Abstract
Skeletal muscle comprises the largest organ in the human body and is the major site for energy expenditure. It exhibits remarkable plasticity in response to physiological stimuli such as exercise. Physical exercise remodels skeletal muscle and enhances its capability to burn calories, which has been shown to be beneficial for many clinical conditions including metabolic syndrome and cancer. Nuclear receptors (NRs) comprise a class of transcription factors found only in metazoans that regulate major biological processes such as reproduction, development, and metabolism. Recent studies have demonstrated crucial roles for NRs and their co-regulators in regulating skeletal muscle energy metabolism and exercise-induced muscle remodeling. While nothing can fully replace exercise, development of exercise mimetics that enhance or even substitute for the beneficial effects of physical exercise would be of great benefit. The unique property of NRs that allows modulation by endogenous or synthetic ligands makes them bona fide therapeutic targets. In this review, we present an overview of the current understanding of NRs and their co-regulators in skeletal muscle oxidative metabolism and summarize recent progress in the development of exercise mimetics that target NRs and their co-regulators.
Introduction
Exercise has been known for its health benefits since ancient times. It is now widely accepted that physical activity positively impacts on a variety of clinical conditions including obesity, type-2 diabetes, metabolic syndrome, neurodegenerative diseases, cardiovascular diseases and cancer (Perseghin, Price et al. 1996) (Grazina and Massano 2013) (Mellett and Bousquet 2013) (Lemanne, Cassileth et al. 2013). On the other hand, physical inactivity poses major negative influences on these disease conditions (Hu, Willett et al. 2004).
How exactly exercise exerts its beneficial effects is not fully understood, however, skeletal muscle is believed to play a vital role (Hamilton and Booth 2000). As the largest organ of our body, skeletal muscle comprises ~40% of total body mass and accounts for ~30% of whole-body energy metabolism during resting (Zurlo, Larson et al. 1990). Upon insulin stimulation, skeletal muscle can be responsible for ~85% of total glucose disposal (Defronzo, Jacot et al. 1981). During peak activity, whole-body energy metabolism can be increased by up to 20 fold, ~90% of which is contributed by skeletal muscle (Zurlo, Larson et al. 1990). Hence muscle is the major site of calorie-burning of energy substrates like glucose and free fatty acids. Exercise training remodels skeletal muscle to more efficiently clear these substrates, whose excess levels negatively impact many tissues.
In mammals, skeletal muscle is a mosaic of heterogeneous myofibers with diverse structural and functional properties (Schiaffino and Reggiani 2011). Based on the expression patterns of different myosin heavy chain (MHC) isoforms, which coincide with various biochemical characteristics, myofibers can be classified into four major groups: slow-twitch type I and fast-twitch type IIa, IIx/d, and IIb. Type I and IIa fibers are red in appearance due to their high myoglobin content. They are rich in mitochondria and predominantly powered by complete oxidation of glucose and fatty acids. These oxidative fibers are also dense with vasculature and resistant to fatigue. In contrast, the glycolytic type IIx/d and IIb fibers are generally white in color, have less myoglobin and mitochondria, mainly rely on glycolysis for energy production, have less vasculature, and fatigue rapidly (Schiaffino and Reggiani 2011). In humans, fiber-type composition is strongly associated with metabolic health, with more glycolytic fibers seen in obese and type-2-diabetic patients (Hickey, Carey et al. 1995).
It has been well documented that skeletal muscle undergoes a series of physiological and biochemical adaptations upon exercise training (Hamilton and Booth 2000), of which the most intriguing is fiber-type transformation. Many human and animal studies have clearly demonstrated that prolonged exercise induces the glycolytic type IIb and IIx/d fibers to transform to the more oxidative type IIa fibers (Gollnick, Armstrong et al. 1973) (Foster, Costill et al. 1978) (Wu, Rothermel et al. 2001). Although some professional athletes have an increased proportion of type I fibers (Gollnick, Saltin et al. 1972), it remains unclear whether exercise training can switch type II fibers completely to type I. While exercise has a positive effect on the glycolytic-to-oxidative fiber-type transformation, physical inactivity and obesity usually has the opposite effect and leads to the reverse transformation (Bergouignan, Rudwill et al. 2011). During fiber-type transformation, not only is the expression of MHC isoforms switched, but other fiber-type specific properties, such as mitochondrial density, oxidative phosphorylation (OXPHOS) activity, vasculature, and fatigue resistance, are also changed accordingly (Yan, Okutsu et al. 2011).
Skeletal muscle adaptation during exercise involves numerous transcriptional and epigenetic changes, which are regulated by multiple signaling pathways (Bassel-Duby and Olson 2006) (Barres, Yan et al. 2012). In addition to the widely known Calcineurin/NFAT and HDAC/MEF pathways, it has been recently shown that nuclear receptors and their co-regulatory factors also play important roles in skeletal muscle adaptation.
Nuclear receptors (NRs) are ligand-modulated transcription factors that respond to a variety of hydrophobic molecules including hormones, lipids, steroids, retinoids, and xenobiotics. All NRs share similar modular domains, including a highly conserved DNA binding domain (DBD), a ligand binding domain (LBD), variable N-terminal and C-terminal domains, and a hinge domain between the DBD and LBD (Mangelsdorf, Thummel et al. 1995). The DBD is characterized by a zinc finger motif which recognizes the hormone response element (HRE) on target chromatin and the LBD by a hydrophobic ligand binding pocket. Upon ligand binding, NRs undergo conformational changes, which alter their interactions with other proteins and trigger epigenetic chromatin changes and downstream transcriptional regulation (Wurtz, Bourguet et al. 1996).
A major goal of exercise science is to find substitutes for physical exercise that achieve its beneficial effects in people unable to exercise. The ability of nuclear receptors to sense and respond to small-molecule ligands makes them ideal pharmacological targets. This review focuses on the roles of NRs and their co-regulatory factors in regulating skeletal muscle functions, including fiber-type determination, mitochondrial biogenesis, vasculature development, and fatigue resistance, with the goal to shed some light on developing the ‘exercise in a pill’.
The PPAR Subfamily
The peroxisome proliferator-activated receptor (PPAR) subfamily of NRs is composed of three members: PPARα, PPARδ (also referred to as PPARβ), and PPARγ. PPARα was the first PPAR identified during a screen for the molecular target of fibrates, a class of cholesterol-lowering compounds that increase hepatic fatty acid oxidation and peroxisome proliferation (hence the name) (Issemann and Green 1990). Based on sequence homology, PPARδ and PPARγ, which do not induce peroxisome proliferation, were later cloned from mouse (Zhu, Alvares et al. 1993) (Kliewer, Forman et al. 1994).
PPARs are predominantly localized in the nucleus. They form heterodimers with retinoid X receptors (RXRs) and can be activated by both PPAR ligands and RXR ligands. In the absence of ligand, the PPAR/RXR heterodimers bind to PPAR response elements (PPREs) in association with transcriptional co-repressors such as NCoR and SMRT. Ligand binding leads to a conformational change, and recruitment of co-activators such as PGC-1α and PGC-1β to replace the co-repressors, resulting in activation of downstream target gene expression. PPARs play essential roles in lipid metabolism regulation. They sense and respond to free fatty acids and their derivatives to regulate genes involved at almost all levels of lipid metabolism, including lipid import/export, synthesis, storage, breakdown, and oxidation (Evans, Barish et al. 2004). While the PPAR subfamily shares certain common target genes, PPARα and PPARδ are typically involved in regulating lipid catabolism and oxidation, while PPARγ is responsible for adipogenesis and lipid synthesis. All three PPARs are expressed in skeletal muscle (Muoio, MacLean et al. 2002) (Amin, Mathews et al. 2010), and over the last decade, both gain- and loss-of-function studies have contributed significantly to our understanding of their roles in muscle.
PPARδ is the most abundant PPAR in skeletal muscle (Muoio, MacLean et al. 2002) (Amin, Mathews et al. 2010) and plays important roles in regulating fiber-type determination, mitochondrial function, lipid metabolism, and fatigue resistance (Figure 1). It is expressed relatively higher in oxidative fibers than in glycolytic fibers. Exercise, in both acute and prolonged forms (Watt, Southgate et al. 2004) (Luquet, Lopez-Soriano et al. 2003), induces Pparδ expression in skeletal muscle. Similar to exercise, fasting also triggers a fuel-source switch in skeletal muscle from glucose to fatty acid utilization. Consistently, 6 to 48 hours of fasting dramatically increases Pparδ expression in skeletal muscle (de Lange, Farina et al. 2006).
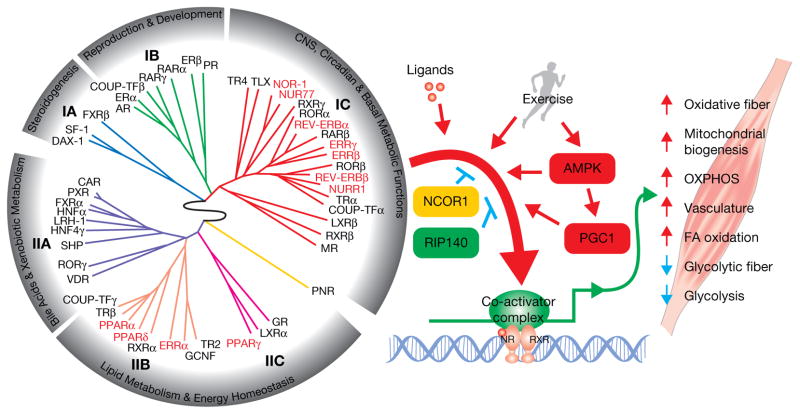
Figure 1.
NR regulation of energy metabolism and remodeling in skeletal muscle. On the left is the nuclear receptor ring of physiology (Bookout, Jeong et al. 2006). It clusters 49 mouse NRs into 6 groups based on their tissue distribution patterns. The NRs that have been found to play crucial roles in skeletal muscle function (highlighted in red/bold) are clustered mainly in two groups: group IC whose members are selectively expressed in highly metabolic tissues and are involved in CNS, circadian and basal metabolic functions, including NOR-1, NUR77, NURR1, ERRβ, ERRγ, REV-ERBα, and REV-ERBβ; and groups IIB and IIC whose members are broadly expressed and are linked to lipid metabolism and energy homeostasis, including PPARα, PPARδ, PPARγ, and ERRα. These NRs work in concert with exercise and co-regulators to regulate many aspects of skeletal muscle physiology. Synthetic ligands targeting NRs and their co-regulators can enhance or replace the physiological benefits induced by exercise, which is of great value to public health.
Two independent studies showed that skeletal muscle over-expression of Pparδ induces a glycolytic-to-oxidative fiber-type transformation (Luquet, Lopez-Soriano et al. 2003) (Wang, Zhang et al. 2004). Mice over-expressing wild-type Pparδ have more oxidative fibers, higher OXPHOS enzyme activities, and more uncoupling proteins. These transgenic mice also have reduced fat content with smaller adipocyte size, similar to what is seen in exercised animals (Luquet, Lopez-Soriano et al. 2003). Mice expressing a constitutively active form of Pparδ were nicknamed ‘marathon mice’ as they can run for up to twice the distance of their wild-type littermates. They have more type I and less type II fibers, have increased mitochondrial biogenesis and uncoupling, are resistant to diet-induced-obesity, and have improved glucose tolerance (Wang, Zhang et al. 2004). Conversely, conditional knockout of Pparδ in skeletal muscle leads to an oxidative-to-glycolytic fiber-type switch. The knockout muscle has lower expression of genes involved in fatty acid catabolism and oxidation, as well as reduced OXPHOS activities (Schuler, Ali et al. 2006). Upon high-fat-diet challenge, the mutant mice gain more weight mainly due to increased fat content and are more susceptible to developing insulin resistance and glucose intolerance (Schuler, Ali et al. 2006). Therefore, Pparδ appears necessary for the maintenance of oxidative fibers and their oxidative functions in skeletal muscle. However, it remains to be demonstrated whether Pparδ is required for exercise induced muscle remodeling.
Pparα is abundantly expressed in tissues with high fatty acid catabolism, such as liver and heart (Figure 1) (Braissant, Foufelle et al. 1996), where it is activated by free fatty acids and promotes fatty acid oxidation (Kersten, Seydoux et al. 1999). Pparα is also expressed at significant levels in skeletal muscle. Both Pparα and Pparδ regulate fatty acid catabolism and share common target genes. Similar to Pparδ, over-expression of Pparα in skeletal muscle also induces the expression of genes involved in fatty acid catabolism, the tricarboxylic acid cycle (TCA cycle), and mitochondrial OXPHOS. As a result, fatty acid oxidation is increased in the transgenic muscle and the mice are resistant to diet-induced-obesity (Finck, Bernal-Mizrachi et al. 2005). However, the transgenic mice are more prone to developing insulin resistance and glucose intolerance due to reduced expression of genes involved in glucose uptake and glycolysis (Finck, Bernal-Mizrachi et al. 2005). Thus, although Pparα has a positive role in regulating fatty acid oxidation in skeletal muscle, its activity needs to be finely regulated to balance glucose and fatty acid metabolism.
In addition to their different roles in metabolic regulation, Pparα also functions distinctly from Pparδ in fiber-type determination. In contrast to Pparδ, over-expression of Pparα in skeletal muscle does not increase endurance but rather reduces it by more than 50% (Gan, Burkart-Hartman et al. 2011). Consistently, an oxidative to glycolytic fiber-type switch is found in these mice, as shown by the expression MHC genes, metachromatic ATPase staining, and MHC immunohistochemistry staining (Gan, Rumsey et al. 2013). The opposing functions of PPARα and PPARδ to induce glycolytic and oxidative fiber-type transformations respectively, seem to be mediated by a miRNA network involving two specific miRNAs, miR-208b and miR-499 (Gan, Rumsey et al. 2013), which play important roles in fiber-type determination by activating the oxidative and repressing the glycolytic myofiber gene program (van Rooij, Quiat et al. 2009). In contrast to the over-expression model, knockout of Pparα in skeletal muscle induces a glycolytic-to-oxidative fiber-type switch (Gan, Rumsey et al. 2013). Therefore, endogenous PPARα counteracts PPARδ to maintain a proper fiber-type composition of skeletal muscle.
Pparγ is expressed most highly in adipose tissues, where it plays an essential role in adipogenesis and whole body lipid homeostasis (Figure 1). Its ablation in adipose tissues leads to severe lipodystrophy and elevated levels of blood triglycerides and free fatty acids. The knockout mice are more susceptible to diet induced insulin resistance. However, treatment with thiazolidinediones (TZDs), a class of PPARγ specific ligands, can still improve insulin sensitivity in these knockout mice, suggesting that PPARγ in non-adipose tissues also contributes to its regulation of lipid homeostasis and insulin sensitivity (He, Barak et al. 2003). The strongest evidence showing a positive role for muscle PPARγ in metabolic regulation comes from the generation of a mouse model with Pparγ specifically deleted in skeletal muscle (Hevener, He et al. 2003). These knockout mice develop glucose intolerance and insulin resistance. Moreover, they are less responsive to TZD-induced skeletal muscle insulin sensitization, while the effects of TZDs in liver and adipose tissues remain unaffected (Hevener, He et al. 2003). A similar study seems to draw a different conclusion, showing that the knockout mice only have mild insulin resistance and respond normally to TZD treatment (Norris, Chen et al. 2003). However, the two studies were performed on mice with different genetic backgrounds, one being a pure C57BL/6J (Hevener, He et al. 2003) and the other a mixed 129/sv, C57BL/6, and FVB background, which might account for the different phenotypes observed. In addition to the knockout models, over-expression of Pparγ in skeletal muscle also demonstrated its importance in metabolic regulation (Amin, Mathews et al. 2010). These transgenic mice are protected from diet-induced insulin resistance and glucose intolerance. Interestingly, these mice produce significant amounts of adiponectin in skeletal muscle, despite their reduced intramuscular adiposity. Furthermore, activation of AMPK, a known adiponectin target, in the transgenic muscle suggests that the increased adiponectin functions locally. Similar to Pparδ, over-expression of Pparγ induces a glycolytic-to-oxidative fiber-type switch and an increase in mitochondrial gene expression, which may be a secondary effect from the activated AMPK pathway (Amin, Mathews et al. 2010). Therefore, PPARγ is required in skeletal muscle for glucose and lipid homeostasis. In addition, its role in generating muscle adiponectin provides another layer of metabolism regulation.
The ERR Subfamily
The estrogen-related receptor (ERR) subfamily includes three members: ERRα, ERRβ, and ERRγ. ERRα was the first to be identified based on its high sequence homology with the estrogen receptor α (ERα) (Giguere, Yang et al. 1988). ERRβ was cloned in the same study using Errα cDNA as a probe (Giguere, Yang et al. 1988). Last but not least, ERRγ was discovered in three independent studies using different strategies (Eudy, Yao et al. 1998) (Hong, Yang et al. 1999) (Heard, Norby et al. 2000). Although all three ERRs share high structural similarities with ERs at both the DNA and protein levels, they are distinct from ERs in both their functions and their regulation of target gene transcription (Eichner and Giguere 2011).
All three ERRs are believed to be constitutively active and to date, no natural ligand(s) has been identified (Eichner and Giguere 2011). Instead, the transcriptional activities of ERRs are regulated by a number of co-regulatory factors, the most studied of which include the steroid receptor co-activators (SRC1, 2, and 3) (Hong, Yang et al. 1999; Xie, Hong et al. 1999; Zhang and Teng 2000), the peroxisome proliferator-activated receptor γ co-activators (PGC-1α and β) (Huss, Kopp et al. 2002) (Kamei, Ohizumi et al. 2003), and the nuclear receptor co-repressors RIP140 and NCoR1 (Sanyal, Matthews et al. 2004) (Perez-Schindler, Summermatter et al. 2012).
Extensive studies in the past decade have clearly established a central role of ERRs in regulating energy metabolism (Eichner and Giguere 2011), which is further supported by their tissue expression patterns. Errα is the most abundant of the three. It is ubiquitously expressed but peaks in tissues with high energy needs including brain, heart, muscle, and kidney (Figure 1) (Giguere, Yang et al. 1988) (Bookout, Jeong et al. 2006). Errβ and Errγ have similar tissue distribution patterns. Both are selectively expressed in metabolically active tissues such as retina, spinal cord, heart, muscle, and kidney, with Errγ generally expressed at a higher level (Figure 1) (Bookout, Jeong et al. 2006). All three ERRs are highly expressed in skeletal muscle and their roles in regulating muscle energy metabolism have been explored in both gain- and loss-of-function studies (Luo, Sladek et al. 1997) (Luo, Sladek et al. 2003) (Huss, Torra et al. 2004) (Wende, Huss et al. 2005) (Alaynick, Kondo et al. 2007) (Chinsomboon, Ruas et al. 2009) (Rangwala, Wang et al. 2010) (Narkar, Fan et al. 2011) (Gan, Rumsey et al. 2013) (Matsakas, Yadav et al. 2013).
Studies of ERRα in skeletal muscle have mainly focused on its synergistic interaction with PGC-1α in target gene regulation. No phenotypic change in skeletal muscle is observed after whole body Errα ablation, possibly due to a compensatory induction of Pgc-1α (Luo, Sladek et al. 2003) (Huss, Torra et al. 2004). ERRα seems to play a role in regulating fatty acid metabolism and fuel selection in skeletal muscle as its over-expression induces the expression of Pparα, a key regulator of fatty acid metabolism, and Pdk4, the mitochondrial gate keeper for pyruvate oxidation. Over-expression of its co-activator PGC-1α can further enhance the expression of these genes (Huss, Torra et al. 2004) (Wende, Huss et al. 2005). Such regulation is mediated by the direct binding of ERRα to the ERR response element (ERRE) on the promoters of Pparα and Pdk4 (Huss, Torra et al. 2004) (Wende, Huss et al. 2005). In addition, ERRα also regulates myocyte differentiation. Over-expression of Errα in C2C12 myoblasts accelerates myotube formation while Errα null primary myocytes show delayed myogenesis and mitochondrial dysfunction (Murray and Huss 2011). Although ERRα positively regulates lipid metabolism and mitochondrial OXPHOS in cooperation with PGC-1α in heart and brown adipose tissue (Dufour, Wilson et al. 2007) (Villena, Hock et al. 2007), its physiological function in skeletal muscle remains to be elucidated.
Similar to ERRα, ERRγ also plays an important role in regulating energy metabolism. Errγ null mice die within the first week of life, possibly from heart failure due to disrupted mitochondrial energy production (Alaynick, Kondo et al. 2007). The importance of ERRγ in energy metabolism is also indicated by its distribution in skeletal muscle, where it is exclusively expressed in oxidative muscles such as soleus and red gastrocnemius but not in glycolytic muscles like white gastrocnemius or quadriceps (Narkar, Fan et al. 2011). Transgenic mice with muscle-specific over-expression of Errγ have a remarkable conversion of glycolytic to oxidative fibers, with all white muscles appearing red (Narkar, Fan et al. 2011). The transgenic mice are fatigue resistant and can run about twice the distance of the controls. They also have a higher energy expenditure rate and a lower respiratory exchange ratio (RER), indicating a fuel preference for fatty acids. Mitochondrial biogenesis and vascularization are both induced. Gene expression analysis further revealed a gene signature change from glycolytic to oxidative muscle, including the induction of genes involved in lipid metabolism, TCA cycle, angiogenesis, and mitochondrial OXPHOS (Narkar, Fan et al. 2011) (Rangwala, Wang et al. 2010). In addition, the over-expression of ERRγ also alleviates symptoms of Duchenne muscular dystrophy and promotes muscle recovery from ischemia damage (Matsakas, Yadav et al. 2012) (Matsakas, Yadav et al. 2013). Therefore, genetic activation of ERRγ can induce an exercise-like phenotype in skeletal muscle with positive impacts on muscle diseases. However, its endogenous roles in regulating skeletal muscle function and exercise-induced muscle remodeling remain to be demonstrated.
Unlike ERRα and ERRγ, little is known about whether and how ERRβ regulates energy metabolism. Loss-of-function studies have demonstrated the crucial roles of ERRβ in placental development (Luo, Sladek et al. 1997), germ cell development (Mitsunaga, Araki et al. 2004), inner ear development (Chen and Nathans 2007), and retinal photoreceptor survival (Onishi, Peng et al. 2010). In skeletal muscle, it has been briefly shown that both ERRβ and ERRγ are required to maintain type I muscle fibers in the oxidative/glycolytic mixed muscle gastrocnemius but not in the mostly oxidative muscle soleus (Gan, Rumsey et al. 2013). However, the extent of functional redundancy between ERRβ and ERRγ in skeletal muscle is unclear and more work is needed to fully understand the role of ERRβ in regulating energy metabolism and skeletal muscle function.
The NR4A Subfamily
The NR4A subfamily of nuclear receptors consists of three closely related members: NR4A1 (NUR77), NR4A2 (NURR1), and NR4A3 (NOR1). Similar to ERRs, the NR4As are also orphan receptors that do not bind to any natural agonist (Pearen and Muscat 2010). They are constitutively active and their transcriptional activities appear to be primarily regulated by their abundance and post-translational modifications (Chao, Wroblewski et al. 2012).
Based on their tissue expression patterns, the NR4As are clustered in the same group as ERRβ and ERRγ; they are preferentially expressed in tissues with high energy needs such as brain, muscle, and brown adipose tissue (Figure 1) (Bookout, Jeong et al. 2006). While little is known about the function of NURR1 in skeletal muscle, both NUR77 and NOR1 have been clearly shown to play important roles in regulating skeletal muscle metabolism (Maxwell, Cleasby et al. 2005) (Chao, Zhang et al. 2007) (Chao, Wroblewski et al. 2012) (Pearen, Myers et al. 2008) (Pearen, Eriksson et al. 2012).
In skeletal muscle, Nur77 is selectively expressed in glycolytic versus oxidative muscles, suggesting a positive role in regulating glucose metabolism (Chao, Zhang et al. 2007). NUR77 expression can be significantly induced by β-adrenergic signaling from the sympathetic nervous system to regulate muscle energy metabolism (Maxwell, Cleasby et al. 2005). Contrarily, skeletal muscle denervation reduces the expression of NUR77 as well as a subset of glucose metabolism genes, which is restored by the ectopic expression of Nur77 in denervated muscle (Chao, Zhang et al. 2007). The importance of NUR77 in regulating glucose metabolism can be further demonstrated by the over-expression of Nur77 in C2C12 cells, which not only induces glucose metabolism genes but also enhances cellular glucose transport (Chao, Zhang et al. 2007). Despite its role in regulating glucose metabolism, muscle-specific over-expression of Nur77 induces an oxidative fiber-type switch, similar to Pparδ and Errγ (Chao, Wroblewski et al. 2012). The transgenic muscle has typical characteristics of oxidative fibers such as increased fatty acid oxidation, higher mitochondrial OXPHOS activity, and fatigue resistance. However, glycogen, which is usually high in glycolytic fibers and low in oxidative fibers, is increased in the Nur77 transgenic muscle, suggesting a different working model for its fiber-type determination compared to PPARδ and ERRγ. More detailed analysis in fiber type composition, endurance performance, and gene expression profiling will be required to understand the mechanism of muscle remodeling induced by NUR77. In addition, the endogenous role of NUR77 in the β-adrenergic signaling cascade remains to be elucidated.
Similar to Nur77, Nor1 is also induced by β-adrenergic signaling in skeletal muscle (Pearen, Myers et al. 2008). However, NOR1 seems to participate more in regulating fatty acid metabolism rather than glucose. Knockdown of NOR1 in C2C12 cells reduces fatty acid oxidation and mitochondrial OXPHOS, but induces glycolysis (Pearen, Myers et al. 2008). Over-expression of an active form of Nor1 in skeletal muscle leads to a fiber-type switch from glycolytic to oxidative fibers (Pearen, Eriksson et al. 2012). The transgenic mice have increased running endurance, improved insulin sensitivity and glucose tolerance, and higher energy expenditure. Myoglobin expression and mitochondrial activity are both induced in the transgenic muscle. The fiber-type switch phenotype seems to be dependent on muscle groups, with overall more type IIa and IIx fibers but less type I and IIb fibers. This intermediate oxidative fiber-type switch might be due to the enhanced HDAC5 activity, which has been shown to promote oxidative fiber formation (Potthoff, Wu et al. 2007). However, the direct targets of NOR1 remain to be identified. It is also not clear how NOR1 activates HDAC5 and whether or not other pathways are involved in the fiber-type conversion induced by NOR1.
The REV-ERB Subfamily
There are two members in the REV-ERB subfamily of nuclear receptors: REV-ERBα and REV-ERBβ. REV-ERBs were originally discovered as orphan receptors (Miyajima, Horiuchi et al. 1989), but were later ‘adopted’ by the identification of heme as their physiological ligand (Raghuram, Stayrook et al. 2007). Upon heme binding, REV-ERBs recruit co-repressors such as NCoR1 and repress target gene expression (Raghuram, Stayrook et al. 2007) (Yin, Wu et al. 2007). REV-ERBs are active components of the circadian clock (Preitner, Damiola et al. 2002) (Bass 2012) and recent studies have also linked their functions to metabolic regulation in adipose tissues, liver, and muscle (Yang, Downes et al. 2006) (Kumar, Solt et al. 2010) (Cho, Zhao et al. 2012) (Woldt, Sebti et al. 2013). Anatomical profiling of NRs clusters REV-ERBs in the same group as ERRβ, ERRγ, NUR77, and NOR1, all of which are preferentially expressed in metabolically active tissues (Figure 1) (Bookout, Jeong et al. 2006). This further indicates an active role of REV-ERBs in regulating energy metabolism.
While little is known about the function of REV-ERBβ in skeletal muscle, REV-ERBα has recently been shown to positively regulate energy metabolism and mitochondrial OXPHOS function in muscle (Woldt, Sebti et al. 2013). Rev-erbα is expressed at higher levels in oxidative muscles compared to glycolytic muscles and exercise can further induce its expression (Woldt, Sebti et al. 2013). The importance of REV-ERBα in skeletal muscle has been demonstrated in Rev-erbα null mice. These mice have reduced voluntary wheel-running activity, diminished endurance exercise performance, and lower energy expenditure during exercise. The knockout muscle has decreased mitochondrial density, reduced OXPHOS activity, and down-regulated fatty acid metabolism genes (Woldt, Sebti et al. 2013). On the other hand, over-expression of Rev-erbα in C2C12 cells increases mitochondrial biogenesis and OXPHOS activity, accompanied with the induction of fatty acid metabolism genes. The in vivo over-expression of Rev-erbα in muscle via adeno-associated viral (AAV) infection also induces mitochondrial OXPHOS activity. These physiological changes seem to be mediated by the AMPK-Sirt1-PGC-1α signaling pathway which is down-regulated in the knockout muscle but up-regulated in Rev-erbα over-expressing muscle cells. In addition to its roles in regulating mitochondrial biogenesis and OXPHOS activity, muscle REV-ERBα is also involved in modulating mitochondrial autophagy (mitophagy) (Woldt, Sebti et al. 2013). Mitophagy is induced in Rev-erbα knockout muscle but suppressed in over-expressing C2C12 cells. REV-ERBα seems to directly bind to and repress genes in multiple steps of mitophagy, including the mitophagy regulator Park2, the autophagosome initiation factor Ulk1, the autophagosome elongation factors Atg5 and Bnip3, and the lysosomal enzymes Ctsl and Atpase6v1b2. Therefore, REV-ERBα increases mitochondrial number by both inducing mitochondrial biogenesis through the AMPK-Sirt1 pathway and reducing mitochondrial turnover by inhibiting mitophagy. However, it is not clear how AMPK is activated by Rev-erbα over-expression since the level of ATP is much lower in Rev-erbα knockout muscle (Woldt, Sebti et al. 2013), which is usually associated with AMPK activation. Also, the inhibition of mitophagy might be deleterious in the long-term due to the diminished clearance of dysfunctional mitochondria (Narendra, Tanaka et al. 2008) (Jin and Youle 2012).
NR Co-Regulatory Factors
The functions of nuclear receptors are finely modulated by associated co-activators and co-repressors. The abundance of these co-regulators and their post-translational modifications are regulated in response to a variety of physiological stimuli such as exercise and fasting, which then induce conformational changes in the NR-chromatin complexes and regulate their transcriptional activities. Recent studies have demonstrated important roles for NR co-regulators in energy metabolism and fiber-type determination in skeletal muscle.
PGC-1
The peroxisome proliferator-activated receptor γ coactivator 1α and β (PGC-1α and PGC-1β) are probably the best-known and most studied NR co-regulators implicated in energy metabolism. Both are highly expressed in metabolically-active tissues such as brain, heart, muscle, and brown adipose tissue, where they serve as co-activators for a number of transcription factors involved in energy metabolism regulation, including the PPAR and ERR nuclear receptors, and the nuclear respiratory factors 1 and 2 (NRF1 and NRF2/GABPA).
PGC-1α was first identified as a cold-inducible thermogenic factor in brown adipose tissue (Puigserver, Wu et al. 1998). In skeletal muscle, Pgc-1α is predominantly expressed in oxidative muscles like soleus (Wu, Puigserver et al. 1999). The expression of Pgc-1α can be induced by exercise or cold exposure in skeletal muscle (Puigserver, Wu et al. 1998) (Baar, Wende et al. 2002) (Russell, Feilchenfeldt et al. 2003). In addition to expression level, its co-transcriptional activity can also be modulated by a variety of post-translational modifications such as phosphorylation (Puigserver, Rhee et al. 2001) (Jager, Handschin et al. 2007), acetylation (Rodgers, Lerin et al. 2005), and methylation (Teyssier, Ma et al. 2005). When over-expressed in C2C12 muscle cells, Pgc-1α stimulates mitochondrial biogenesis by up-regulating the mitochondrial transcription factor A (Tfam) as well as the mitochondrial regulators Nrf1 and Nrf2. It can further function as a co-activator for NRF1 and NRF2 in up-regulating the expression of mitochondrial genes. In addition to mitochondrial biogenesis, Pgc-1α also stimulates mitochondrial uncoupling by up-regulating the mitochondrial uncoupling protein 2 (Ucp2), to further enhance mitochondrial energy expenditure (Wu, Puigserver et al. 1999). In vivo ectopic expression of Pgc-1α in skeletal muscle not only induces mitochondrial biogenesis and OXPHOS activity but also switches type IIb and IIx/d glycolytic fibers to type I and IIa oxidative fibers (Lin, Wu et al. 2002). As a result, the transgenic mice have improved endurance running performance (Calvo, Daniels et al. 2008). Loss-of-function studies, both whole-body and muscle-specific, show that Pgc-1α is required for proper mitochondrial OXPHOS and energy metabolism in skeletal muscle (Leone, Lehman et al. 2005) (Handschin, Choi et al. 2007). However, fiber-type composition and exercise-induced fiber-type switches are not affected by the knockout of Pgc-1α (Geng, Li et al. 2010). On top of that, a recent study shows that muscle mitochondrial biogenesis can still be induced by exercise without Pgc-1α (Rowe, El-Khoury et al. 2012), suggesting an alternate signaling pathway in remodeling skeletal muscle upon exercise induction.
PGC-1β was identified by its high homology with PGC-1α (Kressler, Schreiber et al. 2002) (Lin, Puigserver et al. 2002). It is also highly involved in regulating mitochondrial function and energy metabolism (Kamei, Ohizumi et al. 2003). In vitro over-expression of Pgc-1β in muscle cells has effects similar to Pgc-1α in terms of promoting mitochondrial biogenesis and oxidative fiber-type transformation (Mortensen, Frandsen et al. 2006). Similarly, skeletal muscle over-expression of Pgc-1β stimulates mitochondrial OXPHOS and fatty acid oxidation, along with oxidative fiber-type transformation (Arany, Lebrasseur et al. 2007). However, instead of a switch towards the most oxidative type I and IIa fibers as seen in the PGC-1α model, PGC-1β induces a more intermediate switch towards type IIx/d fibers (Arany, Lebrasseur et al. 2007), suggesting a different working mechanism. Whole-body or muscle-specific knockout of Pgc-1β causes reduced mitochondrial OXPHOS function in skeletal muscle but does not change fiber-type composition (Lelliott, Medina-Gomez et al. 2006) (Sonoda, Mehl et al. 2007) (Zechner, Lai et al. 2010). It would be expected that PGC-1α and PGC-1β compensate for each other when one is absent. This is true for their contributions in regulating mitochondrial function. Double knockout mice lacking Pgc-1α and Pgc-1β in skeletal muscle have significantly lower mitochondrial OXPHOS activity compared to the single knockout mice. However, the fiber-type composition of the double knockout mice is not different from the wild-type controls (Zechner, Lai et al. 2010). Therefore, PGC-1α and PGC-1β are necessary for mitochondrial OXPHOS function in skeletal muscle but appear dispensable for oxidative fiber-type determination.
RIP140
In addition to nuclear receptor co-activators, their co-repressors also contribute to the regulation of energy metabolism in skeletal muscle, one of which is the receptor-interacting protein 140 (RIP140). It was originally identified as a co-regulatory factor for the estrogen receptors (Cavailles, Dauvois et al. 1995). RIP140 is highly expressed in metabolic tissues such as fat and muscle (Leonardsson, Steel et al. 2004). In skeletal muscle, it is selectively expressed in glycolytic versus oxidative muscles (Seth, Steel et al. 2007), indicating a repressive role in regulating oxidative metabolism. Rip140 null mice show ~70% reduction in total fat content, mainly due to increased fatty acid oxidation and mitochondrial energy consumption in muscle and white adipose tissue (Leonardsson, Steel et al. 2004). The knockout mice have ~25% increase in whole-body energy expenditure and a lower respiratory exchange ratio, suggesting a shift towards fat utilization as energy source. In primarily glycolytic muscles where Rip140 is endogenously expressed, loss of Rip140 induces an oxidative fiber-type switch towards type IIa and IIx/d fibers, as well as increases in myoglobin content and mitochondrial biogenesis. Gene expression profiling further reveals significant induction of genes involved in fatty acid oxidation and mitochondrial OXPHOS in the knockout muscle (Seth, Steel et al. 2007). On the contrary, ectopic expression of Rip140 in oxidative muscles causes a reduction of oxidative fibers and myoglobin content. However, the exercise-induced fiber-type conversion is still retained in these transgenic mice (Seth, Steel et al. 2007). A subset of oxidative genes repressed by RIP140 are known targets of PPARs and ERRs and can be co-activated by PGC-1α, including Mcad, Cidea, Cpt1b, and Fabp3 (Christian, White et al. 2006) (Hallberg, Morganstein et al. 2008). Additionally, RIP140 is recruited to either known or predicted PPAR and ERR response elements at the promoters of these genes (Seth, Steel et al. 2007). Hence, RIP140 and PGC-1 could work in a yin-yang fashion in regulating the transcriptional activity of NRs such as PPARs and ERRs.
NCoR1
The nuclear receptor co-repressor 1 (NCoR1) was first identified as a ligand-independent transcriptional co-repressor for thyroid-hormone receptor (TR) and retinoic-acid receptor (RAR) (Horlein, Naar et al. 1995). It is ubiquitously expressed and is required for normal embryonic development (Jepsen, Hermanson et al. 2000). In skeletal muscle, NCoR1 is expressed at similar levels in oxidative and glycolytic muscles (Schuler, Buhler et al. 1999). However, in conditions when fatty acid metabolism is stimulated, such as during fasting, high-fat-diet challenge, and exercise, its expression in skeletal muscle is significantly reduced (Yamamoto, Williams et al. 2011) (Perez-Schindler, Summermatter et al. 2012), indicating that NCoR1 is involved in the repression of fatty acid metabolism. Muscle-specific deletion of NCoR1 increases muscle mass and exercise endurance (Yamamoto, Williams et al. 2011). The NCoR1 null mice have higher locomotor activity and whole-body energy expenditure. Similar to the over-expression of Pgc-1α or deletion of Rip140, the knockout of NCoR1 induces an oxidative fiber-type switch, associated with increased mitochondrial biogenesis and enhanced oxidative metabolism. In addition, there is a high overlap between the genes induced by the over-expression of Pgc-1α and knockout of NCoR1 or Rip140 in skeletal muscle. Similar to RIP140, NCoR1 functions through PPARs and ERRs in opposition to PGC-1α. It is recruited to PPAR or ERR response elements at their target gene promoters to repress their transcriptional activity, which can be antagonized by PGC-1α (Christian, White et al. 2006) (Perez-Schindler, Summermatter et al. 2012). Thus, the three co-regulatory factors work cooperatively with PPARs and ERRs in regulating skeletal muscle adaptation and energy metabolism. However the abundance of NCoR1 and PGC-1α, but not RIP140, fluctuates in response to exercise, suggesting they both play an important role in exerting exercise-induced muscle remodeling (Wright, Little et al. 2011).
AMPK
The adenosine monophosphate (AMP)-activated protein kinase (AMPK) is a central mediator of metabolism by sensing and regulating cellular energy supplies. It is activated when energy levels are low to restore energy balance by promoting catabolism and inhibiting anabolism (Hardie 2007). In skeletal muscle, the activity of AMPK is significantly higher in oxidative versus glycolytic muscles, indicating its contribution in maintaining the basal oxidative metabolism (Narkar, Fan et al. 2011). This was further confirmed by the in vivo over-expression of an inactive form of AMPK in skeletal muscle, which dramatically reduced endurance exercise capacity and induced insulin resistance and glucose intolerance (Fujii, Seifert et al. 2007) (Fujii, Ho et al. 2008). In addition to the basal oxidative metabolism, the activation of AMPK is also required for exercise-induced mitochondrial biogenesis via PGC-1α (Zong, Ren et al. 2002) (Jager, Handschin et al. 2007), in which AMPK is activated by exercise and directly phosphorylates PGC-1α and up-regulates its co-transcriptional activity (Jager, Handschin et al. 2007) (Narkar, Downes et al. 2008). In some NR genetic models where oxidative fiber-type conversion is induced, such as the muscle-specific over-expression of Pparδ, Pparγ, Errγ, or Rev-erbα, AMPK activity is also significantly increased (Amin, Mathews et al. 2010) (Narkar, Downes et al. 2008) (Narkar, Fan et al. 2011) (Woldt, Sebti et al. 2013). Furthermore, direct interaction between AMPK and PPARδ has been observed to synergistically activate target genes involved in oxidative metabolism (Narkar, Downes et al. 2008) (Gan, Burkart-Hartman et al. 2011). Thus, although AMPK is not a canonical NR co-regulator, it interacts with NRs and is highly involved in their regulation of energy metabolism (Fan, Downes et al. 2011).
Road to Exercise Mimetics
A common feature of NRs and AMPK is that their activities can be modulated by small molecule ligands, which makes them ideal pharmacological targets. Towards this end, a number of synthetic ligands have been developed for NRs including the ones described above. Some of these ligands have already been shown to promote skeletal muscle oxidative metabolism, including the PPARδ agonist GW501516 (Narkar, Downes et al. 2008), ERRβ/γ agonist GSK4716 (Rangwala, Wang et al. 2010), and REV-ERBα/β agonists SR9009 and SR9011 (Woldt, Sebti et al. 2013).
GW501516 was originally developed as a potent and selective PPARδ agonist (Oliver, Shenk et al. 2001). Its activation of PPARδ in cultured C2C12 muscle cells induces the expression of genes involved in fatty acid catabolism, mitochondrial OXPHOS, and cholesterol efflux (Dressel, Allen et al. 2003). GW501516 also works in vivo to enhance oxidative metabolism in skeletal muscle. Oral doses of 5mg/kg/day for 4 weeks significantly up-regulated oxidative genes such as Ucp3, Pdk4, and Cpt1a, similar to that seen with the muscle-specific over-expression of Pparδ (Luquet, Lopez-Soriano et al. 2003) (Wang, Zhang et al. 2004) (Narkar, Downes et al. 2008). The ligand activation of PPARδ alone did not stimulate any oxidative fiber-type switch or mitochondrial biogenesis in skeletal muscle, which is different from the muscle over-expression model. However, when co-administered with exercise training, GW501516 treatment increases the proportion of type I oxidative fibers by ~38% and mitochondrial biogenesis by ~50%, while training alone had little effect. In addition, the pairing of GW501516 treatment with exercise training dramatically increased endurance running performance compared to GW501516 treatment or training alone. Gene expression profiling revealed a unique oxidative gene signature, which is also found in the Pparδ transgenic muscle but not in either GW501516 treatment or training alone (Wang, Zhang et al. 2004) (Narkar, Downes et al. 2008). Thus, in vivo activation of PPARδ by oral administration of GW501516 enhances the effect of exercise training.
GSK4716 was identified as a specific agonist for ERRβ and ERRγ, without any crossover activity with the estrogen receptors (Zuercher, Gaillard et al. 2005). It seems to have good potential for promoting oxidative metabolism in skeletal muscle. In primary mouse myotubes, treatment with GSK4716 leads to up-regulation of all three Err genes and their co-activators Pgc-1α and Pgc-1β. Additionally, it induces the expression of genes involved in fatty acid oxidation, TCA cycle, and mitochondrial OXPHOS, such as Cpt1b, Idh3, and Atp5b. It also stimulates mitochondrial biogenesis as the mitochondrial citrate synthase activity and the amount of cytochrome c are both increased (Rangwala, Wang et al. 2010). However, no in vivo trial has been reported and more functional studies will be needed to fully assess its effect in skeletal muscle.
The synthetic REV-ERB agonists SR9009 and SR9011 were recently developed (Solt, Wang et al. 2012). Treatment with SR9009 or SR9011 increases the transcriptional repression of REV-ERBs on their target genes. In vivo, a single injection of SR9009 or SR9011 resulted in induction of genes involved in glycolysis, fatty acid catabolism, and mitochondrial OXPHOS, including Hk1, Pkm2, Pgc-1α, Cpt1b, Fatp1, and Ucp3. Mice treated with SR9011 for 12 days have increased energy expenditure with no change in RER, indicating that fatty acid and glucose oxidation are both induced. Additionally, 30 days of treatment with SR9009 significantly increased mouse running endurance. In C2C12 myotubes, treatment with SR9009 or SR9011 increased mitochondrial number (Woldt, Sebti et al. 2013). While the effects of these agonists on skeletal muscle seems promising, questions regarding the requirement for skeletal muscle REV-ERBs and how REV-ERBs activate energy metabolism genes remain to be answered.
In addition to the NR ligands, the AMPK activator AICAR also works as an exercise mimetic (Narkar, Downes et al. 2008). AICAR treatment for 4 weeks increases mouse energy expenditure and enhances running endurance by ~40%. It induces the expression of a number of genes linked to oxidative metabolism, including Scd1, Pdk4, Fasn, Lipe, and Dgat, most of which are also induced by the over-expression of Pparδ in skeletal muscle (Wang, Zhang et al. 2004). The stimulation of oxidative genes by AICAR seems to be dependent on PPARδ as AICAR fails to induce these genes in Pparδ null muscle cells. In addition, when administered together, AICAR and GW501516 synergistically activate PPARδ target genes such as Ucp3, Pdk4, and Lpl (Narkar, Downes et al. 2008). Therefore, activation of AMPK by its activator AICAR induces an oxidative gene signature change mediated by PPARδ, which causes skeletal muscle remodeling and enhances endurance. However, the mechanism of how AMPK synergistically activates PPARδ target genes remains to be elucidated.
Conclusions
Studies over the past decade have made it clear that nuclear receptors and their co-regulators are key regulatory components of energy metabolism and exercise-induced remodeling in skeletal muscle. Synthetic ligands targeting NRs and their co-regulators, including GW501516, AICAR, GSK4716, and SR9009/9011, have been developed and proven to be effective in enhancing or mimicking exercise effects. To date, many issues remain with the current generation of exercise mimetics, such as toxicity, side effects, and high dosage, which prevent their immediate clinical applications. However, with advances in our understanding of the molecular mechanism by which NRs regulate skeletal muscle physiology, we are optimistic that the next generation of exercise mimetics is not far away.
Acknowledgments
We thank L. Ong and C. Brondos for administrative assistance. R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute and March of Dimes Chair in Molecular and Developmental Biology. This work was supported by US National Institutes of Health grants (DK057978, DK090962, HL088093, HL105278, CA014195 and ES010337), the Glenn Foundation for Medical Research, the Leona M. and Harry B. Helmsley Charitable Trust, Ipsen/Biomeasure, and the Ellison Medical Foundation.
References
- Alaynick WA, Kondo RP, et al. ERRgamma directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metabolism. 2007;6(1):13–24. doi: 10.1016/j.cmet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Amin RH, Mathews ST, et al. Selective activation of PPARgamma in skeletal muscle induces endogenous production of adiponectin and protects mice from diet-induced insulin resistance. Am J Physiol Endocrinol Metab. 2010;298(1):E28–37. doi: 10.1152/ajpendo.00446.2009. [DOI] [PubMed] [Google Scholar]
- Arany Z, Lebrasseur N, et al. The transcriptional coactivator PGC-1 beta drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metabolism. 2007;5(1):35–46. doi: 10.1016/j.cmet.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Baar K, Wende AR, et al. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. Faseb Journal. 2002;16(14):1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- Barres R, Yan J, et al. Acute Exercise Remodels Promoter Methylation in Human Skeletal Muscle. Cell Metabolism. 2012;15(3):405–411. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Bass J. Circadian topology of metabolism. Nature. 2012;491(7424):348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annual Review of Biochemistry. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- Bergouignan A, Rudwill F, et al. Physical inactivity as the culprit of metabolic inflexibility: evidence from bed-rest studies. Journal of Applied Physiology. 2011;111(4):1201–1210. doi: 10.1152/japplphysiol.00698.2011. [DOI] [PubMed] [Google Scholar]
- Bookout AL, Jeong Y, et al. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126(4):789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braissant O, Foufelle F, et al. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology. 1996;137(1):354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- Calvo JA, Daniels TG, et al. Muscle-specific expression of PPAR gamma coactivator-1 alpha improves exercise performance and increases peak oxygen uptake. Journal of Applied Physiology. 2008;104(5):1304–1312. doi: 10.1152/japplphysiol.01231.2007. [DOI] [PubMed] [Google Scholar]
- Cavailles V, Dauvois S, et al. Nuclear Factor Rip140 Modulates Transcriptional Activation by the Estrogen-Receptor. Embo Journal. 1995;14(15):3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao LC, Wroblewski K, et al. Skeletal muscle Nur77 expression enhances oxidative metabolism and substrate utilization. Journal of Lipid Research. 2012;53(12):2610–2619. doi: 10.1194/jlr.M029355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao LC, Zhang Z, et al. Nur77 coordinately regulates expression of genes linked to glucose metabolism in skeletal muscle. Molecular Endocrinology. 2007;21(9):2152–2163. doi: 10.1210/me.2007-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Nathans J. Estrogen-related receptor beta/NR3B2 controls epithelial cell fate and endolymph production by the stria vascularis. Dev Cell. 2007;13(3):325–337. doi: 10.1016/j.devcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Chinsomboon J, Ruas J, et al. The transcriptional coactivator PGC-1alpha mediates exercise-induced angiogenesis in skeletal muscle. Proc Natl Acad Sci U S A. 2009;106(50):21401–21406. doi: 10.1073/pnas.0909131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Zhao X, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485(7396):123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, White R, et al. Metabolic regulation by the nuclear receptor corepressor RIP140. Trends in Endocrinology and Metabolism. 2006;17(6):243–250. doi: 10.1016/j.tem.2006.06.008. [DOI] [PubMed] [Google Scholar]
- de Lange P, Farina P, et al. Sequential changes in the signal transduction responses of skeletal muscle following food deprivation. FASEB J. 2006;20(14):2579–2581. doi: 10.1096/fj.06-6025fje. [DOI] [PubMed] [Google Scholar]
- Defronzo RA, Jacot E, et al. The Effect of Insulin on the Disposal of Intravenous Glucose - Results from Indirect Calorimetry and Hepatic and Femoral Venous Catheterization. Diabetes. 1981;30(12):1000–1007. doi: 10.2337/diab.30.12.1000. [DOI] [PubMed] [Google Scholar]
- Dressel U, Allen TL, et al. The peroxisome proliferator-activated receptor beta/delta agonist, GW501516, regulates the expression of genes involved in lipid catabolism and energy uncoupling in skeletal muscle cells. Molecular Endocrinology. 2003;17(12):2477–2493. doi: 10.1210/me.2003-0151. [DOI] [PubMed] [Google Scholar]
- Dufour CR, Wilson BJ, et al. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERR alpha and gamma. Cell Metabolism. 2007;5(5):345–356. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Eichner LJ, Giguere V. Estrogen related receptors (ERRs): a new dawn in transcriptional control of mitochondrial gene networks. Mitochondrion. 2011;11(4):544–552. doi: 10.1016/j.mito.2011.03.121. [DOI] [PubMed] [Google Scholar]
- Eudy JD, Yao S, et al. Isolation of a gene encoding a novel member of the nuclear receptor superfamily from the critical region of Usher syndrome type IIa at 1q41. Genomics. 1998;50(3):382–384. doi: 10.1006/geno.1998.5345. [DOI] [PubMed] [Google Scholar]
- Evans RM, Barish GD, et al. PPARs and the complex journey to obesity. Nat Med. 2004;10(4):355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- Fan W, Downes M, et al. Nuclear receptors and AMPK: resetting metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:17–22. doi: 10.1101/sqb.2012.76.010470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck BN, Bernal-Mizrachi C, et al. A potential link between muscle peroxisome proliferator-activated receptor-alpha signaling and obesity-related diabetes. Cell Metabolism. 2005;1(2):133–144. doi: 10.1016/j.cmet.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Foster C, Costill DL, et al. Skeletal muscle enzyme activity, fiber composition and VO2 max in relation to distance running performance. Eur J Appl Physiol Occup Physiol. 1978;39(2):73–80. doi: 10.1007/BF00421711. [DOI] [PubMed] [Google Scholar]
- Fujii N, Ho RC, et al. Ablation of AMP-activated protein kinase alpha2 activity exacerbates insulin resistance induced by high-fat feeding of mice. Diabetes. 2008;57(11):2958–2966. doi: 10.2337/db07-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii N, Seifert MM, et al. Role of AMP-activated protein kinase in exercise capacity, whole body glucose homeostasis, and glucose transport in skeletal muscle - Insight from analysis of a transgenic mouse model. Diabetes Research and Clinical Practice. 2007;77:S92–S98. doi: 10.1016/j.diabres.2007.01.040. [DOI] [PubMed] [Google Scholar]
- Gan Z, Burkart-Hartman EM, et al. The nuclear receptor PPARbeta/delta programs muscle glucose metabolism in cooperation with AMPK and MEF2. Genes Dev. 2011;25(24):2619–2630. doi: 10.1101/gad.178434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Z, Rumsey J, et al. Nuclear receptor/microRNA circuitry links muscle fiber type to energy metabolism. J Clin Invest. 2013;123(6):2564–2575. doi: 10.1172/JCI67652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng TY, Li P, et al. PGC-1 alpha plays a functional role in exercise-induced mitochondrial biogenesis and angiogenesis but not fiber-type transformation in mouse skeletal muscle. American Journal of Physiology-Cell Physiology. 2010;298(3):C572–C579. doi: 10.1152/ajpcell.00481.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguere V, Yang N, et al. Identification of a new class of steroid hormone receptors. Nature. 1988;331(6151):91–94. doi: 10.1038/331091a0. [DOI] [PubMed] [Google Scholar]
- Gollnick PD, Armstrong RB, et al. Effect of training on enzyme activity and fiber composition of human skeletal muscle. J Appl Physiol. 1973;34(1):107–111. doi: 10.1152/jappl.1973.34.1.107. [DOI] [PubMed] [Google Scholar]
- Gollnick PD, Saltin B, et al. Enzyme-Activity and Fiber Composition in Skeletal-Muscle of Untrained and Trained Men. Journal of Applied Physiology. 1972;33(3):312. doi: 10.1152/jappl.1972.33.3.312. [DOI] [PubMed] [Google Scholar]
- Grazina R, Massano J. Physical exercise and Parkinson’s disease: influence on symptoms, disease course and prevention. Rev Neurosci. 2013;24(2):139–152. doi: 10.1515/revneuro-2012-0087. [DOI] [PubMed] [Google Scholar]
- Hallberg M, Morganstein DL, et al. A Functional Interaction between RIP140 and PGC-1 alpha Regulates the Expression of the Lipid Droplet Protein CIDEA. Molecular and Cellular Biology. 2008;28(22):6785–6795. doi: 10.1128/MCB.00504-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton MT, Booth FW. Skeletal muscle adaptation to exercise: a century of progress. Journal of Applied Physiology. 2000;88(1):327–331. doi: 10.1152/jappl.2000.88.1.327. [DOI] [PubMed] [Google Scholar]
- Handschin C, Choi CS, et al. Abnormal glucose homeostasis in skeletal muscle-specific PGC-1alpha knockout mice reveals skeletal muscle-pancreatic beta cell crosstalk. J Clin Invest. 2007;117(11):3463–3474. doi: 10.1172/JCI31785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8(10):774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- He WM, Barak Y, et al. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(26):15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard DJ, Norby PL, et al. Human ERRgamma, a third member of the estrogen receptor-related receptor (ERR) subfamily of orphan nuclear receptors: tissue-specific isoforms are expressed during development and in the adult. Molecular Endocrinology. 2000;14(3):382–392. doi: 10.1210/mend.14.3.0431. [DOI] [PubMed] [Google Scholar]
- Hevener AL, He W, et al. Muscle-specific Pparg deletion causes insulin resistance. Nat Med. 2003;9(12):1491–1497. doi: 10.1038/nm956. [DOI] [PubMed] [Google Scholar]
- Hickey MS, Carey JO, et al. Skeletal muscle fiber composition is related to adiposity and in vitro glucose transport rate in humans. Am J Physiol. 1995;268(3 Pt 1):E453–457. doi: 10.1152/ajpendo.1995.268.3.E453. [DOI] [PubMed] [Google Scholar]
- Hong H, Yang L, et al. Hormone-independent transcriptional activation and coactivator binding by novel orphan nuclear receptor ERR3. Journal of Biological Chemistry. 1999;274(32):22618–22626. doi: 10.1074/jbc.274.32.22618. [DOI] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, et al. Ligand-Independent Repression by the Thyroid-Hormone Receptor-Mediated by a Nuclear Receptor Co-Repressor. Nature. 1995;377(6548):397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Hu FB, Willett WC, et al. Adiposity as compared with physical activity in predicting mortality among women. New England Journal of Medicine. 2004;351(26):2694–2703. doi: 10.1056/NEJMoa042135. [DOI] [PubMed] [Google Scholar]
- Huss JM, Kopp RP, et al. Peroxisome proliferator-activated receptor coactivator-1 alpha (PGC-1 alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma - Identification of novel leucine-rich interaction motif within PGC-1 alpha. Journal of Biological Chemistry. 2002;277(43):40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- Huss JM, I, Torra P, et al. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor at signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Molecular and Cellular Biology. 2004;24(20):9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347(6294):645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- Jager S, Handschin C, et al. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104(29):12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepsen K, Hermanson O, et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102(6):753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- Jin SM, Youle RJ. PINK1-and Parkin-mediated mitophagy at a glance. Journal of Cell Science. 2012;125(4):795–799. doi: 10.1242/jcs.093849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei Y, Ohizumi H, et al. PPAR gamma coactivator 1 beta/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(21):12378–12383. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten S, Seydoux J, et al. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103(11):1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Forman BM, et al. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc Natl Acad Sci U S A. 1994;91(15):7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Schreiber SN, et al. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor alpha. Journal of Biological Chemistry. 2002;277(16):13918–13925. doi: 10.1074/jbc.M201134200. [DOI] [PubMed] [Google Scholar]
- Kumar N, Solt LA, et al. Regulation of Adipogenesis by Natural and Synthetic REV-ERB Ligands. Endocrinology. 2010;151(7):3015–3025. doi: 10.1210/en.2009-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelliott CJ, Medina-Gomez G, et al. Ablation of PGC-1beta results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol. 2006;4(11):e369. doi: 10.1371/journal.pbio.0040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemanne D, Cassileth B, et al. The role of physical activity in cancer prevention, treatment, recovery, and survivorship. Oncology (Williston Park) 2013;27(6):580–585. [PubMed] [Google Scholar]
- Leonardsson G, Steel JH, et al. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(22):8437–8442. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone TC, Lehman JJ, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3(4):e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Puigserver P, et al. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta), a novel PGC-1-related transcription coactivator associated with host cell factor. Journal of Biological Chemistry. 2002;277(3):1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu H, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418(6899):797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Luo J, Sladek R, et al. Placental abnormalities in mouse embryos lacking the orphan nuclear receptor ERR-beta. Nature. 1997;388(6644):778–782. doi: 10.1038/42022. [DOI] [PubMed] [Google Scholar]
- Luo JM, Sladek R, et al. Reduced fat mass in mice lacking orphan nuclear receptor estrogen-related receptor alpha. Molecular and Cellular Biology. 2003;23(22):7947–7956. doi: 10.1128/MCB.23.22.7947-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luquet S, Lopez-Soriano J, et al. Peroxisome proliferator-activated receptor delta controls muscle development and oxidative capability. FASEB J. 2003;17(15):2299–2301. doi: 10.1096/fj.03-0269fje. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83(6):835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsakas A, Yadav V, et al. Revascularization of Ischemic Skeletal Muscle by Estrogen-Related Receptor-gamma. Circulation Research. 2012;110(8):1087–1096. doi: 10.1161/CIRCRESAHA.112.266478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsakas A, Yadav V, et al. Muscle ERRgamma mitigates Duchenne muscular dystrophy via metabolic and angiogenic reprogramming. FASEB J. 2013;27(10):4004–4016. doi: 10.1096/fj.13-228296. [DOI] [PubMed] [Google Scholar]
- Maxwell MA, Cleasby ME, et al. Nur77 regulates lipolysis in skeletal muscle cells -Evidence for cross-talk between the beta-adrenergic and an orphan nuclear hormone receptor pathway. Journal of Biological Chemistry. 2005;280(13):12573–12584. doi: 10.1074/jbc.M409580200. [DOI] [PubMed] [Google Scholar]
- Mellett LH, Bousquet G. Cardiology patient page. Heart-healthy exercise. Circulation. 2013;127(17):e571–572. doi: 10.1161/CIRCULATIONAHA.112.000880. [DOI] [PubMed] [Google Scholar]
- Mitsunaga K, Araki K, et al. Loss of PGC-specific expression of the orphan nuclear receptor ERR-beta results in reduction of germ cell number in mouse embryos. Mechanisms of Development. 2004;121(3):237–246. doi: 10.1016/j.mod.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Miyajima N, Horiuchi R, et al. Two erbA homologs encoding proteins with different T3 binding capacities are transcribed from opposite DNA strands of the same genetic locus. Cell. 1989;57(1):31–39. doi: 10.1016/0092-8674(89)90169-4. [DOI] [PubMed] [Google Scholar]
- Mortensen OH, Frandsen L, et al. PGC-1 alpha and PGC-1 beta have both similar and distinct effects on myofiber switching toward an oxidative phenotype. American Journal of Physiology-Endocrinology and Metabolism. 2006;291(4):E807–E816. doi: 10.1152/ajpendo.00591.2005. [DOI] [PubMed] [Google Scholar]
- Muoio DM, MacLean PS, et al. Fatty acid homeostasis and induction of lipid regulatory genes in skeletal muscles of peroxisome proliferator-activated receptor (PPAR) alpha knock-out mice. Evidence for compensatory regulation by PPAR delta. Journal of Biological Chemistry. 2002;277(29):26089–26097. doi: 10.1074/jbc.M203997200. [DOI] [PubMed] [Google Scholar]
- Murray J, Huss JM. Estrogen-related receptor alpha regulates skeletal myocyte differentiation via modulation of the ERK MAP kinase pathway. Am J Physiol Cell Physiol. 2011;301(3):C630–645. doi: 10.1152/ajpcell.00033.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. Journal of Cell Biology. 2008;183(5):795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narkar VA, Downes M, et al. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134(3):405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narkar VA, Fan W, et al. Exercise and PGC-1alpha-independent synchronization of type I muscle metabolism and vasculature by ERRgamma. Cell Metabolism. 2011;13(3):283–293. doi: 10.1016/j.cmet.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris AW, Chen L, et al. Muscle-specific PPARgamma-deficient mice develop increased adiposity and insulin resistance but respond to thiazolidinediones. J Clin Invest. 2003;112(4):608–618. doi: 10.1172/JCI17305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver WR, Shenk JL, et al. A selective peroxisome proliferator-activated receptor delta agonist promotes reverse cholesterol transport. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(9):5306–5311. doi: 10.1073/pnas.091021198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi A, Peng GH, et al. The orphan nuclear hormone receptor ERRbeta controls rod photoreceptor survival. Proc Natl Acad Sci U S A. 2010;107(25):11579–11584. doi: 10.1073/pnas.1000102107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearen MA, Eriksson NA, et al. The nuclear receptor, Nor-1, markedly increases type II oxidative muscle fibers and resistance to fatigue. Molecular Endocrinology. 2012;26(3):372–384. doi: 10.1210/me.2011-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearen MA, Muscat GEO. Minireview: Nuclear Hormone Receptor 4A Signaling: Implications for Metabolic Disease. Molecular Endocrinology. 2010;24(10):1891–1903. doi: 10.1210/me.2010-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearen MA, Myers SA, et al. The orphan nuclear receptor, NOR-1, a target of beta-adrenergic signaling, regulates gene expression that controls oxidative metabolism in skeletal muscle. Endocrinology. 2008;149(6):2853–2865. doi: 10.1210/en.2007-1202. [DOI] [PubMed] [Google Scholar]
- Perez-Schindler J, Summermatter S, et al. The corepressor NCoR1 antagonizes PGC-1alpha and estrogen-related receptor alpha in the regulation of skeletal muscle function and oxidative metabolism. Molecular and Cellular Biology. 2012;32(24):4913–4924. doi: 10.1128/MCB.00877-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perseghin G, Price TB, et al. Increased glucose transport-phosphorylation and muscle glycogen synthesis after exercise training in insulin-resistant subjects. N Engl J Med. 1996;335(18):1357–1362. doi: 10.1056/NEJM199610313351804. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Wu H, et al. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. Journal of Clinical Investigation. 2007;117(9):2459–2467. doi: 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, et al. The orphan nuclear receptor REV-ERB alpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, et al. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPAR gamma coactivator-1. Molecular Cell. 2001;8(5):971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu ZD, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Raghuram S, Stayrook KR, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERB alpha and REV-ERB beta. Nature Structural & Molecular Biology. 2007;14(12):1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala SM, Wang XM, et al. Estrogen-related Receptor gamma Is a Key Regulator of Muscle Mitochondrial Activity and Oxidative Capacity. Journal of Biological Chemistry. 2010;285(29):22619–22629. doi: 10.1074/jbc.M110.125401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, et al. Nutrient control of glucose homeostasis through a complex of PGC-1 alpha and SIRT1. Nature. 2005;434(7029):113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rowe GC, El-Khoury R, et al. PGC-1 alpha is Dispensable for Exercise-Induced Mitochondrial Biogenesis in Skeletal Muscle. PLoS One. 2012;7(7) doi: 10.1371/journal.pone.0041817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AP, Feilchenfeldt J, et al. Endurance training in humans leads to fiber type-specific increases in levels of peroxisome proliferator-activated receptor-gamma coactivator-1 and peroxisome proliferator-activated receptor-alpha in skeletal muscle. Diabetes. 2003;52(12):2874–2881. doi: 10.2337/diabetes.52.12.2874. [DOI] [PubMed] [Google Scholar]
- Sanyal S, Matthews J, et al. Deoxyribonucleic acid response element-dependent regulation of transcription by orphan nuclear receptor estrogen receptor-related receptor gamma. Molecular Endocrinology. 2004;18(2):312–325. doi: 10.1210/me.2003-0165. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Reggiani C. Fiber Types in Mammalian Skeletal Muscles. Physiological Reviews. 2011;91(4):1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- Schuler M, Ali F, et al. PGC1 alpha expression is controlled in skeletal muscles by PPAR beta, whose ablation results in fiber-type switching, obesity, and type 2 diabetes. Cell Metabolism. 2006;4(5):407–414. doi: 10.1016/j.cmet.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Schuler MJ, Buhler S, et al. Effects of contractile activity and hypothyroidism on nuclear hormone receptor mRNA isoforms in rat skeletal muscle. European Journal of Biochemistry. 1999;264(3):982–988. doi: 10.1046/j.1432-1327.1999.00706.x. [DOI] [PubMed] [Google Scholar]
- Seth A, Steel JH, et al. The transcriptional corepressor RIP140 regulates oxidative metabolism in skeletal muscle. Cell Metabolism. 2007;6(3):236–245. doi: 10.1016/j.cmet.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solt LA, Wang YJ, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485(7396):62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, I, Mehl R, et al. PGC-1beta controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc Natl Acad Sci U S A. 2007;104(12):5223–5228. doi: 10.1073/pnas.0611623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyssier C, Ma H, et al. Activation of nuclear receptor coactivator PGC-1 alpha by arginine methylation. Genes & Development. 2005;19(12):1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Quiat D, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17(5):662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villena JA, Hock MB, et al. Orphan nuclear receptor estrogen-related receptor alpha is essential for adaptive thermogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(4):1418–1423. doi: 10.1073/pnas.0607696104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YX, Zhang CL, et al. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2004;2(10):e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt MJ, Southgate RJ, et al. Suppression of plasma free fatty acids upregulates peroxisome proliferator-activated receptor (PPAR) alpha and delta and PPAR coactivator 1alpha in human skeletal muscle, but not lipid regulatory genes. J Mol Endocrinol. 2004;33(2):533–544. doi: 10.1677/jme.1.01499. [DOI] [PubMed] [Google Scholar]
- Wende AR, Huss JM, et al. PGC-1alpha coactivates PDK4 gene expression via the orphan nuclear receptor ERRalpha: a mechanism for transcriptional control of muscle glucose metabolism. Molecular and Cellular Biology. 2005;25(24):10684–10694. doi: 10.1128/MCB.25.24.10684-10694.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldt E, Sebti Y, et al. Rev-erb-alpha modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med. 2013;19(8):1039–1046. doi: 10.1038/nm.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D, Little J, et al. Reductions in RIP140 are not required for exercise and high fat diet mediated increases in mitochondrial enzymes. Faseb Journal. 2011;25 doi: 10.1152/japplphysiol.00279.2011. [DOI] [PubMed] [Google Scholar]
- Wu H, Rothermel B, et al. Activation of MEF2 by muscle activity is mediated through a calcineurin-dependent pathway. Embo Journal. 2001;20(22):6414–6423. doi: 10.1093/emboj/20.22.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZD, Puigserver P, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98(1):115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Wurtz JM, Bourguet W, et al. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3(2):206. doi: 10.1038/nsb0296-206. [DOI] [PubMed] [Google Scholar]
- Xie W, Hong H, et al. Constitutive activation of transcription and binding of coactivator by estrogen-related receptors 1 and 2. Molecular Endocrinology. 1999;13(12):2151–2162. doi: 10.1210/mend.13.12.0381. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Williams EG, et al. NCoR1 Is a Conserved Physiological Modulator of Muscle Mass and Oxidative Function. Cell. 2011;147(4):827–839. doi: 10.1016/j.cell.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Okutsu M, et al. Regulation of exercise-induced fiber type transformation, mitochondrial biogenesis, and angiogenesis in skeletal muscle. J Appl Physiol. 2011;110(1):264–274. doi: 10.1152/japplphysiol.00993.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XY, Downes M, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126(4):801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, et al. Rev-erb alpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318(5857):1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Zechner C, Lai L, et al. Total skeletal muscle PGC-1 deficiency uncouples mitochondrial derangements from fiber type determination and insulin sensitivity. Cell Metabolism. 2010;12(6):633–642. doi: 10.1016/j.cmet.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZP, Teng CT. Estrogen receptor-related receptor alpha 1 interacts with coactivator and constitutively activates the estrogen response elements of the human lactoferrin gene. Journal of Biological Chemistry. 2000;275(27):20837–20846. doi: 10.1074/jbc.M001880200. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Alvares K, et al. Cloning of a new member of the peroxisome proliferator-activated receptor gene family from mouse liver. Journal of Biological Chemistry. 1993;268(36):26817–26820. [PubMed] [Google Scholar]
- Zong HH, Ren JM, et al. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(25):15983–15987. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuercher WJ, Gaillard S, et al. Identification and structure - Activity relationship of phenolic acyl hydrazones as selective agonists for the estrogen-related orphan nuclear receptors ERR beta and ERR gamma. Journal of Medicinal Chemistry. 2005;48(9):3107–3109. doi: 10.1021/jm050161j. [DOI] [PubMed] [Google Scholar]
- Zurlo F, Larson K, et al. Skeletal-Muscle Metabolism Is a Major Determinant of Resting Energy-Expenditure. Journal of Clinical Investigation. 1990;86(5):1423–1427. doi: 10.1172/JCI114857. [DOI] [PMC free article] [PubMed] [Google Scholar]