Abstract
Binding of proteins to particular DNA sites across the genome is a primary determinant of specificity in genome maintenance and gene regulation. DNA-binding specificity is encoded at multiple levels, from the detailed biophysical interactions between proteins and DNA, to the assembly of multi-protein complexes. At each level, variation in the mechanisms used to achieve specificity has led to difficulties in constructing and applying simple models of DNA binding. We review the complexities in protein–DNA binding found at multiple levels and discuss how they confound the idea of simple recognition codes. We discuss the impact of new high-throughput technologies for the characterization of protein–DNA binding, and how these technologies are uncovering new complexities in protein–DNA recognition. Finally, we review the concept of multi-protein recognition codes in which new DNA-binding specificities are achieved by the assembly of multi-protein complexes.
INTRODUCTION
In the mid 1970s, motivated by early X-ray crystal structures of proteins and DNA, Seeman et al. (1) proposed a protein–DNA recognition code based on hydrogen bonding patterns between amino acids and bases. For example, in the DNA major groove, an arginine side-chain can make two hydrogen bonds with a guanine base, but not with any other base. Therefore, an Arg-Gua residue base pairing provides a mechanism—or code—for proteins to preferentially select for guanines. Although aesthetically pleasing, 30 years of subsequent analyses of protein–DNA complexes and interactions have demonstrated myriad complications with such a simple recognition code. We briefly summarize findings from structural and biochemical studies that explain why simple protein–DNA recognition codes do not exist.
Side-chain flexibility and water molecules
Protein side-chains and water molecules in the binding interface create a flexible network of interactions that can readily adopt new conformations in response to changes in DNA or protein sequence (2–5). Two examples illustrate the relevant complexities. Comparing protein–DNA crystal structures for wild-type and mutant zinc finger protein Zif268/Egr1, it was demonstrated that a single amino acid mutation (Zif268 D20A) could lead to altered protein side-chain conformations and DNA-binding preferences (6). Further, the side-chain re-arrangement occurred without a change in the protein docking geometry, but resulted in a new, positioned water molecule in the binding interface for one of the DNA sequences. Structural comparison of two Cre recombinase variants in complex with different DNA sequences revealed that both DNA and protein differences affect the contacts made in the binding interface (7). In complex with the same LoxM7 DNA sequence, two Cre variants, which differ at only three residue positions, had distinctly different conformations for the common side-chains mediating base contacts. Further, comparison of one of the variants bound to a different LoxP DNA site revealed that recognition of the two DNA sequences, LoxM7 and LoxP, led to altered side-chain conformations and different side-chain and water-mediated contacts. These studies highlight the complex interplay between side-chains, water molecules and DNA bases in the binding interface.
DNA structure and indirect readout
‘Direct readout’ (also known as ‘base readout’) refers to the situation where proteins discriminate different bases in a DNA sequence via direct (or water-mediated) interactions with the DNA bases. In contrast, ‘indirect readout’ (also known as ‘shape readout’) describes a different mechanism of recognition in which the sequence-dependent deformability or structural differences between DNA molecules contribute to their discrimination (8). A recent review categorized different types of shape readout and distinguished between ‘local’ shape readout, in which deviations from B-form DNA are localized along the sequence, and ‘global’ shape readout, in which much of the DNA molecule is deformed or bent (2). In local shape readout, sequence-dependent kinks in the DNA molecule or localized deviations in DNA groove width can lead to preferential binding to the cognate protein. Examples of widely used types of local shape readout are the preferential binding of arginine and histidine residues into narrow DNA minor grooves DNA (9,10) or binding to DNA kinks observed at YpR steps, such as seen for TATA-box binding protein (9,11,12). In global shape readout, larger deformations are involved. For example, a mechanism proposed for DNA binding of the human papillomavirus E2 protein is that DNA sequences in the unbound form adopt global conformations similar to that found in the bound protein–DNA complex (13). A particular DNA sequence that was already ‘pre-bent’ would require less energy to deform and hence be preferentially bound by a protein (2,13). What complicates prediction of the effects of indirect readout on the binding specificity is that DNA deformation is a structural property that involves multiple, distributed interactions (residue–base and base–base) and is sensitive to the conformation adopted in the bound complex. Even local shape readout (e.g., the local narrowing of the DNA minor grove) involves integrating the individual structural propensities over multiple bases (2). Therefore, a recognition code for many proteins will need to include or account for the detailed structure of the bound protein–DNA complex.
Docking geometry and spatial relationships
Energetically favorable residue–base interactions, particularly hydrogen bond interactions, depend on the relative spatial orientation of the protein residue and the DNA base (1,14–16). Subsequently, the docking geometry of a protein–DNA complex, defined by the orientation of the protein backbone relative to the DNA, is critical to establishing favorable amino acid–base interactions (14,15,17). Comparisons of the docking geometry for different protein–DNA complexes have revealed complications to a simple recognition code. First, the docking geometry of different protein folds can differ dramatically; therefore, favorable interactions made by different residue types will depend strongly on the protein fold (i.e. not all arginines will be able to make optimal contacts with a guanine). Second, even structurally homologous proteins can dock on the DNA in different orientations (14,15,17,18). Therefore, the distributed protein–DNA interactions that contribute to the overall docking geometry, both base-specific and non-specific (i.e., with the DNA backbone), can all potentially affect the protein–DNA interactions. In other words, effects are non-local and residues distributed throughout the protein will affect DNA-binding specificity.
Studies have also shown that the docking geometry of the same protein can vary when bound to different DNA sequences (14,18,19). In an illustrative case, crystal structures of the nuclear factor kappa B (NF-κB)-family RelA homodimer in complex with different DNA sequences exhibited markedly different orientations for the dimer subunits. In the structure of RelA homodimer bound to the pseudo-symmetric sequence 5′-GGAA(A)TTTC-3′, one subunit makes a canonical set of hydrogen bond interactions with the 5′-GGAA half-site sequence. In contrast, the other subunit undergoes a large rotation and translation and makes no base-specific contacts with the apposing 5′-GAAA half-site, yet allows the protein to bind well to DNA (20). This contrasts starkly with the structure of RelA bound to the more symmetric 5′-GGAA(T)TTCC-3′ site where the same docking and DNA-base contacts are symmetric for the two subunits (21). Flexibility of the protein docking geometry in response to different DNA sequences presents a major difficulty in predicting the interactions and subsequent DNA-binding specificity of a protein (17,22).
The complexities described here that result from flexible protein–DNA interactions, indirect DNA readout and protein docking geometry make it difficult to establish simple recognition rules that can provide a comprehensive description of protein–DNA binding. Structural and biochemical studies have now identified the major specificity-determining residues for many different protein structural families and subfamilies, providing a detailed picture of the basic mechanisms of DNA recognition used by different proteins (2,3). However, despite these structural and mechanistic descriptions, simple recognition codes have not emerged that can accurately characterize the binding affinities of a protein to the vast number of potential DNA-binding site sequences and has spurred the development of more complex models (23–26).
In the last 10 years, newly developed high-throughput (HT) technologies have revolutionized our ability to characterize protein–DNA binding specificity. Such technologies include: bacterial one-hybrid (B1H) (27,28), protein-binding microarrays (PBMs) (29–33), total internal reflectance fluorescence-PBM (34), mechanically induced trapping of molecular interactions (MITOMI) (35), Bind-n-seq (36), EMSA-seq (37), HT-SELEX/SELEX-seq (38–40), microarray evaluation of genomic aptamers by shift (MEGAshift) (41), cognate site identifier (CSI) (42), and HT sequencing fluorescent ligand interaction profiling (HiTS-FLIP) (43). The rich datasets provided by these new HT technologies are facilitating the development of improved models of protein–DNA recognition (44,45), while at the same time revealing new complications for models of binding. Here we will briefly review the most widely used models and outline features of protein–DNA binding that complicate their development and application.
COMPLEXITIES IN PROTEIN–DNA RECOGNITION
Consensus sequences in which base preferences are represented using letters (e.g., A = Ade, Y = Cyt or Thy) are widely used to represent protein–DNA binding specificity. These models are appropriate for high-specificity DNA-interacting proteins such as restriction enzymes. For example, enzyme HincII will cut DNA sites that match the consensus GTYRAC (R = Ade or Gua), but the affinity for all other DNA sites is orders of lower magnitude (46). However, transcription factor (TF)–DNA binding is much more degenerate and is often characterized by a gradual transition from high to low affinity sites (24,29,35,43,46–48). To account for the sequence degeneracy in TF binding, the concept of position weight matrices (PWMs) was proposed and remains the most widely used representation of TF-binding specificity (46,49). A PWM is a matrix of scores (or weights) for each DNA base pair along a binding site. PWM models can be learned from a variety of data types, from small collections of known binding sites to large datasets generated using HT technologies. PWMs also have the benefit of being easy to visualize as DNA logos, providing an intuitive feel for the TF-binding specificity (50).
One drawback of the PWM formalism is that it makes the implicit assumption that individual base pairs within a binding site contribute independently to the protein–DNA binding affinity. It has been shown that this assumption does not always hold (51–53), and more complex models of protein–DNA specificity have been developed to account for position dependencies within protein–DNA binding sites (52,54–56). Some studies have focused on extending traditional PWM models into ‘higher-order’ models by including contributions from dinucleotides and trinucleotides (45,57–62). A drawback of including non-independent contributions is that the number of model parameters increases significantly, which makes the models harder to learn and prone to overfitting. Thus, selecting the relevant dinucleotides and trinucleotides is critical for building higher-order models that take into account dependencies between positions in TF-binding sites (58,60). We note that other approaches have also been proposed to model DNA-binding specificity, however, a full survey of these various approaches is outside the scope of this review. Beyond inherent sequence degeneracy and positional dependencies, several other aspects of protein–DNA binding complicate the development and application of binding models and are reviewed below.
Low-affinity binding sites
Regardless of how degeneracy is represented in protein–DNA binding models, it is critically important to be able to capture the full breadth of binding sites used in vivo. Complicating this goal is the fact that TFs can specifically utilize low-affinity DNA-binding sites to regulate genes. TF binding to low-affinity DNA sites can provide a mechanism for interpreting both spatial (63–66) and temporal (67,68) TF gradients that often arise during development to control where and when genes are expressed. Analysis of genome-wide binding data has also provided evidence that low-affinity sites are under wide-spread evolutionary selection (69,70) and that their inclusion can greatly improve quantitative models of TF binding and gene regulation used for predicting segmentation patterns during early embryonic development in Drosophila (71). Utilization of sites selected to be lower affinity than an optimal sequence opens the door for functionally relevant sites to deviate strongly from the consensus sequence and may not be well represented by a particular binding model. For example, a comprehensive analysis of DNA binding by NF-κB dimers identified numerous lower affinity, non-traditional sites that differ significantly from the consensus sites and are not captured by the widely used PWMs (37,72). Disagreement with a PWM model may be due to: (i) a protein having multiple binding modes, which will require multiple PWMs (discussed more below) or (ii) poor or biased parameterization of the PWM model. PWMs can capture low-affinity binding sites but must be explicitly parameterized using low-affinity binding data (59). In either case, by focusing only on the highest affinity sites, protein–DNA binding models are likely to miss functionally important interactions that occur through lower affinity sites; however, making binding models too flexible or encompassing, without proper parameterization, runs the risk of increasing the rate of false-positive predictions.
TF-specific preferences
Another complexity highlighted by recent HT protein–DNA binding studies is that closely related TFs often exhibit both common and TF-specific binding preferences (23,24,35,37,40,72–79). In a study of 104 mouse TFs, it was found that even proteins with a high degree of similarity in their DNA-binding domains (DBDs) (as high as 67% amino acid sequence identity) can exhibit distinct DNA-binding profiles (24). In many cases, the highest affinity site is identical for several members of a DBD family, but individual proteins within the family have different preferences for lower affinity sites. For example, mouse TFs Irf4 and Irf5 share a strong preference for sequences containing the 5′-CGAAAC-3′ site, but prefer, albeit with lower affinity, 5′- TGAAAG-3′ or 5′-CGAGAC-3′, respectively. Homeodomain proteins have been extensively studied using HT approaches (23,77,78), revealing rich and diverse sequence preferences for members even within protein subfamilies. For example, the Lhx subfamily binds with high affinity sites containing the core 5′-TAATTA-3′, but different subfamily members have different preferences for medium- and low-affinity sites: Lhx2 prefers 5′-TAATGA-3′ and 5′-TAACGA-3′, whereas Lhx4 prefers 5′-TAACGA-3′ and 5′-TAATCT-3′. These TF-specific preferences cause difficulties for DNA-binding models, as the models must accurately characterize the common binding sites while at the same time capture the sites specific to each TF.
Flanking DNA
Even in the case of TFs with well-defined core DNA binding sites, complexities can still arise from the DNA sequence flanking the core, which can affect binding affinity. Two such examples have been highlighted in recent HT studies (45). The TF Gcn4 binds to the core motif 5′-TGACTCA-3′. Binding measurements of Gcn4 to thousands of sequences containing this core motif revealed a wide range of binding affinities, from no appreciable binding to high-affinity binding (43). Nucleotides immediately adjacent to the core 7-mer were important for binding affinity, but even the two nucleotides flanking the best 9-mer affected affinity by an order of magnitude. Another recent study examined the DNA-binding preferences of the paralogous yeast TFs Cbf1 and Tye7 to hundreds of genomic sequences containing the E-box motif 5′-CACGTG-3′ (45). The study revealed a strong dependence on flanking DNA and computational analyses suggested that the sequence beyond the canonical E-box motif contributes to binding specificity by influencing the three-dimensional structure of the DNA-binding site. Importantly, the additional specificity coming from sequences flanking the core motif could not be described by simply extending the core (45), which is not surprising given that the influence of flanking DNA is likely to be exerted through DNA shape and not through specific DNA contacts. DNA shape is a function of the DNA sequence; however, this relationship is complex (80) and cannot be captured by simple PWM models. Thus, in order to account for the influence of flanking regions on the affinity of DNA-binding sites, future models of binding specificity will likely use DNA shape characteristics explicitly.
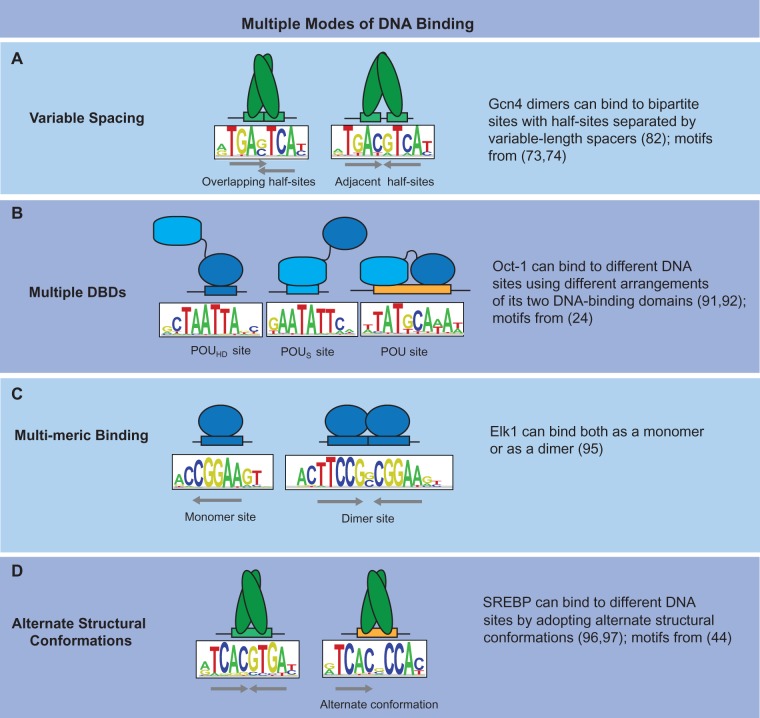
Multiple modes of DNA binding
Complicating a simple model of DNA binding for many proteins is that they can bind DNA using two or more distinct modes (Figure 1). These different modes of interaction can lead to fundamentally different DNA-binding preferences (i.e., motifs), complicating simple binding models that do not explicitly account for these multiple modes. HT studies are increasingly revealing unknown diversity in DNA-binding preferences of numerous proteins, many of which may be the results of different binding modes (29,72,75,81). Here, we classify variable binding modes into four categories (Figure 1) and briefly review each category.
Figure 1.
Multiple modes of DNA binding. Schematized are examples illustrating four mechanistic categories by which proteins recognize different DNA sites via distinct structural modes.
Variable spacing
Some proteins that bind to bipartite DNA motifs (i.e. motifs composed of two half-sites), can recognize distinct classes of motifs in which the half-sites are separated by different numbers of bases (Figure 1A). This phenomenon was first observed more than 20 years ago for basic leucine zipper (bZIP) proteins, which can bind overlapping or adjacent TGAC half-sites (82). Subsequent studies have classified the bZIP family into two subfamilies based on protein sequence: (i) Activator protein 1 (AP-1) proteins, which generally prefer overlapping half-sites, but bind to adjacent half-sites with almost equal affinity and (ii) Activating transcription factor (ATF)/cAMP response element-binding protein (CREB) proteins, which generally prefer adjacent half-sites and bind very poorly to overlapping half-sites (82,83). However, a recent PBM-based study found that at least one ATF/CREB protein in yeast (Yap3) binds with high affinity to both adjacent and overlapping half-sites (74). Therefore, although some of the residues that defined bZIP half-site spacing have been identified (84), additional residues are also involved. Variable half-site spacing has also been well documented for the nuclear receptors (NRs). For example, both the peroxisome proliferator activated receptors (PPARs) and retinoic acid receptors (RARs) form heterodimers with the retinoid X receptor (RXR) and can bind to response elements composed of direct repeats of the 5′-AGGTCA-3′ half-site separated by different length spacers (85). PPAR:RXR dimers bind direct repeats spaced by 1 or 2 bases (i.e. DR1 = 5′-AGGTCANAGGTCA-3′; DR2 = 5′-AGGTCANNAGGTCA-3′), while RAR:RXR dimers bind to repeats spaced by 1 (DR1) or 5 (DR5) bases. The variable spacing has even been shown to affect in vivo function of DNA-bound NR dimers (86). RAR:RXR bound to DR5 elements can activate transcription in response to ligand, but will not activate transcription when bound to the DR1 elements (86).
Multiple DBDs
DNA-binding proteins can contain multiple, independent DBDs (3,87). Multiple DBDs can allow a protein to alternatively recognize different DNA elements using different DBDs. For example, the zinc finger (ZF) protein Evi1 contains 10 ZF domains separated into two autonomously functioning DBDs—an N-terminal seven ZF DBD and a C-terminal three ZF DBD—that recognize completely different DNA motifs (88–90). A still more complicated scenario is seen for the mouse TF Oct-1 that has two DBDs known as the POU (Pit-Oct-Unc) homeodomain (POUHD) and the POU-specific domain (POUS) (Figure 1B) (24,91,92). Oct-1 can bind to three distinct DNA motifs using different combinations of the two DBDs: the POUHD site is recognized by the POUHD domain, the POUS site is recognized by POUS domain and the composite POU site is recognized using both domains.
Multi-meric binding
Selective multimerization provides another means to expand or diversify the DNA-binding specificity of a protein. Proteins such as Oct-1(93) and RXR (94) have been reported to bind DNA as either monomers or homodimers. Recent large-scale studies using HT-SELEX assays (39,95) have revealed that the dimeric mode of binding might be more common than previously appreciated and even proteins that are known to bind DNA primarily as monomers, such as Elk1, (Figure 1C) can also form dimers with specific orientation and spacing preferences. These dimeric sites are enriched within genomic regions bound in vivo, suggesting that the dimeric profiles are biologically relevant. Furthermore, the dimer orientation and spacing preferences can sometimes distinguish individual members of the same TF family (95). For example, T-box factors share a common monomeric binding specificity but show seven distinct dimeric spacing/orientation preferences.
Alternate structural conformations
The recognition of distinct DNA motifs is expected when a protein contains multiple DBDs or binds as a multi-mer; however, some proteins with only a single DBD can also bind to multiple, distinctly different DNA sites. One such example is mouse TF sterol regulatory element-binding factor 1 (SREBF1) (Figure 1D), the ortholog of human TF sterol regulatory element-binding protein 1 (SREBP) (59,96). SREBF/SREBP proteins belong to the basic helix–loop–helix (bHLH) family of TFs that typically recognize symmetric E-boxes (5′-CAnnTG-3′). Unlike most bHLH proteins, SREBF/SREBP can bind the asymmetric sterol regulatory element 5′-ATCACnCCAC-3′. A co-crystal structure of the DNA-binding domain of human SREBP-1a bound to DNA revealed an asymmetric DNA–protein interface with one monomer binding the E-box half-site (5′-ATCAC-3′) using protein–DNA contacts typical of bHLH proteins and the second monomer recognizing the non-E-box half-site (5′-GTGGG-3′) using entirely different protein–DNA contacts (96,97). Thus, in the case of SREBF/SREBP, residues in the DBD are responsible for the multiple modes of DNA binding. Regions outside the DNA-binding domain of the protein can also enable alternate binding modes. For example, a region N-terminal to the basic DBD of yeast TF Hac1 is required for dual recognition modes. Mutations in this region were shown to preferentially reduce binding to one of the modes, whereas mutation of an arginine in the basic domain was crucial for the other mode of binding (98). This indicates that the protein can bind DNA using two distinct conformations, and individual residues (within and outside the DBD) play specific roles within each binding mode (98).
The ability of proteins to utilize different DNA-binding modes presents difficulties or even precludes the construction of simple DNA-binding models for many proteins. The presence of distinct modes usually requires that multiple DNA-binding models are used to represent a protein’s specificity. This can cause difficulties when the data available to construct the DNA-binding model contains a mixture of multiple types of binding motifs, such as in genome-wide chromatin immunoprecipitation (ChIP)-seq datasets. In some situations, the DNA-binding modes can potentially be separated and independently characterized. For example, one could independently characterize the binding of individual DBDs when a protein contains multiple DBDs. However, even these situations can lead to difficulties as illustrated by the case of Oct-1 where the autonomous DBDs can function either independently or together, in which case examining them separately will cause one to miss the binding sites recognized when both DBDs are involved (Figure 1B).
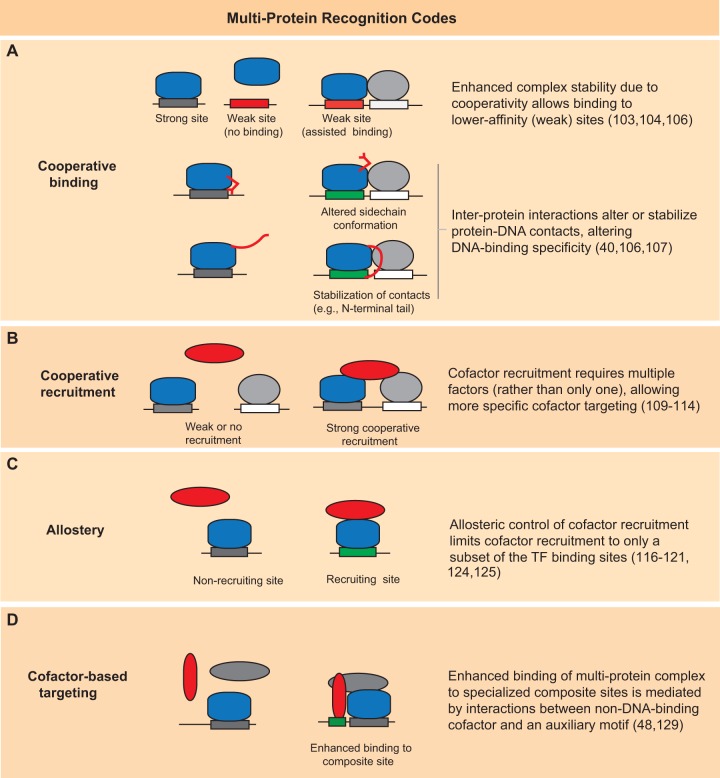
Multi-protein recognition codes
The DNA-binding specificity of a TF is a primary determinant of where it will bind in the genome (99,100). However, transcriptional regulation often involves the assembly of multi-protein complexes on DNA (101,102), and it has been shown that multi-protein complexes can exhibit novel DNA-binding specificities not predictable from measurements of the individual proteins (40,48). Therefore, in efforts aimed at understanding the biophysical determinants of specificity in gene regulation it is of primary importance to also consider how multi-protein complexes can lead to new or enhanced specificities. Here, we will review the varied mechanism by which novel DNA-binding specificities can arise when proteins, both DNA-binding and non-DNA-binding, assemble together. We suggest that these mechanisms represent higher-order ‘multi-protein’ recognition codes—mechanisms by which genomic targeting of regulatory factors is encoded in multi-protein complexes.
Cooperative binding
Cooperative binding of TFs to DNA—usually via protein–protein interactions between adjacently bound TFs—stabilizes the proteins on the DNA and can enhance their individual contributions to transcription of a gene (3,103). Cooperative binding can also affect the DNA-binding specificity and lead to recognition of new binding sites (3) (Figure 2A). One way in which cooperative binding alters specificity is by extending the binding of a TF to include lower affinity sites as a result of the enhanced overall affinity of the cooperative complex for DNA. A well-studied example is the binding of yeast TFs MATα2, MATa1 and Mcm1, which regulate mating-type genes (103,104). MATα2 binds DNA cooperatively with cofactors MATa1 or Mcm1 to repress different sets of genes. MATα2:MATa1 heterodimers bind to sites found in the promoter regions of haploid-specific genes and repress their expression, whereas MATα2:Mcm1 heterotetramers bind different sites found in mating-type a-specific gene promoters and repress gene expression. Both complexes repress gene expression via recruitment of co-repressor proteins Tup1 and Ssn6 by interactions with MATα2. Therefore, MATα2 functions to recruit co-repressors and repress gene expression, but its DNA-binding specificity—and subsequently its target genes—are mediated by cooperative binding with cell-type-specific cofactors. In a recent study, a more subtle form of specificity alteration by cooperative binding was presented in which DNA-binding of one protein can stabilize or destabilize the DNA-binding of another protein via the deformation of the DNA structure (105). Notably, the cooperative binding was not mediated by direct protein contacts, but by an allosteric mechanism where DNA structural deformations propagate along the DNA helix. The cooperative allosteric effects were shown to operate even to ∼16 bp away.
Figure 2.
Multi-protein recognition codes. Schematized are examples illustrating four mechanistic categories by which targeting of proteins to distinct DNA sites involves the assembly of multi-protein complexes.
A second, more active, mechanism has been described where cooperative binding alters the residue–base contacts that a TF can make with the DNA, thereby altering its inherent binding specificity (3,40) (Figure 2A). One example is the binding of the TFs Ets-1 and Pax5, where Pax5 alters the DNA contacts made by Ets-1, making it more permissive to alternate DNA sequences (106). Bound alone to DNA, Ets-1 binds with high-affinity to 5′-GGAA-3′ and with lower affinity to 5′-GGAG-3′. A tyrosine residue (Y395) in the Ets-1 recognition helix interacts with this fourth base position (underlined) and mediates the preference for adenine over guanine. However, in complex with Pax-5, the Y395 residue adopts an alternate conformation and no longer interacts with the DNA at this base position. This has the effect of making the Ets-1 DNA binding permissive for both sequences, and therefore broadens the specificity of the Pax-5:Ets-1 complex to include the suboptimal 5′-GGAG′-3′ Ets-1 half-site. Another example has been elucidated in a series of studies on Drosophila Hox proteins and their cofactor Extradenticle (Exd) (40,107). The Hox protein Sex combs reduced (Scr) binds with the cofactor protein Exd preferentially to a paralog-specific site (fkh250), found in the fork head (fkh) gene, over a consensus site (fkh250con) recognized by other Hox:Exd complexes (107,108). The preferential binding of the Scr:Exd complex to the fkh250 site involves the stabilization of a flexible ‘arm’ region N-terminal to the Scr homeodomain (107). Two of the stabilized residues in the N-terminal arm region (Arg3 and His-12) insert into the fkh250 DNA minor groove, which is narrower than the same region in the fkh250con sequence and more electrostatically favorable for the Arg and His residues. Therefore, the enhanced specificity of the Scr:Exd complex to the fhk250 sequence is due to local DNA shape (i.e., minor groove width) readout by the Scr Arg and His residues when stabilized by the cooperatively bound Exd cofactor. This work highlights how the intrinsic specificity of a monomeric homeodomain (Scr) can be altered or refined through interaction with a protein cofactor (Exd), a characteristic called latent specificity. A recent comprehensive analysis of other Hox proteins extended these results and demonstrated latent specificities for all the Drosophila Hox proteins (40). A HT-SELEX/SELEX-seq assay was used to measure and compare the DNA-binding specificity of all eight Drosophila Hox proteins alone and in complex with Exd. As seen for Scr:Exd, binding with the Exd cofactor revealed latent specificity differences between the Hox proteins, not observable when Hox proteins were examined as monomers.
Cooperative cofactor recruitment
The recruitment of non-DNA-binding cofactors (i.e. coactivators and corepressors) to promoters and enhancers by DNA-binding TFs is central to gene regulation (101). ‘Cooperative’ recruitment occurs when a cofactor can make simultaneous interactions with multiple, DNA-bound TFs; the result is a synergistic enhancement in the cofactor recruitment (109,110). Cooperative cofactor recruitment integrates the contributions from multiple TFs to achieve maximal gene expression and is a powerful mechanism for establishing an integrative, ‘AND-type’ regulatory logic in gene transcription (111–114). At the same time, cooperative recruitment enhances the binding specificity of the cofactor. The genomic location of the cofactor is determined not by the presence of a single TF, but by the less-frequent (i.e., more specific), coincident presence of multiple TFs (Figure 2B).
A paradigm for cooperative cofactor recruitment are multi-protein, enhanceosome complexes in which multiple TFs cooperatively assemble on DNA and coordinately recruit a common cofactor. A well-studied example is the enhanceosome that binds to the Interferon beta (IFNβ) gene promoter and cooperatively recruits the coactivator CREB-binding protein (CBP) (111,112). In response to viral infection, the TFs ATF-2, c-Jun, Irf3, Irf7 and NF-κB, facilitated by the architectural factor Hmga1, bind cooperatively to the IFNβ promoter. Multiple proteins in the DNA-bound complex contribute to the cooperative recruitment of CBP, and operate synergistically to enhance transcription (111). While NF-κB, the ATF-2:c-Jun dimer and Irf factors can recruit CBP individually, studies have demonstrated that removal of the activation domains from IRF or NF-κB factors resulted in complete loss of CBP recruitment, highlighting the cooperative nature of the CBP cofactor recruitment (111). A similar situation occurs in the cooperative recruitment of class II, major histocompatibility complex transactivator (CIITA) to a set of conserved transcriptional control elements found in the promoters of major histocompatibility complex class II (MHC-II) genes (113,115). The control sequences contain four DNA elements termed the W, X, X2 and Y boxes, with a conserved sequence, spacing and orientation across the different promoters. The TFs X-box binding protein (RFX), X2-binding protein (X2BP) and Nuclear factor Y (NF-Y) bind to the X, X2 and Y boxes, respectively, and form a cooperative, nucleoprotein, enhanceosome complex. This multi-protein complex recruits the CIITA to the MHC-II promoters via multiple weak interactions mediated by different components of the enhanceosome complex (113). Removal of interactions from any of these component proteins leads to significant loss of CIITA recruitment.
Allosteric effects in cofactor recruitment
DNA can act as a sequence-specific allosteric ligand that alters the function of a DNA-bound TF (116–121). A common mechanism by which this functions is by DNA sequence-dependent effects on cofactor recruitment. A TF bound to one set of DNA sites will recruit a specific cofactor, whereas the same TF bound to a different set of sites will not recruit the cofactor. Therefore, allosteric control of cofactor recruitment functionally partitions DNA binding sites of a TF and consequently refines binding specificity of the recruited cofactor (Figure 2C).
Allosteric mechanisms have been reported for both the glucocorticoid (GR) and the estrogen (ER) nuclear receptors (116,118). The DNA sequence bound by ER can influence its transcriptional activity and recruitment of LXXLL-containing peptides and p160 cofactors (SRC-1, GRIP1, ACTR) (116). The DNA sequence bound by GR, called the GR response element (GRE), was shown to influence gene expression, but the effect did not correlate with GR-binding affinity, suggesting a mechanism that involves differential recruitment of cofactor proteins (118). Furthermore, knock down of Brahma, a subunit of the SWI/SNF nucleosome remodeling complex, affected GR-dependent gene expression in a GRE sequence-dependent manner, suggesting that the composition of the DNA-bound protein complexes were allosterically regulated. Allosteric control of cofactor recruitment has also been described for different NF-κB family dimers (117,120). Single-base changes in NF-κB binding sites have been demonstrated to affect recruitment of the TF Irf3 protein by DNA-bound p65/Rel-containing dimers (117) as well as the recruitment of the cofactors histone de-acetylase 3 (HDAC3) and Tip60 to a DNA-bound, ternary complex containing p50 homodimers and Bcl3 (120).
The mechanism of allosteric control for GR and NF-κB appears to be moderated by structural differences between TFs bound to different DNA sites. Structural studies of GR bound to different GREs demonstrated that the structure of a ‘lever arm’ loop region, connecting the DNA recognition helix and the ligand binding domain, was highly dependent on the GRE sequence (118). Studies of NF-κB dimers bound to different kB sites have shown that DNA sequences differences can result in structural rearrangements (rotations and translations) of one dimer subunit (19,21,122). Protease protection assays and detailed binding studies have also suggested DNA sequence-dependent changes in protein conformation (72,123).
In contrast to the situation for GR and NF-κB where allostery is mediated by structural differences between the DNA-bound complexes, allostery involving the POU factors operates via larger, more global differences in quaternary structure when bound to different DNA sites. POU factors, such as Oct-1 described above, have two DBDs connected by a flexible linker: POUS and a POUHD (121). POU factors can homo- and heterodimerize to a diverse set of DNA-binding sites in which the relative orientations and spacing of the POUS and POUHD can vary and subsequently lead to differential cofactor recruitment (121,124). The POU factors Oct-1 and Oct-2 can bind as homodimers to two distinct response elements, PORE and MORE, found in the promoters of B-cell-specific genes (121). When bound to the PORE sequence, Oct dimers can recruit the transcriptional coactivator Oct-binding factor 1 (OBF-1); however, dimers bound to the MORE sequence do not recruit OBF-1. Crystal structures of the DNA-bound Oct-1 dimer have revealed that in the MORE-bound dimer the OBF-1 binding site on Oct-1 is blocked due to the relative orientation of the dimer subunits, but in the PORE-bound dimer the site is exposed, allowing OBF-1 recruitment (125,126). A similar situation arises with the Pit-1 POU factor in which binding site differences lead to cell-type-specific expression patterns and differential recruitment of the N-CoR co-repressor complex (124,127,128). Studies revealed that an extra 2 bp between the POUS and POUHD half-sites found in the growth hormone (GH) gene promoter, compared with a similar affinity site in the prolactin gene promoter, altered the quaternary structure of the DNA-bound Pit-1 and enabled the recruitment of the N-CoR co-repressor complex to the GH site (124).
Cofactor-based specificity
Targeting of non-DNA-binding cofactors to DNA is traditionally viewed as being mediated solely via protein–protein interactions with DNA-bound TFs. In other words, the cofactor is ‘recruited’ and interacts with DNA indirectly through the TFs. However, several studies have described a provocative alternative mechanism in which recruited cofactors interact directly with DNA when part of a larger, multi-protein complex (48,129). Further, as part of this larger complex, the non-DNA-binding cofactors mediate sequence-specific interactions that preferentially stabilize the binding to composite DNA sites containing specific auxiliary motifs (Figure 2D). This represents yet another mechanism to enhance the DNA sequence specificity of multi-protein regulatory complexes.
The regulation of sulfur metabolism genes in yeast involves a multi-protein complex composed of a sequence-specific TF Cbf1 and two non-DNA-binding cofactors Met28 and Met4 (130,131). Cbf1 is a bHLH protein and binds as a homodimer to E-box sites with a consensus 5′-CACGTG-3′ core (131). A PBM-based analysis revealed that the Met4:Met28:Cbf1 complex binds preferentially to composite DNA sites in which the Cbf1 E-box is flanked by an adjacent 5′-RYAAT-3′ ‘recruitment’ motif (i.e. 5′-RYAATNNCACGTG-3′) (48). High-affinity binding to the composite sites is highly cooperative and requires the full heteromeric complex. Sequence analysis, modeling and binding assays suggest that the recruitment motif is recognized by the non-DNA-binding Met28 subunit. A highly similar situation has also been described for the multi-protein Oct-1:HCF-1:VP16 complex that regulates expression of the herpes simplex virus immediate-early genes by binding to a composite 5′-TAATGARAT sequence in the gene promoters (129). The complex is composed of the DNA-binding TF Oct-1 and two non-DNA-binding cofactors: the viral activator VP16 and the host cell factor 1 (HCF-1). Oct-1 binds to the 5′-TAAT sequence and a DNA-binding surface of VP16 mediates interactions with the 5′-GARAT sequence. Similar to the situation in yeast, the VP16 cofactor does not interact with DNA well on its own, but when stabilized on DNA as part of a larger multi-protein complex it can adopt a configuration that mediates recognition of the 5′-GARAT subsequence. Therefore, in both situations multi-protein complexes preferentially bind to composite binding sites composed of a recruitment motif (5′-RYAAT-3′ and 5′-GARAT-3′) and an adjacent TF-binding motif (5′-CACGTG-3′ and 5′-TAAT-3′) that are recognized by a non-DNA-binding cofactor (Met28 and VP16) and sequence-specific TF (Cbf1 and Oct-1) subunits, respectively.
SUMMARY
Complexities in DNA-binding operate at multiple levels and complicate efforts to construct simple models, or recognition codes, of DNA-binding. Flexible protein-DNA interactions and non-local structural effects confound simple descriptions of DNA base preferences, and require the use of binding models, such as PWMs, that can account for the resulting sequence degeneracy. The ability of proteins to bind DNA via multiple modes further complicates the situation and leads to the requirement of multiple binding models for many proteins. Fortunately, HT technologies are not only increasing the rate at which DNA-binding proteins are being characterized, but are providing the comprehensive binding data needed to construct models that involve multiple DNA-binding modes (23,24,27,28,35,38–40,42,43,48,59,73,74,79,132–134). An added benefit of the comprehensive nature of certain HT datasets, such as comprehensive k-mer or binding site level data, is that predictions of binding sites in the genome can be performed directly using the measured binding data (23,24,40,43,133).
Multi-protein complexes add tremendously to the DNA-binding diversity of proteins and provide a key mechanism to integrate signals in gene regulation. However, these higher-order multi-protein mechanisms cause difficulties in constructing models, primarily due to the fact that DNA-binding models of proteins examined in isolation may not capture the binding sites utilized in vivo when cofactors are present. Here again, HT technologies are being used successfully to characterize binding of multi-protein complexes, revealing recognition codes mediated by cooperative binding (40,133) and cofactor-mediated targeting (48). The continued application of HT technologies to examine DNA binding of multi-protein complexes will undoubtedly provide increasingly refined binding models and provide new insights into gene targeting in vivo. Furthermore, just as the application of HT technologies has drawn our attention to the prevalence of TFs that exhibit subtle binding preferences or bind DNA via multiple modes, studies examining multi-protein complexes will likely identify wide spread use of higher-order mechanisms such as allostery, cooperative recruitment and cofactor-mediated targeting. Integrating these datasets with whole genome chromatin immunoprecipitation (ChIP) and expression datasets will lead to much more complete and sophisticated descriptions of specificity in gene regulation.
FUNDING
NIH [K22AI093793 to T.S.]; PhRMA Foundation Research Starter Grant (to R.G.); Basil O'Connor Research Award #5-FY13-212 from March of Dimes Foundation (to R.G.). Funding for open access charge: Waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Natl Acad. Sci. USA. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garvie CW, Wolberger C. Recognition of specific DNA sequences. Mol. Cell. 2001;8:937–946. doi: 10.1016/s1097-2765(01)00392-6. [DOI] [PubMed] [Google Scholar]
- 4.Jayaram B, Jain T. The role of water in protein-DNA recognition. Annu. Rev. Biophys. Biomol. Struct. 2004;33:343–361. doi: 10.1146/annurev.biophys.33.110502.140414. [DOI] [PubMed] [Google Scholar]
- 5.Reddy CK, Das A, Jayaram B. Do water molecules mediate protein-DNA recognition? J. Mol. Biol. 2001;314:619–632. doi: 10.1006/jmbi.2001.5154. [DOI] [PubMed] [Google Scholar]
- 6.Miller JC, Pabo CO. Rearrangement of side-chains in a Zif268 mutant highlights the complexities of zinc finger-DNA recognition. J. Mol. Biol. 2001;313:309–315. doi: 10.1006/jmbi.2001.4975. [DOI] [PubMed] [Google Scholar]
- 7.Baldwin EP, Martin SS, Abel J, Gelato KA, Kim H, Schultz PG, Santoro SW. A specificity switch in selected cre recombinase variants is mediated by macromolecular plasticity and water. Chem. Biol. 2003;10:1085–1094. doi: 10.1016/j.chembiol.2003.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otwinowski Z, Schevitz RW, Zhang RG, Lawson CL, Joachimiak A, Marmorstein RQ, Luisi BF, Sigler PB. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988;335:321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- 9.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang YP, Xu M, Machado AC, Yu XJ, Rohs R, Chen XS. Mechanism of origin DNA recognition and assembly of an initiator-helicase complex by SV40 large tumor antigen. Cell Reports. 2013;3:1117–1127. doi: 10.1016/j.celrep.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Werner MH, Gronenborn AM, Clore GM. Intercalation, DNA kinking, and the control of transcription. Science. 1996;271:778–784. doi: 10.1126/science.271.5250.778. [DOI] [PubMed] [Google Scholar]
- 12.Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 13.Rohs R, Sklenar H, Shakked Z. Structural and energetic origins of sequence-specific DNA bending: Monte Carlo simulations of papillomavirus E2-DNA binding sites. Structure. 2005;13:1499–1509. doi: 10.1016/j.str.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Pabo CO, Nekludova L. Geometric analysis and comparison of protein-DNA interfaces: why is there no simple code for recognition? J. Mol. Biol. 2000;301:597–624. doi: 10.1006/jmbi.2000.3918. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki M, Gerstein M, Yagi N. Stereochemical basis of DNA recognition by Zn fingers. Nucleic Acids Res. 1994;22:3397–3405. doi: 10.1093/nar/22.16.3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kortemme T, Morozov AV, Baker D. An orientation-dependent hydrogen bonding potential improves prediction of specificity and structure for proteins and protein-protein complexes. J. Mol. Biol. 2003;326:1239–1259. doi: 10.1016/s0022-2836(03)00021-4. [DOI] [PubMed] [Google Scholar]
- 17.Siggers TW, Honig B. Structure-based prediction of C2H2 zinc-finger binding specificity: sensitivity to docking geometry. Nucleic Acids Res. 2007;35:1085–1097. doi: 10.1093/nar/gkl1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siggers TW, Silkov A, Honig B. Structural alignment of protein–DNA interfaces: insights into the determinants of binding specificity. J. Mol. Biol. 2005;345:1027–1045. doi: 10.1016/j.jmb.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Chen FE, Ghosh G. Regulation of DNA binding by Rel/NF-kappaB transcription factors: structural views. Oncogene. 1999;18:6845–6852. doi: 10.1038/sj.onc.1203224. [DOI] [PubMed] [Google Scholar]
- 20.Chen YQ, Ghosh S, Ghosh G. A novel DNA recognition mode by the NF-kappa B p65 homodimer. Nat. Struct. Biol. 1998;5:67–73. doi: 10.1038/nsb0198-67. [DOI] [PubMed] [Google Scholar]
- 21.Chen YQ, Sengchanthalangsy LL, Hackett A, Ghosh G. NF-kappaB p65 (RelA) homodimer uses distinct mechanisms to recognize DNA targets. Structure. 2000;8:419–428. doi: 10.1016/s0969-2126(00)00123-4. [DOI] [PubMed] [Google Scholar]
- 22.Yanover C, Bradley P. Extensive protein and DNA backbone sampling improves structure-based specificity prediction for C2H2 zinc fingers. Nucleic Acids Res. 2011;39:4564–4576. doi: 10.1093/nar/gkr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat. Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng X, Smith RM, Giesecke AV, Joung JK, Wolfe SA. Counter-selectable marker for bacterial-based interaction trap systems. Biotechniques. 2006;40:179–184. doi: 10.2144/000112049. [DOI] [PubMed] [Google Scholar]
- 28.Noyes MB, Meng X, Wakabayashi A, Sinha S, Brodsky MH, Wolfe SA. A systematic characterization of factors that regulate Drosophila segmentation via a bacterial one-hybrid system. Nucleic Acids Res. 2008;36:2547–2560. doi: 10.1093/nar/gkn048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, 3rd, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bulyk ML, Gentalen E, Lockhart DJ, Church GM. Quantifying DNA-protein interactions by double-stranded DNA arrays. Nat. Biotechnol. 1999;17:573–577. doi: 10.1038/9878. [DOI] [PubMed] [Google Scholar]
- 31.Mukherjee S, Berger MF, Jona G, Wang XS, Muzzey D, Snyder M, Young RA, Bulyk ML. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linnell J, Mott R, Field S, Kwiatkowski DP, Ragoussis J, Udalova IA. Quantitative high-throughput analysis of transcription factor binding specificities. Nucleic Acids Res. 2004;32:e44. doi: 10.1093/nar/gnh042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Field S, Udalova I, Ragoussis J. Accuracy and reproducibility of protein-DNA microarray technology. Adv Biochem. Eng. Biotechnol. 2007;104:87–110. doi: 10.1007/10_2006_035. [DOI] [PubMed] [Google Scholar]
- 34.Bonham AJ, Neumann T, Tirrell M, Reich NO. Tracking transcription factor complexes on DNA using total internal reflectance fluorescence protein binding microarrays. Nucleic Acids Res. 2009;37:e94. doi: 10.1093/nar/gkp424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–237. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 36.Zykovich A, Korf I, Segal DJ. Bind-n-Seq: high-throughput analysis of in vitro protein-DNA interactions using massively parallel sequencing. Nucleic Acids Res. 2009;37:e151. doi: 10.1093/nar/gkp802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong D, Teixeira A, Oikonomopoulos S, Humburg P, Lone IN, Saliba D, Siggers T, Bulyk M, Angelov D, Dimitrov S, et al. Extensive characterization of NF-kappaB binding uncovers non-canonical motifs and advances the interpretation of genetic functional traits. Genome Biol. 2011;12:R70. doi: 10.1186/gb-2011-12-7-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Y, Granas D, Stormo GD. Inferring binding energies from selected binding sites. PLoS Comput. Biol. 2009;5:e1000590. doi: 10.1371/journal.pcbi.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jolma A, Kivioja T, Toivonen J, Cheng L, Wei G, Enge M, Taipale M, Vaquerizas JM, Yan J, Sillanpaa MJ, et al. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 2010;20:861–873. doi: 10.1101/gr.100552.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, Dror I, Zhou T, Rohs R, Honig B, Bussemaker HJ, et al. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell. 2011;147:1270–1282. doi: 10.1016/j.cell.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tantin D, Gemberling M, Callister C, Fairbrother WG. High-throughput biochemical analysis of in vivo location data reveals novel distinct classes of POU5F1(Oct4)/DNA complexes. Genome Res. 2008;18:631–639. doi: 10.1101/gr.072942.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warren CL, Kratochvil NC, Hauschild KE, Foister S, Brezinski ML, Dervan PB, Phillips GN, Jr, Ansari AZ. Defining the sequence-recognition profile of DNA-binding molecules. Proc. Natl Acad. Sci. USA. 2006;103:867–872. doi: 10.1073/pnas.0509843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nutiu R, Friedman RC, Luo S, Khrebtukova I, Silva D, Li R, Zhang L, Schroth GP, Burge CB. Direct measurement of DNA affinity landscapes on a high-throughput sequencing instrument. Nat. Biotechnol. 2011;29:659–664. doi: 10.1038/nbt.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weirauch MT, Hughes TR. Dramatic changes in transcription factor binding over evolutionary time. Genome Biol. 11:122. doi: 10.1186/gb-2010-11-6-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gordan R, Shen N, Dror I, Zhou T, Horton J, Rohs R, Bulyk ML. Genomic regions flanking E-box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape. Cell Reports. 2013;3:1093–1104. doi: 10.1016/j.celrep.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 47.Maniatis T, Ptashne M, Backman K, Kield D, Flashman S, Jeffrey A, Maurer R. Recognition sequences of repressor and polymerase in the operators of bacteriophage lambda. Cell. 1975;5:109–113. doi: 10.1016/0092-8674(75)90018-5. [DOI] [PubMed] [Google Scholar]
- 48.Siggers T, Duyzend MH, Reddy J, Khan S, Bulyk ML. Non-DNA-binding cofactors enhance DNA-binding specificity of a transcriptional regulatory complex. Mol. Syst. Biol. 2011;7:555. doi: 10.1038/msb.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Staden R. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 1984;12:505–519. doi: 10.1093/nar/12.1part2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Workman CT, Yin Y, Corcoran DL, Ideker T, Stormo GD, Benos PV. enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic Acids Res. 2005;33:W389–W392. doi: 10.1093/nar/gki439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Man TK, Stormo GD. Non-independence of Mnt repressor-operator interaction determined by a new quantitative multiple fluorescence relative affinity (QuMFRA) assay. Nucleic Acids Res. 2001;29:2471–2478. doi: 10.1093/nar/29.12.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bulyk ML, Johnson PL, Church GM. Nucleotides of transcription factor binding sites exert interdependent effects on the binding affinities of transcription factors. Nucleic Acids Res. 2002;30:1255–1261. doi: 10.1093/nar/30.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomovic A, Oakeley EJ. Position dependencies in transcription factor binding sites. Bioinformatics. 2007;23:933–941. doi: 10.1093/bioinformatics/btm055. [DOI] [PubMed] [Google Scholar]
- 54.Zhou Q, Liu JS. Modeling within-motif dependence for transcription factor binding site predictions. Bioinformatics. 2004;20:909–916. doi: 10.1093/bioinformatics/bth006. [DOI] [PubMed] [Google Scholar]
- 55.Agius P, Arvey A, Chang W, Noble WS, Leslie C. High resolution models of transcription factor-DNA affinities improve in vitro and in vivo binding predictions. PLoS Comput. Biol. 2010;6 doi: 10.1371/journal.pcbi.1000916. pii: e1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ben-Gal I, Shani A, Gohr A, Grau J, Arviv S, Shmilovici A, Posch S, Grosse I. Identification of transcription factor binding sites with variable-order Bayesian networks. Bioinformatics. 2005;21:2657–2666. doi: 10.1093/bioinformatics/bti410. [DOI] [PubMed] [Google Scholar]
- 57.Gershenzon NI, Stormo GD, Ioshikhes IP. Computational technique for improvement of the position-weight matrices for the DNA/protein binding sites. Nucleic Acids Res. 2005;33:2290–2301. doi: 10.1093/nar/gki519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao Y, Ruan S, Pandey M, Stormo GD. Improved models for transcription factor binding site identification using nonindependent interactions. Genetics. 2012;191:781–790. doi: 10.1534/genetics.112.138685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weirauch MT, Cote A, Norel R, Annala M, Zhao Y, Riley TR, Saez-Rodriguez J, Cokelaer T, Vedenko A, Talukder S, et al. Evaluation of methods for modeling transcription factor sequence specificity. Nat. Biotechnol. 2013;31:126–134. doi: 10.1038/nbt.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mordelet F, Horton J, Hartemink AJ, Engelhardt BE, Gordan R. Stability selection for regression-based models of transcription factor-DNA binding specificity. Bioinformatics. 2013;29:i117–i125. doi: 10.1093/bioinformatics/btt221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siddharthan R. Dinucleotide weight matrices for predicting transcription factor binding sites: generalizing the position weight matrix. PLoS One. 2010;5:e9722. doi: 10.1371/journal.pone.0009722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mathelier A, Wasserman WW. The next generation of transcription factor binding site prediction. PLoS Comput. Biol. 2013;9:e1003214. doi: 10.1371/journal.pcbi.1003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang J, Levine M. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell. 1993;72:741–752. doi: 10.1016/0092-8674(93)90402-c. [DOI] [PubMed] [Google Scholar]
- 64.White MA, Parker DS, Barolo S, Cohen BA. A model of spatially restricted transcription in opposing gradients of activators and repressors. Mol. Syst Biol. 2012;8:614. doi: 10.1038/msb.2012.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scardigli R, Baumer N, Gruss P, Guillemot F, Le Roux I. Direct and concentration-dependent regulation of the proneural gene Neurogenin2 by Pax6. Development. 2003;130:3269–3281. doi: 10.1242/dev.00539. [DOI] [PubMed] [Google Scholar]
- 66.Struhl G, Struhl K, Macdonald PM. The gradient morphogen bicoid is a concentration-dependent transcriptional activator. Cell. 1989;57:1259–1273. doi: 10.1016/0092-8674(89)90062-7. [DOI] [PubMed] [Google Scholar]
- 67.Rowan S, Siggers T, Lachke SA, Yue Y, Bulyk ML, Maas RL. Precise temporal control of the eye regulatory gene Pax6 via enhancer-binding site affinity. Genes Dev. 2010;24:980–985. doi: 10.1101/gad.1890410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- 69.Jaeger SA, Chan ET, Berger MF, Stottmann R, Hughes TR, Bulyk ML. Conservation and regulatory associations of a wide affinity range of mouse transcription factor binding sites. Genomics. 2010;95:185–195. doi: 10.1016/j.ygeno.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tanay A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 2006;16:962–972. doi: 10.1101/gr.5113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature. 2008;451:535–540. doi: 10.1038/nature06496. [DOI] [PubMed] [Google Scholar]
- 72.Siggers T, Chang AB, Teixeira A, Wong D, Williams KJ, Ahmed B, Ragoussis J, Udalova IA, Smale ST, Bulyk ML. Principles of dimer-specific gene regulation revealed by a comprehensive characterization of NF-kappaB family DNA binding. Nat. Immunol. 2012;13:95–102. doi: 10.1038/ni.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gordan R, Murphy KF, McCord RP, Zhu C, Vedenko A, Bulyk ML. Curated collection of yeast transcription factor DNA binding specificity data reveals novel structural and gene regulatory insights. Genome Biol. 2011;12:R125. doi: 10.1186/gb-2011-12-12-r125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nakagawa S, Gisselbrecht SS, Rogers JM, Hartl DL, Bulyk ML. DNA-binding specificity changes in the evolution of forkhead transcription factors. Proc. Natl Acad. Sci. USA. 2013;110:12349–12354. doi: 10.1073/pnas.1310430110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bolotin E, Chellappa K, Hwang-Verslues W, Schnabl JM, Yang C, Sladek FM. Nuclear receptor HNF4alpha binding sequences are widespread in Alu repeats. BMC Genomics. 2011;12:560. doi: 10.1186/1471-2164-12-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chu SW, Noyes MB, Christensen RG, Pierce BG, Zhu LJ, Weng Z, Stormo GD, Wolfe SA. Exploring the DNA-recognition potential of homeodomains. Genome Res. 2012;22:1889–1898. doi: 10.1101/gr.139014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fang B, Mane-Padros D, Bolotin E, Jiang T, Sladek FM. Identification of a binding motif specific to HNF4 by comparative analysis of multiple nuclear receptors. Nucleic Acids Res. 2012;40:5343–5356. doi: 10.1093/nar/gks190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou T, Yang L, Lu Y, Dror I, Dantas Machado AC, Ghane T, Di Felice R, Rohs R. DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res. 2013;41:W56–W62. doi: 10.1093/nar/gkt437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim J, Struhl K. Determinants of half-site spacing preferences that distinguish AP-1 and ATF/CREB bZIP domains. Nucleic Acids Res. 1995;23:2531–2537. doi: 10.1093/nar/23.13.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M. Classification of human B-ZIP proteins based on dimerization properties. Mol. Cell. Biol. 2002;22:6321–6335. doi: 10.1128/MCB.22.18.6321-6335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kuo D, Licon K, Bandyopadhyay S, Chuang R, Luo C, Catalana J, Ravasi T, Tan K, Ideker T. Coevolution within a transcriptional network by compensatory trans and cis mutations. Genome Res. 2010;20:1672–1678. doi: 10.1101/gr.111765.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Khorasanizadeh S, Rastinejad F. Nuclear-receptor interactions on DNA-response elements. Trends Biochem. Sci. 2001;26:384–390. doi: 10.1016/s0968-0004(01)01800-x. [DOI] [PubMed] [Google Scholar]
- 86.Kurokawa R, DiRenzo J, Boehm M, Sugarman J, Gloss B, Rosenfeld MG, Heyman RA, Glass CK. Regulation of retinoid signalling by receptor polarity and allosteric control of ligand binding. Nature. 1994;371:528–531. doi: 10.1038/371528a0. [DOI] [PubMed] [Google Scholar]
- 87.Luscombe NM, Austin SE, Berman HM, Thornton JM. An overview of the structures of protein-DNA complexes. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-1-reviews001. REVIEWS001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Perkins AS, Fishel R, Jenkins NA, Copeland NG. Evi-1, a murine zinc finger proto-oncogene, encodes a sequence-specific DNA-binding protein. Mol. Cell. Biol. 1991;11:2665–2674. doi: 10.1128/mcb.11.5.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Delwel R, Funabiki T, Kreider BL, Morishita K, Ihle JN. Four of the seven zinc fingers of the Evi-1 myeloid-transforming gene are required for sequence-specific binding to GA(C/T)AAGA(T/C)AAGATAA. Mol. Cell. Biol. 1993;13:4291–4300. doi: 10.1128/mcb.13.7.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Funabiki T, Kreider BL, Ihle JN. The carboxyl domain of zinc fingers of the Evi-1 myeloid transforming gene binds a consensus sequence of GAAGATGAG. Oncogene. 1994;9:1575–1581. [PubMed] [Google Scholar]
- 91.Klemm JD, Rould MA, Aurora R, Herr W, Pabo CO. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell. 1994;77:21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- 92.Verrijzer CP, Alkema MJ, van Weperen WW, Van Leeuwen HC, Strating MJ, van der Vliet PC. The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO J. 1992;11:4993–5003. doi: 10.1002/j.1460-2075.1992.tb05606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kemler I, Schreiber E, Muller MM, Matthias P, Schaffner W. Octamer transcription factors bind to two different sequence motifs of the immunoglobulin heavy chain promoter. EMBO J. 1989;8:2001–2008. doi: 10.1002/j.1460-2075.1989.tb03607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kersten S, Gronemeyer H, Noy N. The DNA binding pattern of the retinoid X receptor is regulated by ligand-dependent modulation of its oligomeric state. J. Biol. Chem. 1997;272:12771–12777. doi: 10.1074/jbc.272.19.12771. [DOI] [PubMed] [Google Scholar]
- 95.Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 96.Kim JB, Spotts GD, Halvorsen YD, Shih HM, Ellenberger T, Towle HC, Spiegelman BM. Dual DNA binding specificity of ADD1/SREBP1 controlled by a single amino acid in the basic helix-loop-helix domain. Mol. Cell. Biol. 1995;15:2582–2588. doi: 10.1128/mcb.15.5.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Parraga A, Bellsolell L, Ferre-D'Amare AR, Burley SK. Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 A resolution. Structure. 1998;6:661–672. doi: 10.1016/s0969-2126(98)00067-7. [DOI] [PubMed] [Google Scholar]
- 98.Fordyce PM, Pincus D, Kimmig P, Nelson CS, El-Samad H, Walter P, DeRisi JL. Basic leucine zipper transcription factor Hac1 binds DNA in two distinct modes as revealed by microfluidic analyses. Proc. Natl Acad. Sci. USA. 2012;109:E3084–E3093. doi: 10.1073/pnas.1212457109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, Pritchard JK. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bulyk ML. Computational prediction of transcription-factor binding site locations. Genome Biol. 2003;5:201. doi: 10.1186/gb-2003-5-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 102.Johnson AD. Molecular mechanisms of cell-type determination in budding yeast. Curr. Opin Genet. Dev. 1995;5:552–558. doi: 10.1016/0959-437x(95)80022-0. [DOI] [PubMed] [Google Scholar]
- 103.Wolberger C. Multiprotein-DNA complexes in transcriptional regulation. Annu. Rev. Biophys. Biomol. Struct. 1999;28:29–56. doi: 10.1146/annurev.biophys.28.1.29. [DOI] [PubMed] [Google Scholar]
- 104.Johnson AD. In: Transcriptional Regulation. McKnight SL, Yamamoto KR, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1992. pp. 975–1006. [Google Scholar]
- 105.Kim S, Brostromer E, Xing D, Jin J, Chong S, Ge H, Wang S, Gu C, Yang L, Gao YQ, et al. Probing allostery through DNA. Science. 2013;339:816–819. doi: 10.1126/science.1229223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Garvie CW, Hagman J, Wolberger C. Structural studies of Ets-1/Pax5 complex formation on DNA. Mol. Cell. 2001;8:1267–1276. doi: 10.1016/s1097-2765(01)00410-5. [DOI] [PubMed] [Google Scholar]
- 107.Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, Crickmore MA, Jacob V, Aggarwal AK, Honig B, Mann RS. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131:530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ryoo HD, Mann RS. The control of trunk Hox specificity and activity by Extradenticle. Genes Dev. 1999;13:1704–1716. doi: 10.1101/gad.13.13.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 110.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 111.Merika M, Williams AJ, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol. Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 112.Kim TK, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon-beta enhanceosome. Mol. Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 113.Masternak K, Muhlethaler-Mottet A, Villard J, Zufferey M, Steimle V, Reith W. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 2000;14:1156–1166. [PMC free article] [PubMed] [Google Scholar]
- 114.del Blanco B, Garcia-Mariscal A, Wiest DL, Hernandez-Munain C. Tcra enhancer activation by inducible transcription factors downstream of pre-TCR signaling. J. Immunol. 2012;188:3278–3293. doi: 10.4049/jimmunol.1100271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Benoist C, Mathis D. Regulation of major histocompatibility complex class-II genes: X, Y and other letters of the alphabet. Annu. Rev. Immunol. 1990;8:681–715. doi: 10.1146/annurev.iy.08.040190.003341. [DOI] [PubMed] [Google Scholar]
- 116.Hall JM, McDonnell DP, Korach KS. Allosteric regulation of estrogen receptor structure, function, and coactivator recruitment by different estrogen response elements. Mol. Endocrinol. 2002;16:469–486. doi: 10.1210/mend.16.3.0814. [DOI] [PubMed] [Google Scholar]
- 117.Leung TH, Hoffmann A, Baltimore D. One nucleotide in a kappaB site can determine cofactor specificity for NF-kappaB dimers. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 118.Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mrinal N, Tomar A, Nagaraju J. Role of sequence encoded kappaB DNA geometry in gene regulation by Dorsal. Nucleic Acids Res. 2011;39:9574–9591. doi: 10.1093/nar/gkr672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang VY, Huang W, Asagiri M, Spann N, Hoffmann A, Glass C, Ghosh G. The transcriptional specificity of NF-kappaB dimers is coded within the kappaB DNA response elements. Cell Reports. 2012;2:824–839. doi: 10.1016/j.celrep.2012.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Remenyi A, Scholer HR, Wilmanns M. Combinatorial control of gene expression. Nat. Struct. Mol. Biol. 2004;11:812–815. doi: 10.1038/nsmb820. [DOI] [PubMed] [Google Scholar]
- 122.Chen YQ, Ghosh S, Ghosh G. A novel DNA recognition mode by the NF-kappa B p65 homodimer. Nat. Struct. Biol. 1998;5:67–73. doi: 10.1038/nsb0198-67. [DOI] [PubMed] [Google Scholar]
- 123.Fujita T, Nolan GP, Ghosh S, Baltimore D. Independent modes of transcriptional activation by the p50 and p65 subunits of NF-kappa B. Genes Dev. 1992;6:775–787. doi: 10.1101/gad.6.5.775. [DOI] [PubMed] [Google Scholar]
- 124.Scully KM, Jacobson EM, Jepsen K, Lunyak V, Viadiu H, Carriere C, Rose DW, Hooshmand F, Aggarwal AK, Rosenfeld MG. Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science. 2000;290:1127–1131. doi: 10.1126/science.290.5494.1127. [DOI] [PubMed] [Google Scholar]
- 125.Remenyi A, Tomilin A, Pohl E, Lins K, Philippsen A, Reinbold R, Scholer HR, Wilmanns M. Differential dimer activities of the transcription factor Oct-1 by DNA-induced interface swapping. Mol. Cell. 2001;8:569–580. doi: 10.1016/s1097-2765(01)00336-7. [DOI] [PubMed] [Google Scholar]
- 126.Chasman D, Cepek K, Sharp PA, Pabo CO. Crystal structure of an OCA-B peptide bound to an Oct-1 POU domain/octamer DNA complex: specific recognition of a protein-DNA interface. Genes Dev. 1999;13:2650–2657. doi: 10.1101/gad.13.20.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ingraham HA, Chen RP, Mangalam HJ, Elsholtz HP, Flynn SE, Lin CR, Simmons DM, Swanson L, Rosenfeld MG. A tissue-specific transcription factor containing a homeodomain specifies a pituitary phenotype. Cell. 1988;55:519–529. doi: 10.1016/0092-8674(88)90038-4. [DOI] [PubMed] [Google Scholar]
- 128.Ingraham HA, Flynn SE, Voss JW, Albert VR, Kapiloff MS, Wilson L, Rosenfeld MG. The POU-specific domain of Pit-1 is essential for sequence-specific, high affinity DNA binding and DNA-dependent Pit-1-Pit-1 interactions. Cell. 1990;61:1021–1033. doi: 10.1016/0092-8674(90)90067-o. [DOI] [PubMed] [Google Scholar]
- 129.Babb R, Huang CC, Aufiero DJ, Herr W. DNA recognition by the herpes simplex virus transactivator VP16: a novel DNA-binding structure. Mol. Cell Biol. 2001;21:4700–4712. doi: 10.1128/MCB.21.14.4700-4712.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kuras L, Cherest H, Surdin-Kerjan Y, Thomas D. A heteromeric complex containing the centromere binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J. 1996;15:2519–2529. [PMC free article] [PubMed] [Google Scholar]
- 131.Blaiseau PL, Thomas D. Multiple transcriptional activation complexes tether the yeast activator Met4 to DNA. EMBO J. 1998;17:6327–6336. doi: 10.1093/emboj/17.21.6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wong D, Teixeira A, Oikonomopoulos S, Humburg P, Lone IN, Saliba D, Siggers T, Bulyk M, Angelov D, Dimitrov S, et al. Extensive characterization of NF-KappaB binding uncovers non-canonical motifs and advances the interpretation of genetic functional traits. Genome Biol. 2011;12:R70. doi: 10.1186/gb-2011-12-7-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ferraris L, Stewart AP, Kang J, DeSimone AM, Gemberling M, Tantin D, Fairbrother WG. Combinatorial binding of transcription factors in the pluripotency control regions of the genome. Genome Res. 2011;21:1055–1064. doi: 10.1101/gr.115824.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bolotin E, Liao H, Ta TC, Yang C, Hwang-Verslues W, Evans JR, Jiang T, Sladek FM. Integrated approach for the identification of human hepatocyte nuclear factor 4alpha target genes using protein binding microarrays. Hepatology. 2010;51:642–653. doi: 10.1002/hep.23357. [DOI] [PMC free article] [PubMed] [Google Scholar]