Abstract
Plant MADS-domain transcription factors act as key regulators of many developmental processes. Despite the wealth of information that exists about these factors, the mechanisms by which they recognize their cognate DNA-binding site, called CArG-box (consensus CCW6GG), and how different MADS-domain proteins achieve DNA-binding specificity, are still largely unknown. We used information from in vivo ChIP-seq experiments, in vitro DNA-binding data and evolutionary conservation to address these important questions. We found that structural characteristics of the DNA play an important role in the DNA binding of plant MADS-domain proteins. The central region of the CArG-box largely resembles a structural motif called ‘A-tract’, which is characterized by a narrow minor groove and may assist bending of the DNA by MADS-domain proteins. Periodically spaced A-tracts outside the CArG-box suggest additional roles for this structure in the process of DNA binding of these transcription factors. Structural characteristics of the CArG-box not only play an important role in DNA-binding site recognition of MADS-domain proteins, but also partly explain differences in DNA-binding specificity of different members of this transcription factor family and their heteromeric complexes.
INTRODUCTION
The MADS-domain is a conserved DNA-binding domain present in a eukaryote-wide family of transcription factors (TFs). MADS-domain proteins typically contact their cognate binding site, the CArG-box (consensus: CCW6GG) as dimers (1). Structural analysis of animal and yeast MADS-domain protein dimers revealed that central parts of their MADS-domains form an antiparallel coiled-coil, made of two amphipathic α helices—one from each subunit. This coiled coil lies flat on the DNA minor groove (2). The N-terminal regions penetrate into the minor groove and stabilize bending of the DNA. The C-terminal part of the MADS-domain forms β-sheets that allow protein dimerization (2–4).
The family of MADS-box genes has dramatically expanded during plant evolution, and in particular in flowering plants (5). Two major classes of MADS-domain proteins can be distinguished: type I proteins, which are a heterogeneous group of proteins having only the MADS-domain in common, and type II proteins, which have a highly conserved modular domain architecture (5). In type II proteins, which are also called MIKC-type proteins, the MADS-domain (‘M’)is followed by an intervening (‘I’) domain, which is predicted to form an α helix and contributes to the selection of dimer partners (6). After the I-domain a keratin-like (‘K’) domain is located, which, presumably, assembles into coiled-coil structures enabling dimer and higher-order complex formation. The K-domain is followed by a highly variable C-terminus that has roles in transcriptional regulation (7). MIKC-type genes function as master regulators of developmental phase transitions, meristem and floral organ specification. Their encoded proteins function together in a combinatorial manner, as they interact with each other forming heterodimers and higher-order molecular complexes (8–11) [for review, see (12)].
The function of each MADS-domain protein complex is presumably achieved by regulating partly different sets of target genes through specific binding to their DNA regulatory elements. Although the CArG-box motif is the common DNA-binding consensus sequence of the MADS-domain TF family, several variants of the CArG-box exist that differ in length of the A/T-rich region in the central part of the motif and still can be considered as MADS-domain TF binding sites (13). However, the main in vivo determinants of MADS-domain TF binding site recognition and their DNA-binding specificity remain enigmatic. To understand the various important and specialized roles of MADS-domain TFs in plant development, it is essential to understand the mechanisms of the DNA-binding site recognition by this diverse family of TFs.
The identification of in vivo DNA-binding events of MADS-domain TFs at genome-wide scale provides novel opportunities to study parameters and factors influencing DNA-binding site recognition. Chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) or hybridization to tiling arrays (ChIP-CHIP) has allowed to generate genome-wide binding maps of several MADS-domain TFs involved in floral transition (14,15) and flower development (16–18). Especially a study on the floral MADS-domain TF SEPALLATA3 (SEP3), which acts as a mediator of higher-order interactions among floral MADS-domain proteins, has revealed that the CArG-box consensus sequence (CCW6GG) has only poor predictability for DNA-binding ‘in planta’ (17): only 7.7% of all perfect CArG-boxes are bound by SEP3, and only 17% of the SEP3 binding events identified contain a perfect CArG-box consensus. This indicates that the perfect CArG-box consensus is not an optimal definition for the in vivo DNA binding of MADS-domain proteins
In this article, we analyze the structural properties of DNA regions bound by specific MADS-domain TFs to unravel DNA sequence determinants affecting their binding affinity. Our results show that regions bound by MADS-domain TFs have a tendency to display particular structural properties, and that these structural properties may play a role in determining the DNA-binding specificity of different MADS-domain protein dimers. In particular, our results show that certain structural elements called A-tracts facilitate MADS-domain TF DNA binding when located inside the CArG-box motif and periodically distributed around it.
MATERIALS AND METHODS
Bioinformatic analysis of ChIP experiments
ChIP-seq data sets for SEP3 (17), AP1 (16) and FLC (19), and ChIP-chip data sets for SVP (14) and SOC1 (14) were re-analyzed in this study. For ChIP-seq experiments, sequence reads were mapped to the Arabidopsis thaliana (TAIR9) genome using SOAPv2 (20). Reads mapped to multiple regions or to the mitochondria or chloroplast genome were discarded. We modified the R package CSAR (21) to generate read-enrichment score values at each single-nucleotide position, without performing peak calling. This score represents the ratio between density of reads overlapping a given nucleotide in the IP sample versus the control sample after normalization. For ChIP-chip experiments, probe sequences were remapped to the TAIR9 Arabidopsis genome with the Starr package (22). Only probes that mapped to unique locations were retained. Subsequently, CisGenome (23) was used to detect potential binding regions, using the hidden Markov model to combine intensities of neighboring probes. In this case, the score value ranges between 0 and 1, where 1 is the most significant.
Subsequently, all CArG-box motifs (CCW6GG) were located in the TAIR9 genome. We used three definitions for the CArG-box consensus: (i) perfect CArG-box (CCW6GG); (ii) long CArG-box (CCW7G); and (iii) short CARG-box (CCW4S2GG). No mismatches were allowed. Afterwards, instead of performing a peak calling step directly on the ChIP-seq and ChIP-chip data, we defined regions 250 bp around each 10 bp motif (510 bp in total) and we assigned them a ChIP score with the maximum ChIP-seq or ChIP-chip score in that region. For ChIP-seq analysis, a ChIP-seq threshold was calculated for false discovery rate (FDR) < 0.05 using the function ‘permutatedWinScores’ from the package CSAR. This threshold was used to define a set of bound and unbound regions. We defined regions bound by SEP3 in ‘wild-type’ but not in agamous mutant, as these regions with a SEP3 (wt) ChIP-seq score >4.15 (FDR < 0.05) and a SEP3 (ag mutant) ChIP-seq score <1. Scores of ≤1 indicate that the normalized number of mapped reads in the control sample is equal to or larger than in the IP sample.
Analyzing DNA structural properties
Dinucleotide properties (73 in total) were obtained from the DiProDB database (24). They were used to estimate several properties of the DNA at each dinucleotide step. From these properties, we calculated average differences between the set of regions identified as bound by SEP3 in our ChIP-seq analysis (FDR < 0.05) and the set of regions identified as SEP3 unbound.
A-tract elements were defined with the motif AmTn, where n + m > 3. The length of a consecutive stretch of A followed by T was counted in both cases as the maximum n + m in the consensus AmTn.
DNA conservation studies
The aligned DNA sequences of 81 A. thaliana accessions were obtained from the 1001 genome project (http://www.1001genomes.org/; release 5 December 2010). We associated CArG-box motifs with the SEP3 ChIP-Seq score in the accession Col-0, and we extracted their corresponding sequence in the other accessions. Only sequences where all nucleotides has been identified were considered; sequences containing Ns were removed from the analysis. These regions were classified depending on the presence or absence of an A-tract element in the Col-0 accession. For the conservation analysis, only CArG-box regions that have, at least, one SNP in one ecotype compared with Col-0 on their 10 bp sequences were considered. The proportion of CArG-box regions with conserved A-tract element length was calculated as the ratio between the number of CArG-box regions at a given ChIP-seq score threshold that have exactly the same length for the A-tract element in all ecotypes considered divided by the total number of CArG-box regions at that ChIP-seq score threshold.
Quantitative multiple fluorescence relative affinity (QuMFRA)
QuMFRA experiments were performed as described previously (25). Oligonucleotide sequences used in the ‘AG intron’ experiments (Figure 4 and Supplementary Figure S8) were derived from the first intron of the AG locus and contained a single CArG-box with an A-tract element of length four inside. The probe ‘AG wt’ has the sequence: 5′-TATATATATT(CCAAATAAGG)AAAGTATGGA. The probe ‘AG mut’ represent the same sequence, but the A-tract element inside of the CArG-box was eliminated by the substitution of ApA or ApT steps by TpA, exactly, it has the sequence: 5′-TATATATATT(CCTATATAGG)AAAGTATGGA. CArG-box sequences are represented in bold, and substitutions are underlined.
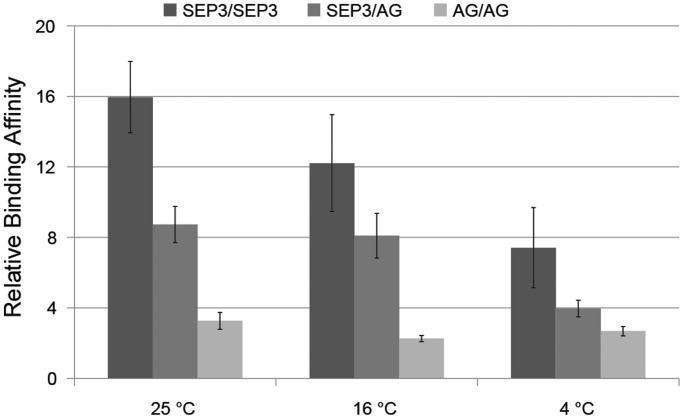
Figure 4.
Temperature-dependent DNA affinity of MADS-domain complexes. Relative binding affinity of three MADS-domain complexes to a probe representing the AGAMOUS intron relative to the affinity to a probe representing the same region but with the A-tract element inside of the CArG-box mutated (see ‘Materials and Methods’ section). The relative affinity was studied at different temperatures by QuMFRA experiments. Error bars indicates standard error calculated out of six replicates. Supplementary Figure S8 shows the images of gels of two replicates from which these affinities were calculated.
Oligonucleotide sequences used in the SOC1 promoter studies (Supplementary Figure S7) were derived from the SOC1 promoter and contained two CArG-boxes [CArG III: −96 bp and CArG-box IV: −125 bp as described by Immink et al., 2012 (15)] separated by four A-tract elements. Probe ‘SOC1 wt’ has the sequence 5′-TTG(CTATTTTTGG)TCCCTCGGATTACTAAAGAAAACGTAACTTAGAAATCCAATAATAATTCAGCTTATCGAACGTCTTGTCTAGCTAGTGGCACCAAAAAAATAT(CCTTTTTTGG)AGA, and probe ‘SOC1 mut1 represents the same sequence but the four A-tract elements were eliminated by the substitution of ApA or ApT steps by TpA. A-tract elements inside of the two CArG-boxes were not modified. Exactly, the sequence is 5′-TTG(CTATTTTTGG)TCCCTCGGATTACTAAAGATATCGTAACTTAGATATCCAATAATATATCAGCTTATCGAACGTCTTGTCTAGCTAGTGGCACCATATATATAT(CCTTTTTTGG)AGA. CArG-boxes are indicated within parentheses, and bold and mutated nucleotides are underlined and bold.
Single-stranded DNA oligonucleotides were commercially synthesized, annealed and inserted into pGEM-T vector (Promega). Double-stranded DNA (dsDNA) fragments were amplified by PCR with infrared 5′-fluorescent-labeled (Dy682 or Dy782) primers specific for pGEM-T vector, gel-purified and their concentration was measured. Electrophoretic mobility shift assays (EMSAs) were performed as described previously (11), with 2 µl of in vitro synthesized proteins (TNT Coupled Wheat Germ Extract, Promega) with an equimolar (75 fmol each) mixture of two different dsDNA sequences each labeled with a different IR-fluorophore. Both the protein–DNA-binding reaction and the EMSA were performed in temperature-controlled environments at 4°C (cold room), 16°C (water bath) and 25°C (incubator). Low voltage of the electrophoresis run (75 V/6.8 cm gel) was applied to avoid temperature change within the gel-running chamber during the run. EMSA gels were scanned with Odyssey Infrared Imaging System (Li-Cor) and the band shift signals were quantified using Odyssey Software v1.2 (Li-Cor) taking ‘Integrated Intensity’ (I.I.) parameter for further quantification process. Relative binding affinity [Kb(D1)/Kb(D2)] of the protein complex considered to probe 1 (D1) compared with probe 2 (D2) was calculated as described previously (25) using the equation Kb(D1)/Kb(D2) = ([P·D1]*[D2])/([P·D2]*[D1]), where [P·Di] is estimated as the intensity of the bound dsDNA probe i (Di), and [Di] is estimated as the intensity of the free dsDNA probe i (Di) within a single EMSA lane after background noise subtraction. The relative binding affinity was measured based on six independent QuMFRA replicates for the AG intron element measurements and four replicates for the SOC1 promoter sequence element. For both experiments, half of the replicates were done with probe 1 labeled with Dy682 and probe 2 with Dy782, and the other half with probe 1 labeled with Dy782 and probe 2 with Dy682.
RESULTS
CArG-boxes bound by SEP3 complexes are defined by particular DNA structural properties
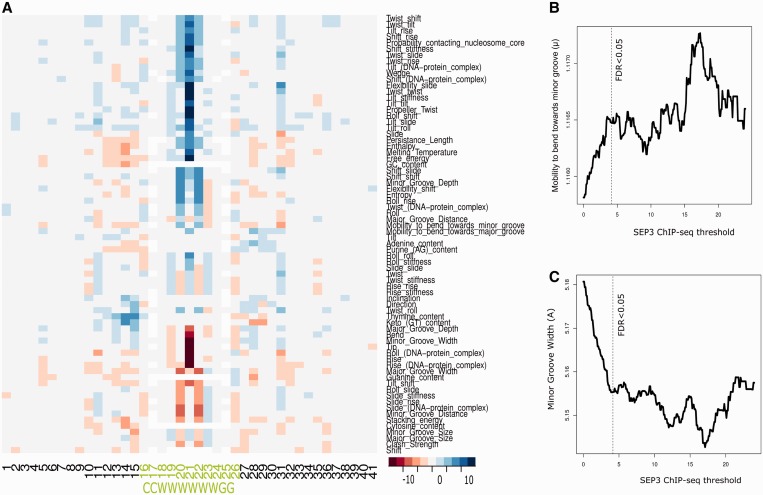
To understand the specificity of SEP3 DNA binding, we identified patterns of DNA structural properties common to a set of ‘functional’ CArG-boxes as identified by SEP3 binding (FDR < 0.05) (17). We focussed on SEP3 because of its ability to form complexes with several other MADS-domain TFs and, therefore, to give a broad picture of the MADS-domain TF binding events. To do so, we estimated DNA structural properties, as defined in the dinucleotide property database [DiProDB (24)], for each dinucleotide step of regions around all (7741) CArG-boxes (CCW6GG) in the Arabidopsis nuclear genome. Figure 1A shows a heatmap representing regions with different structural properties obtained by comparing CArG-box regions bound (FDR < 0.05) versus not bound by SEP3 at each dinucleotide position using a t-test statistic. The three most central dinucleotides and the flanking regions of the CArG-box sequence showed the highest differences when comparing CArG-boxes that are bound versus non-bound by SEP3, indicating that the structural properties of these locations are important for binding (Figure 1A). To identify which properties show the best correlation with the SEP3 ChIP-score, we related the average property value over the 10-bp CArG-box sequence with its associated SEP3 ChIP-seq score threshold value. We observed the strongest correlation with the ‘mobility to bend toward the minor groove’ (µ) property (r = 0.69; pv < 10−21; Figure 1B), which measures the ability of the DNA to be bent toward the minor groove by the Escherichia coli catabolite activator protein measured as the relative complex gel mobility (µ) (26). In addition, among the 10 structural properties with the strongest correlations to the SEP3 ChIP-seq score threshold, we found ‘minor groove width’ (Å) (r = −0.56; pv < 10−21; Figure 1C) and DNA ‘minor groove depth’ (Å) (r = 0.55; pv < 10−21). In summary, properties of the DNA groove and degree of bending seem to correlate with the SEP3 binding event.
Figure 1.
DNA structure properties of CArG-box regions bound by SEP3. (A) Heatmap showing which DNA properties at which location have significantly higher (−log10 P-value; blue color) or lower values (log10 P-value; red color) in the CArG-box regions bound by SEP3 (FDR<0.05) compared with CArG-box regions unbound by SEP3 using a t-test statistic. The three most central dinucleotides of the CArG-box and its flanking regions show the highest differences. The properties ‘mobility to bend toward the minor groove’ (B) and ‘minor groove width’ (C) are among the properties with the highest correlations with SEP3 ChIP-seq score. For panel (B) and (C), values are only plotted till such a ChIP-seq score threshold where the average property is calculated from at least 50 CArG-box regions.
A-tract elements are overrepresented in SEP3-bound CArG-box sequences
The structural properties of functional CArG-boxes (CCW6GG) that were detected in our analysis show striking similarities with the properties of DNA elements known as A-tracts. A-tracts have been defined as 4–8 consecutive A*T base pairs without a TpA step (27). The consensus of one A-tract element can be described with the motif: NiAmTnNj, where m+n > 3 and the total length of the motif being 10 bp. DNA regions containing in-phase A-tract repeats show a narrower minor groove width and higher bendability toward the minor groove than other AT-rich regions (27).
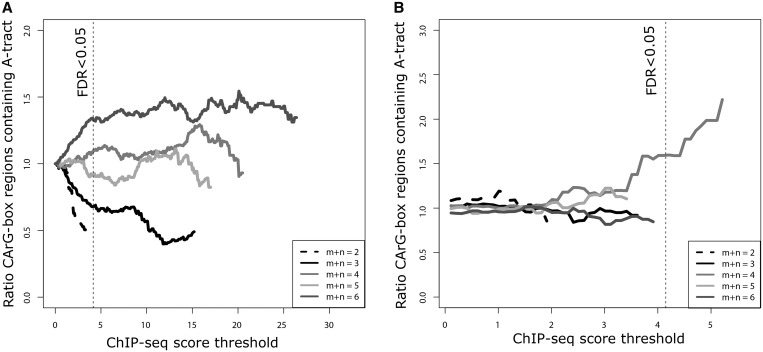
Because of their structural and sequence similarities, we studied how the presence of A-tracts in the CArG-box region relates with the binding of SEP3. Figure 2 shows that the normalized proportion of DNA regions containing an A-tract (m + n > 3) inside the 10 bp CArG-box sequence increases with the ChIP-seq score threshold used, supporting the idea of its positive dependency. In contrast, the proportion of regions without an A-tract inside the CArG-box (m + n < 4) tends to decrease with the threshold used. In particular, for SEP3 wt ChIP-seq (Figure 2A), the Pearson correlation (r) was −0.96 (pv < 2 × 10−16), −0.94 (pv < 2 × 10−16), 0.50 (pv < 2 × 10−14), 0.02 (pv < 0.81), 0.66 (pv < 2 × 10−16) for A-tract length of 2–6, respectively. When we eliminate from this data set the binding events that are also present in the SEP3 ag mutant ChIP-seq experiment, we expect to have an enrichment of binding events of complexes formed mainly by AG and SEP3. This allows to investigate if there is a different pattern of A-tract length enrichment depending on the type of SEP3 MADS-domain complex (Figure 1). Because of the large overlap of these two data sets, the subtraction of common binding sites will decrease the range of score values of the binding sites considered, and this is also the reason why several enrichment curves of Figure 2B do not reach the FDR < 0.05 threshold. The A-tracts of length 4 (pv < 0.06; hypergeometric test) and 6 (pv < 1.3 × 10−7; hypergeometric test) for SEP3 ‘wt’ and of length 4 (pv < 0.006; hypergeometric test) for SEP3 binding events not present in ag mutant showed the highest enrichment at the threshold level of FDR < 0.05 in the SEP3 ChIP-seq experiments studied. A similar pattern of enrichment for A-tracts was also observed for other MADS-domain TF ChIP-seq and -chip experiments and alternative definitions of the CArG-box consensus (Supplementary Figure S1–3). In particular, for FLC, AP1, SOC1 and SCP the A-tract element of length six was the one most strongly enriched.
Figure 2.
Enrichment of A-tract elements in SEP3-bound CArG-box sequences. The proportion of CArG-box motifs with a particular A-tract element inside normalized by the proportion of CArG-boxes with each particular A-tract element at genome-wide level and plotted against the ChIP-seq score threshold used, for (A) SEP3 ChIP-seq; and (B) SEP3 ChIP-seq regions that loose the binding event in the ag mutant. The figure shows an increase in the normalized proportion of CArG-box sequences containing an A-tract (m+n > 3) with the ChIP-seq score used. In contrast, the proportion of CArG-box motifs without an A-tract (m+n<4) decreases with the ChIP-seq score. Values are only plotted until a ChIP-seq score where there are at least 15 CArG-boxes to calculate the ratio. Dashed line indicates the SEP3 ChIP-seq threshold value for FDR < 0.05.
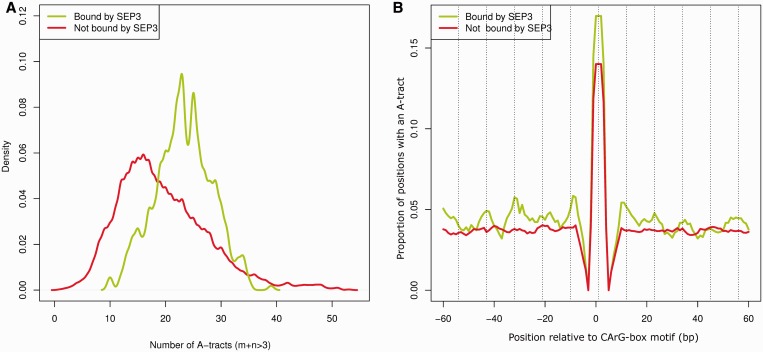
The flanking regions of CArG-boxes (CCW6GG), defined with an arbitrary length of 250 bp at each side (for a total of 510 bp), bound by SEP3 were also characterized by a higher presence of A-tract elements than the flanking regions of non-bound CArG-box regions (Figure 3A). This overrepresentation is not due to a different AT content, as when we eliminated the A-tract sequences from the studied regions the AT-content was almost identical (Supplementary Figure S4). Furthermore, we used a Fisher’s exact g-test (28) to test periodicity of the location of the A-tract elements for each single 510 bp CArG-box containing region. We found that 88% of the SEP3-bound (FDR < 0.05) CArG-box surrounding regions showed a significant periodicity on the location of A-tract elements (pv < 0.05), whereas the percentage for unbound regions containing CArG-boxes was only 67%. The distribution of the P-values for the g-test of bound and unbound regions was markedly different (t-test; pv < 10−15) (Supplementary Figure S5). Moreover, we studied the relative location distribution of the A-tracts elements to the middle position of the CArG-box sequence (Figure 3B and Supplementary Figure S6) and the estimated dominant A-tract location periodicity for the 510 bp regions bound by SEP3 was found to be 22.1 bp. This distance was estimated as the average distance (1/dominant frequency) for each SEP3-bound (FDR < 0.05) CArG-box region that shows a significant (pv < 0.05) periodicity.
Figure 3.
Multiple A-tracts in SEP3-bound CArG-box regions. (A) Distribution of multiple A-tracts (m+n > 3) elements within the 250-bp region at either side of the CArG-box motif (510-bp region in total) bound by SEP3 or not bound by SEP3. (B) Proportion of CArG-box regions with an A-tract element in a particular position. A moving average of length 5 bp was applied to obtain a more smooth representation of the data. Regions with a SEP3 ChIP-seq binding event (FDR < 0.05) are indicated in green, and regions without a binding event are indicated in red. Dashed lines are located each 11 bp from the middle of the CArG-box motif, representing a helical turn. For illustrative reasons only the region −60 to 60 bp is shown, for the 510 bp region see Supplementary Figure S6.
The bioinformatics analyses suggest a role for periodically distributed A-tract elements in DNA binding of MADS-domain protein complexes. It is possible that the flanking regions of CArG-box sequences may facilitate the looping of the DNA by higher-order complexes of MADS-domain proteins. Therefore, next, we experimentally studied the importance of A-tract elements for MADS TF/DNA complex formation. The SOC1 promoter contains two CArG-box sequences where SEP3 is able to bind (15). Each CArG-box sequence contains one A-tract element and they are separated by four A-tract elements. We studied the affinity of SEP3 to a probe representing this region compared with a probe representing the same region but with the A-tract elements between the two CArG-boxes mutated by substitution of the ApA or ApT steps by TpA. The relative affinity of the SEP3 homodimer seemed to be slightly affected by the elimination of the A-tract elements (1.4-fold change for the unmutated probe compared with the mutated; standard error 0.07 over 4 replicates; Supplementary Figure S7). However, the relative affinity of a SEP3 higher order complex to the unmutated probe had an increase of 3.6-fold (standard error 0.26 over 4 replicates; Supplementary Figure S7) when compared with the mutated probe, indicating that the location of A-tract elements between the two CArG-boxes on the SOC1 promoter facilitates the formation of the SEP3 tetramer (or higher-order protein)–DNA complexes.
A-tract DNA curvature plays a role in the DNA-binding specificity of MADS-domain proteins
Our analysis of ChIP-seq data presented previously suggests the importance of A-tracts for DNA binding by MADS-domain proteins. Because A-tract length is related with the degree of curvature of the DNA region where it is located and because several MADS-domain protein homo- and heterodimers bend the DNA in vitro at different degrees (29,30), we analyzed the in vivo preference of MADS-domain protein complexes within CArG-box sequences with different A-tract length. ChIP-seq experiments identify the binding regions of a set of protein complexes targeted by the used antibody. To narrow the specificity to particular protein complexes, one can compare ChIP-seq experiments in mutants lacking some of the potential protein binding partners. In such a way, DNA regions detected by the SEP3 ChIP-seq experiment in wild-type (wt) but not in the ‘agamous’ (ag) mutant (17) are expected to be mainly bound by protein complexes containing SEP3 and AG. These DNA regions are enriched in CArG-boxes with an A-tract of length 4 (Figure 2B), in contrast to the preferences of length 4 and 6 in the wt ChIP-seq experiment (Figure 2A). These results indicate that some MADS-domain protein complexes, e.g. the SEP3-AG heterodimer, have a preference for CArG-boxes with particular A-tract properties.
DNA curvature of regions containing A-tract elements strongly depends on the temperature. Koo et al. (31) found a decrease in bending magnitude when passing from 4°C to room temperature, and Diekmann et al. (32) revealed that the decrease with temperature is monotonic. This property enables us to modulate the DNA curvature of the same DNA sequence fragments, and, therefore, to experimentally study the importance of DNA curvature in the DNA-binding affinity and specificity of MADS-domain proteins. We used QuMFRA experiments at different temperatures to estimate the relative affinities of three MADS-domain protein combinations (only SEP3, SEP3 and AG, and only AG) to a probe representing the AG intron compared with a probe representing the AG intron where the A-tract element inside the CArG-box region was mutated by the introduction of TpA steps. We chose these three combinations because our analysis (Figure 2A and B) showed that the SEP3-AG heterodimer has a preference to A-tract elements of different length than other SEP3 complexes, and we also added the AG-AG homodimer to be sure that the temperature-dependent changes in affinities were not only due to a temperature-dependent change in the proportion of homodimer/heterodimer formed on the mix. The results of the QuMFRA experiments (Figure 4, Supplementary Figure S8) show that the elimination of the A-tract element decreases the affinity of three MADS-domain protein/DNA complexes compared with the ‘wt’ sequence in 2–16-fold depending on the dimer and condition considered. This supports our hypothesis of the importance of A-tract elements inside the CArG-box sequence to facilitate DNA binding. Strikingly, their relative affinity changed with the temperature (Figure 4). Although the DNA binding of the AG homodimer is relatively independent of the temperature, the relative affinity of the SEP3 homodimer and SEP3-AG heterodimer depends more strongly on the temperature.
A-tract length in SEP3 binding sites is conserved among Arabidopsis ecotypes
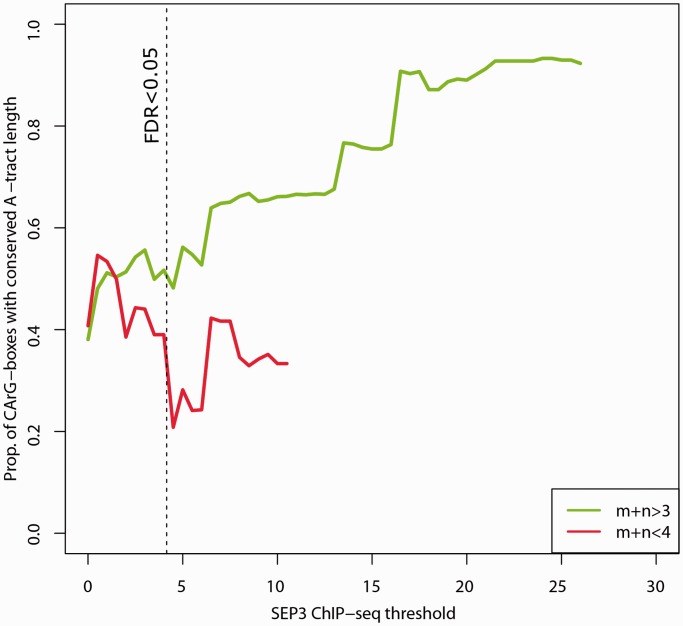
To further assess the functional importance of A-tract length within the central CArG-box core sequence, we analyzed DNA sequence conservation. The proportion of 10 bp Col-0 CArG-box sequences with conserved length of their A-tract among the 81 sequenced Arabidopsis ecotypes (1001 genome project) is higher in regions bound by SEP3 TF complexes than in CArG box sequences without SEP3 binding (Figure 5; Pearson correlation r = 0.97; pv < 2 × 10−16). In contrast, the proportion of CArG-box sequences with conserved length of consecutive A and T base pairs for non A-tracts (m + n < 4) decreases with the SEP3 ChIP-seq score (Pearson correlation r = −0.52; pv < 0.012). This supports not only the functionality of the A-tract inside the CArG-box sequence but also the importance of its length.
Figure 5.
Conservation of the A-tract length in functional CArG-box regions. The average proportion of CArG-box motifs with conserved length of the motif AmTn among the 81 A. thaliana ecotypes (see ‘Material and Methods’ section) are shown as a function of the SEP3 ChIP-seq score threshold. Green, A-tract element with length 4–6; and red, AT-regions with length 2–3 (non–A-tract elements). Only CArG-box motifs with at least one SNP compared with Col-0 inside the 10 bp CArG-box region in at least one ecotype are considered. Proportions are only plotted to such threshold level that at least 15 CArG-box sequences are considered. Vertical dashed line indicates the threshold score value corresponding to FDR < 0.05.
DISCUSSION
The 10 bp DNA sequence motif known as CArG-box represents the DNA-binding consensus of MADS-domain TFs. Previous studies focused on the characterization of the primary DNA sequence of this binding site, largely omitting the importance of structural properties of the DNA. Since the first structural characterization of the DNA-binding domain of an animal MADS-domain TF in 1995 (2), it has been suggested that this family of TFs binds DNA by the interaction of their amino acids mainly with the minor groove side of the DNA. This type of recognition usually relies on structural properties of the DNA more than a specific sequence of DNA bases (33). Here, we studied the importance of the DNA structure as a determinant in the DNA recognition and specificity of MADS-domain TFs.
We studied a set of 73 DNA properties as potential factors that can influence the binding of plant SEP3 TF complexes. Among the most significant properties associated with functional CArG-boxes were those related to the minor DNA groove and bendability of DNA. Genomic regions bound by SEP3 complexes were also found to be associated with the presence of periodically distributed A-tracts elements. These elements are known to confer a particularly high level of curvature and narrow minor groove width to the DNA regions where they are periodically located. Interestingly, previous in vitro studies have shown that some MADS-domain TFs are able to bend the DNA at different degrees [e.g. 53° by AP1, 70° by AG; (29)]. We hypothesize that the affinity of MADS-domain TFs could be related with the energy needed to modify the DNA conformation to the one observed on binding, and therefore, DNA-binding affinity will depend on a priori structural properties of the DNA. This mechanism of DNA-binding recognition has been already proposed for the human protein NF-κB (34), where DNA bending in the binding site of this factor in the bound state is similar to the bending already present in its free state. This particular bent conformation seems to be facilitated by the presence of A-tract elements. Suggesting a similar mechanism for the MADS-domain TF binding event, we found a positive association of A-tracts inside CArG-box sequences and in vivo MADS-domain TF binding. Our in vitro experiments on a sequence representing the AG intron also support the importance of A-tract elements inside of the CArG-box motif, as its presence increases the relative affinity of SEP3 complexes between 2–16-fold, depending on the dimer and temperature considered. Additionally, MADS-domain TFs can form quaternary protein complexes that loop the DNA around two CArG-box elements (8,9,11,35–37). We hypothesize that our observed periodicity of A-tracts in CArG-box flanking regions could be associated with the need of looping the DNA by higher order complexes in vivo. In fact, our in vitro experiment on the SOC1 promoter shows that the elimination of A-tract elements between two CArG-boxes with the simple substitution of ApA or ApT steps by TpA decreases the in vitro binding of SEP3 higher order complexes to this sequence. This result supports the hypothesis that A-tract elements in flanking regions may facilitate the looping of the DNA on binding of some MADS-domain TFs (i.e SEP3), and it is tempting to speculate that they may contribute to the DNA-binding specificity of higher order complexes.
Because several MADS-domain protein dimers are able to bend the DNA at different degrees, this structural property can play a role in the specificity of different dimers. For example, the mammalian MADS-domain factor Myocyte enhancer factor 2A (MEF2A), which hardly induces DNA bending, has the consensus binding motif CTAW4TAG, whereas the serum response factor, with the standard CCW6GG consensus binding motif (13,38), induces a dramatic DNA bending on binding. In fact, West and Sharrocks (39) already speculated about a possible link between DNA-bending and DNA-binding specificity of MADS-domain TFs. By exploiting the temperature-dependent curvature of A-tract elements, we obtained confirmation for this hypothesis. Changing the temperature will not modify the primary DNA sequence, but it will affect the curvature of the DNA containing an A-tract (31,40). We observed that the relative in vitro affinity of the SEP3 and SEP3-AG dimers changes with the temperature, supporting the influence of the DNA curvature in the in vitro DNA-binding specificity of these two dimers. Meanwhile the affinity of AG homodimers only shows minimal changes. Additionally, we found that DNA regions bound by different SEP3 dimers in vivo show an overrepresentation of A-tracts of different length (Figure 2). The curvature induced by short A-tract elements in vitro is lower than for long A-tracts (27), which supports the hypothesis that the DNA curvature-dependent specificity of MADS-domain TFs may be also important in vivo. The fact that the length of A-tract elements is conserved among the Arabidopsis ecotypes for regions bound by MADS-domain TFs also indicates the evolutionary importance of this structural property.
The fact that temperature may affect differentially the DNA-binding affinity of particular MADS-domain dimers opens the door to new possibilities of how temperature can affect transcriptional regulation by MADS-domain TFs. Several MADS-domain TFs act in processes that are temperature-dependent, such as floral transition, flower maturation and fruit ripening (5). There is a large overlap among the target genes of several MADS-domain dimers (16). Therefore, it is tempting to speculate that temperature can be partially sensed by the plant via modification of the DNA-binding affinity of various dimers competing for binding common regulatory regions. This would provide a way to activate or repress the downstream pathways of target genes affected by these regulatory regions depending on the activity of the dimers. A similar mechanism of temperature sensing has been observed in bacteria, where temperature-dependent changes in DNA curvature in promoter regions containing A-tract elements play an important role in temperature-controlled gene expression (40,41). In eukaryotes, the TATA binding protein also shows a temperature-dependent binding affinity; Kuddus et al. (42) propose that this could be related with the fact that TATA binding protein affinity is dictated by the conformational flexibility of its DNA target (43). Recently, Lee et al. (44) and Posé et al. (45) unravelled various aspects of temperature-dependent activity of MADS-domain SVP-FLM complexes, including temperature-dependent degradation of SVP (44) and temperature-dependent alternative splicing of FLM (45). Future studies need to reveal the biological importance of the various mechanisms of temperature-dependent binding and regulation ‘in planta’.
SUPPLEMENTAL DATA
Supplementary Data are available at NAR Online.
FUNDING
Wageningen UR IPOP Systems Biology programme and NWO-NGI Horizon Breakthrough project [93519015 to A.D.J.vD.], NWO-VIDI project (to K.K.) and NGI-Netherlands Proteomics Centre (NPC) project (to G.C.A. and C.S.). Funding for open access charge: NWO-NGI Horizon Breakthrough project 93519015.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Schwarz-Sommer Z, Huijser P, Nacken W, Saedler H, Sommer H. Genetic control of flower development by homeotic genes in Antirrhinum majus. Science (New York, N.Y.) 1990;250:931–936. doi: 10.1126/science.250.4983.931. [DOI] [PubMed] [Google Scholar]
- 2.Pellegrini L, Tan S, Richmond TJ. Structure of serum response factor core bound to DNA. Nature. 1995;376:490–498. doi: 10.1038/376490a0. [DOI] [PubMed] [Google Scholar]
- 3.Huang K, Louis JM, Donaldson L, Lim FL, Sharrocks AD, Clore GM. Solution structure of the MEF2A-DNA complex: structural basis for the modulation of DNA bending and specificity by MADS-box transcription factors. EMBO J. 2000;19:2615–2628. doi: 10.1093/emboj/19.11.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan S, Richmond TJ. Crystal structure of the yeast MATalpha2/MCM1/DNA ternary complex. Nature. 1998;391:660–666. doi: 10.1038/35563. [DOI] [PubMed] [Google Scholar]
- 5.Smaczniak C, Immink RG, Angenent GC, Kaufmann K. Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development. 2012;139:3081–3098. doi: 10.1242/dev.074674. [DOI] [PubMed] [Google Scholar]
- 6.van Dijk AD, Morabito G, Fiers M, van Ham RC, Angenent GC, Immink RG. Sequence motifs in MADS transcription factors responsible for specificity and diversification of protein-protein interaction. PLoS Comput. Biol. 2010;6:e1001017. doi: 10.1371/journal.pcbi.1001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaufmann K, Melzer R, Theissen G. MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene. 2005;347:183–198. doi: 10.1016/j.gene.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 8.Egea-Cortines M, Saedler H, Sommer H. Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 1999;18:5370–5379. doi: 10.1093/emboj/18.19.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honma T, Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001;409:525–529. doi: 10.1038/35054083. [DOI] [PubMed] [Google Scholar]
- 10.de Folter S, Immink RG, Kieffer M, Parenicova L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, et al. Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell. 2005;17:1424–1433. doi: 10.1105/tpc.105.031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smaczniak C, Immink RG, Muino JM, Blanvillain R, Busscher M, Busscher-Lange J, Dinh QD, Liu S, Westphal AH, Boeren S, et al. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl Acad. Sci. USA. 2012;109:1560–1565. doi: 10.1073/pnas.1112871109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Immink RG, Kaufmann K, Angenent GC. The ‘ABC’ of MADS domain protein behaviour and interactions. Semin. Cell Dev. Biol. 2010;21:87–93. doi: 10.1016/j.semcdb.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Nurrish SJ, Treisman R. DNA binding specificity determinants in MADS-box transcription factors. Mol. Cell Biol. 1995;15:4076–4085. doi: 10.1128/mcb.15.8.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tao Z, Shen L, Liu C, Liu L, Yan Y, Yu H. Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J. 2012;70:549–561. doi: 10.1111/j.1365-313X.2012.04919.x. [DOI] [PubMed] [Google Scholar]
- 15.Immink R, Pose D, Ferrario S, Ott F, Kaufmann K, Leal Valentim F, De Folter S, Van der Wal F, van Dijk AD, Schmid M, et al. Characterisation of SOC1's central role in flowering by the identification of its up- and downstream regulators. Plant Physiol. 2012;160:433–449. doi: 10.1104/pp.112.202614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaufmann K, Wellmer F, Muino JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueno F, Krajewski P, Meyerowitz EM, et al. Orchestration of floral initiation by APETALA1. Science. 2010;328:85–89. doi: 10.1126/science.1185244. [DOI] [PubMed] [Google Scholar]
- 17.Kaufmann K, Muino JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC. Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 2009;7:e1000090. doi: 10.1371/journal.pbio.1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wuest SE, O'Maoileidigh DS, Rae L, Kwasniewska K, Raganelli A, Hanczaryk K, Lohan AJ, Loftus B, Graciet E, Wellmer F. Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc. Natl Acad. Sci. USA. 2012;109:13452–13457. doi: 10.1073/pnas.1207075109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng W, Ying H, Helliwell CA, Taylor JM, Peacock WJ, Dennis ES. FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc. Natl Acad. Sci. USA. 2011;108:6680–6685. doi: 10.1073/pnas.1103175108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 21.Muino JM, Kaufmann K, van Ham RC, Angenent GC, Krajewski P. ChIP-seq analysis in R (CSAR): an R package for the statistical detection of protein-bound genomic regions. Plant Methods. 2011;7:11. doi: 10.1186/1746-4811-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zacher B, Kuan PF, Tresch A. Starr: simple tiling array analysis of affymetrix ChIP-chip data. BMC Bioinformatics. 2010;11:194. doi: 10.1186/1471-2105-11-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedel M, Nikolajewa S, Suhnel J, Wilhelm T. DiProGB: the dinucleotide properties genome browser. Bioinformatics. 2009;25:2603–2604. doi: 10.1093/bioinformatics/btp436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Man TK, Stormo GD. Non-independence of Mnt repressor-operator interaction determined by a new quantitative multiple fluorescence relative affinity (QuMFRA) assay. Nucleic Acids Res. 2001;29:2471–2478. doi: 10.1093/nar/29.12.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gartenberg MR, Crothers DM. DNA sequence determinants of CAP-induced bending and protein binding affinity. Nature. 1988;333:824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- 27.Stefl R, Wu H, Ravindranathan S, Sklenar V, Feigon J. DNA A-tract bending in three dimensions: solving the dA4T4 vs. dT4A4 conundrum. Proc. Natl Acad. Sci. USA. 2004;101:1177–1182. doi: 10.1073/pnas.0308143100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher RA. Tests of significance in harmonic analysis. Proc. R. Soc. Lond. A Math. Phys. Character. 1929;125:54–59. [Google Scholar]
- 29.Riechmann JL, Wang M, Meyerowitz EM. DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 1996;24:3134–3141. doi: 10.1093/nar/24.16.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.West AG, Shore P, Sharrocks AD. DNA binding by MADS-box transcription factors: a molecular mechanism for differential DNA bending. Mol. Cell Biol. 1997;17:2876–2887. doi: 10.1128/mcb.17.5.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koo HS, Wu HM, Crothers DM. DNA bending at adenine.thymine tracts. Nature. 1986;320:501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- 32.Diekmann S. Temperature and salt dependence of the gel migration anomaly of curved DNA fragments. Nucleic Acids Res. 1987;15:247–265. doi: 10.1093/nar/15.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang DB, Phelps CB, Fusco AJ, Ghosh G. Crystal structure of a free kappaB DNA: insights into DNA recognition by transcription factor NF-kappaB. J. Mol. Biol. 2005;346:147–160. doi: 10.1016/j.jmb.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 35.Melzer R, Theissen G. Reconstitution of ‘floral quartets’ in vitro involving class B and class E floral homeotic proteins. Nucleic Acids Res. 2009;37:2723–2736. doi: 10.1093/nar/gkp129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melzer R, Verelst W, Theissen G. The class E floral homeotic protein SEPALLATA3 is sufficient to loop DNA in ‘floral quartet'-like complexes in vitro. Nucleic Acids Res. 2009;37:144–157. doi: 10.1093/nar/gkn900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mendes MA, Guerra RF, Berns MC, Manzo C, Masiero S, Finzi L, Kater MM, Colombo L. MADS domain transcription factors mediate short-range dna looping that is essential for target gene expression in Arabidopsis. Plant Cell. 2013;25:2560–2572. doi: 10.1105/tpc.112.108688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pollock R, Treisman R. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 1991;5:2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- 39.West AG, Sharrocks AD. MADS-box transcription factors adopt alternative mechanisms for bending DNA. J. Mol. Biol. 1999;286:1311–1323. doi: 10.1006/jmbi.1999.2576. [DOI] [PubMed] [Google Scholar]
- 40.Prosseda G, Mazzola A, Di Martino ML, Tielker D, Micheli G, Colonna B. A temperature-induced narrow DNA curvature range sustains the maximum activity of a bacterial promoter in vitro. Biochemistry. 2010;49:2778–2785. doi: 10.1021/bi902003g. [DOI] [PubMed] [Google Scholar]
- 41.Katayama S, Matsushita O, Jung CM, Minami J, Okabe A. Promoter upstream bent DNA activates the transcription of the Clostridium perfringens phospholipase C gene in a low temperature-dependent manner. EMBO J. 1999;18:3442–3450. doi: 10.1093/emboj/18.12.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuddus R, Schmidt MC. Effect of the non-conserved N-terminus on the DNA binding activity of the yeast TATA binding protein. Nucleic Acids Res. 1993;21:1789–1796. doi: 10.1093/nar/21.8.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parvin JD, McCormick RJ, Sharp PA, Fisher DE. Pre-bending of a promoter sequence enhances affinity for the TATA-binding factor. Nature. 1995;373:724–727. doi: 10.1038/373724a0. [DOI] [PubMed] [Google Scholar]
- 44.Lee JH, Ryu HS, Chung KS, Pose D, Kim S, Schmid M, Ahn JH. Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science. 2013;342:628–632. doi: 10.1126/science.1241097. [DOI] [PubMed] [Google Scholar]
- 45.Pose D, Verhage L, Ott F, Yant L, Mathieu J, Angenent GC, Immink RG, Schmid M. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature. 2013;503:414–417. doi: 10.1038/nature12633. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.