Abstract
The CRISPR-Cas-derived RNA-guided Cas9 endonuclease is the key element of an emerging promising technology for genome engineering in a broad range of cells and organisms. The DNA-targeting mechanism of the type II CRISPR-Cas system involves maturation of tracrRNA:crRNA duplex (dual-RNA), which directs Cas9 to cleave invading DNA in a sequence-specific manner, dependent on the presence of a Protospacer Adjacent Motif (PAM) on the target. We show that evolution of dual-RNA and Cas9 in bacteria produced remarkable sequence diversity. We selected eight representatives of phylogenetically defined type II CRISPR-Cas groups to analyze possible coevolution of Cas9 and dual-RNA. We demonstrate that these two components are interchangeable only between closely related type II systems when the PAM sequence is adjusted to the investigated Cas9 protein. Comparison of the taxonomy of bacterial species that harbor type II CRISPR-Cas systems with the Cas9 phylogeny corroborates horizontal transfer of the CRISPR-Cas loci. The reported collection of dual-RNA:Cas9 with associated PAMs expands the possibilities for multiplex genome editing and could provide means to improve the specificity of the RNA-programmable Cas9 tool.
INTRODUCTION
Editing genomes using the RNA-guided DNA targeting principle of CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeats-CRISPR associated proteins) immunity has been exploited widely over the past few months (1–13). The main advantage provided by the bacterial type II CRISPR-Cas system lies in the minimal requirement for programmable DNA interference: an endonuclease, Cas9, guided by a customizable dual-RNA structure (14). As initially demonstrated in the original type II system of Streptococcus pyogenes, trans-activating CRISPR RNA (tracrRNA) (15,16) binds to the invariable repeats of precursor CRISPR RNA (pre-crRNA) forming a dual-RNA (14–17) that is essential for both RNA comaturation by RNase III in the presence of Cas9 (15–17), and invading DNA cleavage by Cas9 (14,17–19). As demonstrated in Streptococcus, Cas9 guided by the duplex formed between mature activating tracrRNA and targeting crRNA (14–16) introduces site-specific double-stranded DNA (dsDNA) breaks in the invading cognate DNA (14,17–19). Cas9 is a multi-domain enzyme (14,20,21) that uses an HNH nuclease domain to cleave the target strand (defined as complementary to the spacer sequence of crRNA) and a RuvC-like domain to cleave the non-target strand (14,22,23), enabling the conversion of the dsDNA cleaving Cas9 into a nickase by selective motif inactivation (2,8,14,24,25). DNA cleavage specificity is determined by two parameters: the variable, spacer-derived sequence of crRNA targeting the protospacer sequence (a protospacer is defined as the sequence on the DNA target that is complementary to the spacer of crRNA) and a short sequence, the Protospacer Adjacent Motif (PAM), located immediately downstream of the protospacer on the non-target DNA strand (14,18,23,26–28).
Recent studies have demonstrated that RNA-guided Cas9 can be employed as an efficient genome editing tool in human cells (1,2,8,11), mice (9,10), zebrafish (6), drosophila (5), worms (4), plants (12,13), yeast (3) and bacteria (7). The system is versatile, enabling multiplex genome engineering by programming Cas9 to edit several sites in a genome simultaneously by simply using multiple guide RNAs (2,7,8,10). The easy conversion of Cas9 into a nickase was shown to facilitate homology-directed repair in mammalian genomes with reduced mutagenic activity (2,8,24,25). In addition, the DNA-binding activity of a Cas9 catalytic inactive mutant has been exploited to engineer RNA-programmable transcriptional silencing and activating devices (29,30).
At present, RNA-guided Cas9 from S. pyogenes, S. thermophilus** and Neisseria meningitidis have been developed into tools for genome manipulation (1–13,24,25,31–34). Here, we explore the possibilities of expanding the RNA-programmable Cas9 toolbox to additional orthologous systems. We investigated the diversity and interchangeability of dual-RNA:Cas9 in eight representatives of phylogenetically defined type II CRISPR-Cas groups. The results of this work not only introduce a wider range of Cas9 enzymes, dual-RNA structures and associated specific PAMs but also enlighten the evolutionary aspects of type II CRISPR-Cas systems, including coevolution and horizontal transfer of the system components.
MATERIALS AND METHODS
Bacterial strains and culture conditions
Supplementary Table S1 lists bacterial strains used in this study. Streptococcus pyogenes, Streptococcus mutans, Campylobacter jejuni, N. meningitidis, Escherichia coli and Francisella novicida were grown as previously described (15,16). Brain Heart Infusion (BHI, Becton Dickinson) agar and BHI broth medium supplemented with 1% glucose and 1% lactose were used to culture S. thermophilus at 42°C in a 5% CO2 environment (16). Pasteurella multocida and Staphylococcus aureus were grown at 37°C on BHI agar plates and in BHI broth with shaking. Cell growth was monitored by measuring the optical density of cultures at 620 nm (OD620) using a microplate reader (BioTek PowerWave).
Bacterial transformation
Escherichia coli was transformed with plasmid DNA according to standard protocols (35). Transformation of S. pyogenes was performed as previously described (36) with some modifications. Streptococcus pyogenes pre-cultures were diluted 1:100 in fresh THY medium and grown at 37°C, 5% CO2 until OD620 reached 0.3. Glycine was added to the medium to 10% final concentration and growth was maintained for an additional hour. Cells were spun down at 4°C at 2500 × g and washed three times with electroporation buffer (5 mM KH2PO4, 0.4 M D-sorbitol, 10% glycerol, pH 4.5), finally suspended in the same buffer and equalized to the same OD620. For electroporation, 1 µg of plasmid was incubated with the competent cells on ice for 10 min. The conditions were 25 µF, 600 Ω and 1.5 V using 1 mm electroporation cuvettes (Biorad). After a regeneration time of 3 h, bacteria were spread on agar medium supplemented with kanamycin (300 µg/ml). Transformation assays were performed at least three times independently with technical triplicates. The efficiencies were calculated as colony-forming units (CFU) per µg of plasmid DNA. Positive and negative control transformations were done with backbone plasmid pEC85 and sterile H2O, respectively.
DNA manipulations
DNA manipulations including DNA preparation (QIAprep Spin MiniPrep Kit, Qiagen), polymerase chain reaction (PCR) (Phusion® High-Fidelity DNA Polymerase, Finnzyme), DNA digestion (restriction enzymes, Fermentas), DNA ligation (T4 DNA ligase, Fermentas), DNA purification (QIAquick PCR Purification Kit, Qiagen) and agarose gel electrophoresis were performed according to the standard techniques or manufacturers’ protocols with some modifications (35). Site-directed mutagenesis was done using QuikChange II XL kit (Stratagene) or PCR-based mutagenesis (37). Synthetic oligonucleotides (Sigma-Aldrich and Biomers) and plasmids used and generated in this study are listed in Supplementary Table S1. The integrity of all constructed plasmids was verified by enzymatic digestion and sequencing at LGC Genomics.
Construction of plasmids for complementation studies in S. pyogenes
The backbone shuttle vector pEC85 was used for complementation study (38,39). The RNase-III encoding genes (rnc genes) of S. pyogenes, S. mutans, S. thermophilus, C. jejuni, N. meningitidis, P. multocida, F. novicida, E. coli and S. aureus, and the genes encoding truncated and inactive RNase III variants (truncated and inactive (D51A) rnc mutants) of S. pyogenes were cloned in pEC483 (pEC85 containing the native promoter of S. pyogenes rnc) using NcoI and EcoRI restriction sites (Supplementary Table S1, Supplementary Figure S6). The orthologous and mutant cas9 genes were cloned in pEC342 (pEC85 containing a sequence encoding tracrRNA-171 nt (16) and the native promoter of the S. pyogenes cas operon) using SalI and SmaI restriction sites (Supplementary Table S1). Note that in a previous study, we observed low abundance of tracrRNA in the cas9 deletion mutant. For this reason, plasmids used in cas9 complementation studies were designed to encode tracrRNA in addition to cas9 (16). The generated rnc and cas9 recombinant plasmids were introduced in S. pyogenes Δrnc and Δcas9 deletion strains, respectively (Supplementary Table S1). Plasmid integrity in all complemented strains was checked by plasmid DNA extraction and digestion.
Construction of plasmids for transformation studies in S. pyogenes
Plasmid pEC85 was used as backbone vector for transformation studies. A DNA fragment containing WT speM protospacer sequence was cloned in the PstI site of plasmids containing coding sequences of WT or mutated cas9 from S. pyogenes (Supplementary Table S1).
Construction of plasmids for protein purification
The overexpression vector pET16b (Novagen) was modified by inserting three additional restriction sites (SalI, SacI, NotI) into the NdeI restriction site, generating pEC621. The genes coding for the orthologous Cas9 proteins were PCR amplified from genomic DNA of the corresponding strains using primers containing a SalI and a NotI restriction site (Supplementary Table S1). The S. pyogenes cas9 mutant genes were PCR amplified from the complementation plasmids mentioned above. All orthologous and mutant cas9 genes were cloned into the SalI and NotI sites of pEC621.
Construction of substrate plasmids for in vitro cleavage assays
Plasmid pEC287 that contains the speM protospacer sequence was used as a vector to construct all substrate plasmids. The PAM sequence located in 3′ just next to the crRNA-targeted sequence of the speM protospacer (GGG on this plasmid) was modified by PCR-mediated site-directed mutagenesis (37) using one standard oligonucleotide (OLEC3140 or OLEC3194) that either introduced or removed a XbaI restriction site for screening purposes, and a second mutagenic oligonucleotide to exchange the protospacer adjacent sequence (Supplementary Table S1).
RNA preparation
Total RNA from S. pyogenes SF370 WT, deletion mutants and complemented strains was prepared from culture samples collected at the mid-logarithmic phase of growth using TRIzol (Sigma-Aldrich). The total RNA samples were treated with DNase I (Fermentas) according to the manufacturer’s instructions. The concentration of RNA in each sample was measured using NanoDrop.
Northern blot analysis
Northern blot analysis was carried out essentially as described previously (40–42). Total RNA was separated on 10% polyacrylamide 8 M urea gels and further processed for blotting on nylon membranes (Hybond™ N+, GE healthcare; Trans-Blot® SD semi-dry transfer apparatus, Biorad; 1X TBE, 2 h at 10 V/cm), chemical cross-linking with EDC (1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride) (41) and prehybridization (Rapid-hyb buffer, GE healthcare; 1 h at 42°C). Oligonucleotide probes (40 pmol) were labeled with 32P (20 μCi) using the T4-polynucleotide kinase (10 U, Fermentas) and purified using G-25 columns (GE Healthcare) prior use. Visualization of the radioactive signal was done using a phosphorimager. 5S rRNA served as loading control.
Protein purification
Escherichia coli Rosetta2(DE3) and E. coli NiCo21(DE3) (New England Biolabs) were transformed with overexpression plasmids coding for S. pyogenes WT and mutant or orthologous Cas9, respectively. Cells were grown at 37°C to reach an OD600 of 0.7–0.8, protein expression was induced by adding IPTG to a final concentration of 0.5 mM and cultures were further grown at 13°C overnight. The cells were harvested by centrifugation and the pellet was resuspended in lysis buffer (20 mM HEPES, pH 7.5, 500 mM KCl [1 M for S. thermophilus* Cas9], 0.1% Triton X-100, 25 mM imidazole) and lysed by sonication. The lysate was cleared by centrifugation (>20 000 × g) and incubated with Ni-NTA (Qiagen) for 1 h at 4°C. After washing the Ni-NTA with lysis buffer and wash buffer (20 mM HEPES, pH 7.5, 300 mM KCl, 0.1% Triton X-100, 25 mM imidazole), the recombinant protein was eluted with elution buffer (20 mM HEPES, pH 7.5, 150 mM KCl, 0.1 mM DTT, 250 mM imidazole, 1 mM (EDTA)) and the fractions were analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). In the case of S. pyogenes Cas9 WT and mutants, the protein containing eluates were pooled and further purified via HiTrap SP FF (GE Healthcare) cation-exchange chromatography. Briefly, the protein was loaded on the column equilibrated with buffer A (20 mM HEPES pH 7.5, 100 mM KCl) using an FPLC system (Äkta, GE Healthcare). Cas9 was eluted with a gradient of buffer B (20 mM HEPES pH 7.5, 1 M KCl) over 12 ml. 1 ml fractions were collected and analyzed by (SDS–PAGE). The protein containing fractions were pooled and dialyzed overnight (20 mM HEPES, pH 7.5, 150 mM KCl, 50% glycerol). For Cas9 orthologs, the eluates from Ni-NTA purification were checked for purity by SDS–PAGE. In case of contaminants, a second purification over chitin beads was performed as described in the manual for NiCo21(DE3) cells from New England Biolabs. Briefly, 1 ml chitin beads (New England Biolabs) equilibrated with buffer A was incubated with the Ni2+-IMAC eluates for 1 h at 4°C. Afterwards, the beads were added onto a column and the Cas9 containing flowthroughs were collected and again checked for purity by SDS–PAGE (Supplementary Figure S1). The purified proteins were dialyzed overnight. The protein concentration was calculated by measuring the OD280 using the extinction coefficient. The detailed characteristics of purified proteins are summarized in Supplementary Figure S1A.
In vitro transcription
RNA for in vitro DNA cleavage assays was generated by in vitro transcription using the AmpliScribe™ T7-Flash™ Transcription Kit (Epicentre) according to the manufacturer’s instructions. PCR products or synthetic oligonucleotides used as templates are listed in Supplementary Table S1. The synthesized tracrRNA and repeat region of crRNA from each bacterial species correspond to the mature forms of RNAs as determined by deep RNA sequencing (15) or bioinformatics predictions. The spacer region of all crRNAs used in this study targets the speM protospacer (encoding superantigen; targeted by spacer 2 of S. pyogenes SF370 CRISPR array, Spyo1h_002 (16)). Transcribed RNAs were precipitated and further purified from 10% polyacrylamide 8 M urea denaturing gel. The RNA concentration was determined by measuring the OD260 and the molarity was calculated. Equimolar amounts of crRNA and tracrRNA were mixed in 5X RNA annealing buffer (1 M NaCl, 100 mM HEPES, pH 7.5), heated up to 95°C for 5 min and slowly cooled to room temperature before use.
In vitro DNA cleavage assays
For the cleavage assays using Cas9 mutant proteins, 25 nM of Cas9 were incubated with equimolar amounts of prehybridized S. pyogenes dual-RNA in cleavage buffer (20 mM HEPES, pH 7.5, 150 mM KCl, 10 mM MgCl2, 0.5 mM DTT, 0.1 mM EDTA) for 15 min at 37°C. Plasmid DNA (5 nM) containing speM (NGG PAM) was added and further incubated for 1 h at 37°C. The reaction was stopped by addition of 5X loading buffer (250 mM EDTA, 30% glycerol, 1.2% SDS, 0.1% (w/v) bromophenol blue) and analyzed by 1% agarose gel electrophoresis in 1X TAE. Cleavage products were visualized by ethidium bromide staining. All other cleavage assays were carried out using the same conditions with the following modifications: KGB (43) (100 mM potassium glutamate, 25 mM Tris/acetate, pH 7.5, 10 mM Mg-acetate, 0.5 mM 2-mercaptoethanol, 10 µg/ml bovine serum albumin) was used as cleavage buffer and different concentrations of dual-RNA:Cas9 complex were analyzed. The concentration of plasmid DNA was kept constant in all experiments, i.e. 5 nM.
Search for PAM motifs
Spacer sequences of the selected bacterial species were extracted from the CRISPRdatabase (http://crispr.u-psud.fr/crispr/) and used to find cognate protospacer candidates using megaBLAST (http://blast.ncbi.nih.gov/Blast). Protospacer candidates were defined as containing a sequence with ≥90% similarity to the crRNA spacer sequence and originating from phage, plasmid or genomic DNA related to the bacterial species of the targeting CRISPR-Cas. For the investigated CRISPR-Cas loci, the orientation of transcription was determined previously by RNA sequencing or northern blot analysis (15,16). It was also shown before that in type II CRISPR-Cas, the PAM sequence is located in 3′ of the protospacer, juxtaposed to the sequence targeted by cognate crRNA on the non-target strand (14,18,23,44). To identify possible PAMs in each bacterial species, 10 nt sequences on the non-target strand directly downstream of each protospacer sequence were aligned. A logo plot (http://weblogo.berkeley.edu/) showing the most abundant nucleotides was created and PAM sequences were predicted. In the cases of CRISPR-Cas loci for which no suitable protospacer sequences could be identified (S. mutans UA159, C. jejuni NCTC 11168, P. multocida Pm70, F. novicida U112), closely related strains of the same species were selected (Supplementary Table S2). The spacer contents of the type II CRISPR arrays in selected strains were analyzed (http://crispr.u-psud.fr/Server/). The spacer sequences were then used to select cognate protospacer sequences as described above.
Protein sequence analysis
Position-Specific Iterated (PSI)-BLAST program (45) was used to retrieve orthologs of the Cas9 family in the NCBI nr database. Sequences shorter than 800 amino acids were discarded. The BLASTClust program (46) set up with a length coverage cutoff of 0.8 and a score coverage threshold (bit score divided by alignment length) of 0.8 was used to cluster the remaining sequences (Supplementary Table S2). This procedure produced 82 clusters. In the case of sequences reported in this study, one or several representatives from each cluster were selected and aligned using the MUSCLE program (47) with default parameters, followed by a manual correction on the basis of local alignments obtained using PSI-BLAST (45) and HHpred programs (48). The confidently aligned blocks (Supplementary Figure S2) with 285 informative positions were used for maximum likelihood tree reconstruction using the FastTree program (49) with the default parameters: JTT evolutionary model, discrete gamma model with 20 rate categories. The same program was used to calculate the bootstrap values. Cas1 sequences were selected from the corresponding cas operons (Supplementary Table S2). A few incomplete sequences were substituted by other Cas1 sequences from the same Cas9 cluster (Supplementary Table S2). Several Cas1 proteins from subtypes I-A, B, C and E were included as an outgroup. Cas1 sequences were aligned using the same approach described above and 252 informative positions (Supplementary Figure S3) were used for maximum likelihood tree reconstruction using the FastTree program. RNase III multiple sequence alignment was prepared using the MUSCLE program.
RNA sequence and structure analysis
RNA duplex secondary structures were predicted using RNAcofold of the Vienna RNA package (50,51) and RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/). The structure predictions were then visualized using VARNA (52).
RESULTS
Diversity of Cas9 orthologs
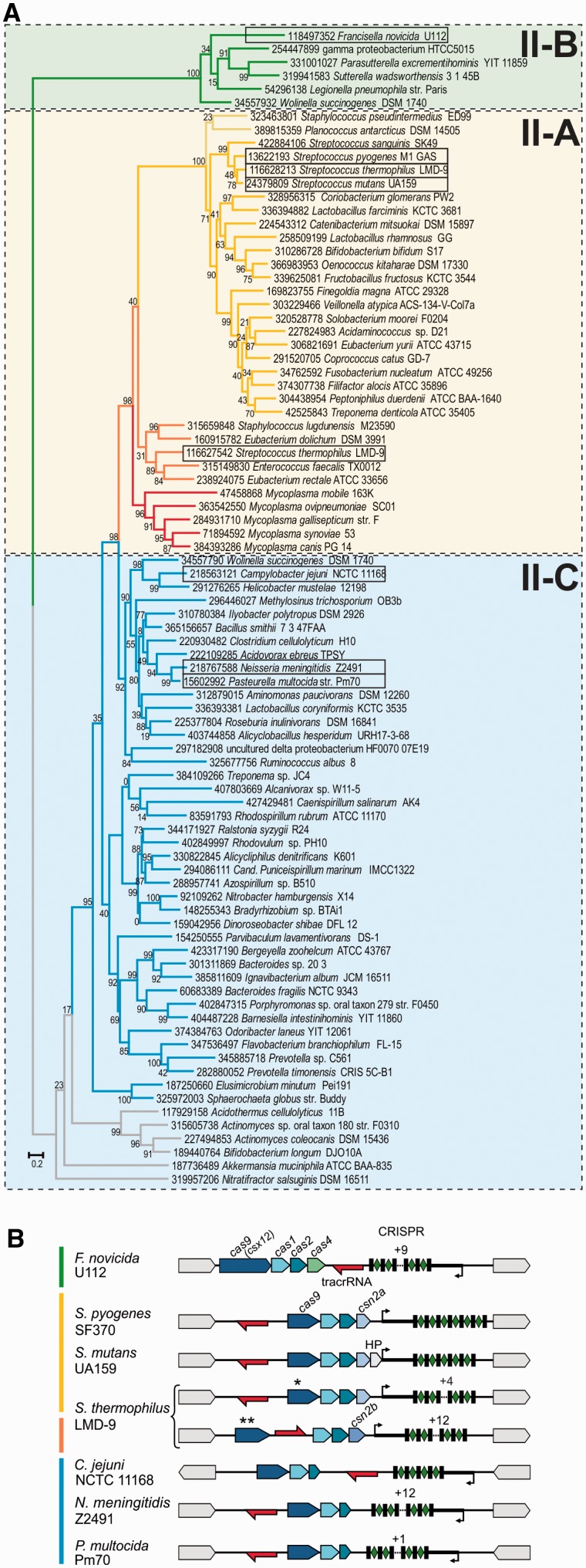
To investigate the evolution and diversity of dual-RNA:Cas9 systems, we subjected publicly available genomes to multiple rounds of BLAST search using previously retrieved Cas9 sequences as queries (15). Cas9 orthologs were identified in 653 bacterial strains representing 347 species (Supplementary Table S2). After removing incomplete or highly similar sequences, we selected 83 diverse, representative Cas9 orthologs for multiple sequence alignment and phylogenetic tree reconstruction (Figure 1A, Supplementary Table S2, Supplementary Figures S2 and S4, see ‘Materials and Methods’ section). The Cas9 tree topology largely agrees with the phylogeny of the corresponding Cas1 proteins (Supplementary Table S2, Supplementary Figures S3 and S4) and fully supports the previously described classification of type II CRISPR-Cas into three subtypes, II-A (specified by csn2), II-B (characterized by long and most diverged cas9 variants (formerly csx12) and cas4) and II-C (three-cas gene operon) (15).
Figure 1.
Phylogeny of representative Cas9 orthologs and schematic representation of selected bacterial type II CRISPR-Cas systems. (A) Phylogenetic tree of Cas9 reconstructed from selected, informative positions of representative Cas9 orthologs multiple sequence alignment is shown (Supplementary Figure S2 and Supplementary Table S2). The Cas9 orthologs of the subtypes classified as II-A, II-B and II-C are highlighted with shaded boxes. The colored branches group distinct proteins of closely related loci with similar locus architecture (15). Each protein is represented by the GenInfo (GI) identifier followed by the bacterial strain name. The bootstrap values are given for each node (see ‘Materials and Methods’ section). Note that the monophyletic clusters of subtypes II-A and II-B are supported by high bootstrap values. The scale bar for the branch length is given as the estimated number of amino acid substitution per site. (B) Genetic loci of type II (Nmeni/CASS4) CRISPR-Cas in Streptococcus pyogenes SF370, Streptococcus mutans UA159, Streptococcus thermophilus LMD-9 *(CRISPR3), **(CRISPR1), Campylobacter jejuni NCTC 11168, Neisseria meningitidis Z2491, Pasteurella multocida Pm70 and Francisella novicida U112. Red arrow, transcription direction of tracrRNA; blue arrows, cas genes; black rectangles, CRISPR repeats; green diamonds, spacers; thick black line, leader sequence; black arrow, putative pre-crRNA promoter; HP, Hypothetical Protein. The colored bars represented on the left correspond to Cas9 tree branches colors. The transcription direction and putative leader position of C. jejuni and N. meningitidis pre-crRNAs were derived from previously published RNA sequencing data (15). The CRISPR-Cas locus architecture of P. multocida was predicted based on its close similarity to that of N. meningitidis and further confirmed by bioinformatics prediction of tracrRNA based on a strongly predicted promoter and a transcriptional terminator as described in (15). Type II CRISPR-Cas loci can differ in the cas gene composition, mostly with cas9, cas1 and cas2 being the minimal set of genes (type II-C, blue), sometimes accompanied with a fourth gene csn2a/b (type II-A, yellow and orange) or cas4 (type II-B, green). The CRISPR array can be transcribed in the same (type II-A, yellow and orange) or in the opposite (types II-B and C, blue and green) direction of the cas operon. The location of tracrRNA and the direction of its transcription differ within the groups (compare type II-A of S. thermophilus** with type II-A from the other species indicated here (yellow) and compare type II-C of C. jejuni with type II-C of N. meningitidis and P. multocida (blue)).
Analysis of the composition of cas genes, transcription direction of the CRISPR arrays with respect to that of the cas operon, and location and orientation of tracrRNAs resulted in the division of subtypes into groups with distinct locus characteristics, especially within the subtype II-A (Figure 1, clusters marked with different colors) (15). We selected Cas9 enzymes representative of the major type II groups. Cas9 orthologs of S. pyogenes, S. thermophilus* (CRISPR3) and S. mutans were chosen for type II-A systems associated with shorter, ∼220 amino acid Csn2 variants (Csn2a). Cas9 of S. thermophilus** (CRISPR1) represents a distinct group of type II-A sequences associated with longer, ∼350 amino acid version of Csn2 orthologs (Csn2b). Cas9 of F. novicida was selected for type II-B. The closely related Cas9 orthologs of P. multocida and N. meningitidis and the distinct, short Cas9 of C. jejuni were chosen for type II-C (Figure 1B). Expression of associated tracrRNAs and crRNAs in S. pyogenes, S. mutans, F. novicida, N. meningitidis and C. jejuni was already validated by deep RNA sequencing (15,16). The RNAs in S. thermophilus and P. multocida were predicted bioinformatically based on the sequences from related species within the same type II group. Figure 1B shows the organization of the eight selected type II CRISPR-Cas loci and highlights our previous findings demonstrating that the type II loci architectures are highly variable among subtypes, yet conserved within each group (15). These variations are in good agreement with the clustering derived from the Cas9 and Cas1 phylogenetic trees (Figure 1A, Supplementary Figure S4).
Bacterial RNases III are interchangeable in dual-RNA maturation
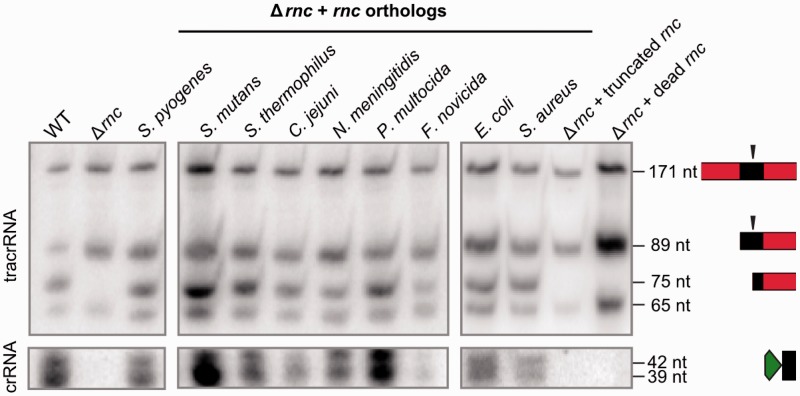
As described in S. pyogenes and S. thermophilus, RNase III plays an essential role in the biogenesis of dual-RNA:Cas9 systems by coprocessing tracrRNA and pre-crRNA at the level of antirepeat:repeat duplexes (16,17). We analyzed the interchangeability of S. pyogenes RNase III with RNases III from selected bacterial species in the coprocessing of S. pyogenes tracrRNA:pre-crRNA, including strains that lack type II CRISPR-Cas (S. aureus COL, E. coli TOP10). Northern blot analysis shows that all RNases III studied here can coprocess the RNA duplex (Figure 2, Supplementary Figure S5), indicating that there is no species-specificity for tracrRNA:pre-crRNA cleavage by RNase III. Multiple sequence alignment of RNase III orthologs demonstrates conservation of the catalytic aspartate residue and the dsRNA binding domain (Figure 2, Supplementary Figure S6) that are both required for RNA coprocessing (Figure 2, Supplementary Figure S5). These data imply that the conservation of tracrRNA:pre-crRNA coprocessing by bacterial RNase III provides a degree of flexibility allowing the functionality of dual-RNA:Cas9 systems in multiple species upon horizontal transfer.
Figure 2.
RNase III is a general executioner of tracrRNA:pre-crRNA processing in type II CRISPR-Cas. Northern blot analysis of total RNA from S. pyogenes WT, Δrnc and Δrnc complemented with rnc orthologs or mutants (truncated rnc and inactivated (dead) (D51A) rnc) probed for tracrRNA (top) and crRNA repeat (bottom). RNA sizes in nucleotide and schematic representations of tracrRNA (red-black) and crRNA (green-black) are indicated on the right (16). The vertical black arrows indicate the processing sites. tracrRNA-171 nt and tracrRNA-89 nt forms correspond to primary tracrRNA transcripts. The presence of tracrRNA-75 nt and crRNA 39-42 nt forms indicates tracrRNA and pre-crRNA co-processing. S. pyogenes tracrRNA and pre-crRNA are coprocessed by all analyzed RNase III orthologs. The truncated version and catalytic inactive mutant of S. pyogenes RNase III are both deficient in tracrRNA:pre-crRNA processing.
Cas9 HNH and split RuvC domains are the catalytic moieties for DNA interference
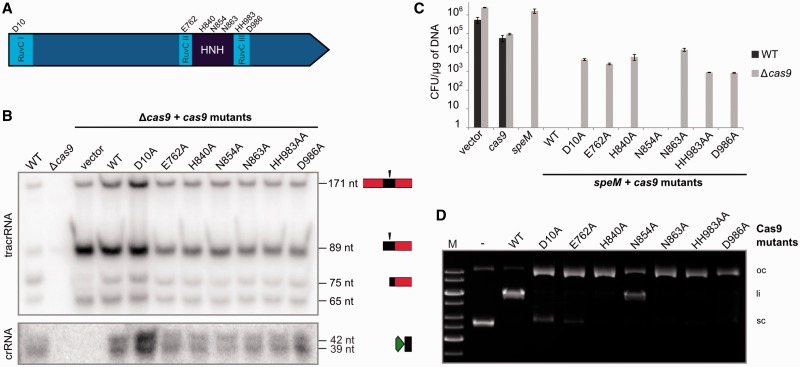
Comparison of Cas9 sequences revealed high diversity in amino acid composition and length (984 amino acid for C. jejuni to 1648 amino acids for F. novicida), especially in the linker sequence between the highly conserved N-terminal RuvC and central RuvC-HNH-RuvC regions and in the C-terminal extension (Supplementary Figure S2). Several studies demonstrated the importance of the nuclease motifs for dsDNA cleavage activity by mutating one aspartate in the N-terminal motif of the RuvC domain and one or several residues in the predicted catalytic motif of the HNH domain of the Cas9 enzyme (14,22,23). To investigate the relevance of all catalytic motifs for tracrRNA:pre-crRNA processing and/or DNA interference, alanine substitutions of selected residues were created (Figure 3A). In addition to the already published catalytic amino acids, we created Cas9 point mutants of conserved amino acid residues in the central RuvC motifs (14) (Figure 3A, Supplementary Figure S2). Northern blot analysis of S. pyogenes cas9 deletion mutant complemented with each of the cas9 point mutants revealed the presence of mature tracrRNA and crRNA forms, demonstrating that none of the catalytic motifs is involved in dual-RNA maturation by RNase III. This is in agreement with previous data showing that RNase III is the enzyme that specifically cleaves tracrRNA:pre-crRNA duplex (16). Cas9 seems to have a stabilizing function on dual-RNA. We show that the catalytic motifs are not involved in RNA duplex stabilization (Figure 3B, Supplementary Figure S7).
Figure 3.
Conserved motifs of Cas9 are required for DNA interference but not for dual-RNA processing by RNase III. (A) Schematic representation of S. pyogenes Cas9. The conserved HNH and splitted RuvC motifs and analyzed amino acids are indicated. (B) Northern blot analysis of total RNA from S. pyogenes WT, Δcas9 and Δcas9 complemented with pEC342 or pEC342 containing cas9 WT or mutant genes, probed for tracrRNA and crRNA repeat. Maturation of tracrRNA and pre-crRNA generating tracrRNA-75 nt and crRNA-39-42 nt forms is observed in all Δcas9 strains complemented with the cas9 mutants. (C) In vivo protospacer targeting. Transformation assays of S. pyogenes WT and Δcas9 with pEC85 (vector), pEC85Ωcas9 (cas9), pEC85ΩspeM (speM), and pEC85ΩtracrRNA-171 nt plasmids containing speM and cas9 mutants. The CFUs per µg of plasmid DNA were determined in at least three independent experiments. The results ±SD of technical triplicates of one representative experiment are shown. Cas9 N854A is the only mutant that did not tolerate the protospacer plasmid as observed for WT Cas9, indicating that this residue is not involved in DNA interference. (D) In vitro plasmid cleavage. Agarose gel electrophoresis of plasmid DNA (5 nM) containing speM protospacer (pEC287) incubated with 25 nM Cas9 WT or mutants in the presence of equimolar amounts of dual-RNA-speM (see ‘Materials and Methods’ section). Cas9 WT and N854A generated linear cleavage products while the other Cas9 mutants created only nicked products. M, 1 kb DNA ladder (Fermentas); oc: open circular, li: linear; sc: supercoiled.
To investigate the involvement of the conserved motifs of Cas9 in DNA interference in vivo, we used a previously described plasmid-based read-out system that mimics infection with invading protospacer-containing DNA elements (16). Transformation assays were done in S. pyogenes WT or a cas9 deletion mutant using plasmids containing the speM protospacer gene (complementary to the second spacer of S. pyogenes SF370 type II CRISPR array (16)) and WT or mutant cas9 (Figure 3C). In this assay, Cas9 expressed following plasmid delivery in bacterial cells catalyzes its own vector cleavage, when active. Control experiments showed that the speM protospacer-containing plasmid was not tolerated in WT S. pyogenes, demonstrating activity of WT CRISPR-Cas. Similarly, a plasmid containing the speM protospacer and encoding WT Cas9 could not be maintained in the cas9 deletion mutant, demonstrating that Cas9 is able to cleave the plasmid from which it is expressed. Except for Cas9 N854A, all plasmids encoding Cas9 mutants were tolerated in the cas9 deletion strain, indicating abrogation of Cas9 interference activity for these variants.
The in vivo DNA targeting data were confirmed with in vitro DNA cleavage assays. Purified WT and mutant Cas9 proteins were incubated with tracrRNA:crRNA targeting speM and subjected to cleavage of plasmid DNA containing the speM protospacer. WT and N854A Cas9 show dsDNA cleavage activity, whereas the other Cas9 mutants cleave only one strand of the dsDNA substrate, yielding nicked open circular plasmid DNA (Figure 3D). This corroborates the results obtained in vivo showing the importance of the conserved nuclease motifs for DNA interference by Cas9. In addition to the previously published data demonstrating the importance of the N-terminal RuvC motif and the catalytic motif of HNH, we thus defined new catalytic residues in the central RuvC motifs.
Only Cas9 from closely related CRISPR-Cas systems can substitute for S. pyogenes Cas9 in tracrRNA-directed pre-crRNA maturation by RNase III
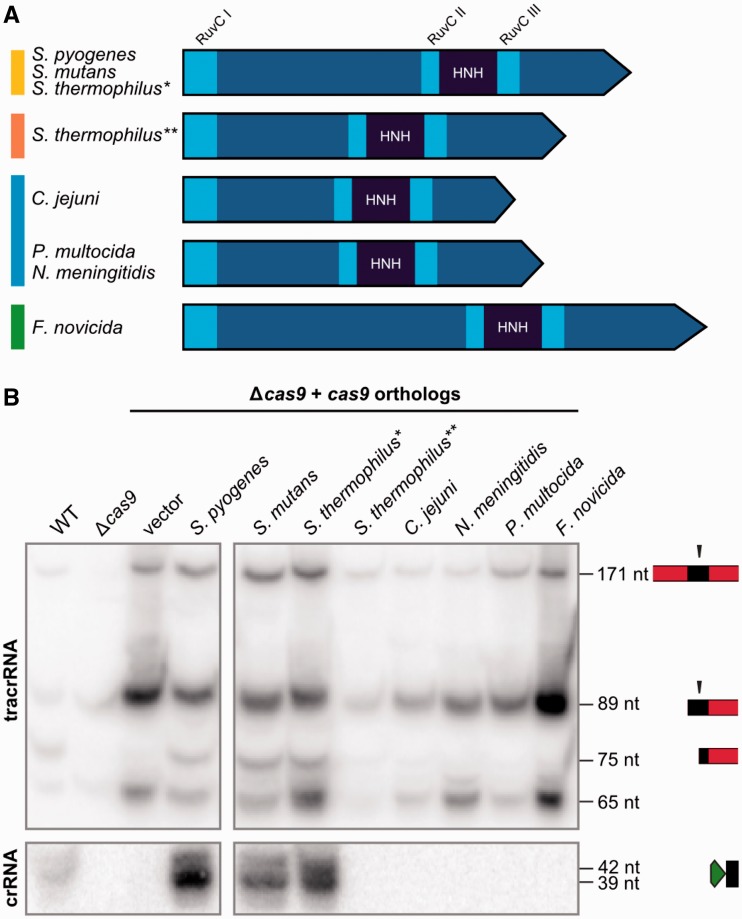
Beside the conservation of the HNH and split RuvC domains involved in DNA cleavage (14,15), the length of Cas9 orthologs and the amino acid sequences of Cas9 are highly variable among the different groups of type II CRISPR-Cas systems (Figure 4A, Supplementary Figure S2). Hence, we investigated whether this variability plays a role in the specificity of Cas9 with regard to tracrRNA:pre-crRNA duplex and mature crRNA stabilization. We complemented S. pyogenes cas9 deletion mutant with Cas9 from selected bacterial species representative of the various type II groups and analyzed tracrRNA:pre-crRNA processing by northern blot. Cas9 proteins from S. mutans and S. thermophilus* can substitute for the stabilizing role of S. pyogenes Cas9 in RNA processing by RNase III (Figure 4B, Supplementary Figure S8). In contrast, Cas9 from S. thermophilus**, C. jejuni, N. meningitidis, P. multocida and F. novicida could not complement the lack of RNA processing in the cas9 mutant of S. pyogenes. In these strains, the 75-nt processed form of tracrRNA is observed as a very weak signal of background level of dual-RNA processed by RNase III in the absence of Cas9. Overall, only Cas9 from closely related systems of S. pyogenes in the type II-A cluster can substitute endogenous Cas9 role in dual-RNA stabilization and subsequent maturation by RNase III.
Figure 4.
Cas9 from closely related CRISPR-Cas systems can substitute the role of S. pyogenes Cas9 in RNA processing by RNase III. (A) Schematic representation of Cas9 from selected bacterial species. The protein sizes and distances between conserved motifs (RuvC and HNH) are drawn in scale. See Supplementary Figure S2. (B) Northern blot analysis of total RNA extracted from S. pyogenes WT, Δcas9 and Δcas9 complemented with pEC342 (backbone vector containing tracrRNA-171 nt and the cas operon promoter from S. pyogenes) or pEC342-based plasmids containing cas9 orthologous genes, probed for tracrRNA and crRNA repeat. Mature forms of S. pyogenes tracrRNA and pre-crRNA are observed only in the presence of S. pyogenes Cas9 WT or closely related Cas9 orthologs from S. mutans and S. thermophilus*.
Cas9 orthologs require their specific PAM sequence for DNA cleavage activity
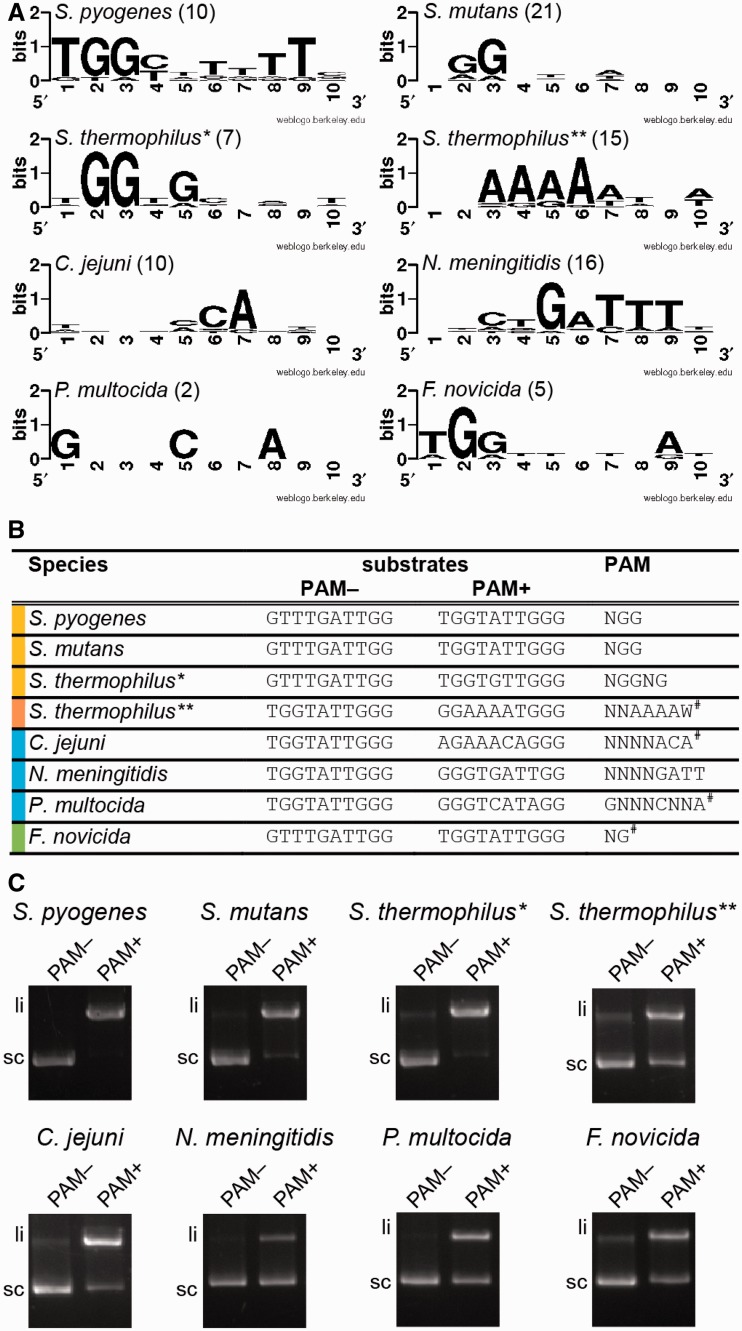
In S. pyogenes and S. thermophilus* types II-A, PAMs were identified as NGG and NGGNG, respectively. In these two species, mutating the PAM abrogates DNA interference by dual-RNA:Cas9 (14,22,23). To identify the functional PAMs for Cas9 from bacterial species other than S. pyogenes and S. thermophilus, we searched for potential protospacers matching spacer sequences in the selected CRISPR arrays using BLAST. For S. mutans UA159, C. jejuni NCTC 11168, P. multocida Pm70 and F. novicida U112, we were unable to identify potential protospacers. Therefore, we searched for strains that harbor a closely related variant of Cas9 (Supplementary Table S2) and analyzed their spacer sequences following the same approach (Supplementary Table S3). We aligned the identified 10 nt sequences located directly downstream of the protospacer sequence and delineated the most common nucleotides that could represent PAM sequences. Based on the data visualized as a logo plot (Figure 5A), we designed plasmid DNA substrates containing the speM protospacer followed by different adjacent sequences either comprising the predicted PAM or not (Figure 5B). The Cas9 orthologous proteins were purified (Supplementary Figure S1) and dual-RNA orthologs were designed based on deep RNA sequencing data (15), with the spacer sequence of crRNA targeting speM. To determine the protospacer-adjacent sequences critical for efficient DNA targeting, the purified Cas9 orthologs and their cognate dual-RNAs were used in DNA cleavage assays with different plasmid substrates (Figure 5C, Supplementary Figure S9). The previously published PAMs for Cas9 from S. pyogenes (NGG), S. mutans (NGG), S. thermophilus* (NGGNG) and N. meningitidis (NNNNGATT) (27,28,53,54) were confirmed by multiple sequence alignments and in vitro cleavage assay, validating our approach. However, dual-RNA guided Cas9 from S. thermophilus* could efficiently cleave target DNA in the presence of only NGG instead of NGGNG (Supplementary Figure S9). This is in contrast to data obtained in vivo, where mutation of the third G abrogates interference by Cas9 of S. thermophilus* (23). For S. thermophilus**, the PAM was published as NNAGAAW (27), which differs by one base from the sequence that we derived (NNAAAAW). In vitro cleavage assays with these two sequences demonstrate that the DNA substrate with the ‘NNAAAAW’ PAM is cleaved more efficiently by Cas9 of S. thermophilus** compared to the ‘NNAGAAW’ PAM (Supplementary Figure S9). Using the same approach, we also validated the PAM activity of the most common protospacer-downstream sequences for C. jejuni, F. novicida and P. multocida by in vitro cleavage assays, resulting in the most probable PAM sequences being NNNNACA (C. jejuni), GNNNCNNA (P. multocida) and NG (F. novicida) (Figure 5C, Supplementary Figure S9). Analysis of the protospacer-adjacent sequence from C. jejuni shows the same frequency of C and A (‘NNNNCCA’ or ‘NNNNACA’) at position 5 downstream of the protospacer (Supplementary Table S3). Hence, we tested both substrates for cleavage activity by C. jejuni dual-RNA:Cas9. Only the DNA target containing A at this position was cleaved efficiently (Supplementary Figure S9). This result could be explained by the origin of the protospacer, with the ‘NNNNCCA’ PAM being mostly found in genomic DNA or prophages of Campylobacter strains. In this case, the mutated PAM sequence on the chromosomally located protospacer prevents self-targeting. The P. multocida PAM requires further verification given that the multiple sequence alignment was derived from only two protospacer sequences. Thus, we identified a series of specific PAMs that enable dsDNA cleavage by dual-RNA:Cas9 complexes from different bacterial species in vitro. For gene editing purposes, it would be advisable to test a range of potential motifs to select those PAMs that would allow efficient targeting with limited off-site effect.
Figure 5.
Cas9 orthologs cleave DNA in the presence of their cognate dual-RNA and specific PAM in vitro. (A) Logo plot of protospacer adjacent sequences derived from BLAST analysis of spacer sequences for selected bacterial species. The logo plot gives graphical representation of most abundant nucleotides downstream of the protospacer sequence. The numbers in brackets correspond to the number of analyzed protospacers. (B) DNA substrates designed for specific PAM verification. Based on the logo plot for each species, plasmid DNA substrates were designed to contain the speM protospacer and the indicated sequence downstream, either comprising (PAM+) or not (PAM−) the proposed PAM. The predicted PAMs were verified by cleavage assays narrowing down the necessary nucleotides for activity (data not shown); therefore the sequence used differs slightly from the logoplot shown in (A). The high abundance of other nucleotides not being part of the PAM can be explained by redundancy of the coding sequences containing the protospacers, and by the limited number of found protospacer targets. The last column shows the PAM sequence for each species, which was already published (no symbol) or derived from this work (#). (C) In vitro plasmid cleavage assays by dual-RNA:Cas9 orthologs on plasmid DNA with the 10-bp protospacer adjacent sequence (summarized in (B)). Each Cas9 ortholog in complex with its cognate dual-RNA cleaves plasmids containing the corresponding species-specific PAM (PAM+). No cleavage is observed with plasmids that did not contain the specific PAM (PAM−). li: linear cleavage product, sc: supercoiled plasmid DNA.
Phylogenetic clustering of Cas9 defines dual-RNA:Cas9 exchangeability
As described above, clustering of Cas9 orthologs correlates with the ability to substitute for the RNA-stabilizing role of S. pyogenes Cas9 in tracrRNA:pre-crRNA processing by RNase III in vivo (Figure 4B). We investigated the exchangeability between Cas9 and dual-RNA in closely related CRISPR-Cas systems at the level of DNA interference. Plasmid cleavage assays were performed using S. pyogenes Cas9 complexed with dual-RNAs from selected CRISPR-Cas systems representative of the clustering of the type II CRISPR-Cas systems. As shown in Figure 6A (upper panel), S. pyogenes Cas9 can cleave target DNA in the presence of dual-RNAs from S. mutans and S. thermophilus* (type II-A, yellow subcluster), but not from any other tested species. The same result was observed when the dual-RNA from S. pyogenes was incubated with Cas9 orthologs from different bacteria (Figure 6A, lower panel). Cleavage assays were also performed with all Cas9 orthologs incubated with cognate and non-cognate dual-RNAs on their PAM-specific plasmid DNA. Only the combinations of Cas9 and dual-RNA within the same type II subcluster conferred dsDNA cleavage activity (Figure 6B, Supplementary Figure S10). More striking was the gradient of activity dependent on how closely related the species are in the corresponding type II group. This effect can be observed for C. jejuni Cas9 that is able to cleave DNA in the presence of dual-RNA from P. multocida and N. meningitidis, but not as efficient as with its own RNA (type II-C, blue subcluster). This finding is in good agreement with the phylogenetic tree of Cas9 (Figure 1A) showing that all three Cas9 orthologs belong to type II-C but C. jejuni Cas9 clusters more distantly from P. multocida and N. meningitidis Cas9. This effect was even greater for S. thermophilus** Cas9, which belongs to type II-A together with S. pyogenes, S. mutans and S. thermophilus*. However, none of the dual-RNAs from the three latter loci could direct DNA cleavage by S. thermophilus** Cas9. This result supports the recent findings demonstrating the lack of exchangeability between Cas9 from CRISPR1 and CRISPR3 of S. thermophilus DGCC7710 with regard to dual-RNA binding (17). We conclude that Cas9 and tracrRNA:crRNA interchangeabilty directly results from Cas9 coevolution with dual-RNA and follows the Cas9 phylogeny that may differ from the phylogeny of the respective bacterial species due to horizontal transfer.
Figure 6.
Cas9 and dual-RNA coevolved. (A) In vitro plasmid cleavage assays using S. pyogenes Cas9 in complex with orthologous dual-RNA (upper panel) and orthologous Cas9 enzymes in complex with S. pyogenes dual-RNA (lower panel). Plasmid DNA containing protospacer speM and S. pyogenes PAM (NGG) was incubated with different dual-RNAs in complex with S. pyogenes Cas9. tracrRNA and crRNA-repeat sequences of the dual-RNAs are from the indicated bacterial species, with crRNA spacer targeting speM. In the lower panel, plasmid DNA containing speM protospacer and the specific PAM was incubated with Cas9 orthologs in complex with S. pyogenes dual-RNA. S. pyogenes Cas9 can cleave plasmid DNA only in the presence of dual-RNA from S. pyogenes, S. mutans and S. thermophilus* (yellow). Dual-RNA from S. pyogenes can mediate DNA cleavage only with Cas9 from S. pyogenes, S. mutans and S. thermophilus* (yellow). li: linear cleavage product; sc: supercoiled plasmid DNA. (B) Summary of Cas9 and dual-RNA orthologs exchangeability. Specific PAM sequences were used according to Figure 5. The color code reflects the type II CRISPR-Cas subgroups (Figure 1). +++: 100–75% cleavage activity; ++: 75–50% cleavage activity; +: 50–25% cleavage activity; −: 25–0% cleavage activity observed under the conditions tested. Cas9 and dual-RNA duplexes from the same type II group can be interchanged and still mediate plasmid cleavage providing that the PAM sequence is specific for Cas9. See also Supplementary Figure S10.
DISCUSSION
In this work, we identified and characterized dual-RNA and PAM requirements for eight Cas9 orthologous enzymes representative of the Cas9 phylogenetic grouping. To evaluate dual-RNA:Cas9 diversity, we performed bioinformatics analysis of type II CRISPR-Cas systems from available genomes and identified Cas9 orthologs in a plethora of bacterial species that belong to 12 phyla and were isolated from diverse environments (Supplementary Tables S2 and S4). Most of the strains that harbor type II CRISPR-Cas systems (and accordingly Cas9) are pathogens and commensals of vertebrates. A majority of these strains were isolated from gastrointestinal tracts and feces of mammals, fish and birds, but also from wounds, abscesses and spinocereberal fluid of septicaemia patients. We also identified strains isolated from invertebrates and environmental samples, including fresh and sea water, plant material, soil and food, the latter comprising species used in fermentation processes. Cas9 is also present in species from extreme environments such as deep sea sediments, hot springs and Antarctic ice, further demonstrating the wide spread of type II CRISPR-Cas systems in bacteria. A comparison of the taxonomy and habitats of representative strains with the phylogenetic clustering of Cas9 sequences shows little correlation (Supplementary Figure S11). In particular, we identified clusters of Cas9 genes from taxonomically distant bacteria that were isolated from similar habitats. Examples include diverse Firmicutes, Molicutes, Spirochaete and Fusobacteria, that were all isolated from gastrointestinal tracts of mammals, and members of different Proteobacteria, Firmicutes and Fusobacteria families mostly found in environmental samples (Supplementary Figure S11, clusters 1 and 3). A few exceptions involve grouping of Cas9 genes from closely related species isolated from diverse habitats such as Actinobacteria isolated from human and dog specimens but also from hot springs (Supplementary Figure S11, clusters 2, 4 and 5). This complex distribution of Cas9 across bacterial genomes indicates that evolution of dual-RNA:Cas9 systems in bacteria occurs both vertically and horizontally (55).
To investigate the basis for the horizontal dissemination of CRISPR-Cas modules among bacteria, we assessed the specificity of RNase III utilized by type II CRISPR-Cas for dual-RNA maturation. Complementation analysis shows that RNase III from a variety of species, including bacteria that lack type II CRISPR-Cas, can process S. pyogenes tracrRNA:pre-crRNA, suggesting that type II CRISPR-Cas systems can exploit any double-stranded RNA cleavage activity. This finding is consistent with the observation of S. pyogenes dual-RNA maturation in human cells which is apparently mediated by host RNases (2).
Dual-RNA and Cas9 sequences have widely evolved in bacteria (15). However, despite the high sequence variability among Cas9 sequences, certain motifs are conserved. In addition to the previously identified central HNH and N-terminal RuvC catalytic motifs (20,21,44,56), we show that the two middle RuvC motifs are required for interference activity in vivo and in vitro. In agreement with previous findings, deactivation of either one of the catalytic motifs (RuvC or HNH) results in nicking activity of Cas9 originating from the other motif (2,8,24,25). None of the mutations introduced in these conserved motifs affected the role of Cas9 in tracrRNA:pre-crRNA maturation by RNase III in vivo. The specificity of Cas9 towards different dual-RNAs might be explained by the high variability among Cas9 orthologs from different type II groups. To test this hypothesis, we selected representative bacterial species from the three major type II subtypes, namely S. pyogenes, S. mutans and S. thermophilus (II-A), F. novicida (II-B) and C. jejuni, N. meningitidis and P. multocida (II-C). Substitution of orthologs from the selected species for the endogenous S. pyogenes Cas9 shows that only Cas9 proteins from the S. pyogenes subcluster are capable of assisting tracrRNA:pre-crRNA processing by RNase III. This result indicates that the less-conserved inter-motif regions, which are the basis for the Cas9 subgrouping, could be responsible for Cas9 specificity for certain dual-RNAs.
To investigate the interchangeability between type II subgroups at the level of DNA interference, we first determined the PAMs specific for each of the eight selected Cas9 orthologs (28). By aligning potential crRNA-targeted sequences, we identified conserved motifs adjacent to the protospacers in all selected species. We then showed that these motifs are essential for DNA interference activity of the cognate dual-RNA:Cas9 complex in vitro. The interchangeability between dual-RNA and Cas9 from different subclusters was tested using plasmid cleavage assays. Only closely related Cas9 proteins can exchange their cognate dual-RNAs and still exert cleavage activity when using the Cas9 specific PAM. The specificity of Cas9 towards dual-RNAs is highly sensitive to the Cas9 sequence relatedness. This sensitivity is observed with Cas9 from C. jejuni that displays full cleavage activity with its cognate dual-RNA but reduced activity with dual-RNAs from N. meningitidis or P. multocida which belong to different subclusters of type II-C. We hypothesize that Cas9 possesses specificity for the secondary structure of dual-RNAs, given that bioinformatics predictions suggest similar structures of repeat:antirepeat duplexes in closely related CRISPR-Cas systems (Supplementary Figure S12).
This work provides the first experimental evidence in support of tracrRNA:crRNA duplex and Cas9 protein coevolution. Previously, we investigated the diversity of type II CRISPR-Cas system with respect to the loci architecture, tracrRNA sequence and position, and showed their correlation with the phylogenetic grouping of Cas9 (15). Here, the biological relevance of similarities within the Cas9 tree subclusters is demonstrated by limited exchangeability of the RNA and protein components. Thus, Cas9 and dual-RNA coevolved to maintain the functionality of the type II system. This finding also indicates that in bacteria harboring two distinct type II CRISPR-Cas systems (e.g S. thermophilus (18,57) or Wolinella succinogenes (15)), each system functions independently, with its own repeat-spacer array.
While this article was in preparation, a study by Hou et al. (34) showed the successful application of Cas9 from N. menigitidis for gene targeting in mammalian cells using the same PAM sequence as described here. This finding demonstrates that the various dual-RNA and Cas9 orthologs with their associated PAM sequences presented in this work have the potential to substantially enhance this novel genome editing tool by offering increased versatility and possibly specificity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [58–59].
FUNDING
Swedish Research Council [K2010-57X-21436-01-3, K2013-57X-21436-04-3, 621-2011-5752-LiMS to E.C.]; the Kempe Foundation to E.C.; Umeå University [Dnr: 223- 2728-10, Dnr: 223-2836-10, Dnr: 223-2989-10 to E.C.]; the Laboratory for Molecular Infection Medicine Sweden to E.C. and the Helmholtz Association to E.C., K.S.M. and E.V.K. are supported by intramural funds of the US Department of Health and Human Services (to the National Library of Medicine). K.C. was a fellow of the Austrian Doctoral Program in RNA Biology. Funding for open access charge: Helmholtz Centre for Infection Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Martin Jinek for his gift of plasmid pMJ833 and Nikola Zlatkov Kolev for technical help.
REFERENCES
- 1.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 2.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O'Connor-Giles KM. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194:1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P, Yang L, Esvelt KM, Aach J, Guell M, Dicarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, Zhang X, Zhang P, Huang X. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23:720–723. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLIFE. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013;31:688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:691–693. doi: 10.1038/nbt.2655. [DOI] [PubMed] [Google Scholar]
- 14.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chylinski K, Le Rhun A, Charpentier E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013;10:726–737. doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karvelis T, Gasiunas G, Miksys A, Barrangou R, Horvath P, Siksnys V. crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biol. 2013;10:841–851. doi: 10.4161/rna.24203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 19.Magadan AH, Dupuis ME, Villion M, Moineau S. Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3-Cas system. PLoS One. 2012;7:e40913. doi: 10.1371/journal.pone.0040913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl Acad. Sci. USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. Cas9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, et al. Double nicking by RNA-Guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 29.Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charpentier E, Doudna JA. Biotechnology: rewriting a genome. Nature. 2013;495:50–51. doi: 10.1038/495050a. [DOI] [PubMed] [Google Scholar]
- 32.Horvath P, Barrangou R. RNA-guided genome editing a la carte. Cell Res. 2013;23:733–734. doi: 10.1038/cr.2013.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Oost J. Molecular biology. New tool for genome surgery. Science. 2013;339:768–770. doi: 10.1126/science.1234726. [DOI] [PubMed] [Google Scholar]
- 34.Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl Acad. Sci. USA. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Caparon MG, Scott JR. Genetic manipulation of pathogenic streptococci. Methods Enzymol. 1991;204:556–586. doi: 10.1016/0076-6879(91)04028-m. [DOI] [PubMed] [Google Scholar]
- 37.Kirsch RD, Joly E. An improved PCR-mutagenesis strategy for two-site mutagenesis or sequence swapping between related genes. Nucleic Acids Res. 1998;26:1848–1850. doi: 10.1093/nar/26.7.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siller M, Janapatla RP, Pirzada ZA, Hassler C, Zinkl D, Charpentier E. Functional analysis of the group A streptococcal luxS/AI-2 system in metabolism, adaptation to stress and interaction with host cells. BMC Microbiol. 2008;8:188. doi: 10.1186/1471-2180-8-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mangold M, Siller M, Roppenser B, Vlaminckx BJ, Penfound TA, Klein R, Novak R, Novick RP, Charpentier E. Synthesis of group A streptococcal virulence factors is controlled by a regulatory RNA molecule. Mol. Microbiol. 2004;53:1515–1527. doi: 10.1111/j.1365-2958.2004.04222.x. [DOI] [PubMed] [Google Scholar]
- 40.Herbert S, Barry P, Novick RP. Subinhibitory clindamycin differentially inhibits transcription of exoprotein genes in Staphylococcus aureus. Infect. Immun. 2001;69:2996–3003. doi: 10.1128/IAI.69.5.2996-3003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 42.Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McClelland M, Hanish J, Nelson M, Patel Y. KGB: a single buffer for all restriction endonucleases. Nucleic Acids Res. 1988;16:364. doi: 10.1093/nar/16.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wheeler D, Bhagwat M. BLAST QuickStart: example-driven web-based BLAST tutorial. Methods Mol. Biol. 2007;395:149–176. [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bernhart SH, Tafer H, Muckstein U, Flamm C, Stadler PF, Hofacker IL. Partition function and base pairing probabilities of RNA heterodimers. Algorithms Mol. Biol. 2006;1:3. doi: 10.1186/1748-7188-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 52.Darty K, Denise A, Ponty Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Heidrich N, Ampattu BJ, Gunderson CW, Seifert HS, Schoen C, Vogel J, Sontheimer EJ. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol. Cell. 2013;50:488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takeuchi N, Wolf YI, Makarova KS, Koonin EV. Nature and intensity of selection pressure on CRISPR-associated genes. J. Bacteriol. 2012;194:1216–1225. doi: 10.1128/JB.06521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol. Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 58.Sun W, Li G, Nicholson AW. Mutational analysis of the nuclease domain of Escherichia coli ribonuclease III. Identification of conserved acidic residues that are important for catalytic function in vitro. Biochemistry. 2004;43:13054–13062. doi: 10.1021/bi049258i. [DOI] [PubMed] [Google Scholar]
- 59.Sun W, Jun E, Nicholson AW. Intrinsic double-stranded-RNA processing activity of Escherichia coli ribonuclease III lacking the dsRNA-binding domain. Biochemistry. 2001;40:14976–14984. doi: 10.1021/bi011570u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.