86.4) | 243 | (77.9) | 213 | (98.6) |
| 10–20 | 58 | (11.0) | 56 | (17.9) | 2 | (0.5) |
| > 20 | 14 | (2.6) | 13 | (4.2) | 1 | (0.9) |
| Gleason Sum | ||||||
| 6 | 231 | (43.7) | 25 | (8.0) | 206 | (95.4) |
| 7 | 255 | (48.3) | 245 | (78.5) | 10 | (4.6) |
| 8–9 | 42 | (8.0) | 42 | (13.5) | 0 | (0.0) |
| Ethnicity | ||||||
| African-American | 23 | (4.4) | 14 | (4.5) | 9 | (4.2) |
| Asian | 29 | (5.5) | 17 | (5.4) | 12 | (5.5) |
| Caucasian | 470 | (89.0) | 279 | (89.4) | 191 | (88.4) |
| Native American | 1 | (0.2) | 0 | (0.0) | 1 | (0.5) |
| Other | 5 | (0.9) | 2 | (0.6) | 3 | (1.4) |
| T-stage | ||||||
| T1a | 2 | (0.4) | 1 | (0.3) | 1 | (0.4) |
| T1c | 425 | (80.5) | 241 | (77.2) | 184 | (85.2) |
| T2a | 77 | (14.6) | 46 | (14.7) | 31 | (14.4) |
| T1b or Other T2 | 24 | (4.5) | 24 | (7.7) | 0 | (0.0) |
| Upstaged | 97 | (18.4) | 82 | (26.3) | 15 | (6.9) |
| Upgraded | 121 | (23.0) | 31 | (9.9) | 90 | (42.1) |
NCCN AS eligibility criteria are defined as follows. Low risk: T1-T2a, Gleason ≤ 6, PSA <10, OR
Intermediate risk: for < 10 years of life expectancy, T2b-T2c or Gleason 7 or PSA 10–20. NCCN recommends AS for very low risk patients; these patients are also eligible for AS and are contained in the low risk group as defined by NCCN.
EPS Collection
Consented patients underwent an attentive digital rectal examination prior to surgery. This is described as sweeping the index finger three times over the right and left lobes, and then three times from the apex to the base of the prostate with attentive pressure during a maximum of 30 seconds manipulation. EPS specimens were collected by milking the urethra, and immediately placed on ice for transport to the laboratory within 1 hour of collection.
Expressed Volume Measurement
Upon arrival in the laboratory, specimens were spun to the bottom of the collection tube in a low speed centrifuge. The fluid volume was measured by drawing it up into a micropipette (Gilson, Middleton, WI).
RNA Preparation
Once the volume was measured, the specimen was suspended in 3ml Phosphate Buffered Saline. Cells and debris were collected by centrifugation and both the pellet and supernatant fluid were frozen and stored at −80° C. RNA was prepared from the pellet with the RNEasy® Mini Kit (Qiagen®, Hilden Germany). RNA concentrations were measured with a Nanodrop 2000 Spectrophotometer (Thermo Scientific, Wilmington, DE).
cDNA Preparation
cDNA was prepared with the Invitrogen™ SuperScript® VILO™ cDNA Synthesis Kit. Synthesis of PCR amplifiable product DNA was determined to be linear over the range 0 to 200ng of input RNA. A maximum of 200ng input RNA from each specimen was used in cDNA synthesis.
qPCR Methods
Levels of PSA-mRNA, Type II and Type VI TMPRSS2:ERG-mRNA, TXNRD1-mRNA, and PCA3-RNA in EPS were determined by qPCR as previously described20, 25. Primer and probe sequences, as well as cycling times were those described in Clark et al20 with the exception of the TXNRD1. This qPCR used 5′-TGCAGCTGCGCTCAAATGTGGA-3′ as forward primer, 5′-TCAGCAGCCAGCCTGGAGGA-3′ as reverse primer and 5′FAM-TGACCAAGCCAAGCGCTCTGGGGCCA-BHQ-3′ as probe. The activation step was 95°C for 10 minutes followed by 50 cycles of 95°C for 15 sec, 56°C for 30 sec and 72°C for 30 sec. Methods used to prepare cloned standards appropriate to each reaction were as previously described25. qPCR reactions were carried out using Rotor Gene qPCR cyclers (Qiagen®, Hilden Germany). The Ct method was used to determine the number of copies detected in a 25μL reaction volume by comparison to the reference standard curve corresponding to the desired amplicon. Copy numbers were taken to be zero if amplification was not observed during the 50 cycle qPCR reaction. Data normalized to the measured RNA concentration used in preparing the cDNA reaction was expressed as copy number observed per ng RNA.
Statistical Analysis
Our approach was to use a two-stage selection scheme to classify patients into “AS eligible” and “AS ineligible” based on their risk of being subsequently upstaged. In the first selection step, all subjects were stratified into a single group that met the NCCN guidelines for AS eligibility. As provided by the guidelines, this group included all patients classified as very low risk or low risk regardless of age, and patients classified as intermediate risk with a life expectancy of less than 10 years (Table 1). We then used backward elimination to fit multivariate logistic regression models for predicting both upstaged (defined as an increase in clinical stage from T1-T2c to pathology stage pT3) and upgraded (defined as an increase in Gleason Sum after surgery) patients. The independent variables considered appear in Table 2. For the regression analysis, we examined each model’s convergence and tested for lack-of-fit. ROC curves were used to determine cutoff values for predicted risk of upstaging after surgery. Cutoff values, derived by identifying the point on the ROC curve closest to coordinates (0,1), were used to determine sensitivity and specificity of each model. For model validation, we performed 5-fold cross validation. Statistical analyses were performed in JMP version 10.0.2 (SAS, Cary, NC) or R version 2.15.1. (GNU Project, www.gnu.org/software/r/).
Table 2.
Variables Tested
| Age | Pre-Biopsy PSA |
|---|---|
| Total ng into cDNA Reaction* | Total ng RNA in specimen* |
| Expressed EPS Volume (uL)* | Gleason Sum |
| TMPRSS2 Fusion Average** | ln(TMPRSS2 Fusion Average)§ |
| TMPRSS2 Fusion Average/PSA-mRNA¶ | ln(TMPRSS2 Fusion Average/PSA-mRNA)§ |
| TMPRSS2 Fusion Average/RNA† | ln(TMPRSS2 Fusion Average/RNA)§ |
| TXNDR1-mRNA Average** | ln(TXNDR1-mRNA Average)§ |
| TXNDR1-mRNA Average/PSA-mRNA¶ | ln(TXNDR1-mRNA Average/PSA-mRNA)§ |
| TXNDR1-mRNA Average/RNA† | ln(TXNDR1-mRNA Average/RNA)§ |
| PCA3-mRNA Average** | ln(PCA3-mRNA Average)§ |
| PCA3-mRNA Average/PSA-mRNA¶ | ln(PCA3-mRNA Average/PSA-mRNA)§ |
| PCA3-mRNA Average/RNA† | ln(PCA3-mRNA Average/RNA)§ |
| PSA-mRNA Average** | ln(TXNDR1-mRNA Average)§ |
| PSA-mRNA Average/RNA† | ln(PSA-mRNA Average/RNA)§ |
Raw value
Average of two replicate determinations
Normalized to ng of RNA substrate input in cDNA reaction.
Normalized to PSA-mRNA Average
Non-zero natural log transformed
Choice of Included Variables
In previous work we studied PSA mRNA levels in EPS specimens obtained from patients undergoing biopsy for PCa. In that work20, 26, we noted that patients who were subsequently diagnosed with PCa had elevated levels of PSA-mRNA in EPS relative to the levels in EPS specimens from patients subsequently diagnosed as benign. We also noted20, 26 that this effect tended to diminish the performance of biomarkers like the TMPRSS2:ERG fusions in EPS specimens when they were normalized to PSA-mRNA. Our results suggested that raw values for each mRNA expression variable were often most informative; hence these values were included in our set of variables (Table 2). When normalized mRNA variables were included, normalization was to the amount of RNA used in the cDNA reaction. Finally, given that anatomical damage to the prostate gland and impaired secretory function are expected to be linked, we included two measures of secretion capacity as variables in our analysis: total recovered EPS RNA and total recovered EPS volume (Table 2).
Results
Effect of Primary Patient Selection for AS with NCCN Guidelines
The primary selection based on NCCN guidelines reduced upstaging by approximately 3.8 fold from 26% in the NCCN AS-ineligible cohort to 6.9% in the NCCN AS-eligible cohort. However, the application of the guidelines increased the potential for upgrading by approximately 4.4 fold from 9.9% in the NCCN AS-ineligible cohort to 42% in the NCCN AS-eligible cohort (Table 1).
Variable Elimination and Secondary-Selection Model Performance
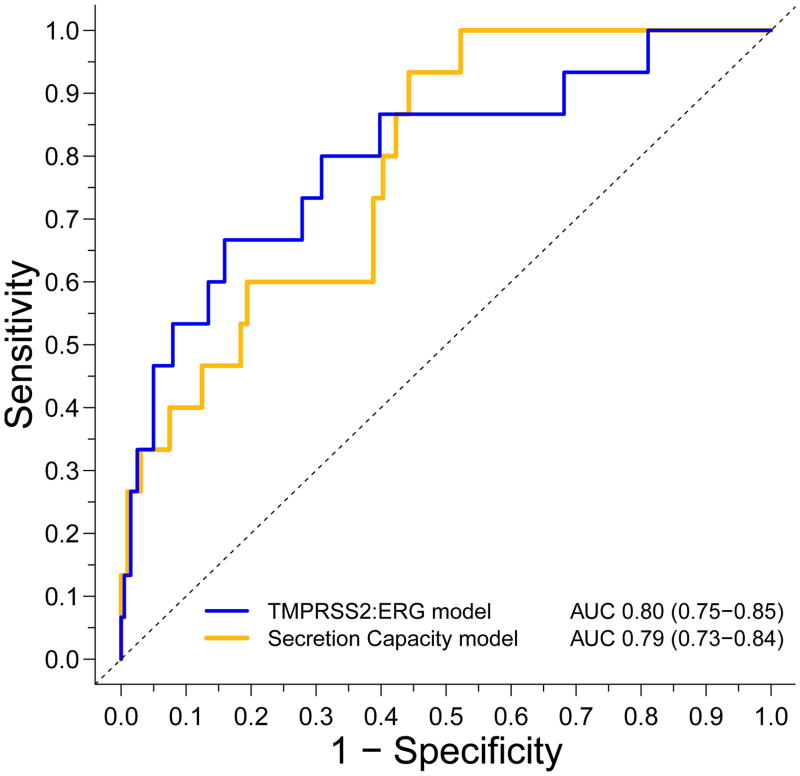
Using the variable set listed in Table 2, variable selection by backward elimination yielded two models for detecting upstaging in the NCCN AS Group prior to surgery (Figure 1). The TMPRSS2:ERG model includes three variables: Serum PSA, coupled with raw and RNA normalized values for total levels of Type III and Type VI TMRPSS2:ERG-mRNAs in EPS. The secretion capacity model includes serum PSA, total recovered EPS RNA and total recovered EPS volume. The AUC value for the TMPRSS2 model was 0.80 with a 95% confidence interval of [0.75–0.85]. It showed almost complete overlap with the secretion capacity model at 0.79 with a 95% confidence interval of [0.73–0.84] indicating roughly equivalent performance based on ROC analysis. The binary cross-validation results for both models were greater than 90%, indicating high internal validity.
Figure 1. Relative Performance ROC curves.
Multivariable logistic regression generated ROC curves with baseline serum PSA level coupled with either secretion capcity biomarkers: total expressed Vol (μL) and total RNA (ng) (secretion capacity model,yellow line); non-zero natural log transformed values for the average level of Type III and Type VI TMPRSS2:ERG expression and the average level of Type III and Type VI TMPRSS2:ERG expression normalized to the amount of RNA used in the cDNA reaction (TMPRSS2:ERG model, blue line).
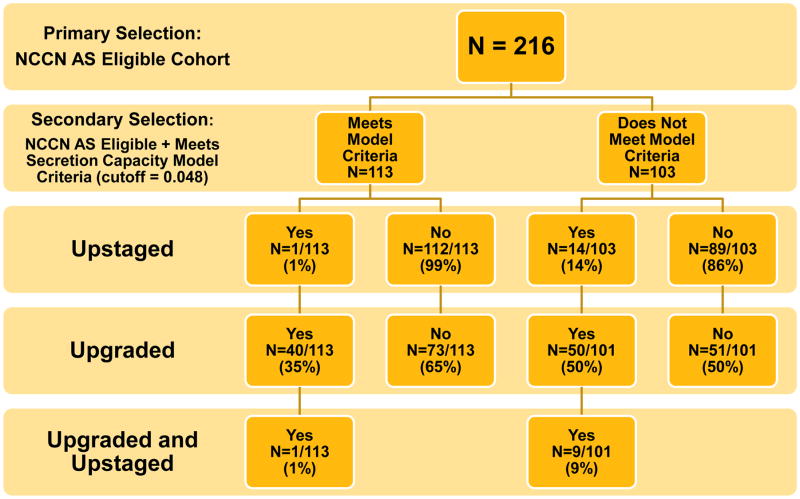
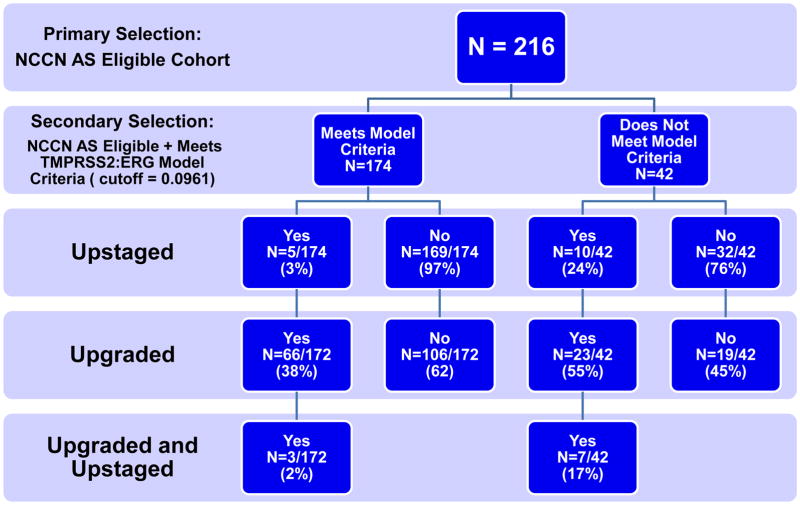
Flow charts depicting the effects of the secondary selection models in minimizing upstaging within the NCCN AS-eligible cohort are given in Figure 2. While the TMPRSS2:ERG model had a higher specificity than the secretion capacity model: 84% vs 56%; the secretion capacity model had a higher sensitivity than the TMPRSS2:ERG model: 93% vs 66%. This resulted in an improved negative predictive value for the secretion capacity model: 99% vs 97%.
Figure 2. Model Segregation Flow Charts.
(A) The flow and residual risk factors associated with a positive or negative test in the secretion capacity model. (B) The flow and residual risk factors associated with a positive or negative test in the TMPRSS2:ERG model.
The net effect of the two models on the NCCN AS-eligible cohort appear in Table 3. A negative test from the TMPRSS2:ERG model reduced the number of upstaged AS patients from 6.9% in the NCCN cohort to 2.9% (p=0.1041), for a reduction of about 2.4-fold. The residual number of AS patients who were both upstaged and upgraded was reduced from 4.6% to 1.7% (p=0.1576) by a negative TMPRSS2:ERG test for a reduction of 2.7-fold. Conversely, a negative test in the secretion capacity model reduced the number of upstaged patients from 6.9% in the NCCN cohort to 0.9% (p=0.0141), for a reduction of about 7.8-fold, as it reduced the residual number of AS patients who were upstaged and upgraded from 4.6% to 0.9% (p=0.1054) for a reduction of 5.2-fold. Neither test significantly reduced the elevated number of upgraded AS patients selected by the NCCN guidelines. A summary of performance statistics for the two models is given in Table 4.
Table 3.
AS selection model enrichment measured by patients with subsequent risk misclassifications. Fold changes are the resulting improvements in misclassification rates after applying our tests to the NCCN AS selection.
| AS Candidate Selection Criteria | ||||
|---|---|---|---|---|
| Cohort | Entire Cohort (N = 528) | NCCN Selection (N = 216) | NCCN + negative test on TMPRSS2:ERG model | NCCN + negative test on Secretion Capacity model |
| Upstaged | 18% | 6.9% | 2.9% (2.4)* | 0.9% (7.8) |
| Upgraded | 23% | 42% | 38% (1.1) | 35% (1.2) |
| Upstaged and Upgraded | 4% | 4.6% | 1.7% (2.7) | 0.9% (5.2) |
Values in parentheses indicate the fold change improvement in misclassification over NCCN selection
Table 4.
Performance Statistics for Secondary Selection Models of Upstaging.
| Performance Statistics* | AS Candidate Selection Model | |
|---|---|---|
| NCCN + TMPRSS2:ERG | NCCN + Secretion Capacity | |
| Sensitivity | 66.67 % (95% CI: 38.41 % to 88.05 %) | 93.33 % (95% CI: 67.98 % to 98.89 %) |
| Specificity | 84.08 % (95% CI: 78.27 % to 88.85 %) | 55.72 % (95% CI: 48.56 % to 62.71 %) |
| Positive Predictive Value | 23.81 % (95% CI: 12.07 % to 37.45 %) | 13.59 % (95% CI: 7.64 % to 21.76 %) |
| Negative Predictive Value | 97.13 % (95% CI: 93.42 % to 99.05 %) | 99.12 % (95% CI: 95.15 % to 99.85 %) |
| 5-fold cross validation | 92.8% (95% CI: 91.3 to 94.3%) | 92.9% (95% CI: 91.4 to 94.4%) |
Parameters determined at ROC cutoff value for each model: 0.0961 for NCCN + TMPRSS2:ERG model, and 0.048 for the NCCN + Secretion Capacity Model.
Discussion
On reflection, it seems obvious that an extensive tumor will impair the secretory function of the prostate gland. Aside from the loss of glandular anatomy, androgen demand from the tumor may reduce the levels of key enzymes (e.g. Monoamine Oxidase A, ornithine decarboxylase, IGF-1) in surrounding normal tissue that are required for the biosynthesis of the prostatic secretions (e.g. spermine, and spermidine). Moreover, impaired Zn2+ uptake by the tumor may induce a broad decrease in the amount of citrate available for secretion. These considerations and the data presented here support the hypothesis that secretion capacity biomarkers like collected EPS volume and total RNA obtained from the EPS specimen might be biomarkers of aggressive tumors.
The variables considered in this analysis included the well known prostate tumor biomarkers PCA3 and TMPRSS2:ERG, as well as the recently identified biomarker TXNRD1 known to be expressed in prostate tumor adjacent stromal tissue27. Neither PCA3 nor TXNRD1 contributed to either model, even though the normalized and raw measures of these parameters were tested. The elimination of TXNRD1 is perhaps not unexpected since we have not been able to detect significant numbers of stromal cells in Ficoll separated EPS preparations (Linehan et al. unpublished). However, the elimination of PCA3 was unexpected given that PCA3 values obtained from post DRE urine specimens have been reported to predict ECE in a 72 patient cohort. That cohort had a rather high prevalence of ECE (29%) and was taken from the general population28 and not from a group preselected for AS, where PCA3 has been shown to have poor predictive ability29 However, the. failure of PCA3 to contribute to the most significant models is consistent with its minor contribution to TMPRSS2:ERG dominated models for the prediction of biopsy outcome in both EPS20, 26 and post DRE urine22.
It is interesting that the two models cannot be further reduced, that is to say that the raw level of Type III and Type VI TMPRSS2:ERG expression in the TMPRSS2 model or the total recovered EPS RNA quantity in the secretion capacity model connot be omitted without abolishing the effectiveness of the models. This suggests that the models are actually linked by the recovered RNA values. Among patients subsequently found to be upstaged, total recovered RNA was lower than it was among those who were not upstaged. Thus, the raw value for the level of Type III and Type VI TMPRSS2:ERG expression embeds RNA recovery because low values of recovered RNA often required RNA inputs less than the maximum 200ng value used in the cDNA reaction. Serum PSA was also a required parameter in each model. This underscores its importance in AS eligibility, and suggests that future guidelines may need to address exact values in addition to the current ranges.
In deciding which model is the most effective, a number of considerations must be taken into account. Both models are examples of evidence-driven approaches to AS12 and both tests have their strengths. In terms of patient acceptance negative predictive value provides the strongest support for secondary selection by the tests. In this regard the serum capacity test was the most effective in eliminating upstaging from the NCCN AS-eligible cohort, reducing the risk by nearly 8 fold. In terms of over-treatment, only 113 or 21% of the original 528 evaluated patients would now be permitted to enter AS. Given the trend toward over-treatment3, 5, 30, permitting the additional risk associated with choosing the TMPRSS2:ERG test could be warranted by the larger percentage of patients (174 or 33% of the original 528) who could enter AS.
In terms of clincal time and expense invested in the performance of the test itself, the secretion capacity test is the most cost effective. This is because the information required to evaluate a patient with the secretion capacity model can be obtained without performing time-consuming cDNA preparation and DNA or RNA amplification.
Neither test reduces the residual level of upgrading brought in with the primary selection under NCCN guidelines. While this cannot be used as a basis for discriminating between the two tests, it is consistent with a recent report on the general prevalence of upgrading after surgery13 and suggests that further research on biomarkers is required in this area. Moreover, given the predominance of GS 6 in our NCCN AS-eligible cohort (more than 95%), and the relatively short time between diagnosis and surgery in our cohort (mean 9.5 months), it appears that transrectal ultrasound-guided biopsy misclassifies a significant number of GS 6 patients.
Neither model can be performed on post-DRE urine. This is because the raw value for the level of Type III and Type VI TMPRSS2:ERG expression will be altered by urine catch in the TMPRSS2:ERG test, and because the total recovered RNA will be altered by the urine catch in the secretion capacity model. Moreover, EPS volume cannot be measured in post-DRE Urine.
Both models were able to segregate patients who are likely to be upstaged into a group with much higher probability of upstaging. For example nearly 24% of the patients with a positive test in the TMPRSS2:ERG model and 14% of the patients with a positive test in the secretion capacity model harbor residual upstaging risk factors (Figure 2). This represents an increase of nearly 3.5-fold for the TMPRSS2:ERG model and about 2-fold for the secretion capacity model over the 6.9% in the untested NCCN group. These results suggest that a positive test in either model might influence a choice for nerve sparing surgery or a recommendation for image-guided biopsies in this group.
Finally, the study has several limitations. First, the relatively small sample size of 216 patients may not extrapolate to the population of men eligible for AS. Second, the complete list of available biomarkers was not tested, so there may be biomarkers which could improve the modeling results reported here. And third, a randomized prospective study that compares outcomes for model-eligible patient groups versus NCCN AS-eligible AS patient group is needed to quantify the utility of the approach.
Conclusion
Our results suggest that non-invasive EPS testing may improve patient acceptance of AS by dramatically reducing the presence of residual risk factors among patients eligible for AS under NCCN guidelines.
Acknowledgments
This study was supported in part by grants CA102521 and CA136055 from the US National Cancer Institute of the National Institutes of Health to S.S.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.National Cancer Institute. [Accessed April 2008]; http://www.cancer.gov/cancertopics/types/prostate.
- 2.O'Dowd GJ, Miller MC, Orozco R, et al. Analysis of repeated biopsy results within 1 year after a noncancer diagnosis. Urology. 2000;55:553. doi: 10.1016/s0090-4295(00)00447-7. [DOI] [PubMed] [Google Scholar]
- 3.Welch HG, Albertsen PC. Prostate cancer diagnosis and treatment after the introduction of prostate-specific antigen screening: 1986–2005. J Natl Cancer Inst. 2009;101:1325. doi: 10.1093/jnci/djp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schroder FH, Hugosson J, Roobol MJ, et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 5.Esserman L, Shieh Y, Thompson I. Rethinking screening for breast cancer and prostate cancer. JAMA. 2009;302:1685. doi: 10.1001/jama.2009.1498. [DOI] [PubMed] [Google Scholar]
- 6.Loeb S, Roehl KA, Helfand BT, et al. Complications of open radical retropubic prostatectomy in potential candidates for active monitoring. Urology. 2008;72:887. doi: 10.1016/j.urology.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 7.D'Amico AV, Whittington R, Malkowicz SB, et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA. 1998;280:969. doi: 10.1001/jama.280.11.969. [DOI] [PubMed] [Google Scholar]
- 8.Choo R, Klotz L, Danjoux C, et al. Feasibility study: watchful waiting for localized low to intermediate grade prostate carcinoma with selective delayed intervention based on prostate specific antigen, histological and/or clinical progression. J Urol. 2002;167:1664. [PubMed] [Google Scholar]
- 9.Klotz L, Zhang L, Lam A, et al. Clinical results of long-term follow-up of a large, active surveillance cohort with localized prostate cancer. J Clin Oncol. 2010;28:126. doi: 10.1200/JCO.2009.24.2180. [DOI] [PubMed] [Google Scholar]
- 10.Smaldone MC, Cowan JE, Carroll PR, et al. Eligibility for active surveillance and pathological outcomes for men undergoing radical prostatectomy in a large, community based cohort. J Urol. 2010;183:138. doi: 10.1016/j.juro.2009.08.152. [DOI] [PubMed] [Google Scholar]
- 11.Epstein JI, Walsh PC, Carmichael M, et al. Pathologic and clinical findings to predict tumor extent of nonpalpable (stage T1c) prostate cancer. JAMA. 1994;271:368. [PubMed] [Google Scholar]
- 12.Carter HB. Management of low (favourable)-risk prostate cancer. BJU Int. 2011;108:1684. doi: 10.1111/j.1464-410X.2010.10489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nazmy M, Lau C, Wilson T, et al. Gleason Score Upgrading In NCCN Very Low Risk Classification Prostate Cancer Patients. Journal of Urology. 2012;187:e284. [Google Scholar]
- 14.Tosoian J, Loeb S. PSA and beyond: the past, present, and future of investigative biomarkers for prostate cancer. Scientific World Journal. 2010;10:1919. doi: 10.1100/tsw.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sokoll LJ, Sanda MG, Feng Z, et al. A prospective, multicenter, National Cancer Institute Early Detection Research Network study of [-2]proPSA: improving prostate cancer detection and correlating with cancer aggressiveness. Cancer Epidemiol Biomarkers Prev. 2010;19:1193. doi: 10.1158/1055-9965.EPI-10-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson IM, Ankerst DP. Prostate-specific antigen in the early detection of prostate cancer. CMAJ. 2007;176:1853. doi: 10.1503/cmaj.060955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross AE, Loeb S, Landis P, et al. Prostate-specific antigen kinetics during follow-up are an unreliable trigger for intervention in a prostate cancer surveillance program. J Clin Oncol. 2010;28:2810. doi: 10.1200/JCO.2009.25.7311. [DOI] [PubMed] [Google Scholar]
- 18.Lechevallier E, Eghazarian C, Ortega JC, et al. Effect of digital rectal examination on serum complexed and free prostate-specific antigen and percentage of free prostate-specific antigen. Urology. 1999;54:857. doi: 10.1016/s0090-4295(99)00239-3. [DOI] [PubMed] [Google Scholar]
- 19.Aubin SM, Reid J, Sarno MJ, et al. PCA3 molecular urine test for predicting repeat prostate biopsy outcome in populations at risk: validation in the placebo arm of the dutasteride REDUCE trial. J Urol. 2010;184:1947. doi: 10.1016/j.juro.2010.06.098. [DOI] [PubMed] [Google Scholar]
- 20.Clark JP, Munson KW, Gu JW, et al. Performance of a Single Assay for both Type III-Type VI TMPRSS2:ERG fusions in Non-Invasive Prediction of Prostate Biopsy Outcome. Clinical Chemistry. 2008;54:2007. doi: 10.1373/clinchem.2008.108845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laxman B, Morris DS, Yu J, et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68:645. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomlins SA, Aubin SM, Siddiqui J, et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med. 2011;3:94ra72. doi: 10.1126/scitranslmed.3001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalgo ML, Pavlovich CP, Lee SM, et al. Prostate cancer detection by GSTP1 methylation analysis of postbiopsy urine specimens. Clin Cancer Res. 2003;9:2673. [PubMed] [Google Scholar]
- 24.Crocitto LE, Korns D, Kretzner L, et al. Prostate cancer molecular markers GSTP1 and hTERT in expressed prostatic secretions as predictors of biopsy results. Urology. 2004;64:821. doi: 10.1016/j.urology.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 25.Munson K, Clark J, Lamparska-Kupsik K, et al. Recovery of Bisulfite-Converted Genomic Sequences in the Methylation-Sensitive QPCR. Nucleic Acids Research. 2007;35:2893. doi: 10.1093/nar/gkm055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whelan C, Crocitto L, Kawachi M, Chan K, Smith D, Wilson T, Smith SS. The influence of PSA-RNA yield on the analysis of expressed prostatic secretions (EPS) for prostate cancer diagnosis. Canadian Journal of Urology. 2013;20:6597. [PMC free article] [PubMed] [Google Scholar]
- 27.Singer EM, Crocitto LE, Weiss LM, et al. Biomarker Identification with Ligand-Targeted Nucleoprotein Assemblies Nanomedicine. 2011;6:659. doi: 10.2217/nnm.11.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitman EJ, Groskopf J, Ali A, et al. PCA3 score before radical prostatectomy predicts extracapsular extension and tumor volume. J Urol. 2008;180:1975. doi: 10.1016/j.juro.2008.07.060. [DOI] [PubMed] [Google Scholar]
- 29.Tosoian JJ, Loeb S, Kettermann A, et al. Accuracy of PCA3 measurement in predicting short-term biopsy progression in an active surveillance program. J Urol. 2010;183:534. doi: 10.1016/j.juro.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Wilt TJ, Brawer MK, Jones KM, et al. Radical prostatectomy versus observation for localized prostate cancer. N Engl J Med. 2012;367:203. doi: 10.1056/NEJMoa1113162. [DOI] [PMC free article] [PubMed] [Google Scholar]