Abstract
A number of genome-wide analyses have revealed that estrogen receptor α binding to and regulation of its target genes correlate with binding of FOXA1, a pioneer factor, to nearby DNA sites in MCF-7 breast cancer cells. The enhancer element-specific histone H3K4me1/2 mark is enriched at the specific FOXA1/ERα recruitment sites in chromatin, but the mechanism by which these enhancer marks are established in chromatin before hormone treatment is unclear. Here, we show that mixed-lineage leukemia 1 (MLL1) protein is a key determinant that maintains permissive chromatin structure of the TFF1 enhancer region. MLL1 occupies the TFF1 enhancer region and methylates H3K4 before hormone stimulation. In vitro, MLL1 binds directly to the CpG-rich region of the TFF1 enhancer, and its binding is dependent on hypomethylation of DNA. Furthermore, the depletion of MLL1 in MCF-7 cells results in a dramatic decrease of chromatin accessibility and recruitment of FOXA1 and ERα to the enhancer element. Our study defines the mechanism by which MLL1 nucleates histone H3K4 methylation marks in CpG-enriched regions to maintain permissive chromatin architecture and allow FOXA1 and estrogen receptor α binding to transcriptional regulatory sites in breast cancer cells.
INTRODUCTION
Enhancers are distal cis-acting elements that regulate gene expression in a highly controlled manner. It is generally thought that the enhancer region is looped out or organized to permit enhancer–promoter interaction. This flexibility of enhancer element position relative to the transcription start site, however, impedes the comprehensive assignment of enhancer elements to the target genes they control. Recent genome-wide studies from independent laboratories have revealed several characteristic chromatin ‘signatures’ that define the enhancer elements in genes. For enhancer elements that are actively engaged or poised for activation, these signatures consist of high levels of p300 occupancy and H3K4me1 and a low level of H3K4me3 (1–4). A recent study of embryonic stem cells suggested three classes of enhancers: active (H3K4me1+, H3K27ac+), intermediate (H3K4me1+, H3K27−) and poised (H3K4me1+, H3K27me3+) (4,5). However, the mechanisms by which these histone marks are maintained and cooperate with other epigenetic factors, such as DNA methylation and pioneer factors, remain unclear.
Mixed lineage leukemia (MLL) proteins are SET1 family members that play critical roles in HOX gene activation (6–8). In mammals, six SET1 family members have been identified: SET1A, SET1B and four MLL family H3K4 methyltransferases, MLL1, MLL2, MLL3 and MLL4. Ash2, WDR5 and RBBP5 have been reported to form the core components of MLL1, MLL2, MLL3 and SET1A (7,9–11). Recent studies have also demonstrated that MLLs act as coactivators for nuclear receptor-mediated activation (12–15). Recently, we found that MLL1 plays a critical role in estrogen-dependent gene expression, possibly through the establishment of a histone methylation mark at the enhancer region of target genes (16), suggesting a potential role of MLL1 or other H3K4-specific histone methyltransferases in maintaining the local chromatin structure in the enhancer region.
The pioneer factor FOXA1 is a transcription factor that binds to thousands of sites across the human genome in breast and prostate cancer cells (17–19). FOXA1 is believed to scan chromatin for enhancers with forkhead motifs and can actively initiate chromatin decompaction (20–22). FOXA1 functions as a permissive or pioneer factor for estrogen receptor (ER) α-chromatin interactions and transcriptional activity in diverse target tissues (18). In particular, FOXA1 binds to genomic regions showing locally high levels of H3K4me1 and DNA hypomethylation, and its cell-type-specific recruitment to chromatin is inversely correlated with the differential DNA methylation levels of its corresponding binding sites (17,23). Selective DNA hypomethylation is also associated with the formation of DNase I hypersensitive chromatin regions (24–26) at potential glucocorticoid receptor (GR) binding sites, which are enriched in CpG dinucleotides compared with the surrounding genomic regions (27). Although these studies imply that DNA hypomethylation, H3K4me and FOXA1 binding are present in enhancer elements found within a pre-existing open chromatin structure, a comprehensive knowledge of the functional relationships among these enhancer marks is still lacking.
Considering that MLL1 binds to specific clusters of CpG residues within the HOXA1 locus and regulates the expression of multiple transcripts (28), we hypothesized that MLL1 plays a role in maintaining the pre-existing open chromatin structure of ERα-target genes by establishing methylation marks. We determined that MLL1 binds directly to the CpG-rich region of the TFF1 enhancer, and that its binding is dependent on DNA hypomethylation. Using formaldehyde-assisted isolation of regulatory elements (FAIRE)-qPCR to assess chromatin conformation and nucleosome occupancy methylome–sequencing (NOMe-seq) to monitor nucleosome positioning, we investigated how the depletion of MLL1 affects the establishment of open (active) chromatin and the binding of the pioneer factor, FOXA1. These findings provide the first evidence of the molecular mechanisms of MLL1, which nucleates histone H3K4 methylation marks at CpG islands to maintain chromatin accessibility for binding of ERα and its pioneer factor FOXA1 in breast cancer cells.
MATERIALS AND METHODS
Plasmids and cell culture
The pSG5–2 × FLAG expression vectors encoding the following proteins were constructed by inserting the appropriate cDNA coding region into EcoRI and XhoI sites: pSG5–2 × FLAG-MLL1(CX) (a.a. 1067–1432) and pSG5–2 × FLAG-MLL1 (CXPB) (a.a. 1067–1989). The pTriEX-His-MLL1(CX) expression vector encoding His-tagged MLL1 (a.a. 1067–1432) was constructed by inserting the appropriate cDNA coding region into NcoI and XhoI sites. Point mutations in MLL1 were introduced by QuikChange site-directed mutagenesis kit (Stratagene). pF-MLL1 was kindly provided by Jay Hess (University of Michigan). MCF-7, MDA-MB231 and 293T cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum.
RNA interference and qRT-PCR
Small interfering RNA experiments were performed according to previously published methods (29). Transfection of the MCF-7 cells was performed with Oligofectamine (Invitrogen) according to the manufacturer’s protocol. The sequences for siRNAs are available in Supplementary data. Total RNA was isolated from MCF-7 cells with Trizol (Invitrogen) after hormone treatment as indicated and subjected to reverse transcription (RT) by iScript cDNA synthesis kit (Bio-Rad) in a total volume of 50 µl. In all, 2 µl of RT product was used for qPCR analysis with the primers shown in Supplementary data. Relative expression levels were normalized to GAPDH mRNA levels. Results shown are mean and range of variation of duplicate PCR reactions performed on the same cDNA sample; the results are from a single experiment that is representative of at least two independent experiments.
Cell proliferation assay
MCF-7 cells were plated into six-well plates (2 × 105 cells/well). The next day, the cells were transfected with siRNA and then cultured for 3 days in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Cells were trypsinized on indicated days, and total live cells were counted after Trypan blue staining.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed according to previously described protocols (16,29). Briefly, MCF-7 cells were transfected with siRNA and then cultured for 3 days in phenol red-free Dulbecco’s modified Eagle’s medium supplemented with 5% charcoal-dextran-stripped fetal bovine serum. At ∼90% confluency, cells were treated with 100 nM estradiol (E2) or ethanol for the indicated time. After cross-linking with formaldehyde, the cell extracts were prepared from control and E2-treated MCF-7 cells. Immunoprecipitation of sonicated chromatin solutions was conducted by overnight incubation at 4°C with anti-ERα (Santa Cruz Biotechnology) anti-MLL1 (Bethyl Laboratories) or anti-FOXA1 (Abcam) antibody. Cross-linking was reversed by heating, and immunoprecipitated DNA was purified by phenol-chloroform extraction and ethanol precipitation. The purified DNA was dissolved in 100 µl of TE buffer [10 mM Tris–HCl (pH 8.0), 1 mM EDTA] and analyzed by quantitative PCR using Roche LightCycler 480 II system with SYBRGreen dye. For the Transfection-ChIP experiment, MCF-7 cells were transiently transfected with 2 × FLAG-tagged MLL1 for 3 days before E2 treatment. Immunoprecipitation was conducted with anti-FLAG antibody (Sigma Aldrich). Results shown are mean and range of variation of duplicate PCR reactions performed on the same DNA sample. Results were expressed as percentage of input chromatin (before immunoprecipitation) and were derived from a single experiment that is representative of at least two independent experiments.
Bisulfite genomic sequencing
Genomic DNA was purified by phenol/chloroform extraction and ethanol precipitation. Bisulfite conversion was performed using the EZ DNA methylation kit (Zymo Research), and molecules were cloned using the Topo TA Kit (Invitrogen) according to the manufacturers’ instructions.
DNA pull down assay and immunoblotting
FLAG-tagged MLL proteins were expressed in 293T cells, and total cell lysates were used for the binding assay. The 6 × His-tagged proteins were expressed in Escherichia coli strain BL21(DE3), purified by incubation with Ni-NTA agarose beads and eluted with phosphate-buffered saline (PBS) containing 250 mM imidazole. Point mutations in MLL1 were introduced by QuikChange site-directed mutagenesis kit (Stratagene). The CpG-rich enhancer (approximately −10 kb from TSS) or control region (approximately −5 kb from TSS) of the TFF1 gene was amplified using 5′-biotinylated oligonucleotide primers. Biotinylated PCR products were immobilized on streptavidin-agarose beads and incubated with MLL1 proteins. Immunoblotting was performed as described previously (29), using the following antibodies: anti-His (Santa Cruz Biotechnology); anti-FLAG (Sigma Aldrich); anti-ERα, anti-tubulin (Santa Cruz Biotechnology); anti-MLL1 (Bethyl Laboratories); and anti-FOXA1 (Abcam).
FAIRE-qPCR
FAIRE-qPCR was performed as previously described (30). Briefly, MCF-7 cells were transfected with siRNA and then cultured for 3 days in phenol red-free Dulbecco’s modified Eagle’s medium supplemented with 5% charcoal-dextran-stripped fetal bovine serum. At ∼90% confluency, the cells were treated with 100 nM E2 or ethanol for 60 min. After cross-linking with formaldehyde, cell extracts were prepared from control and E2-treated MCF-7 cells. Cross-linked chromatin was sonicated and open chromatin (FAIRE DNA) was purified from the aqueous phase by phenol-chloroform extraction and ethanol precipitation. For input control (Input DNA), cross-linking was reversed by heating before extraction. The purified DNA was dissolved in 100 µl of TE buffer and analyzed by the Roche LightCycler 480 II system with SYBRGreen dye. Results shown are mean and range of variation of duplicate PCR reactions performed on the same DNA sample. Results were expressed as percentage of input chromatin (Input DNA) and were derived from a single experiment that is representative of at least two independent experiments.
Nucleosome occupancy methylome-sequencing (NOMe-seq)
NOMe-seq analysis of enhancers was performed as previously described (31), with minor modifications. Briefly, MCF-7 cells in 15-cm dishes were transfected with siRNA and then cultured for 3 days in phenol red-free Dulbecco’s modified Eagle’s medium supplemented with 5% charcoal-dextran-stripped fetal bovine serum. Chromatin was isolated from 200 000 cells and treated for 60 min with E2 or ethanol. Isolated chromatin was incubated for 7.5 min with 200 U of GpC-specific enzyme, M.CviPI (New England BioLabs), followed by another 7.5 min of incubation with 100 U of M.CviPI. Enhancer regulatory regions were PCR amplified from DNA and cloned using the TOPO TA cloning kit (Invitrogen). Colonies were screened for positive clones, and at least 15 positive clones per amplicon were sequenced per experiment. Accessible regions (green bubbles) and inaccessible regions (white bubbles, pink bar) were plotted as bubble charts. Statistical analyses (chi-square) of the ethanol and E2 treatments were performed for one GpC site every 100 bp. The percentage of inaccessible molecules was calculated for every GpC site in the amplicon [percentage of inaccessibility = (number of inaccessible molecules/total number of molecules) × 100], and the results were averaged over a distance of 100 bp and plotted.
RESULTS
MLL1 mediates ERα-dependent transcriptional activity
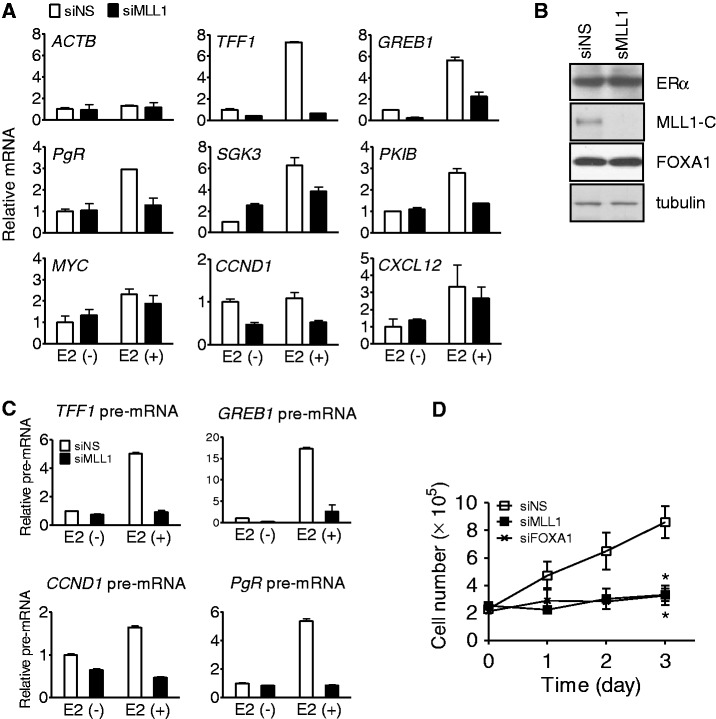
To understand the function of MLL1 in ERα-mediated transcription, we monitored the expression of several endogenous ERα target genes in MCF-7 breast cancer cells. Compared with cells transfected with non-specific siRNA (siNS), the estradiol (E2)-induced expression of TFF1, GREB1, PgR, SGK3 and PKIB was compromised (Figure 1A) when MLL1 was depleted from MCF-7 cells by siRNA duplexes (Figure 1B and Supplementary Figure S1B). MLL1 cleavage by taspase generates N-terminal 320 kDa and C-terminal 180 kDa fragments, which form a stable complex that localizes to a subnuclear compartment (32). The levels of E2-induced MYC and CXCL12 mRNAs were marginally reduced, and the basal expression of CCND1 was affected by the depletion of MLL1, and thus MLL1 is critically required for E2-induced expression of most of the ERα target genes examined in this study.
Figure 1.
Requirement of MLL1 for the expression of endogenous ERα target genes and cell proliferation. (A) Effect of reduced MLL1 on the expression of estrogen-responsive genes. MCF-7 cells were transfected with siRNA against MLL1 (siMLL1) or non-specific siRNA (siNS), and treated with E2 (10 nM) for 16 h before harvest. Total RNA was analyzed by qRT-PCR. Levels of all mRNAs were normalized to that of GAPDH mRNA; β-actin mRNA served as a control that was unaffected by E2 or by siMLL1. (B) Depletion of MLL1 protein by siRNA transfection. MCF-7 cells were transfected with siMLL1 or siNS as in (A) and grown in hormone-free media for 72 h. Levels of ERα, tubulin and C-terminal cleavage of MLL1 (MLL1-C) were assessed by immunoblotting. (C) The pre-mRNA levels in the MCF-7 cells transfected with siNS or siMLL1 were measured by qRT-PCR using primers spanning the 3′ end of exon 1 and the 5′ end of intron 1 of each gene. Cells were treated for 8 h with 10 nM E2. Results shown are the mean and range of variation of duplicate PCR reactions performed on the same cDNA sample; the results are from a single experiment that is representative of at least two independent experiments. (D) Cell proliferation was performed in MCF-7 cells transfected with siNS or siMLL1 and then cultured for 3 days in normal media supplemented with 10% fetal bovine serum. Cells transfected with siFOXA1 were used as a positive control. The data are the mean of independent replicates ± S.D.
We also measured the pre-mRNA levels of four ERα target genes, which is a more reliable reflection of the rapid changes in transcriptional rates, by using PCR primers spanning an exon/intron junction. E2 treatment increased the pre-mRNA levels of TFF1, GREB1, CCND1 and PgR after 8 h, and most of this increase was eliminated by depletion of MLL1 (Figure 1C), indicating that MLL1 is critically involved in the transcriptional regulation of endogenous ERα target genes. Furthermore, silencing of endogenous MLL1 by siRNA in MCF-7 cells growing in normal media containing fetal bovine serum, which is equivalent to growth in E2, resulted in significant growth inhibition (P < 0.01) (Figure 1D), confirming that hormone-dependent expression of ERα-target genes and the growth of MCF-7 cells require MLL1. Similar results were obtained on the levels of mRNA (Supplementary Figure S1) and pre-mRNA (data not shown) of ERαtarget genes by using a second siRNA that targeted a different part of the MLL1 coding region.
MLL1 is recruited to the hypomethylated CpG-rich enhancer region of the TFF1 gene
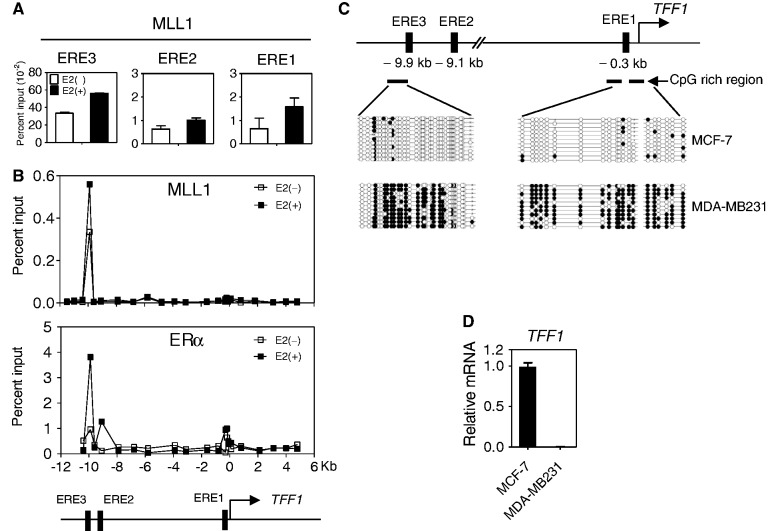
Although MLL1 is necessary for ERα-mediated transcription, the specific genomic regions occupied by MLL1 are not known. TFF1 has three EREs, and ERE3 at −9.9 kb upstream of transcriptional start site (TSS) is the major ER binding site and considered to be the major enhancer element. Our chromatin immunoprecipitation (ChIP) assay showed that the enhancer region of the TFF1 gene (ERE3, −9.9 kb) was highly occupied by MLL1 and that the recruitment of MLL1 to this region was further enhanced after E2 treatment in MCF-7 cells (Figure 2A). We validated these findings by a ChIP scanning assay, which surveyed from −10 kb upstream through 5 kb downstream of the TSS of the TFF1 gene. The distal enhancer region of TFF1 was found to be highly occupied by MLL1. Importantly, the enhancer region of TFF1 gene was occupied by MLL1 even before E2 treatment (Figure 2B).
Figure 2.
Recruitment of MLL1 to the hypomethylated CpG-rich enhancer region of the TFF1 gene. (A) ChIP assays were performed with MCF-7 cells treated with E2 (100 nM) or ethanol for 1 h. The amount of the indicated regions surrounding the three TFF1 ER binding sites precipitated by MLL1 antibody was determined by qPCR. Results shown are the mean and range of variation of duplicate PCR reactions performed on the same DNA sample. Results were expressed as percent of input chromatin (before immunoprecipitation) and were derived from a single experiment, which is representative of at least two independent experiments. (B) ChIP scanning of the TFF1 locus. DNA precipitated with the indicated antibody was analyzed by qPCR with primer sets spaced at 1-kb intervals and spanning the region from −10 to +5 kb relative to the TSS (designated by a horizontal arrow in the diagram) of the TFF1 gene. Primer sequences for ChIP scanning are available on request. (C) The TFF1 gene displays cell type-specific methylation patterns. The DNA methylation of the TFF1 promoter and enhancer regions in MCF-7 and MDA-MB231 cells was analyzed by bisulfite sequencing. Filled and open circles represent methylated and unmethylated CpG sites, respectively. Schematic diagrams of the TFF1 promoter and enhancer regions are shown. CpG-rich regions were determined by CpG Island Searcher. (D) mRNA levels of TFF1 was determined in MCF-7 and MDA-MB231 cells grown in normal media.
Although MLL1 is capable of localizing to the enhancer region in the absence of E2 (Figure 2B), and has a CXXC motif that is known to bind to the unmethylated CpG region of the HOXA9 gene (28,33), we investigated whether the enhancer region of TFF1 contains hypomethylated CpG sites. The enhancer region to which MLL1 binding occurred has a high CpG content and is not methylated in MCF-7 cells that actively express the TFF1 gene (Figure 2C and D). In contrast, the enhancer region of TFF1 in MDA-MB231 cells, which are ERα-negative and do not express the TFF1 gene, was highly methylated.
MLL1 binds directly to the unmethylated CpG region of TFF1 gene
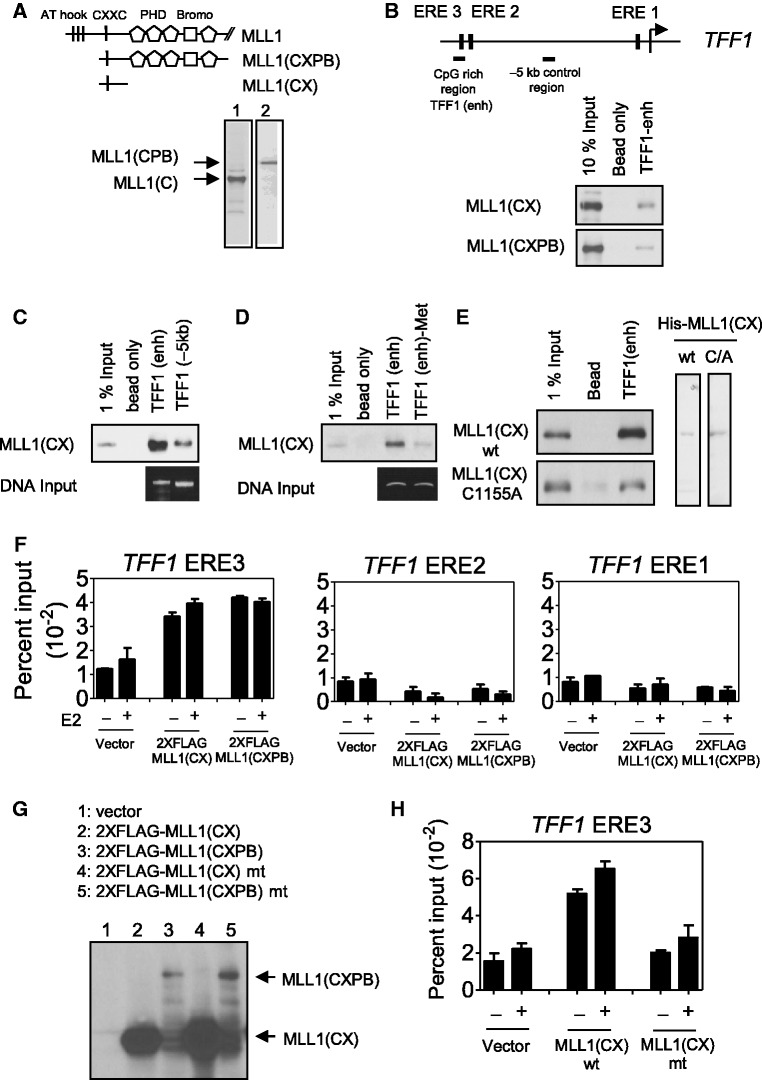
As the enhancer region of the TFF1 gene occupied by MLL1 is hypomethylated, we also assessed the possible effects of the CXXC motif on MLL1 binding to the unmethylated TFF1 enhancer region. We first identified which protein domain of MLL1 binds specifically to the unmethylated or methylated CpG region of the TFF1 gene. The MLL1-CXPB fragment (amino acids 1067–1989; containing a CXXC motif, four PHD finger domains and a bromodomain) bound to the CpG-rich ERE3-region (Figure 3A and B). In addition, a 40-kDa N-terminal fragment of MLL1 that contained only the CXXC motif (MLL1-CX) was sufficient for binding to the enhancer. However, MLL1-CX bound much more weakly to the −5 kb control region where the CpG level was low (Figure 3C), suggesting that the interaction was CpG-specific. In vitro methylation of the CpG-rich region by the CpG-specific methyltransferase (M.SssI) inhibited most of the binding of MLL1-CX (Figure 3D). Mutation of the CXXC motif of MLL1-CX to AXXC (Cys-1155 changed to Ala) eliminated most of the binding to the enhancer of the TFF1 gene (Figure 3E), indicating that the CXXC motif is involved in the interaction between MLL1 and the unmethylated TFF1 enhancer. When FLAG-tagged MLL1-CX or MLL1-CXPB was overexpressed in MCF-7 cells by transient transfection, quantitative ChIP assays performed with anti-FLAG antibody demonstrated the occupancy by both MLL1-CX and MLL1-CXPB on ERE3 (but not ERE2 or ERE1) of the TFF1 gene, indicating that the 40 kDa MLL1-CX fragment is sufficient for recruitment to the enhancer region in vivo (Figure 3F) with the same genomic binding specificity as wild-type MLL1 (Figure 2B). Although both MLL1-CX and MLL1-CXPB showed similar occupancy to ERE3 (Figure 3F), the expression level of MLL1-CX was higher than that of MLL1-CXPB (Figure 3G), suggesting that MLL1-CXPB molecules occupy ERE3 more efficiently than MLL1-CX molecules, probably due to extra DNA interaction or structural stabilization through other domains. Furthermore, when FLAG-tagged wild-type or mutant MLL1-CX were overexpressed at similar levels in MCF-7 cells, only wild-type MLL1-CX, but not mutant MLL1-CX, was associated with the CpG-rich region of the TFF1 enhancer (Figure 3H). Thus, these results indicate that the interaction of CXXC motif with the unmethylated CpG-rich enhancer region of the TFF1 gene is required for MLL1 occupancy.
Figure 3.
Direct interaction of MLL1 with the unmethylated CpG region of TFF1 gene. (A) Expression of truncated MLL1 fragment containing CXXC motif only (MLL1-CX), or CXXC motif, three PHD domains and one Bromodomain (MLL1-CXPB) in 293T cells. (B) The DNA-pull down assay was performed using total cell lysate of 293T cells transfected with FLAG-tagged MLL1-CX or MLL1-CXPB and incubated with biotinylated genomic PCR product of the CpG-rich region of the TFF1 enhancer region bound to streptavidin-Sepharose beads. Bound proteins were analyzed by immunoblotting with anti-FLAG antibody. (C) FLAG-tagged MLL1-CX was incubated with biotinylated genomic PCR product of the CpG-rich TFF1 enhancer region or the −5 kb control region bound to streptavidin-Sepharose beads. (D) The biotinylated CpG-rich enhancer region of TFF1 gene was methylated in vitro using the CpG Methyltransferase (M.SssI), immobilized on streptavidin-agarose beads and then incubated with MLL1-CX proteins. (E) Recombinant wild-type (wt) MLL1-CX or MLL1-CX with mutation (Cys1155 to Ala) in CXXC motif were tested for binding to the biotinylated CpG-rich region of TFF1 enhancer. (F) FLAG-tagged MLL1-CX or MLL1-CXPB was transiently expressed in the MCF-7 cells, which were then grown in hormone-free media for 48 h and then treated with 100 nM E2 for 1 h before performing the ChIP assay with anti-FLAG antibody. Precipitated DNA was analyzed by qPCR with primers representing the ER binding sites of the TFF1 genes. (G) The expression of MLL1-CX or MLL1-CXPB in the MCF-7 cells was monitored by immunoblotting using antibodies against the FLAG epitope. (H) FLAG-tagged wild-type MLL1-CX or AXXC mutant was transiently expressed in the MCF-7 cells. After E2 treatment for 1 h, the ChIP assay was performed with anti-FLAG antibody. The precipitated DNA was analyzed by qPCR using primers representing the ERE3 site of the TFF1 gene.
MLL1 is required for binding of FOXA1 and ERα to the TFF1 enhancer
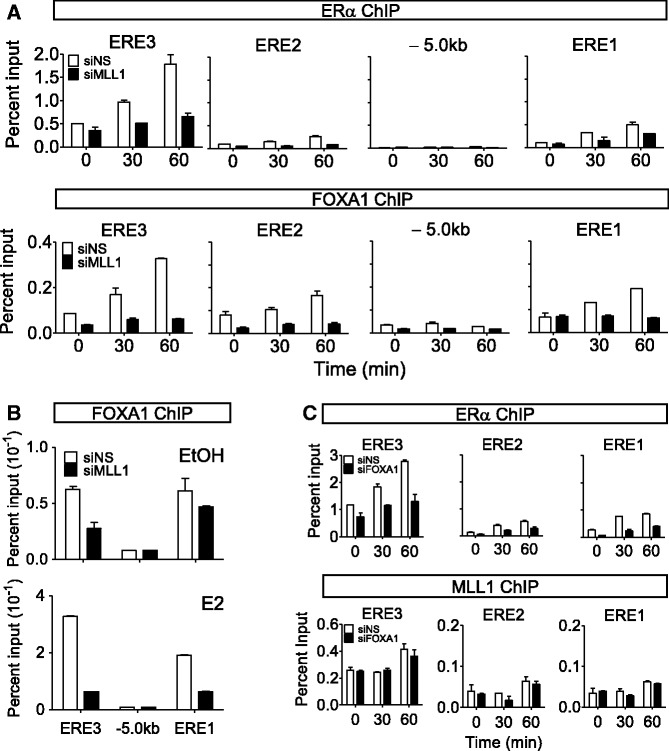
In MCF-7 cells, H3K4me1 and me2 are detected at estrogen responsive enhancers before E2 stimulation and ERα binding, reminiscent of FOXA1 recruitment (17). We have also shown that MLL1 occupies ERE3 of TFF1 before estrogen treatment (Figure 2A) and is responsible for the histone H3K4 methylation marks at the promoter and enhancer regions of the TFF1 gene before E2 treatment (16). We hypothesized that MLL1 may positively regulate the binding of FOXA1 by establishing the H3K4me signature at the enhancer region of TFF1. To investigate whether MLL1 is required for ERα and FOXA1 binding, we assessed their occupancies at EREs in MLL1-depleted MCF-7 cells. Depletion of MLL1 caused dramatic decreases in the occupancy of ERα and FOXA1 at ERE3 (Figure 4A), indicating that both ERα and FOXA1 recruitment are dependent on MLL1. Similar results were obtained by using a second siRNA that targeted a different part of the MLL1 coding region (Supplementary Figure S2).
Figure 4.
Effect of MLL1 depletion on the recruitment of ERαand FOXA1 to TFF1 EREs. (A-C) ChIP assays were performed as detailed in Figure 2 after transfection with siNS, siMLL1 or siFOXA1. The MCF-7 cells were treated with E2 (100 nM) or ethanol for the indicated times (A, C) or 60 min (B). The amount of the indicated region of the TFF1 gene precipitated by ERα, MLL1 or FOXA1 antibodies was determined by qPCR.
In the absence of E2, FOXA1 occupancy at ERE3 was 7.8-fold higher than that in the −5 kb control region, and increased 5.2-fold after estrogen treatment in MCF-7 cells. Surprisingly, specific silencing of MLL1 resulted not only in a 2-fold decrease of the pre-existing FOXA1 occupancy (Figure 4B, upper panel) but also in a dramatic inhibition of E2-induced FOXA1 recruitment (Figure 4B, lower panel), confirming the absolute requirement of MLL1 for the binding of FOXA1 to ERE3 of the TFF1 gene. We validated these findings in independent experiments (Supplementary Figure S3). In contrast, depletion of FOXA1 by specific siRNA did not affect MLL1 occupancy of TFF1 ERE3 (Figure 4C lower panel and Supplementary Figure S4B), even though E2-induced ERα binding to ERE3 was almost eliminated (Figure 4C, upper panel and Supplementary Figure S4A). These data support the notion that MLL1 is the key molecule that regulates chromatin occupancy by FOXA1 both before and after E2 stimulation, and also regulates the subsequent E2-induced ERα recruitment to the TFF1 enhancer.
Role of MLL1 in chromatin accessibility
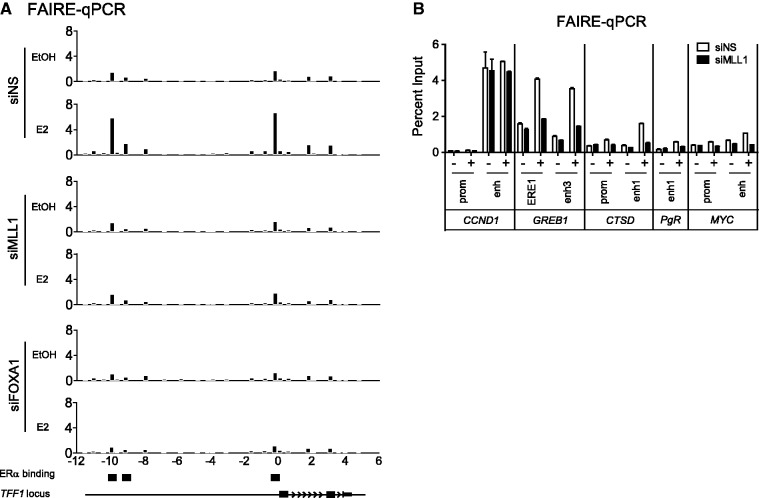
Given that the enhancer region of the TFF1 gene requires MLL1 for the binding of FOXA1 and ERα and for transcriptional activity (Figures 1 and 4), we investigated whether MLL1 influences the chromatin accessibility of the enhancer region of the TFF1 gene. We performed formaldehyde-assisted isolation of regulatory elements (FAIRE) coupled with qPCR. Hormone-deprived MCF-7 cells were transfected with siNS or siMLL1 and treated with ethanol or E2 for 1 h. siNS-transfected and ethanol-treated MCF-7 cells showed weak FAIRE signals at the proximal and distal ERα binding sites (Figure 5A). We observed a several-fold increase in the FAIRE signal after estrogen treatment at the ERα binding sites (ERE1 and 3). Surprisingly, specific silencing of MLL1 by siRNA dramatically decreased the E2-induced FAIRE signal of the enhancer and promoter regions of the TFF1 gene that were observed in siNS-transfected cells (Figure 5A), suggesting reduced chromatin accessibility to EREs caused by MLL1 depletion. The silencing of FOXA1 also reduced the estrogen-induced FAIRE signal. Similarly, depletion of MLL1 altered the chromatin accessibility of the promoter and enhancer regions of other ERα-mediated genes (GREB1, CTSD, PgR and MYC) (Figure 5B). In addition, ERα loading onto the enhancer region of these genes appears to be correlated with MLL1 occupancy. For example, ERα binding to TFF1, GREB1, CTSD and MYC is dependent on MLL1 (Supplementary Figure S5); in contrast, ERα binding to the c11orf49 gene that is not occupied by MLL1 was not affected by MLL1 depletion. These data suggest that MLL1 is required to maintain the optimal chromatin structure at the enhancers of many estrogen target genes.
Figure 5.
MLL1 is required for open chromatin structure. (A) Chromatin accessibility at the TFF1 locus was assessed by FAIRE-qPCR analysis using chromatin samples prepared from the MCF-7 cells transfected with siNS, siMLL1 or siFOXA1. Data are normalized against non–cross-linked genomic DNA for each primer pair. (B) The FAIRE-qPCR analysis was performed at the promoter and enhancer regions of CCND1, GREB1, CTSD, PgR and MYC in MLL1-depleted or control MCF-7 cells treated with E2 or ethanol for 60 min.
Enhancer chromatin structure is maintained by MLL1
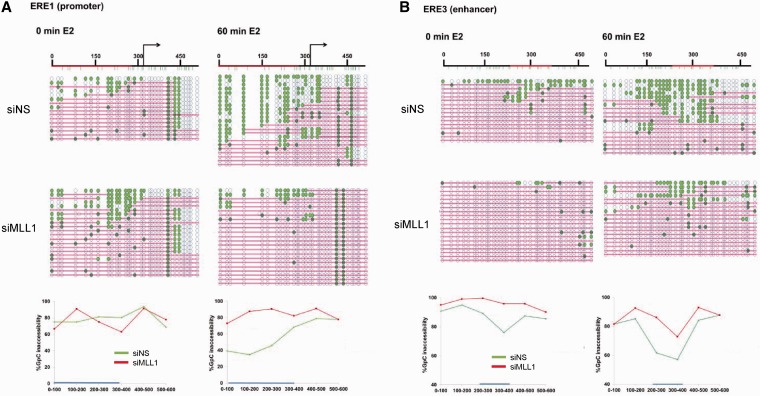
Although MLL1 is localized to the TFF1 enhancer even before hormone stimulation, we were unable to observe any significant changes in the FAIRE signal by MLL1 silencing in ethanol-treated cells. A possible explanation for this may be the limited resolution level of the assay. To further understand the local nucleosome dynamics, we performed NOMe-seq, a method that measures the methylation of GpC in isolated chromatin by M.CviPI DNA methyltransferase, which allows high-resolution determination of nucleosome occupancy (Figure 6), as this enzyme only methylates DNA that is unoccupied by nucleosomes and other proteins. Although the region upstream of the TSS of the TFF1 promoter is largely occupied by nucleosome (indicated by pink bars showing regions inaccessible to the GpC methyltransferase), it is somewhat accessible to enzyme (teal circles indicate methylation by the M.CviPI enzyme), indicating the presence of a minimal nucleosome-depleted region (NDR) in some of the cells that may allow the binding of basal transcription factors (Figure 6A). After 1 h of estrogen treatment, the TFF1 promoter region is open more broadly and is open in a majority of the cells, generating a large NDR at the regulatory site of the ERα target gene. Similarly, a significant increase in accessibility was observed at the enhancer region after 60 min of E2-treatment (Figure 6B), suggesting that the increase of the NDR at both the promoter and enhancer regions is important for gene expression. However, in MLL1-depleted cells, the majority of the promoter region remained closed even after estrogen stimulation, and the estrogen-induced increase of the NDR at the enhancer region was remarkably reduced (Figure 6B), confirming that MLL1 appears to be important for hormone-induced nucleosome repositioning and chromatin accessibility. Importantly, NOMe-seq data in estrogen-starved MCF-7 cells revealed a substantial decrease in chromatin openness at the enhancer (ERE3) region in MLL1-depleted cells compared with control (siNS) cells (Figure 6B), indicating a role for MLL1 in facilitating active chromatin architecture prior to E2 treatment. Together, these data indicate that MLL1 plays a critical role in maintaining nucleosome positioning and chromatin configurations, which allow for establishment of active chromatin states required for gene activation.
Figure 6.
Nucleosome occupancy of the TFF1 enhancer is regulated by MLL1. (A and B) Nucleosome occupancy at the TFF1 promoter and enhancer was analyzed by NOMe-seq in siNS transfected or MLL1-depleted MCF-7 cells. Blue circles represent GpC sites of the DNA (unfilled blue circles represent GpC sites that are inaccessible to GpC methyltransferase; teal-filled circles represent cytosines accessible to GpC methyltransferase). Pink bars represent regions of inaccessibility large enough to accommodate a nucleosome (∼150 bp).
DISCUSSION
Site-specific binding of MLL1 to TFF1 enhancer
Activation or enhancement of transcription in response to a signal apparently involves scores or perhaps even hundreds of coregulator proteins that are recruited by the DNA-bound transcriptional activator protein and subsequently modulate local chromatin conformation and assembly and/or activation of a transcription complex involving RNA polymerase II. Most recently, the Encyclopedia of DNA Elements (ENCODE) project has systematically mapped regions of transcription, chromatin structure and histone modification, providing new insights into the mechanisms of gene regulation (http://encodeproject.org). One of the most highly studied gene systems is the TFF1 gene and its response to estradiol; the timing of specific occupancy by many coregulators and histone modifications has been carefully documented (34,35). However, the mechanisms by which the highly choreographed sequence of events is coordinated are unknown. Here, we defined the role of MLL1, an H3K4-specific histone methyltransferase, in maintaining the local chromatin structure of an enhancer that is required for transcription initiation. We demonstrated how CpG hypomethylation, histone H3K4 methylation, FOXA1 binding and ERα recruitment are coordinated.
The CXXC motif is found in proteins that recognize the cytosine methylation status in genomic regions containing CpG dinucleotides; examples are DNMT1, methyl-CpG-binding protein (MBD), and human CpG-binding protein (hCGBP) (36–40). In MLL1, the CXXC motif binds to the unmethylated promoter of the HOX gene and regulates its expression (28,33,41). We have shown that the CXXC motif in MLL1 can also bind directly to the hypomethylated enhancer region of the TFF1 gene (Figure 3). Introducing a mutation into the CXXC motif revealed that the DNA binding function of the CXXC motif is indispensable for MLL1 recruitment to the TFF1 gene (Figure 3E and H). Previously, multiple MLL1 domains were shown to be necessary for the recruitment of MLL1 to the HOXA9 gene in vivo (6). A fragment containing the CXXC region alone or PHD fingers 1–4 alone failed to associate with HOXA9 in Mll−/− mouse embryonic fibroblast cells. In contrast, we observed that the CXXC domain alone (MLL1-CX) was sufficient to bind to the CpG region of the TFF1 gene in vitro and in vivo, although the inclusion of PHD fingers and bromodomain further increased MLL1 binding efficiency (Figure 3F and G). The binding of MLL1 to the enhancer appears to be guided by hypomethylated CpG. MLL1-CX does not bind efficiently to the −5 kb control region in which CpG content is low (Figure 3D), and in vitro methylation of the CpG-rich TFF1 enhancer fragment by the CpG Methyltransferase (M.SssI) reduced the binding of MLL1-CX (Figure 3D). Thus, the interaction of the CXXC motif in MLL1 with the unmethylated CpG-rich enhancer region of the TFF1 gene is required for occupancy by MLL1 (Figure 3); and the MLL1 occupancy of the ER binding sites associated with many ER target genes that require MLL1 for E2-induced expression (Figure 2 and Supplementary Figure S5) suggests that MLL1 occupancy is required for the transcriptional activity of MLL1 in ERα-mediated gene expression (Figure 1).
Requirement of MLL1 for ERα and FOXA1 binding
Previous studies have shown that FOXA1 is colocalized to the majority of ERα binding sites in MCF-7 cells (17,19). Moreover, introduction of FOXA1 into U2OS cells that stably express ERα alters the ERα binding pattern (18), suggesting that one of the primary roles of FOXA1 is to aid ERα binding to chromatin (42). In addition, significant enrichment of histone H3K4me1/2 marks has been observed at the FOXA1 recruitment site (17). Differential binding of FOXA1 to chromatin is dependent on the distribution of H3K4 dimethylation, although how H3K4me1/2 guides FOXA1 recruitment remains uncertain (17). Recently, it was also reported that FOXA1 binds to genomic regions showing local DNA hypomethylation, and histone H3K4 methylation at the enhancers may stabilize FOXA1 binding during P19 cell neural differentiation (23,43). Importantly, during cellular differentiation, activation of FOXA1-dependent enhancers is accompanied by a decrease in DNA methylation (23), suggesting a strong correlation between DNA hypomethylation and FOXA1 binding.
In this study, we showed that both FOXA1 and ERα binding required MLL1, which preferentially occupies the CpG-rich region of the TFF1 enhancer region (Figure 4). Knockdown of MLL1 resulted in dramatic decreases in ERα and FOXA1 occupancy at ERE3 (Figure 4A). Given that MLL1 occupies the enhancer regions of TFF1 and other E2-induced genes before estrogen treatment (Figure 2A and Supplementary Figure S5) and establishes the histone H3K4 methylation marks (16), our results indicate that MLL1 is a key molecule that regulates chromatin occupancy by FOXA1 by maintaining the histone methylation mark in the absence of E2. Thus, MLL1 facilitates binding of ERα after E2 stimulation by maintaining permissive chromatin architecture that allows FOXA1 occupancy.
Interestingly, FOXA1 occupancy at the enhancer of TFF1 was increased by ∼5.2-fold after E2 stimulation. The most likely explanation is that chromatin remodeling triggered by the SWI/SNF or other chromatin remodeling complexes facilitated the further occupancy of FOXA1 on its binding site. Similarly, the increase of FOXA1 occupancy by E2 has been observed at several ERα binding sites (ERBS-1, ERBS-3, ERBS-5) in the RET gene locus (44). Another possible explanation for this observation may be that FOXA1 became more accessible to the ChIP antibody after E2 stimulation through the formation of relaxed open chromatin structure caused by SWI/SNF.
Role of MLL1 in the chromatin structure of TFF1 enhancer
Hormone-deprived MCF-7 cells showed weak FAIRE signals at proximal and distal ERα binding sites (Figure 5A), but were somewhat accessible to the M.CviPI enzyme, suggesting a minimal NDR that may allow for the binding of basal transcription factors. The FAIRE signal increased significantly after estrogen treatment at ERα binding sites (ERE1–3). Thus, estrogen binding sites (ERE1–3) seem to be partially opened in the absence of E2 (Figures 5A and 6). Surprisingly, specific silencing of MLL1 by siRNA dramatically inhibited the E2-induced increases in the FAIRE-signal and M.CviPI DNA methyltransferase accessibility of the enhancer and promoter regions of the TFF1 gene compared with siNS-transfected cells (Figures 5A and 6).
The correlation between the presence of a hypomethylated CpG region and chromatin hypersensitivity has been previously reported (45–48). Most recently, it was proposed that DNA methylation status may also play a role in the formation of open chromatin regions and in the maintenance of enhancer elements (27,49,50). Cells create and maintain a pre-programmed accessible chromatin architecture at active and potentially active transcriptional regulatory sites that includes specific histone modifications and, at many sites, is also characterized by enriched-CpG hypomethylation. However, the mechanism by which the transcriptionally permissive chromatin architecture (including H3K4 methylation) at DNA hypomethylation sites is maintained remains unclear. We demonstrate here that both the estrogen-induced increase of NDR extent and frequency at enhancer regions and the basal chromatin accessibility were remarkably reduced by MLL1 depletion. These results suggest that MLL1 plays a critical role in maintaining nucleosome positioning as well as epigenetic marks and possibly other aspects of chromatin configurations before and after hormone stimulation, thus facilitating the establishment of active chromatin states by H3K4 methylation in MCF-7 breast cancer cells. It is not clear to us at this time how the H3K4 methylation by MLL1 coordinates FOXA1 binding to TFF1 enhancer, but much work remains to be done to understand the relationships among epigenetic marks, chromatin architecture, nucleosome positioning and occupancy by transcription factors at enhancer elements. However, the observation that the depletion of MLL1 reduced pre-existing FOXA1 occupancy serves as an intriguing finding for further investigation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [DK043093 to M.R.S., R37CA 082422 to P.A.J.]; National Cancer Institute provided the Cancer Center Support Grant [P30CA014089 to the University of Southern California]. Funding for open access charge: NIH [DK043093].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dan Gerke and Kelly Chang (University of Southern California) for expert technical assistance and Dr. Jay Hess (University of Michigan) for providing plasmid pF-MLL.
REFERENCES
- 1.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 3.Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2010;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milne TA, Kim J, Wang GG, Stadler SC, Basrur V, Whitcomb SJ, Wang Z, Ruthenburg AJ, Elenitoba-Johnson KS, Roeder RG, et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol. Cell. 2010;38:853–863. doi: 10.1016/j.molcel.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 8.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 9.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 10.Crawford BD, Hess JL. MLL core components give the green light to histone methylation. ACS Chem. Biol. 2006;1:495–498. doi: 10.1021/cb600367v. [DOI] [PubMed] [Google Scholar]
- 11.Steward MM, Lee JS, O'Donovan A, Wyatt M, Bernstein BE, Shilatifard A. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat. Struct. Mol. Biol. 2006;13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 12.Dreijerink KM, Mulder KW, Winkler GS, Hoppener JW, Lips CJ, Timmers HT. Menin links estrogen receptor activation to histone H3K4 trimethylation. Cancer Res. 2006;66:4929–4935. doi: 10.1158/0008-5472.CAN-05-4461. [DOI] [PubMed] [Google Scholar]
- 13.Lee S, Kim DH, Goo YH, Lee YC, Lee SK, Lee JW. Crucial roles for interactions between MLL3/4 and INI1 in nuclear receptor transactivation. Mol. Endocrinol. 2009;23:610–619. doi: 10.1210/me.2008-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mo R, Rao SM, Zhu YJ. Identification of the MLL2 complex as a coactivator for estrogen receptor alpha. J. Biol. Chem. 2006;281:15714–15720. doi: 10.1074/jbc.M513245200. [DOI] [PubMed] [Google Scholar]
- 15.Ansari KI, Hussain I, Shrestha B, Kasiri S, Mandal SS. HOXC6 Is transcriptionally regulated via coordination of MLL histone methylase and estrogen receptor in an estrogen environment. J. Mol. Biol. 2011;411:334–349. doi: 10.1016/j.jmb.2011.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong KW, Kim K, Situ AJ, Ulmer TS, An W, Stallcup MR. Recognition of enhancer element-specific histone methylation by TIP60 in transcriptional activation. Nat. Struct. Mol. Biol. 2011;18:1358–1365. doi: 10.1038/nsmb.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 20.Zaret K. Developmental competence of the gut endoderm: genetic potentiation by GATA and HNF3/fork head proteins. Dev. Biol. 1999;209:1–10. doi: 10.1006/dbio.1999.9228. [DOI] [PubMed] [Google Scholar]
- 21.Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 2006;20:2513–2526. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sekiya T, Muthurajan UM, Luger K, Tulin AV, Zaret KS. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 2009;23:804–809. doi: 10.1101/gad.1775509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Metivier R, et al. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res. 2011;21:555–565. doi: 10.1101/gr.111534.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomassin H, Flavin M, Espinas ML, Grange T. Glucocorticoid-induced DNA demethylation and gene memory during development. EMBO J. 2001;20:1974–1983. doi: 10.1093/emboj/20.8.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim ST, Fields PE, Flavell RA. Demethylation of a specific hypersensitive site in the Th2 locus control region. Proc. Natl Acad. Sci. USA. 2007;104:17052–17057. doi: 10.1073/pnas.0708293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santangelo S, Cousins DJ, Winkelmann N, Triantaphyllopoulos K, Staynov DZ. Chromatin structure and DNA methylation of the IL-4 gene in human T(H)2 cells. Chromosome Res. 2009;17:485–496. doi: 10.1007/s10577-009-9040-3. [DOI] [PubMed] [Google Scholar]
- 27.Wiench M, John S, Baek S, Johnson TA, Sung MH, Escobar T, Simmons CA, Pearce KH, Biddie SC, Sabo PJ, et al. DNA methylation status predicts cell type-specific enhancer activity. EMBO J. 2011;30:3028–3039. doi: 10.1038/emboj.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erfurth FE, Popovic R, Grembecka J, Cierpicki T, Theisler C, Xia ZB, Stuart T, Diaz MO, Bushweller JH, Zeleznik-Le NJ. MLL protects CpG clusters from methylation within the Hoxa9 gene, maintaining transcript expression. Proc. Natl Acad. Sci. USA. 2008;105:7517–7522. doi: 10.1073/pnas.0800090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeong KW, Lee YH, Stallcup MR. Recruitment of the SWI/SNF chromatin remodeling complex to steroid hormone-regulated promoters by nuclear receptor coactivator flightless-I. J. Biol. Chem. 2009;284:29298–29309. doi: 10.1074/jbc.M109.037010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon JM, Giresi PG, Davis IJ, Lieb JD. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat. Protoc. 2012;7:256–267. doi: 10.1038/nprot.2011.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miranda TB, Kelly TK, Bouazoune K, Jones PA. Methylation-sensitive single-molecule analysis of chromatin structure. Curr. Protoc. Mol. Biol. 2010 doi: 10.1002/0471142727.mb2117s89. Chapter 21, Unit 21 17 1–Unit 21 17 16. [DOI] [PubMed] [Google Scholar]
- 32.Hsieh JJ, Ernst P, Erdjument-Bromage H, Tempst P, Korsmeyer SJ. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol. Cell Biol. 2003;23:186–194. doi: 10.1128/MCB.23.1.186-194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cierpicki T, Risner LE, Grembecka J, Lukasik SM, Popovic R, Omonkowska M, Shultis DD, Zeleznik-Le NJ, Bushweller JH. Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia. Nat. Struct. Mol. Biol. 2010;17:62–68. doi: 10.1038/nsmb.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 35.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 36.Fujita N, Takebayashi S, Okumura K, Kudo S, Chiba T, Saya H, Nakao M. Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms. Mol. Cell Biol. 1999;19:6415–6426. doi: 10.1128/mcb.19.9.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujita N, Shimotake N, Ohki I, Chiba T, Saya H, Shirakawa M, Nakao M. Mechanism of transcriptional regulation by methyl-CpG binding protein MBD1. Mol. Cell Biol. 2000;20:5107–5118. doi: 10.1128/mcb.20.14.5107-5118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cross SH, Meehan RR, Nan X, Bird A. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nat. Genet. 1997;16:256–259. doi: 10.1038/ng0797-256. [DOI] [PubMed] [Google Scholar]
- 39.Voo KS, Carlone DL, Jacobsen BM, Flodin A, Skalnik DG. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Mol. Cell Biol. 2000;20:2108–2121. doi: 10.1128/mcb.20.6.2108-2121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 41.Ansari KI, Mishra BP, Mandal SS. Human CpG binding protein interacts with MLL1, MLL2 and hSet1 and regulates Hox gene expression. Biochim. Biophys. Acta. 2008;1779:66–73. doi: 10.1016/j.bbagrm.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 42.Cheung E, Ruan Y. Determination of transcription factor binding. Nat. Genet. 2011;43:11–12. doi: 10.1038/ng0111-11. [DOI] [PubMed] [Google Scholar]
- 43.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan SK, Lin ZH, Chang CW, Varang V, Chng KR, Pan YF, Yong EL, Sung WK, Cheung E. AP-2gamma regulates oestrogen receptor-mediated long-range chromatin interaction and gene transcription. EMBO J. 2011;30:2569–2581. doi: 10.1038/emboj.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xi H, Shulha HP, Lin JM, Vales TR, Fu Y, Bodine DM, McKay RD, Chenoweth JG, Tesar PJ, Furey TS, et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Auerbach RK, Euskirchen G, Rozowsky J, Lamarre-Vincent N, Moqtaderi Z, Lefrancois P, Struhl K, Gerstein M, Snyder M. Mapping accessible chromatin regions using Sono-Seq. Proc. Natl Acad. Sci. USA. 2009;106:14926–14931. doi: 10.1073/pnas.0905443106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 49.He HH, Meyer CA, Chen MW, Jordan VC, Brown M, Liu XS. Differential DNase I hypersensitivity reveals factor-dependent chromatin dynamics. Genome Res. 2012;22:1015–1025. doi: 10.1101/gr.133280.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.