Abstract
The nucleotide excision repair pathway removes ultraviolet (UV) photoproducts from the human genome in the form of short oligonucleotides ∼30 nt in length. Because there are limitations to many of the currently available methods for investigating UV photoproduct repair in vivo, we developed a convenient non-radioisotopic method to directly detect DNA excision repair events in human cells. The approach involves extraction of oligonucleotides from UV-irradiated cells, DNA end-labeling with biotin and streptavidin-mediated chemiluminescent detection of the excised UV photoproduct-containing oligonucleotides that are released from the genome during excision repair. Our novel approach is robust, with essentially no signal in the absence of UV or a functional excision repair system. Furthermore, our non-radioisotopic methodology allows for the sensitive detection of excision products within minutes following UV irradiation and does not require additional enrichment steps such as immunoprecipitation. Finally, this technique allows for quantitative measurements of excision repair in human cells. We suggest that the new techniques presented here will be a useful and powerful approach for studying the mechanism of human nucleotide excision repair in vivo.
INTRODUCTION
Ultraviolet (UV) light induces the formation of DNA adducts, including cyclobutane pyrimidine dimers (CPDs), pyrimidine (6–4) pyrimidone photoproducts [(6–4)PPs] and Dewar photoproducts, that can only be removed from the genome in humans by the nucleotide excision repair (excision repair) system (1–3). Studies in vitro and in vivo have shown that the UV damage is removed from the genome in the form of an oligonucleotide ∼24 to 32 nt in length (4–6). Six core excision repair factors are necessary and sufficient for the general excision process in humans, which include XPA, RPA, XPC, TFIIH (8–10 proteins, including XPB and XPD), XPG and XPF-ERCC1 (7). Mutations in many of these factors are found in patients with the human disease xeroderma pigmentosum, which is associated with an extreme sensitivity to sunlight and a vastly elevated risk of skin cancer (8,9).
Although nucleotide excision repair has been investigated extensively, and many assays have been developed for studying its molecular mechanism, these techniques often require great technical expertise, lack sensitivity or only indirectly measure excision repair events in vivo. We recently reported a method to extract and immunoprecipitate the short UV photoproduct-containing oligonucleotides from UV-irradiated human cells and then visualize the oligomers by radiolabeling, gel electrophoresis and phosphorimaging (6). This work allowed the first detection of the primary UV photoproduct-containing oligonucleotide products of excision repair in vivo and validated several of the mechanistic insights into excision repair that had previously only been demonstrated in vitro.
In this study, to avoid safety issues associated with radioactivity and time-consuming steps related to immunoprecipitation, we now describe a convenient and powerful non-radioisotopic methodology for detecting UV photoproduct-containing oligomers in UV-irradiated human cells. Importantly, these oligomers are only observed following UV irradiation and in cells with a functional nucleotide excision repair system. We further show that the biotinylation/chemiluminescence method that we present here is comparable in sensitivity with the radiolabeling method. Finally, our methodology allows for accurate and precise measurements of excision repair activity to be made within minutes of exposure of human cells to UV radiation.
MATERIALS AND METHODS
Cell lines and cell culture
HeLa cells were obtained from the Korean Cell Line Bank of Seoul National University (Seoul, Korea). The Chinese hamster ovarian (CHO) cell line AA8 (wild-type) and its derivative UV135 (XPG-deficient) were purchased from the American Type Culture Collection (Rockville, MD, USA). The human melanoma cell line A375 was used as previously described (10). All cell lines were cultured in Dulbecco’s modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 humidified incubator.
Preparation of cell lysates
Cells grown to near confluency in 100 mm or 150 mm diameter plates were washed with phosphate-buffered saline (PBS, Gibco) and then exposed to the indicated doses of UV using a germicidal lamp that emits primarily 254 nm light. UV doses were determined by using a portable digital radiometer (UVX Digital Radiometer UVP). After UV irradiation, the cells were replenished with cultured medium and returned to the incubator. At the indicated time points, the cells were washed with ice-cold PBS and harvested with a cell scraper. The cells were pelleted by centrifugation and then subjected to the modified Hirt procedure (11) or where indicated, cell lysis with a mild NP-40 lysis buffer (10 mM HEPES, pH 7.6, 60 mM KCl, 1 mM EDTA, 0.075% NP-40 and 1 mM DTT), radioimmunoprecipitation (RIPA) buffer (25 mM Tris–HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate and 0.1% SDS), a mild Triton X-100 lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl and 0.1% Triton X-100) or a strong Triton X-100 lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA and 1% Triton X-100). For the modified Hirt procedure, the cell pellets were resuspended in a 10× packed cell volume of lysis buffer (50 mM Tris-Cl, pH 8.0, 10 mM EDTA, 1.2% SDS and 100 μg/ml RNase A) and incubated for 15 min at room temperature. Following addition of a one-fourth volume of 5 M NaCl, the mixtures were gently mixed and incubated on ice for 12–16 h. After centrifugation at maximum speed (∼20 000 × g) for 1 h, the supernatants were gently transferred to new tubes. For cell lysis using the other lysis buffers, the cell pellets were resuspended in 10× packed cell volume of lysis buffer, incubated for 15 min on ice and centrifuged. The supernatants were then transferred to new tubes and processed as described later in the text.
Purification of oligonucleotides containing UV damage
Cell lysates were treated with 0.1 mg/ml of proteinase K (Qiagen) at 55°C for 30 min and then extracted with phenol/chloroform. Following ethanol precipitation, the pellets were washed with 70% ethanol and resuspended in 100 μl of Tris-EDTA (TE) buffer (10 mM Tris-Cl, pH 8.0, 1 mM EDTA). To completely remove any trace of contaminating RNA from the preparations, 2 μl of 5 M NaCl and 50 μg of RNase A were added to the DNA preparations and incubated at 37°C for 30 min. The RNA-free DNA molecules were then extracted by phenol/chloroform following treatment with 50 μg of proteinase K in the presence of 0.4% SDS. Following ethanol precipitation, the pellets were resuspended in TE buffer and then subjected to labeling with terminal deoxynucleotidyl transferase.
Labeling of purified DNAs
For non-radioisotopic labeling, the total extracted DNA was incubated with 20 U of terminal deoxynucleotidyl transferase (New England Biolabs) and 10 µM of biotin-11-UTP or -dUTP (Biotium, Inc.) in 50 µl of 1× Terminal Transferase Reaction Buffer (New England Biolabs) and 0.25 mM CoCl2 at 37°C for 30 min. For radioisotopic labeling, 1 μCi of [α-32P]-3′-deoxyadenosine 5′-triphosphate (cordycepin 5′-triphosphate, Perkin Elmer Life Sciences) was used in place of the biotinylated nucleotide. The reactions were stopped by addition of 1 µl of 0.5 M EDTA and then incubated with 20 µg of proteinase K in the presence of 0.4% SDS. Samples were then extracted with phenol/chloroform and precipitated in ethanol. Identical labeling reactions were used to prepare 100 ng of 10 bp DNA ladder (Invitrogen) for use as a DNA ladder during gel electrophoresis. Quantitative measurements of excised oligonucleotide generation used a 20-mer oligomer standard that was similarly biotinylated or radiolabeled.
Detection of labeled oligomers
The labeled DNAs were loaded onto 10% or 12% TBE-urea gels (8 × 8 cm) and run in 1× Tris-Borate-EDTA (TBE) at 150 V. After gel electrophoresis, the DNAs were transferred to a modified nylon membrane that has a strong positive potential (Zeta-Probe membranes by Bio-Rad or Biodyne membrane by Pierce) in 0.5× TBE buffer at 300 mA for 1 h using a wet-transfer apparatus (Mini Trans-Blot Cell Assembly, Bio-Rad). DNAs were then fixed to the membrane by UV cross-linking (1500 J/m2 of UV-C). Detection of biotin-labeled DNA was achieved using the Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific). Briefly, the membranes were blocked with blocking buffer and incubated with horseradish peroxidase (HRP)-conjugated streptavidin that was diluted in blocking buffer. Membranes were then washed with Wash Buffer or 1× PBS containing 0.5% SDS and then incubated in substrate equilibration buffer or a buffer containing 100 mM Tris-Cl (pH 9.8) and 5 mM MgCl2. Chemiluminescent detection was achieved with Luminol/Enhancer Solution (Thermo Scientific) or Amersham ECL Select reagent (GE Healthcare Life Sciences). Membranes were exposed to X-ray films or imaged with a Bio-Rad ChemiDoc system or an ImageQuant LAS 4000 Mini apparatus (GE Healthcare Life Sciences). The relative intensities of the bands were analyzed by using ImageQuant TL Software (GE Healthcare Life Sciences). Radiolabeled excised oligonucleotides were similarly electrophoresed by urea-PAGE. Gels were then exposed to a phosphorimager screen and visualized with a GE Typhoon phosphorimager.
Immunoprecipitation of excised oligonucleotides containing CPDs or (6–4)PPs
Purified DNAs from the modified Hirt lysate procedure described earlier in the text were subjected to immunoprecipitation (IP) with anti-CPD or anti-(6–4) PP antibodies as reported previously (6). Briefly, protein G Dynabeads slurry (Invitrogen) and anti-rabbit Dynabeads slurry (Invitrogen) were mixed (5 µl each) and washed with Wash Buffer I (20 mM Tris-Cl, pH 8.0, 2 mM EDTA, 150 mM NaCl, 1% Triton X-100 and 0.1% SDS). The beads were resuspended in 20 µl of IP buffer (20 mM Tris-Cl, pH 8.0, 2 mM EDTA, 150 mM NaCl, 1% Triton X-100 and 0.5% sodium deoxycholate) containing rabbit anti-mouse IgG (Abcam) and anti-CPD (Kamiya Biomedical) or anti-(6–4)PP (Cosmo Bio Co.) antibody (1 µl each). After incubation at 4°C for 3 h, the beads were isolated and incubated with the purified DNA in 100 µl of IP buffer overnight at 4°C. The beads were then subjected to sequential washes with Wash Buffer I, Wash Buffer II (20 mM Tris-Cl pH8.0, 2 mM EDTA, 500 mM NaCl, 1% Triton X-100 and 0.1% SDS), Wash Buffer III (10 mM Tris-Cl pH 8.0, 1 mM EDTA, 150 mM LiCl, 1% NP-40 and 1% sodium deoxycholate) and Wash Buffer IV (100 mM Tris-Cl pH 8.0, 1 mM EDTA, 500 mM LiCl, 1% NP-40 and 1% sodium deoxycholate). The beads were additionally washed twice with TE, and the oligonucleotides containing UV photoproducts were eluted with 100 µl of Elution Buffer (50 mM NaHCO3, 1% SDS and 20 µg/ml of glycogen) at 65°C for 15 min, followed by phenol/chloroform extraction and ethanol precipitation. The isolated oligonucleotides were 3′-end labeled with biotin, separated on 10% TBE-urea gels, transferred to a nylon membrane and detected with HRP-conjugated streptavidin using chemiluminescence reagents as described earlier in the text.
RESULTS
Development of a chemiluminescence-based method for the detection of excised UV photoproduct-containing oligonucleotides
We recently reported a radioisotopic method for the detection of the short UV photoproduct-containing oligonucleotides that are released from the genome during nucleotide excision repair in UV-irradiated human cells (6). To expand on the technique and its potential applications, we have now adapted the labeling method to allow for the chemiluminescent detection of the excised DNA oligomers. A schematic of the methodology is presented in Figure 1A. After exposure to a UV light source, cells are harvested and then lysed using the method of Hirt (11), which allows for the extraction of small DNA fragments away from most of the high molecular weight genomic DNA in the cell. After extensive treatment with RNase and protease to remove RNA and proteins, respectively, the DNAs are 3′-end labeled with terminal transferase and the nucleotide analog biotin-11-dUTP (or biotin-11-UTP) (12). The biotinylated DNAs are then separated by denaturing urea-PAGE and transferred onto a nylon membrane. The DNAs are cross-linked to the membrane and then visualized by incubation of the membrane with HRP-conjugated streptavidin and addition of a chemiluminescent substrate. As will be shown later in the text, this labeling protocol is as sensitive as radiolabeling and is highly specific for the detection of the UV photoproduct-containing oligonucleotide products of nucleotide excision repair.
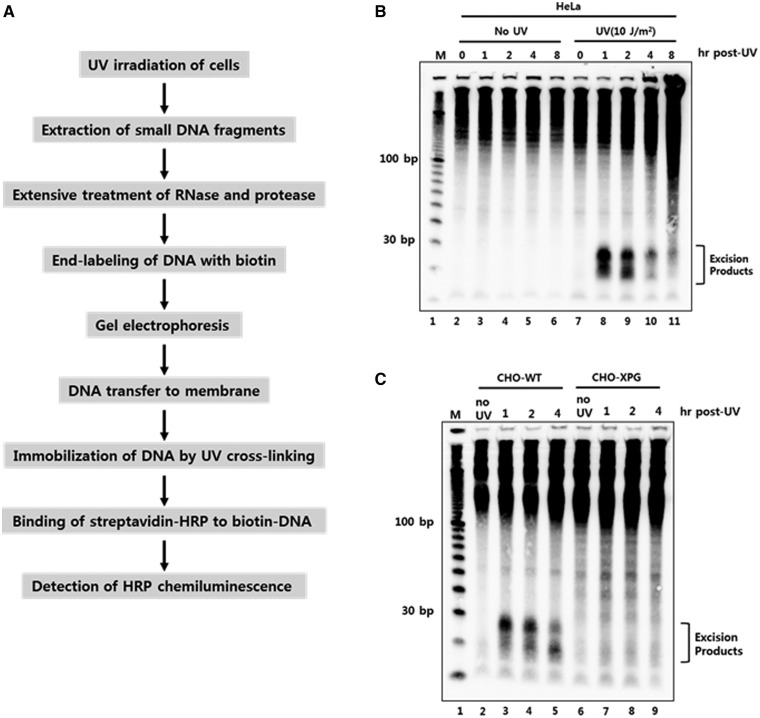
Figure 1.
Development of a novel non-radioisotopic method for the detection of excised oligonucleotide products of nucleotide excision repair in vivo. (A) Schematic representation for the detection of excised oligonucleotides containing UV damage in human cells. UV-irradiated cells are harvested and subjected to DNA extraction to isolate low molecular weight DNAs. After extensive treatment with RNase and protease, the short DNA molecules are labeled with biotin at the 3′-end using terminal deoxynucleotidyl transferase (TdT). The biotin-labeled oligonucleotides are separated by gel electrophoresis and then transferred to a nylon membrane. The transferred biotin-DNA molecules are immobilized by UV cross-linking, incubated with HRP-conjugated streptavidin, and detected with a chemiluminescence reagent. (B) HeLa cells were exposed to 10 J/m2 of UV-C and then harvested at different time points. The cells were subjected to the modified Hirt lysis and labeling procedures as in A. The DNA marker (M) is a 10 bp DNA ladder that was labeled with biotin and loaded onto the gel. (C) CHO wild-type (AA8) and XPG-deficient (UV135) cells were harvested at different times after UV exposure to 10 J/m2 of UV-C. Excised oligonucleotides containing UV damage were then extracted and processed as described in A.
An example of this labeling approach is shown in Figure 1B. DNA oligomers in the range of ∼20 to 30 nt in length are readily observed for several hours following the exposure of HeLa cells to 10 J/m2 of UV radiation (Figure 1B, lanes 7–11). Importantly, DNAs within this size range are not found in non-irradiated cells (Figure 1B, lanes 2–6). Higher molecular weight DNA species ∼150 nt and larger are observed regardless of irradiation and likely reflect contaminating fragments of genomic DNA that are generated during cell lysis (11). At late time points following irradiation (Figure 1B, lane 11), these larger DNAs begin to form a smear down the gel that is likely due to the induction of apoptosis in the UV-irradiated cells.
To demonstrate that these short 20–30–nt DNA oligomers are dependent on the nucleotide excision repair system, we compared the generation of these oligonucleotides in the normal excision repair-proficient hamster cell line AA8 and the excision repair-deficient cell line UV135, which lacks the excision repair factor XPG. These results are shown in Figure 1C. Although the small oligonucleotides products are clearly observed in the AA8 cell line, they are completely absent in the UV135 cells. We conclude from these results that the exposure of mammalian cells to UV results in the generation of small oligonucleotides that are produced in a nucleotide excision repair-dependent manner and which can be readily extracted from cells and visualized by biotinylation and subsequent chemiluminescent detection.
Importantly, although we have routinely used in our study the Hirt method to extract the small oligonucleotides from UV-irradiated cells, we note that our labeling and detection methodology is compatible with the use of a variety of other cell lysis buffers. As shown in Supplementary Figure S1, the lysis of cells with the Hirt method or with lysis buffers containing various combinations of non-ionic and ionic detergents yield similar patterns of oligonucleotides ∼20 to 30 nt in length in a UV-dependent manner. We conclude from these results that there are multiple methods for extracting excised oligomers from UV-irradiated human cells. This flexibility should facilitate further analyses of the function and fate of excised oligomers during nucleotide excision repair.
UV dose- and time-dependent generation of excised oligonucleotides
We next monitored the generation of excised oligonucleotides as a function of UV dose to determine the range of UV doses over which the excised oligomers can be detected. HeLa cells were exposed to varying doses of UV and then harvested 30 min later. Excised oligomers were extracted from the cells, biotinylated and then detected with chemiluminescence. As shown in Figure 2A, even with a dose as low as 2.5 J/m2, excised oligonucleotides were readily apparent in the cell lysates. Quantification of the dose dependence is shown in Figure 2B. These results show that the amount of excised oligonucleotide products that are generated in HeLa cells following UV irradiation is linearly correlated with the UV dose up until ∼10 J/m2, at which point the generation of excised oligomers becomes saturated. Although the lowest dose that we used in these experiments was 2.5 J/m2, lower UV doses also yield detectable signals (data not shown). Thus, the method that we present here for detecting excised oligonucleotides following UV irradiation can be used at low non-toxic doses of UV.
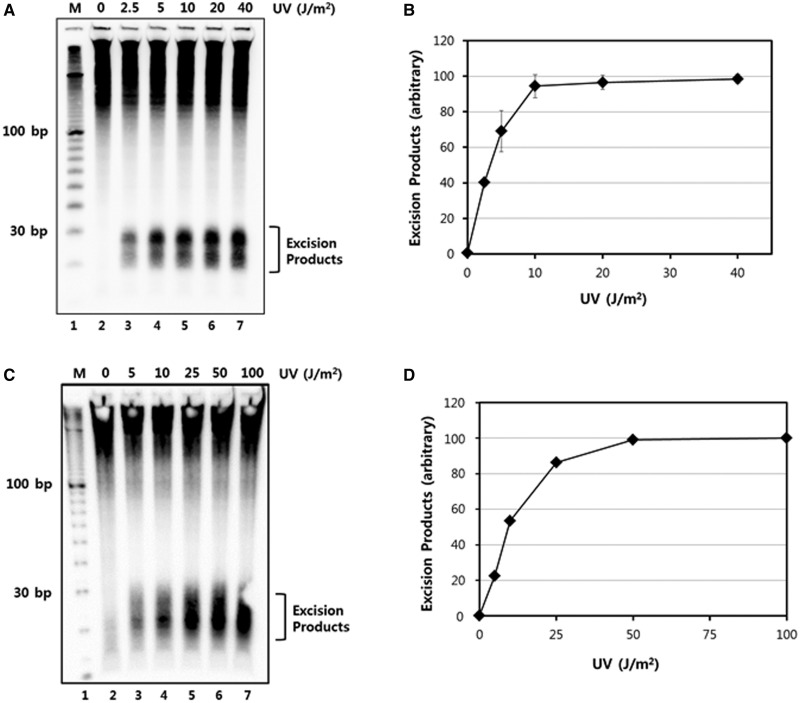
Figure 2.
UV dose-dependent generation of excised oligonucleotides. (A) HeLa cells were treated with the indicated doses of UV-C radiation and then harvested after incubation for 30 min. Excised oligonucleotides containing UV damage were then extracted, 3′ end-labeled with biotin, separated on a 10% TBE-urea gel, transferred to a nylon membrane, and detected with streptavidin-HRP conjugate using chemiluminescence reagents. (B) Quantitative analysis of the excision repair products from three independent experiments as in A are presented as means ± standard deviation (SD). The maximum values were set to 100 and the other values are presented relative to that value. (C) A375 cells were treated with the indicated doses of UV-C radiation and then harvested 1 h later. Excised oligonucleotides were analyzed as in A, except with a 12% TBE-urea gel. (D) Quantitative analysis of the excised oligonucleotide repair products in A375 cells.
To further validate our methodology and the UV dose-dependent generation of excised oligomers in another human cell line, we performed a UV dose response experiment in A375 cells, which is a human melanoma cell line with robust nucleotide excision repair activity (6,10). In this cell line, the generation of excised oligonucleotides did not become saturated until doses >25 J/m2 were used (Figure 2C and D). Together with the experiments with HeLa cells, these results suggest that maximal generation of excised oligonucleotides occurs between 10 and 25 J/m2 of UV-C. Importantly, this saturation range is consistent with other approaches that have been used to measure excision repair activity in human cells (13–15). These results further demonstrate that our method for the direct detection of excised oligonucleotides may be a convenient and robust approach for assaying nucleotide excision repair activity and capacity in a variety of human cell types and lines, including at low non-toxic doses of UV.
We next performed time course experiments to monitor the presence and fate of excised oligonucleotides as a function of time following UV irradiation. As shown in Figure 3A, excised oligomers are readily observed within 6 min following UV irradiation of HeLa cells, which indicates that the excision repair system is able to respond rapidly to UV-induced DNA damage and that our methodology is sensitive enough to detect these early repair events. Moreover, we observe two distinct classes of excision products over time. At early time points following irradiation, the excised oligomers that are generated are exclusively the primary excision products ∼25 to 30 nt in length, which is in agreement with in vitro data that human nucleotide excision repair generates 24–32-nt long oligomers by dual incisions (4–6,16). Within 30 min to 1 h, however, a second peak of oligonucleotide products emerges that are ∼20 nt in length. Based on previous studies, these latter products represent excised oligomers that are partially degraded by cellular nucleases and become bound to the single-stranded DNA binding protein RPA (6,16).
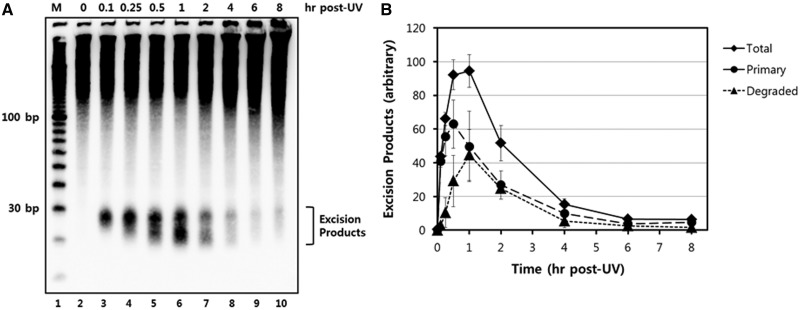
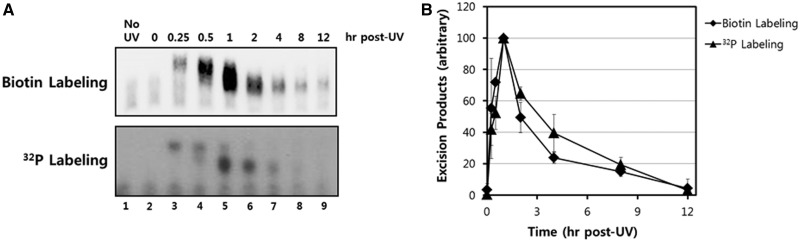
Figure 3.
Time course analysis of UV-induced excision products. (A) HeLa cells were exposed to 10 J/m2 of UV-C and then harvested at various time points. Excised oligonucleotides containing UV damage were then extracted, 3′ end-labeled with biotin, separated on a 10% TBE-urea gel, transferred to a nylon membrane, and detected with HRP-conjugated streptavidin and a chemiluminescent reagent. (B) Quantitative analysis of the excised oligonucleotide repair products. The results from three independent experiments were quantified and are plotted (means ± standard deviation) relative to the maximum.
Both qualitative and quantitative analyses of time course experiments show that the peak time point for detecting excised oligonucleotdies is ∼1 h following UV irradiation (Figure 3B). At later time points, a significant reduction in signal is observed among all of the oligonucleotides in the range of 20–30 nt in length. Because nucleotide excision repair on UV-damaged genomic DNA continues at a near linear rate for at least 12 h following UV irradiation (10), the significant reduction in excised oligomers that is observed at later time points following irradiation is not due to a significant reduction in activity or capacity of the excision repair system to process on UV-damaged DNA in chromatin. Rather, these results indicate that the processing and degradation of the excised oligonucleotides may be more efficient at later time points following UV irradiation. We also note that the absence of detectable oligonucleotides less than ∼20 nt in length may be due to the reduced ability of short oligonucleotides to be precipitated in ethanol (17) or due to processing of the excised oligonucleotides by cellular nucleases that make the oligomers refractory to biotinylation by terminal transferase. Nonetheless, excised oligonucleotides are readily observed from a few minutes to several hours after irradiation. Thus, the methodology described here should be regarded as a powerful approach for studying nucleotide excision repair in human cells.
Excised oligonucleotides contain CPDs and (6–4) photoproducts
To verify that the excised oligonucleotides that are extracted from cells following UV irradiation contain genuine UV photoproducts, we immunoprecipitated the extracted DNAs with anti-CPD and anti-(6–4)PP antibodies as previously reported (6) before biotinylation and chemiluminescent detection. A schematic of this approach is provided in Figure 4A. As shown in Figure 4B, both the anti-CPD and anti-(6–4)PP antibodies were capable of immunoprecipitating the excised oligomers following UV irradiation of HeLa cells. Consistent with the ability of cells to recognize and repair (6–4)PPs more rapidly than CPDs (10,18,19), we observed significantly more (6–4)PP-containing oligonucleotides than CPD-containing oligonucleotides at early time points following irradiation (Figure 4B and C). Furthermore, the lengths of the (6-4)PP-containing oligonucleotides showed a strong time-dependent change, such that the primary full-length excision products were exclusively observed at the early 15 min time point, and only degraded oligomers were observed after 2 h (Figure 4B, Supplementary Figure S2). By 4 h following irradiation, when (6–4)PP removal from genomic DNA is complete (10), we were barely able to detect excised (6–4)PP-containing oligomers. These results indicate that the oligomers that had been excised from the genome have been processed or degraded to the extent that they are no longer detectable with our labeling methodology.
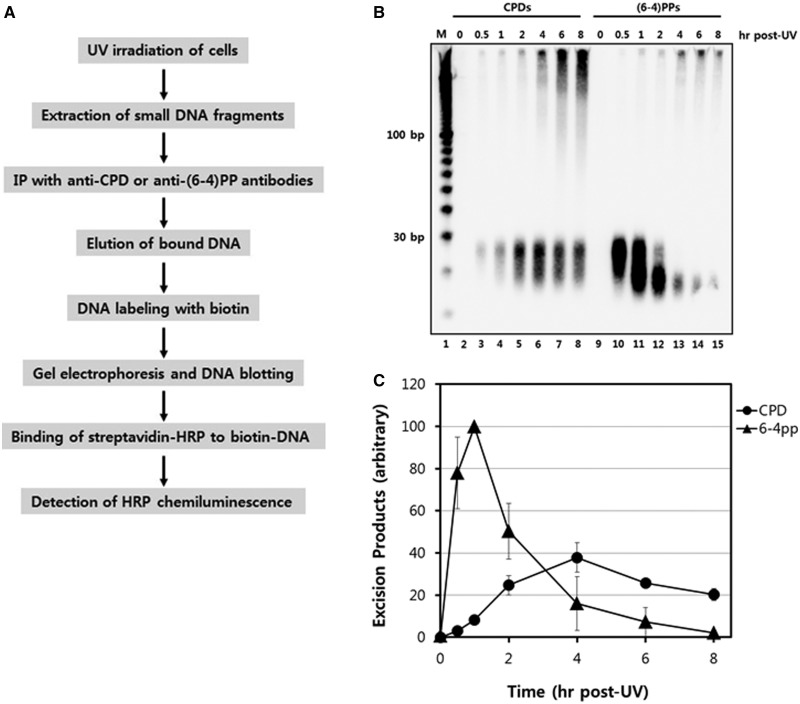
Figure 4.
Detection of specific excised oligonucleotides containing CPDs or (6–4)PPs. (A) Scheme for the detection of excised oligonucleotides containing UV photoproduct. (B) HeLa cells were harvested at various time points after UV exposure to 10 J/m2 of UV-C. The cells were subjected to the modified Hirt procedure, and the extracted oligonucleotides were then immunoprecipitated with anti-(6–4)PPs or anti-CPD antibodies. Purified oligonucleotides 3′-end labeled with biotin-11-UTP were separated on a 10% TBE-urea gel, transferred to a nylon membrane, and detected with HRP-conjugated streptavidin using a chemiluminescent reagent. (C) Quantitative analysis of the CPD- and (6–4)PP-containing excised oligonucleotides products as a function of time following UV irradiation. The results from three independent experiments were quantified and are plotted (means ± standard deviation) relative to the maximum.
In contrast, the pattern of excised CPD-containing oligomers was markedly different than the (6–4)PP-containing oligomers. Though CPD-containing oligonucleotides were detectable 15 min following UV irradiation, the relative abundance of CPD-containing oligonucleotides continued to increase until a peak time point at 4 h post-UV. The slower generation of CPD-containing oligomers that we observe with our approach is consistent with other methods that show that CPDs are repaired at a much slower rate than (6–4)PPs (10,18,19). Furthermore, the distribution of CPD-oligomer lengths was much more uniform throughout the 8 h time period following UV irradiation, which indicates that the rate of generation and degradation of the CPD-containing oligomers is closer to equilibrium for at least 8 h following UV irradiation (Supplementary Figure S2).
We conclude from these experiments that the short 20–30-nt oligomers that can be directly extracted from human cells following UV irradiation contain UV photoproducts. Taken together, these results show that our methodology can be used to monitor specific UV photoproduct-containing excised oligonucleotides that are released from the genome during nucleotide excision repair in UV-irradiated human cells.
Comparison of the radiolabeling and chemiluminescent detection techniques
Because we previously reported that excised UV photoproduct-containing oligonucleotides can be detected by immunoprecipitation and radiolabeling (6), and we have shown here that the oligomers can also be observed by direct extraction and non-radioisotopic labeling (Figures 1–4), we next wanted to directly compare the two labeling methods. We therefore carried out time course experiments in which excised oligomers were directly isolated from A375 cells at various time points following UV irradiation and then either radiolabeled or biotinylated. As shown in Figure 5, we found that the two labeling methods yielded nearly identical patterns of excised oligonucleotides. These results show that both the radiolabeling and biotinylation/chemiluminescence approaches can be used to detect excised oligomers with comparable efficiency. We would also note that there are a few advantages and disadvantages of each of the specific labeling methods. For example, strong chemiluminescent signals can be visualized within minutes of addition of the chemiluminescent substrate, whereas the comparable radioactive signals may take several hours to be visualized depending on the activity of the radiolabel. However, the chemiluminescent detection method requires additional steps following gel electrophoresis, including transfer and immobilization of the DNA on a nylon membrane and incubation with HRP-conjugated streptavidin. Nonetheless, in addition to potential safety issues related to the handling of radioactivity during labeling and subsequent processing steps, the biotinylation and chemiluminescence methodology that we have developed here can be considered to be both sensitive and convenient.
Figure 5.
Sensitivity comparison of the biotinylation and radiolabeling methods. (A) Excised oligonucleotides containing UV damage were recovered from A375 cells at various time points after exposure to 10 J/m2 of UV-C. The purified oligonucleotides were then labeled with biotin-11-dUTP or cordycepin 5′-triphosphate at the 3′-end using terminal deoxynucleotidyl transferase (TdT). Biotin-labeled DNAs were separated on a 12% TBE-urea gel, transferred to a nylon membrane, and detected with HRP-conjugated streptavidin-HRP using a chemiluminescence reagent. Radiolabeled DNAs were separated on a 12% TBE-urea gel and detected by exposing the gel to a phosphorimager screen. (B) Quantitative analysis of the excised repair products. The results from three independent experiments were quantified and are plotted (means ± standard deviation) relative to the maximum.
To further examine the sensitivity and detection ranges of the two labeling methods, we next exposed A375 cells to 10 J/m2 of UV and then isolated the excised oligonucleotides 1 h following irradiation. We then prepared labeling reactions with excised oligonucleotides from varying amounts of Hirt lysate, terminal transferase and either α-[32P]-3′-deoxyadenosine 5′-triphosphate or biotin-11-dUTP. The products of the labeling reactions are shown in Figure 6A. To calculate the absolute amount of excised oligonucleotides that are generated during repair and that can be visualized with each labeling technique, we included defined amounts of a 20-mer oligonucleotide standard in our analyses that had similarly been either biotinylated or radiolabeled.
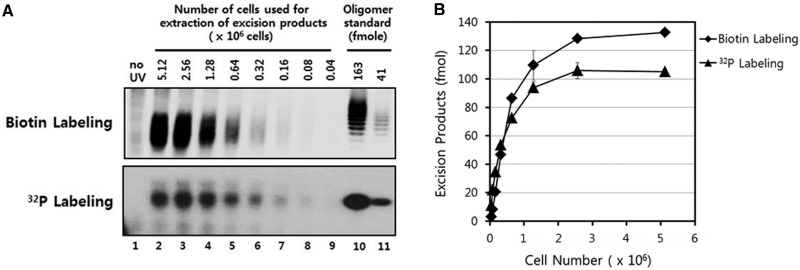
Figure 6.
Absolute quantification of UV photoproducts in vivo. (A) Excised oligonucleotides containing UV damage were recovered from A375 cells 1 h following exposure to 10 J/m2 of UV-C. The purified oligonucleotides were then either biotinylated or radiolabeled as in Figure 5 using Hirt lysate-purified DNA from a defined number of cells. As a standard for quantification of the excised oligonucleotides, a 20-nt long single-stranded DNA oligonucleotide was either biotinylated or radiolabeled and then electrophoresed and analyzed with the UV photoproduct-containing excised oligomer repair products. (B) Absolute quantification of the excised repair products in vivo. The results from two independent experiments were quantified and are plotted (means ± standard deviation).
We found that both labeling methods yielded similar results and showed a linear range of detection when Hirt lysate-extracted DNAs from between ∼1 × 105 and 1 × 106 cells were used in the labeling reactions. When DNA from >1 × 106 cells was used in the labeling reaction, there was no longer an increase in the signal that was observed. These results indicate that a maximum of 100–130 fmol of excised oligomers can be labeled and visualized in a standard reaction. When excised oligonucleotides from fewer than ∼1 × 105 cells were used, the signals were difficult to detect. We conclude from these results that both the radiolabeling and biotinylation/chemiluminescence approaches can be used with comparable effectiveness for the detection and quantification of the excised UV photoproduct-containing oligomer products of nucleotide excision repair in human cells.
Importantly, the quantitative measurements of excision product generation in Figure 6 also allow us to determine the absolute number of excised oligomers that can be detected in UV-irradiated human cells. For example, by using data within the linear range of detection in Figure 6, we calculate that there are ∼80 000 excised oligomers per A375 cell that are recoverable and detectable with our methodology 1 h following exposure to 10 J/m2 of UV (Supplementary Figure S3A). Using this value as a standard, we can convert the data in time course data in Figure 5 into the actual number of molecules of UV photoproduct-containing oligonucleotides that are released and that are recoverable with our methodology at each time point following irradiation (Supplementary Figure S3B). Based on our previous measurements of CPD and (6–4)PP repair in this cell line by immunoslot blot analysis (10), the amount of genomic DNA that is present in each aneuploid A375 cell (15 pg), the number of UV photoproducts that are generated in the genome with 10 J/m2 of UV [1 photoproduct per 10 kb of DNA; 80% CPDs and 20% (6–4)PPs] (20), we can then calculate the cumulative number of UV photoproduct-containing oligonucleotides that are actually released from the genome following irradiation (Supplementary Figure S3C). We can then directly compare these values with the numbers of excised oligomers that we can detect with our new labeling methodology at each time point following irradiation (Supplementary Figure S3D). Interestingly, although >90% of the excised oligonucleotides that are released from the genome within 15 min of UV irradiation are recoverable and detectable with our Hirt extraction and labeling technique, this recovery rapidly decreases as a function of time (Supplementary Figure S3E), such that by 2 h following irradiation we can detect <20% of the excised oligomers that have actually been released from the genome. This low recovery is presumably due to the relatively rapid rate at which excised oligomers are degraded by cellular nucleases following UV irradiation (Figures 3–5), which may become more efficient following irradiation. These quantitative results therefore suggest that the methods we present here for detecting the excised oligomer products of nucleotide excision repair should be a useful technique for both qualitative and quantitative measurements of excision repair in human cells, particularly at early time points following irradiation.
DISCUSSION
A variety of methods have been developed over the past few decades for studying human nucleotide excision repair. Radionucleotides have been widely used both in vitro and in vivo for monitoring the release of UV photoproducts from DNA (4,6,21–27) and for measuring the DNA synthesis that takes place to fill in the gap in the DNA that is left following excision of the damage (8,28–31). Though chemiluminescence-based assays using biotinylated nucleotides have been developed for measuring repair synthesis (29,32), biotin labeling and chemiluminescence detection have not previously been exploited for directly following the excised oligonucleotide products of excision repair. Since the development of specific antibodies for CPDs and (6–4)PPs, immunological approaches (ELISA, slot blot and fluorescence microscopy) have become some of the most routinely used methods for monitoring UV excision repair in vivo (10,19,33). These approaches offer a number of advantages, including the ability to use high-throughput and microplate formats (34). However, a limitation of the classical immunological methods is that they are dependent on monitoring the loss of strong UV photoproduct signals from genomic DNA as a function of repair time. Even relatively low doses of UV generate hundreds of thousands of UV photoproducts throughout the genome, and these immunochemical-based DNA repair assays lack the sensitivity to measure the relatively small number of repair events that take place within minutes following irradiation. These techniques also do not directly demonstrate that UV photoproducts are removed from the genome in the form of ∼30-nt-long oligonucleotides.
Our new methodology to directly visualize the small excised UV photoproduct-containing oligonucleotide products of excision repair therefore offers a number of advantages that should facilitate its addition to the repertoire of available DNA repair assays. First, the labeling methods that we presented here yield virtually no background signal in the absence of UV irradiation (Figure 1B) or in the absence of nucleotide excision repair function (Figure 1C). Moreover, the amount of excised oligonucleotides that are generated in our assays is directly correlated with biologically relevant doses of UV (Figure 2), and the saturation of excised oligomer generation that we observe occurs at the same UV doses that other methods have also shown to saturate the excision repair system (13–15). Importantly, the generation of excision products following UV irradiation is also remarkably rapid, such that a robust clearly detectable signal is observable with minutes following irradiation (Figure 3). The ability to immunoprecipitate oligonucleotides that contain specific UV photoproducts proves that the oligonucleotide products contain genuine photoproducts (Figure 4), which should allow any differences that exist in CPD, (6–4)PP and Dewar isomer repair product processing to be investigated in greater detail in the future. Similarly, we envision that the sequencing of these excised oligonucleotides will allow for the mapping of excised oligomers to their initial locations in the genome and for examining the different repair rates of unique bipyrimidine photoproduct-containing sequences (35). Lastly, our data show that the radiolabeling and chemiluminescence detection methods are quite comparable in sensitivity (Figure 5) and allow for quantitative measurements of nucleotide excision repair to be made in a variety of different in human cell lines (Figure 6). Though we focused on the repair of UV photoproducts here, we would note the chemiluminescence excision assay does not depend on the availability of damage-specific antibodies and can likely be used for any damage that is repaired by the nucleotide excision repair system, including DNA adducts induced by cisplatin and benzo[a]pyrene. We therefore conclude that there are a number of features of our methodology that should facilitate its application for the study of human DNA excision repair in vivo alongside other commonly used techniques.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM32833 to A.S.] and the Korea Research Institute of Standards and Science under the project ‘Development of bio-clinical measurement standards’ [3011018 to J-H.C.]. Funding for open access charge: Korea Research Institute of Standards and Science [3011018].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Sancar A. DNA excision repair. Annu. Rev. Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 2.Wood RD. Nucleotide excision repair in mammalian cells. J. Biol. Chem. 1997;272:23465–23468. doi: 10.1074/jbc.272.38.23465. [DOI] [PubMed] [Google Scholar]
- 3.Reardon JT, Sancar A. Nucleotide excision repair. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 4.Huang JC, Svoboda DL, Reardon JT, Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5' and the 6th phosphodiester bond 3' to the photodimer. Proc. Natl Acad. Sci. USA. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Svoboda DL, Taylor JS, Hearst JE, Sancar A. DNA repair by eukaryotic nucleotide excision nuclease. Removal of thymine dimer and psoralen monoadduct by HeLa cell-free extract and of thymine dimer by xenopus laevis oocytes. J. Biol. Chem. 1993;268:1931–1936. [PubMed] [Google Scholar]
- 6.Hu J, Choi JH, Gaddameedhi S, Kemp MG, Reardon JT, Sancar A. Nucleotide excision repair in human cells: fate of the excised oligonucleotide carrying DNA damage in vivo. J. Biol. Chem. 2013;288:20918–20926. doi: 10.1074/jbc.M113.482257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mu D, Park CH, Matsunaga T, Hsu DS, Reardon JT, Sancar A. Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 8.Cleaver JE. Defective repair replication of DNA in xeroderma pigmentosum. Nature. 1968;218:652–656. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- 9.DiGiovanna JJ, Kraemer KH. Shining a light on xeroderma pigmentosum. J. Invest. Dermatol. 2012;132:785–796. doi: 10.1038/jid.2011.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaddameedhi S, Kemp MG, Reardon JT, Shields JM, Smith-Roe SL, Kaufmann WK, Sancar A. Similar nucleotide excision repair capacity in melanocytes and melanoma cells. Cancer Res. 2010;70:4922–4930. doi: 10.1158/0008-5472.CAN-10-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 12.Riley LK, Marshall ME, Coleman MS. A method for biotinylating oligonucleotide probes for use in molecular hybridizations. DNA. 1986;5:333–337. doi: 10.1089/dna.1986.5.333. [DOI] [PubMed] [Google Scholar]
- 13.Setlow RB, Regan JD, German J, Carrier WL. Evidence that xeroderma pigmentosum cells do not perform the first step in the repair of ultraviolet damage to their DNA. Proc. Natl Acad. Sci. USA. 1969;64:1035–1041. doi: 10.1073/pnas.64.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cleaver JE, Trosko JE. Absence of excision of ultraviolet-induced cyclobutane dimers in xeroderma pigmentosum. Photochem. Photobiol. 1970;11:547–550. doi: 10.1111/j.1751-1097.1970.tb06025.x. [DOI] [PubMed] [Google Scholar]
- 15.Ye N, Bianchi MS, Bianchi NO, Holmquist GP. Adaptive enhancement and kinetics of nucleotide excision repair in humans. Mutat. Res. 1999;435:43–61. doi: 10.1016/s0921-8777(99)00022-1. [DOI] [PubMed] [Google Scholar]
- 16.Kemp MG, Reardon JT, Lindsey-Boltz LA, Sancar A. Mechanism of release and fate of excised oligonucleotides during nucleotide excision repair. J. Biol. Chem. 2012;287:22889–22899. doi: 10.1074/jbc.M112.374447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cleaver JE, Boyer HW. Solubility and dialysis limits of DNA oligonucleotides. Biochim. Biophys. Acta. 1972;262:116–124. doi: 10.1016/0005-2787(72)90224-9. [DOI] [PubMed] [Google Scholar]
- 18.Reardon JT, Sancar A. Recognition and repair of the cyclobutane thymine dimer, a major cause of skin cancers, by the human excision nuclease. Genes Dev. 2003;17:2539–2551. doi: 10.1101/gad.1131003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell DL. The relative cytotoxicity of (6-4) photoproducts and cyclobutane dimers in mammalian cells. Photochem. Photobiol. 1988;48:51–57. doi: 10.1111/j.1751-1097.1988.tb02785.x. [DOI] [PubMed] [Google Scholar]
- 20.Vreeswijk MP, van Hoffen A, Westland BE, Vrieling H, van Zeeland AA, Mullenders LH. Analysis of repair of cyclobutane pyrimidine dimers and pyrimidine 6-4 pyrimidone photoproducts in transcriptionally active and inactive genes in chinese hamster cells. J. Biol. Chem. 1994;269:31858–31863. [PubMed] [Google Scholar]
- 21.Boyce RP, Howard-Flanders P. Release of ultraviolet light-induced thymine dimers from dna in E. coli K-12. Proc. Natl Acad. Sci. USA. 1964;51:293–300. doi: 10.1073/pnas.51.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Setlow RB, Carrier WL. The disappearance of thymine dimers from dna: an error-correcting mechanism. Proc. Natl Acad. Sci. USA. 1964;51:226–231. doi: 10.1073/pnas.51.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Regan JD, Trosko JE, Carrier WL. Evidence for excision of ultraviolet-induced pyrimidine dimers from the DNA of human cells in vitro. Biophys. J. 1968;8:319–325. doi: 10.1016/S0006-3495(68)86490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.La Belle M, Linn S. In vivo excision of pyrimidine dimers is mediated by a DNA N-glycosylase in micrococcus luteus but not in human fibroblasts. Photochem. Photobiol. 1982;36:319–324. doi: 10.1111/j.1751-1097.1982.tb04381.x. [DOI] [PubMed] [Google Scholar]
- 25.Weinfeld M, Gentner NE, Johnson LD, Paterson MC. Photoreversal-dependent release of thymidine and thymidine monophosphate from pyrimidine dimer-containing DNA excision fragments isolated from ultraviolet-damaged human fibroblasts. Biochemistry. 1986;25:2656–2664. doi: 10.1021/bi00357a055. [DOI] [PubMed] [Google Scholar]
- 26.Reardon JT, Sancar A. Purification and characterization of Escherichia coli and human nucleotide excision repair enzyme systems. Methods Enzymol. 2006;408:189–213. doi: 10.1016/S0076-6879(06)08012-8. [DOI] [PubMed] [Google Scholar]
- 27.Evdokimov A, Petruseva I, Tsidulko A, Koroleva L, Serpokrylova I, Silnikov V, Lavrik O. New synthetic substrates of mammalian nucleotide excision repair system. Nucleic Acids Res. 2013;41:e123. doi: 10.1093/nar/gkt301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood RD, Robins P, Lindahl T. Complementation of the xeroderma pigmentosum DNA repair defect in cell-free extracts. Cell. 1988;53:97–106. doi: 10.1016/0092-8674(88)90491-6. [DOI] [PubMed] [Google Scholar]
- 29.Salles B, Provot C. In vitro chemiluminescence assay to measure excision repair in cell extracts. Methods Mol. Biol. 1999;113:393–401. doi: 10.1385/1-59259-675-4:393. [DOI] [PubMed] [Google Scholar]
- 30.Salles B, Frit P, Provot C, Jaeg JP, Calsou P. In vitro eukaryotic DNA excision repair assays: an overview. Biochimie. 1995;77:796–802. doi: 10.1016/0300-9084(96)88198-3. [DOI] [PubMed] [Google Scholar]
- 31.Sibghatullah, Husain I, Carlton W, Sancar A. Human nucleotide excision repair in vitro: repair of pyrimidine dimers, psoralen and cisplatin adducts by HeLa cell-free extract. Nucleic Acids Res. 1989;17:4471–4484. doi: 10.1093/nar/17.12.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salles B, Provot C, Calsou P, Hennebelle I, Gosset I, Fournie GJ. A chemiluminescent microplate assay to detect DNA damage induced by genotoxic treatments. Anal. Biochem. 1995;232:37–42. doi: 10.1006/abio.1995.9964. [DOI] [PubMed] [Google Scholar]
- 33.Wani AA, D'Ambrosio SM, Alvi NK. Quantitation of pyrimidine dimers by immunoslot blot following sublethal UV-irradiation of human cells. Photochem. Photobiol. 1987;46:477–482. doi: 10.1111/j.1751-1097.1987.tb04798.x. [DOI] [PubMed] [Google Scholar]
- 34.Nishinaga M, Kurata R, Onishi K, Kuriyama K, Wakasugi M, Matsunaga T. Establishment of a microplate-formatted cell-based immunoassay for rapid analysis of nucleotide excision repair ability in human primary cells. Photochem. Photobiol. 2012;88:356–362. doi: 10.1111/j.1751-1097.2012.01073.x. [DOI] [PubMed] [Google Scholar]
- 35.Mouret S, Charveron M, Favier A, Cadet J, Douki T. Differential repair of UVB-induced cyclobutane pyrimidine dimers in cultured human skin cells and whole human skin. DNA Repair. 2008;7:704–712. doi: 10.1016/j.dnarep.2008.01.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.