Abstract
Y-family DNA polymerases play a crucial role in translesion DNA synthesis. Here, we have characterized the binding kinetics and conformational dynamics of the Y-family polymerase Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4) using single-molecule fluorescence. We find that in the absence of dNTPs, the binary complex shuttles between two different conformations within ∼1 s. These data are consistent with prior crystal structures in which the nucleotide binding site is either occupied by the terminal base pair (preinsertion conformation) or empty following Dpo4 translocation by 1 base pair (insertion conformation). Most interestingly, on dNTP binding, only the insertion conformation is observed and the correct dNTP stabilizes this complex compared with the binary complex, whereas incorrect dNTPs destabilize it. However, if the n+1 template base is complementary to the incoming dNTP, a structure consistent with a misaligned template conformation is observed, in which the template base at the n position loops out. This structure provides evidence for a Dpo4 mutagenesis pathway involving a transient misalignment mechanism.
INTRODUCTION
Genomic DNA constantly undergoes a wide variety of damage that can lead to cellular death or carcinogenesis. DNA repair pathways minimize the impact of these modifications by restoring the original DNA sequence (1–3). However, damaged DNA that escapes the repair machinery can stall replication carried out by high-fidelity polymerases (4–6). The blocked replisome is unstable and can cause DNA double-strand breaks (7), compromising chromosome stability and increasing the risk of cancer. A specialized variety of DNA polymerases is recruited for the purpose of replicating across from and past damaged bases (translesion DNA synthesis) (7,8). Y-family polymerases perform the majority of translesion DNA syntheses and are generally specialized for a specific lesion type (9–11). These polymerases are characterized by low processivity and fidelity, and therefore can cause increased mutagenesis at or near the adduct site (10,12).
High-fidelity and Y-family polymerases show similar overall structures, presenting a characteristic shape that resembles a right hand (13,14). In addition to the ‘fingers’, ‘thumb’ and ‘palm’ domains, Y-family polymerases possess an additional domain termed the ‘little finger’. The little finger domain is the least conserved of the four domains in the Y-family polymerases, and its variability may determine the different biochemical behavior presented by the various bypass polymerases (15). Another major structural difference between high-fidelity and Y-family polymerases is in the active site. To accommodate bulky DNA lesions or distorted DNA structures, they have a wider active site that makes fewer contacts with its substrate (12). However, the catalytic core remains highly conserved among the Y-family polymerases and presents a tricarboxylate moiety akin to polymerases from other families.
Given the structural analogy between polymerases from different families, it is not surprising that the general mechanism for DNA elongation shows similar features among all DNA polymerases. A minimum kinetic pathway has been proposed based on structural and kinetic studies for high-fidelity polymerases (16), in which a key step takes place on correct dNTP binding. For high-fidelity polymerases, this binding causes the fingers region in the ternary complex (Pol•DNA•dNTP) to undergo a transition that repositions the DNA in the active site, leading to a catalytically active closed complex (17). Using a conformationally sensitive fluorophore bound to the fingers domain of T7 DNA polymerase, a structural state was identified when an incorrect nucleotide was bound that resulted in a faster dissociation of the nucleotide from the active site (18). We have recently presented evidence suggesting that nucleotide selection by the model high-fidelity polymerase Escherichia coli DNA polymerase I [Klenow fragment (KF)] occurs predominantly in the open conformation (19).
However, most bypass polymerases do not undergo a major conformational rearrangement on correct dNTP binding (20,21). Consistent with this, structural studies have shown that the fingers in the binary complex approximate a closed ready-for-catalysis conformation (22). Interestingly, the terminal base pair of the primer template in this structure was found to occupy the dNTP binding site (preinsertion complex) (23), so that the DNA must translocate 1 base pair (insertion complex) before the incoming dNTP can bind in the active site to initiate catalysis. Kinetic studies on Dpo4 using fluorophore reporters have identified various conformational changes that take place during the elongation cycle. By monitoring fluorescence from a unique tryptophan in the protein, it has been found that a prechemistry rearrangement takes place only when an incoming dNTP forms a correct base pair with the template (24). In agreement with this idea, ensemble-averaged FRET experiments have identified two prechemistry conformational changes that occur on correct dNTP binding: a DNA translocation event followed by a subtle and synchronized repositioning of all the polymerase domains (25).
Here, we studied Dpo4 dynamics using single-molecule Förster resonance energy transfer (smFRET). By monitoring smFRET in real time for DNA constructs with primers of different lengths, we found that the Dpo4–DNA binary complex samples two different states, consistent with conformations in which the nucleotide binding site is either occupied by the terminal base pair (preinsertion complex) or empty following Dpo4 translocation by 1 base pair (insertion complex). We have quantified the partition between these two states at different dNTPs concentrations, and provided evidence supporting a 1-base pair translocation that forms the catalytically active structure. Furthermore, incorrect dNTPs were found to destabilize this complex unless the n+1 template base was complementary to the dNTP present, in which case a structure consistent with a slipped misaligned template conformation was observed. Such a structure provides further direct structural support for the transient misalignment mutational mechanism (26,27).
MATERIALS AND METHODS
Dpo4 purification and labeling
Dpo4 was purified by a modification of the method described by Woodgate (28). An overnight culture (1 L) of E. coli RW382 transformed with a plasmid containing the Dpo4 gene (obtained from Roger Woodgate, NICHD) was grown in LB media containing 100 μg/ml carbenicillin. The cells were harvested by centrifugation at 4000 × g, and the pellets were suspended in buffer A [20 mM Tris-HCl, pH 7.5, 25 mM NaCl, 1 mM dithiothreitol (DTT), 0.1 mM ethylenediaminetetraacetic acid] and lysed by sonication followed by heating at 75°C for 15 min. Precipitated materials were removed by centrifugation at 18 000 × g, and DNA was removed from supernatant by dropwise addition of 30% (w/v) streptomycin sulfate with constant stirring at 4°C (1% final concentration) and was stirred for an additional hour at 4°C. The precipitate was removed by centrifugation at 18 000 × g, the supernatant filtered through a 0.2-μm filter, loaded onto a 1-ml SP column (GE healthcare) and then eluted with a linear gradient of 0–1 M NaCl in buffer A. Fractions containing Dpo4 were dialyzed into 20 mM KHPO4, pH 7.0, 25 mM NaCl, 1 mM DTT, loaded onto a 3-ml hydroxyapatite column (Biorad) and eluted with a linear gradient of 100–1000 KHPO4, pH 7.0, containing 25 mM NaCl and 1 mM DTT. Fractions containing Dpo4 were concentrated using Amicon Ultra 30 K centrifugal filter units and stored at −20°C in 40 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.2 mM ethylenediaminetetraacetic acid, 1 mM DTT and 50% (v/v) glycerol. Dpo4 purity was >95% based on sodium dodecyl sulphate-polyacrylamide gel electrophoresis analysis.
Wild-type Dpo4, which contains only one native cysteine, was labeled as described (19). Briefly, Dpo4 was incubated in 50 mM Tris-HCl buffer, pH 7.0, 120 μM Tris(2 carboxyethyl)phosphine, with a 5–10-fold molar excess of Cy5 maleimide (GE healthcare) for 45 min at room temperature. The reaction was stopped with 10 mM DTT, and the enzyme was separated from free dye on a polyacrylamide Bio-Gel P6 spin column. Labeling yield was ∼90% based on ultraviolet and mass spectral analysis. Enzyme activity was measured by primer extension (Supplementary Table S1) analysis on a 20% polyacrylamide gel (Supplementary Figure S1).
DNA oligonucleotide purification and labeling
Custom DNA oligonucleotides were synthesized by Eurofins MWG Operon and purified by high-performance liquid chromatography using an analytical C18 column. Purified oligonucleotides were desalted, and their purity was assessed by MALDI-TOF MS. Exact sequences are shown in Supplementary Table S1. Amino-modified C6-dT 28-mer templates and primers for DNA synthesis gel analysis were labeled with Cy3 NHS ester (GE healthcare) as described (2).
Single-molecule measurements
Microscope quartz slides were prepared as previously described (29), except minor changes were introduced during surface passivation. Amino silanization step: Slides and cover slides were incubated 15 min, sonicated 3 min and incubated another 15 min in 100 ml of methanol, 5 ml of acetic acid and 1.5 ml of N-(2-aminoethyl)-3-aminopropyltrimethoxysilane. Slides and cover slides were rinsed twice with distilled water and dried with air. Surface PEGylation: ∼40 mg of mPEG-succinimidylvalerate MW 5000 and ∼10 mg of and biotin-PEG-succinimidylvalerate MW 3400 (Laysan Bio) were dissolved in 360 µl of PEGylation buffer (filter-sterilized 100 mM sodium bicarbonate, pH 8.4). Seventy microliters of this PEGylation reaction mixture was added onto each slide, sandwiched with a cover slip and incubated in a dark humid chamber for ∼4 h. Subsequent steps were not modified and are described elsewhere (29). DNA was surface-immobilized via biotin–streptavidin–biotin linkage. Briefly, the slide was incubated 10 min with 0.02 mg/ml streptavidin, washed and incubated another 10 min with 20 pM biotinylated primer-template duplex. After a final washing step, Dpo4 was introduced in the solution. Measurements were carried out in 50 mM Tris-HCl, pH 7.5, 3.5 mM CaCl2/MgCl2 and 50 mg/ml bovine serum albumin in the presence of an oxygen scavenging system/antiblinking cocktail (protocatechuate dioxygenase from Pseudomonas sp., 5 mM 3,4-dihydroxybenzoic acid and 1 mM Trolox). Fluorescence intensity was monitored using a home-built prism-based total internal reflection microscope (30). Measurements were performed at 80 ms time resolution at 21°C.
Calcium ions were used in almost all experiments to prevent DNA synthesis in the presence of dNTPs (Supplementary Figure S6) because the absence of a 3′-OH group at the primer terminus inhibits a dNTP-induced conformational rearrangement known to precede catalysis in Dbh (31), a close Dpo4 homolog, and because the substrate orientation is different for deoxy and dideoxy-terminated primers (32). Calcium ions do not affect Dpo4 binding to DNA (33,34). Previous studies with KF and other polymerases have used dideoxy-terminated primers (19,35–38). Others have shown that many enzymes, including Dpo4 (33), that require magnesium for binding and catalysis are able to specifically bind to DNA in the presence of calcium (34).
Data analysis
Apparent FRET efficiencies were calculated as FRET = IA/(ID+IA), where ID and IA are the donor and acceptor intensities, respectively. Only fluorescence trajectories with one single photobleaching event were used to ensure single-molecule imaging, which also enabled accurate background subtraction. Time trajectories were smoothed with a 3–5-point moving average. FRET trajectories for 8Cy3 were further analyzed using a hidden Markov model (HMM) (39). smFRET histograms were created from several tens of single-molecule time trajectories. Dwell-time and smFRET histograms were fit to mono-exponential decays and Gaussian functions, respectively, using Igor.
RESULTS
Dpo4 binds a DNA primer template in two conformations
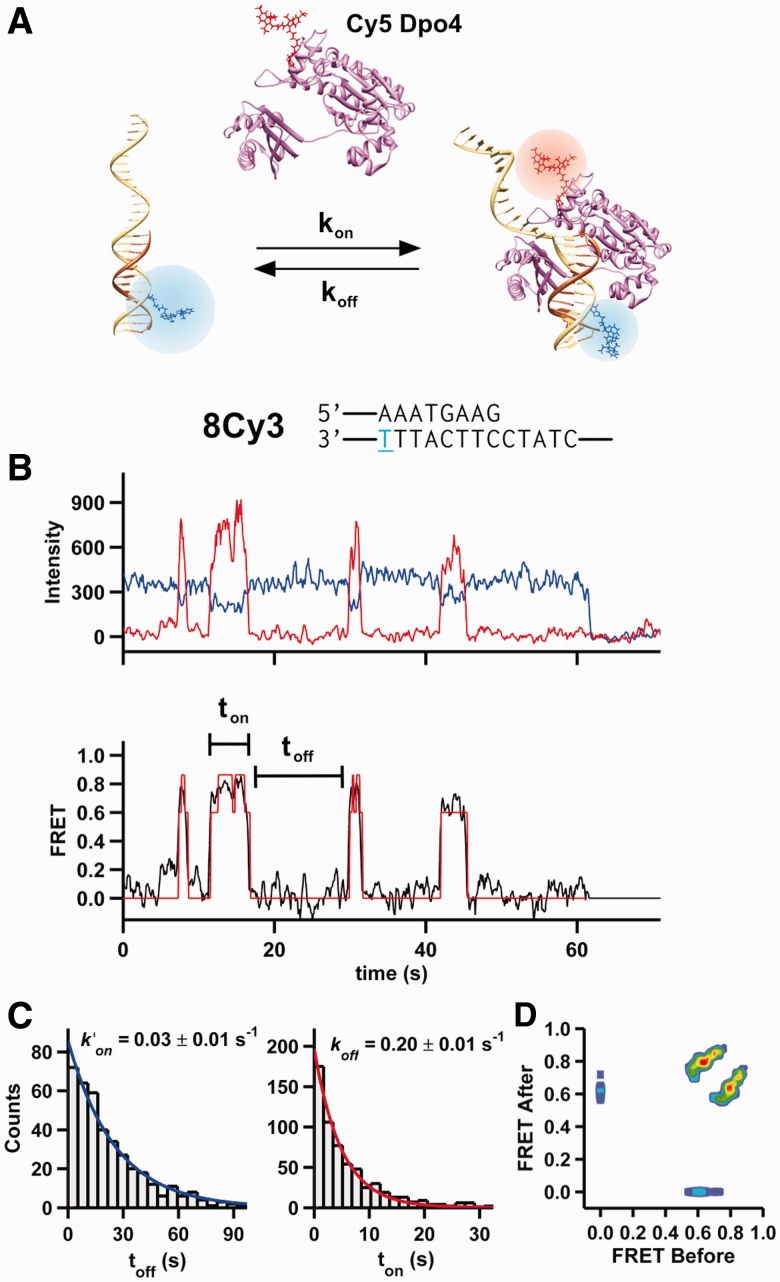
To monitor the conformation and dynamics of Dpo4 bound to a DNA primer template, we surface-immobilized Cy3-labeled DNA (8Cy3, Supplementary Table S1) onto passivated quartz slides and introduced Cy5-labeled Dpo4 in solution (Figure 1A). The labels did not noticeably affect polymerase activity (Supplementary Figure S1). The donor (Cy3) and acceptor (Cy5) fluorescence intensities from individual complexes reveal characteristic anticorrelated fluctuations, indicative of Dpo4 binding, and one-step photobleaching (Figure 1B). The apparent FRET efficiency fluctuates between 0.0 FRET (DNA only) and a high FRET value (Dpo4–DNA binary complex). A dwell time analysis in the 0 and high FRET values (Figure 1C) yields the pseudo-first order binding and dissociation rate constants, k′on = 0.03 s−1 and koff = 0.2 s−1, and a binding affinity KD = 30 ± 10 nM, similar to previously published values (40).
Figure 1.
Observation of time-resolved Dpo4 binding to DNA. (A) A Cy3-labeled (blue) DNA primer-template surface immobilized onto a microscope slide is incubated with a solution containing Cy5-labeled (red) Dpo4. On binding, the polymerase acceptor fluoresces and the donor is quenched. (B) Shown at the top is the primer-template DNA sequence used for the single-molecule experiment shown below. The 8Cy3 notation indicates that there are eight nucleotides between the primer terminus and the T (blue) bearing the Cy3 fluorophore. Below, characteristic time trajectory of a single Dpo4-DNA complex. On binding, the donor fluorescence (blue) decreases with an anticorrelated increase in acceptor fluorescence (red). The apparent FRET (black line, bottom) for the same trajectory is shown. A single photobleaching event is observed at ∼62 s. An HMM fit of the FRET trace is shown in red. (C) On and off dwell time histograms (left and right, respectively) for 5 nM Dpo4 binding to 8Cy3. Both distributions were fit with mono-exponential decay functions. Off dwell times were measured as the time Dpo4 is bound to the DNA (ton), whereas the on dwell times correspond to the time between binding events (toff). (D) Transition density plot for Dpo4 binding to 8Cy3. The transition density plot displays all the FRET transitions calculated by the HMM fit. The initial FRET value is shown at the x-axis, and the final FRET value (after the transition) is shown at the y-axis.
Unlike our prior experiments with KF, the Dpo4 binary complex appears to sample two possible conformations (FRET = 0.78 and 0.65, Figure 1B). We used an HMM to analyze the transitions between these two states (Figure 1B and D). A transition density plot (Figure 1D) clearly shows that the polymerase binds and dissociates primarily from the lower FRET state (0.65) but shuttles between the two states. A dwell time analysis in these two states yields the shuttling rate constants for transitions from the low to high FRET states kh = 2.7 ± 0.1 s−1, while the reverse transition rate kl = 1.1 ± 0.1 s−1 (Supplementary Figure S2), indicating that, in the absence of nucleotides, the 0.78 FRET state is the most stable of the two binary complexes.
Binary complex dynamics reveal Dpo4 translocation on the template
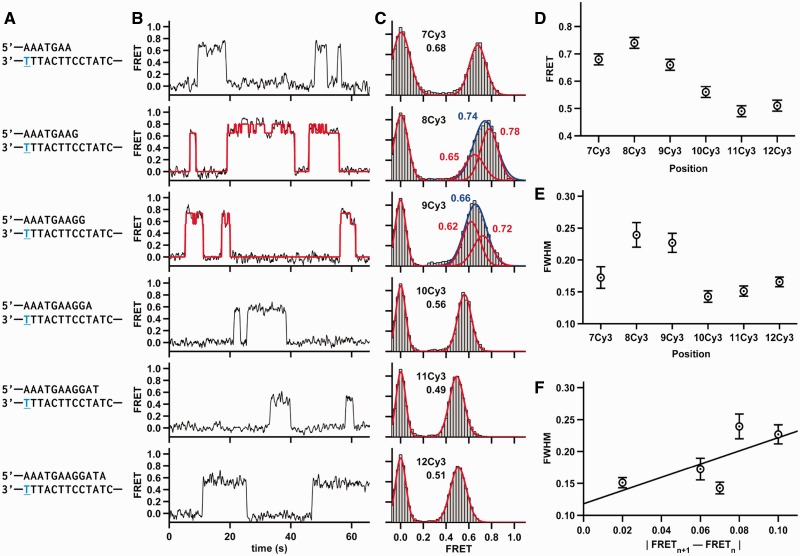
We then varied the primer length from 7 to 12 bp between the Cy3 label and the primer-template junction (termed 7Cy3 through 12Cy3, Figure 2A, Supplementary Table S1). All the single-molecule FRET trajectories reveal the binding and dissociation of individual Dpo4 to the DNA as stochastic jumps in FRET (Figure 2B). FRET histograms were calculated by time binning 40–80 time trajectories for each construct (Figure 2C). As previously observed (19), an increase in primer length typically results in lower FRET efficiency, indicating that the polymerase binds further away from the template label (Figure 2D). Only 8Cy3 and 12Cy3 exhibit a small FRET increase, likely due to the helical pitch of the DNA (41).
Figure 2.
Dpo4 binding different DNA constructs. (A) The sequence of DNA duplex is shown. The same template was used in all DNA constructs, whereas the lengths of the DNA primers varied. The position of the conjugated Cy3 dye is shown in blue and underlined. (B) Characteristic FRET trace for Dpo4 binding to each duplex. The 8Cy3 and 9Cy3 traces shown were fit by an HMM (red line). (C) SmFRET histograms for Dpo4 binding to each duplex with the Gaussian fits shown in red. 8Cy3 and 9Cy3 FRET distributions were fit with one Gaussian (shown in blue) and with two Gaussian functions (shown in red). (D) SmFRET efficiencies from (C) plotted as a function of the number of nucleotides between the Cy3 and the primer-template terminus. (E) Full width at half maximum (FWHM) plotted as a function of the number of nucleotides between the Cy3 and the primer-template terminus. The FWHM for 8Cy3 was calculated by fitting the 8Cy3 binary complex data to a single Gaussian function. Errors in FRET are estimated to be ±0.02, and the errors for the FWHM are the errors of the fit. (F) FWHM plotted versus the absolute value of the FRET difference between (n+1) Cy3 and (n) Cy3.
The binary complex FRET distributions for 8Cy3 and 9Cy3 DNA appear substantially wider than the other distributions (Figure 2E and C). This width increase is consistent with the two FRET states observed in the trajectories (Figure 1B). Two distinct FRET states could also be observed for 9Cy3 binary complex but not for the others (Figure 2B), presumably because the difference between the two FRET states is too small to distinguish them (see later). The 8Cy3 and 9Cy3 FRET distributions could be clearly fit to two Gaussian functions corresponding to the two conformations. This asymmetry in the FRET distribution for 8Cy3 was more obvious in 3.5 mM MgCl2 (Supplementary Figure S3) or with a longer template having a different sequence (Supplementary Figure S4).
As noted earlier, the other DNA constructs did not show two clearly differentiated FRET states for the binary complex. One possible explanation is that the two states are present but their FRET values are too close to be distinguished above the experimental noise. The increase in distribution width correlates well with the FRET difference between adjacent primers (Figure 2F); when the FRET difference for sequential primer templates (FRETn+1 – FRETn) is large, the distribution tends to be wider, whereas a small FRET difference results in a narrower distribution. This correlation suggests that the two FRET states represent the translocation of the polymerase by one nucleotide down the DNA template to the n+1 position, thus opening the nucleotide binding site. This hypothesis is further supported by the crystal structures of the binary and ternary complex, which reveal preinsertion binary and insertion ternary conformations (23). This translocation model predicts that the low FRET state represents the insertion conformation, as this structure should position the polymerase one nucleotide further from the Cy3 donor, and that binding of the correct nucleotide should stabilize this low FRET state.
Dpo4 ternary complex presents only one conformation
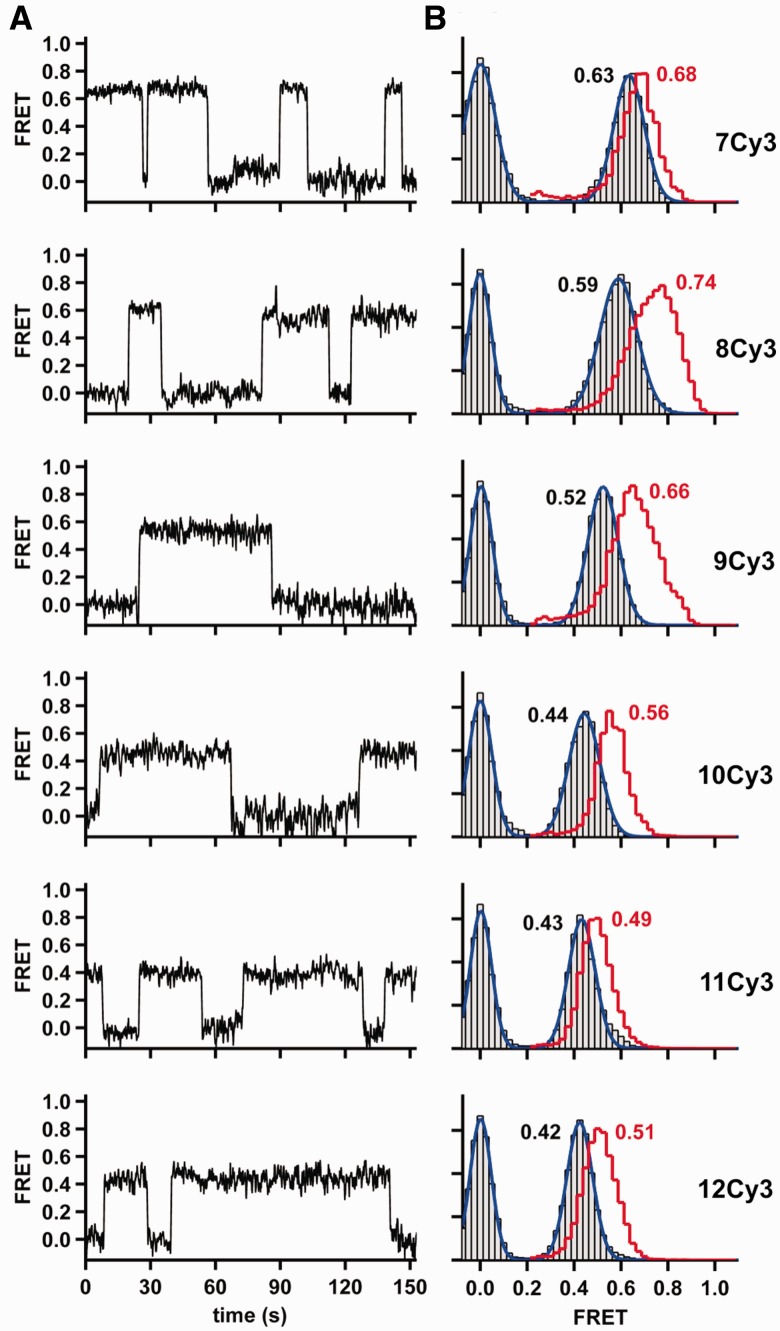
To test this hypothesis, we measured the conformational changes associated with binding of the correct nucleotide by carrying out smFRET experiments in the presence of the complementary dNTP for 7Cy3 through 12Cy3. The resulting time trajectories reveal the presence of a unique FRET state (Figure 3A), suggesting that the complementary dNTP causes Dpo4 binding in a unique well-defined orientation. The associated histograms confirm narrower FRET distributions for all primer templates, especially for 8Cy3 and 9Cy3 (Figure 3B). As predicted by the translocation model, all FRET distributions appear shifted predominantly to lower FRET values and this shift is greatest for both 8Cy3 and 9Cy3. A shift to lower FRET values in the presence of correct dNTPs was also observed with a longer template (Supplementary Figure S4).
Figure 3.
Conformational changes associated with correct dNTP binding. (A) Representative FRET trace for Dpo4 binding to each DNA duplex in the presence of the next correct nucleotide. (B) SmFRET histograms for Dpo4 binding to each DNA duplex in the presence of the correct nucleotide. Binary complex data are the same as in Figure 2C and are shown as a red stairs line. Ternary complex is shown as gray bars (experimental data) and blue line (Gaussian fit). Concentration of the correct nucleotide is 200 μM in all cases.
The presence of the next correct nucleotide also results in moderately longer binding events (Supplementary Figure S5). A dwell time histogram analysis for 8Cy3 shows that the dissociation rate constant decreases by 2.5-fold (Supplementary Figure S5) corresponding to a stabilization of the insertion ternary complex by ∼0.54 kcal/mol.
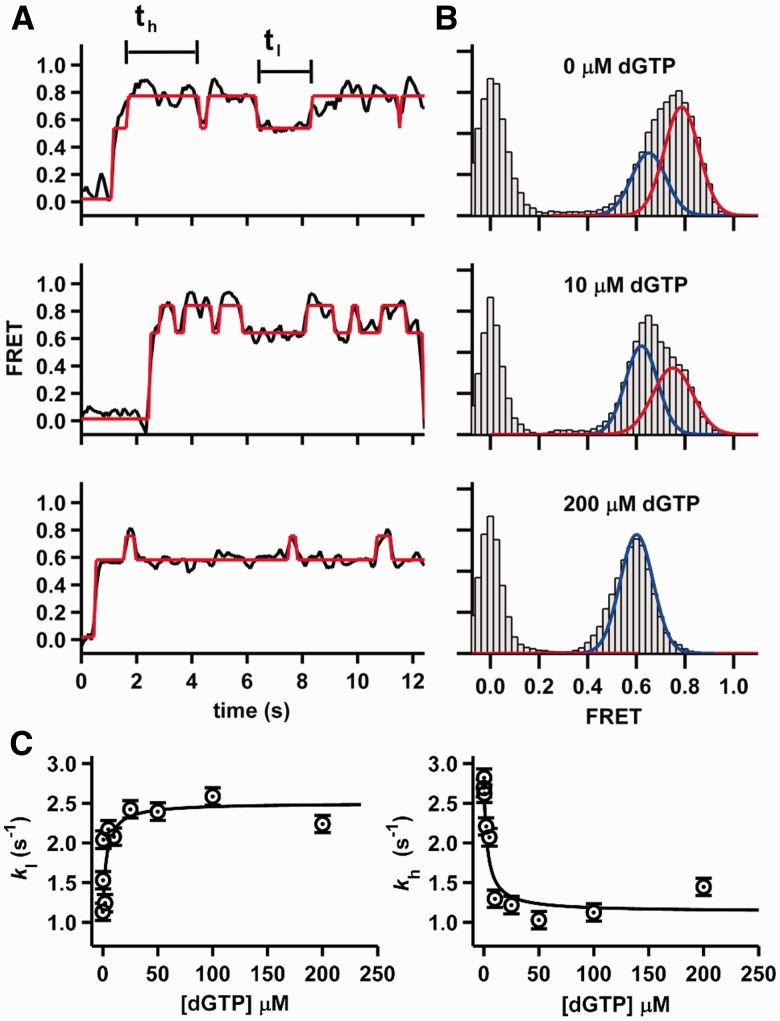
To further evaluate the effect of correct nucleotide on binary complex dynamics, we performed smFRET experiments with 8Cy3 at increasing dGTP concentrations because this construct exhibits the largest FRET difference on nucleotide binding (Figure 3B). The resulting time trajectories (Figure 4A) clearly reveal that increasing dGTP concentrations result in shorter excursions to the high FRET state (preinsertion complex) and longer ones in the low FRET state (insertion complex). The corresponding FRET histograms also confirm that the high FRET population decreases, whereas the low FRET one increases as the dGTP concentration increases (Figure 4B). Dwell time analysis in the high (th) and low FRET (tl) states shows that the shuttling rate constants kl and kh, respectively, increase and decrease by ∼2.5-fold as the dGTP concentration increases (Supplementary Figure S2). A global fit of both titrations to a Langmuir binding isotherm yields a dissociation constant for dGTP of KdGTP = 4 ± 2 μM (Figure 4C).
Figure 4.
Nucleotide binding dynamics. (A) Representative smFRET trajectories for Dpo4 binding the 8Cy3 duplex at different dGTP concentrations. The HMM fit is shown in red. (B) SmFRET histograms for Dpo4 binding to 8Cy3 primer template at different dGTP concentrations. Experimental data (gray bars) were fitted with one or two Gaussian functions (shown as red and blue lines, respectively). (C) Rate constants for the conversion of the high to low FRET state (left) and low to high FRET state (right) at increasing dGTP concentrations. The black lines are fits to a normal binding equation (see Supplementary Information).
Incorrect dNTPs can induce template misalignment
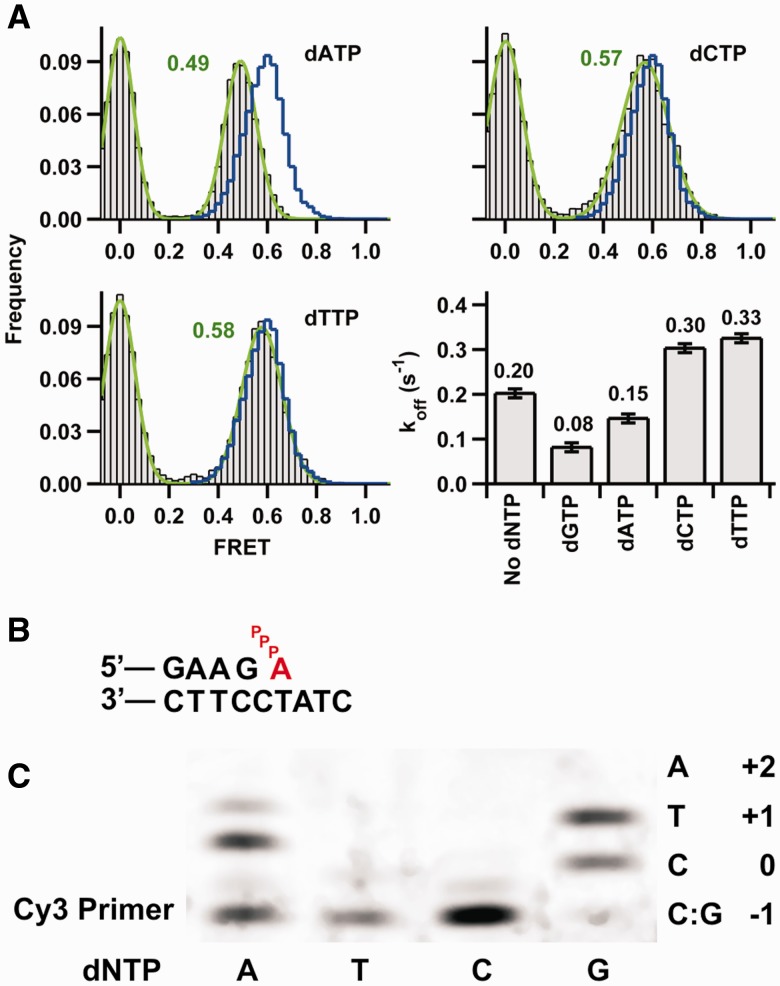
To determine whether incorrect dNTPs also affect the binary complex dynamics, we performed smFRET experiments in the presence of each of the three non-complementary dNTPs using 8Cy3 (Figure 5). The data show that both dCTP and dTTP induced a low FRET shift identical to that observed for the next correct nucleotide, dGTP (Figure 5A), suggesting that either nucleotide induces the formation of a complex with a similar structure to the tertiary complex.
Figure 5.
Conformational changes associated with incorrect nucleotide binding. (A) SmFRET histograms for Dpo4 binding 8Cy3 duplex in the presence of each of the three incorrect nucleotides shown in gray. Ternary complex histograms with the correct nucleotide dGTP are shown as a blue line and are the same as in Figure 3. Dissociation constant koff (calculated as in Figure 1C) in the absence and presence of the indicated dNTP. The concentration of correct nucleotide dGTP was 200 μM, and concentration of the incorrect dNTPs was 1 mM. The reported errors were determined from the mono-exponential fits to the dwell-time distributions. (B) Proposed misaligned template arrangement for the ternary complex with an incoming dATP pairing with the +1 T in the template. (C) Single-nucleotide incorporations by Dpo4. Reactions were carried out at 50°C in buffer containing 50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 50 mM NaCl, 0.025 mg/ml bovine serum albumin, 5 nM primer template (24mer/33mer) and 10 nM Dpo4. Reactions were initiated by the addition of dNTPs (final concentration 500 µM) and incubated for 10 min. The local template sequence is shown to the right of the gel.
In the presence of dATP, however, a different FRET state is observed (Figure 5A). The new FRET value (0.49) resembles that of 9Cy3 in the presence of the next correct nucleotide (dATP). A possible explanation for this unexpected result is that the presence of dATP leads to a misaligned template structure in which the incoming dATP base pairs with the T at the +1 position in the template (Figure 5B). This type of structure has previously been observed in the Dpo4 Type II complex that showed an unpaired template G and the next template base, C in this case, correctly pairing with the incoming dGTP (14).
To estimate the overall stability of the ternary complex in the presence of incorrect dNTPs, we measured their dissociation rate constants (Supplementary Figure S5 and Figure 5A). It is interesting that even though dCTP and dTTP cause a FRET shift similar to dGTP (Figure 5A), the off rates showed a significant increase compared with the binary complex. Based on the off rates, dCTP and dTTP ternary complexes are destabilized 0.24 and 0.29 kcal/mol, respectively, relative to the binary complex. However, the presence of dATP decreases the off rate, indicating a ternary complex stabilization of 0.17 kcal/mol relative to the binary complex.
The misaligned template model also predicts that Dpo4 will incorporate dATP in this template more efficiently than dTTP or dCTP. To test this, we performed single-nucleotide extension assays on the same primer-template sequence used in the single-molecule experiments (Figure 5C). As expected, dGTP is incorporated across the template C and, under these reaction conditions, the primer is extended by one additional nucleotide to the +1 position, possibly caused by a G looping out in the primer or a G:T wobble pair. As predicted by the misaligned template model, neither dCTP nor dTTP results in any significant primer extension, whereas dATP is readily incorporated. Under the latter reaction conditions, the primer is also extended by one additional nucleotide to the +1 position. A possible explanation for this additional extension is that the primer snaps back forming an A:C mispair setting up the T as the next template base.
DISCUSSION
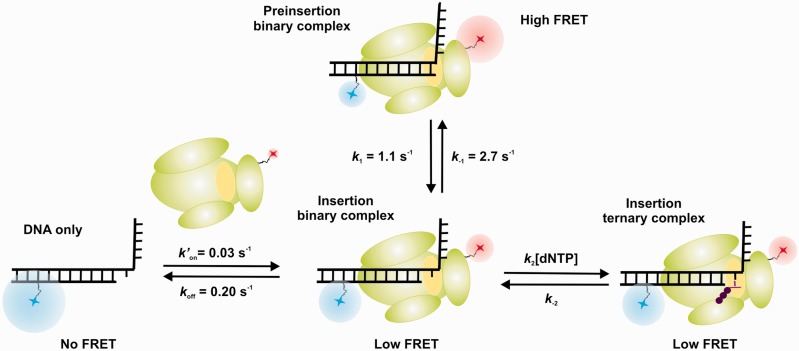
The crystal structure of the Dpo4 binary complex shows that the terminal base pair of the primer template occupies the nucleotide binding site (23), suggesting that the DNA must translocate 1 base pair before an incoming dNTP can be accommodated. Although this conformational change must happen before catalysis, it cannot be readily observed in a static structure. Here, we have characterized this process using smFRET by taking advantage of the fact that this technique can provide not only distance and/or orientation information about the DNA-polymerase complex but also the inter-conversion dynamics between two or more polymerase binding states. Our data clearly show that the binary complex exists in two conformations in dynamic equilibrium. We propose that one of these corresponds to the one captured in the crystal structure (23) with the terminal base pair of the primer template occupying the nucleotide binding site where catalysis would occur (Figure 6, preinsertion complex). The other FRET state can best be explained by the polymerase translocating 1 base pair, such that the nucleotide binding site is now unoccupied (Figure 6, insertion complex). Interestingly, the time trajectories and the transition density plot show that the initial binding occurs in the insertion complex conformation, followed by a translocation to the preinsertion complex conformation.
Figure 6.
Different conformations for Dpo4 binding to DNA shown schematically. Cy3 fluorophore (blue) is on the DNA template, and the Cy5 fluorophore (red) is on the polymerase. Dpo4 active site is shown in yellow. For the preinsertion binary complex, the primer terminus base is sitting in the polymerase active site at the position that an incoming nucleotide binds. The insertion binary complex has a free nucleotide binding site. In the insertion ternary complex, the incoming nucleotide is bound at the active site.
It is generally accepted for high-fidelity polymerases that translocation takes place at the end of the catalytic cycle, following successful nucleotide incorporation (42). Our prior results with the high-fidelity polymerase KF binary complex reveal a single FRET state (19), presumably the one in which the nucleotide binding site is not occupied, as observed in the corresponding crystal structures (43). However, our data here provide direct evidence that the Dpo4 binary complex is able to shuttle between the preinsertion and insertion complex conformations. It is not known whether this behavior occurs with other Y-family polymerases, but the observation that Dpo4 is not always in a nucleotide binding-competent conformation may partially account for the slow polymerization rates observed for Y-family polymerases compared with processive high-fidelity DNA polymerases.
We also find that ternary complex formation on dNTP binding leads to two measurable changes, consistent with the expected prechemistry steps in the DNA polymerase cycle. First, the preinsertion complex disappears in favor of the insertion complex (Figure 6). The fact that the ternary complex FRET ratio is close to the translocated insertion complex suggests that the DNA occupies a similar position in the polymerase active site. Second, the presence of the correct dNTP leads to a significant stabilization of the polymerase–DNA complex, suggesting that a structural change in the polymerase must occur, as previously proposed (44). Although it is likely that Y-family polymerases do not undergo the same large conformational change that occurs with high-fidelity polymerases (19,20,35), ensemble FRET studies have suggested that a subtle but synchronized reposition of all Dpo4 domains takes place on dNTP binding (25). The magnitude of the stabilization observed for Dpo4 was modest compared with high-fidelity polymerases (19), consistent with the fact that Dpo4 does not form as tight a binding pocket around the nascent base pair.
It is well established that non-complementary dNTPs cause a destabilization of high-fidelity polymerase–DNA complexes (19,35). Here, we also observe a destabilizing effect in the presence of either dCTP or dTTP, which do not base pair with the template bases at positions 0 or +1 (Figure 1B). Based on the dissociation rate constants (Figure 5A), the magnitude of this destabilization is comparable with the high-fidelity polymerase KF (19,35). However, the presence of the dATP, which can base pair with the template at the n+1 position, results in a slight stabilization instead. Interestingly, a Dpo4 ternary complex structure (Type II structure) has shown nucleotide base pairing with the +1 nucleotide on the template (14). The ability of Dpo4 to form this misaligned template structure is consistent with the FRET value observed in the presence of dATP, which is the same as found with a 1-nt-longer primer (9Cy3), and the fact that dATP can be readily incorporated by Dpo4 in this primer template (Figure 5C). The formation of this type of structure has been used to account for the single base and frameshift error spectrum observed for Dpo4 (27).
A final question regarding the nucleotide binding mechanism is whether dNTPs can only bind to the insertion complex, where the nucleotide binding site is unoccupied, or whether there is a pathway that allows dNTP association with the preinsertion complex before template movement to form the insertion complex. The most intuitive minimal mechanism consistent with our results is one in which an incoming nucleotide can only bind the insertion binary complex (Figure 6). However, our data do not permit us to exclude two additional scenarios (Supplementary Scheme S1). In these scenarios, the incoming nucleotide could either bind only to the preinsertion complex or to both complexes. These three scenarios are consistent with the observed (dNTP) dependence of the shuttling constants (kh and kl, Figure 4C).
In summary, we have characterized the conformational dynamics of the Y-family DNA polymerase Dpo4 on DNA in real time using smFRET. We observe two different binary complexes consistent with DNA translocation in the polymerase active site. Ternary complex formation locks the polymerase and its substrate in the catalytically active conformation. However, if the incoming nucleotide is complementary to the n+1 template base, a misaligned template structure is observed, in which the 5′-nt becomes the templating base. Our observation of this conformation provides further support for a mutagenic pathway involving this structure that likely plays a role for the high error rate and error spectrum observed for this polymerase.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [CA40605 to L.J.R. and GM085116 to D.R.]; National Science Foundation CAREER Award [MCB0747285 to D.R.] and a Fundación Barrié Fellowship to A.B. Funding for open access charge: NIH.
Conflict of interest statement. The authors declare no competing financial interests.
Supplementary Material
ACKNOWLEDGEMENTS
A.B., L.R. and D.R. designed the research plan. R.M. purified Dpo4. A.B. performed the experiments and analyzed the data. A.B., L.R. and D.R. wrote the manuscript.
REFERENCES
- 1.Hakem R. DNA-damage repair; the good, the bad, and the ugly. EMBO J. 2008;27:589–605. doi: 10.1038/emboj.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 3.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 4.Schmitt MW, Matsumoto Y, Loeb LA. High fidelity and lesion bypass capability of human DNA polymerase delta. Biochimie. 2009;91:1163–1172. doi: 10.1016/j.biochi.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alekseyev YO, Dzantiev L, Romano LJ. Effects of benzo[a]pyrene DNA adducts on Escherichia coli DNA polymerase I (Klenow fragment) primer-template interactions: evidence for inhibition of the catalytically active ternary complex formation. Biochemistry. 2001;40:2282–2290. doi: 10.1021/bi002245u. [DOI] [PubMed] [Google Scholar]
- 6.Zang H, Harris TM, Guengerich FP. Kinetics of nucleotide incorporation opposite polycyclic aromatic hydrocarbon-DNA adducts by processive bacteriophage T7 DNA polymerase. Chem. Res. Toxicol. 2005;18:389–400. doi: 10.1021/tx049683c. [DOI] [PubMed] [Google Scholar]
- 7.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat. Rev. Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl Acad. Sci. USA. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 11.Pata JD. Structural diversity of the Y-family DNA polymerases. Biochim. Biophys. Acta. 2010;1804:1124–1135. doi: 10.1016/j.bbapap.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broyde S, Wang L, Rechkoblit O, Geacintov NE, Patel DJ. Lesion processing: high-fidelity versus lesion-bypass DNA polymerases. Trends Biochem. Sci. 2008;33:209–219. doi: 10.1016/j.tibs.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 14.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 15.Boudsocq F, Kokoska RJ, Plosky BS, Vaisman A, Ling H, Kunkel TA, Yang W, Woodgate R. Investigating the role of the little finger domain of Y-family DNA polymerases in low fidelity synthesis and translesion replication. J. Biol. Chem. 2004;279:32932–32940. doi: 10.1074/jbc.M405249200. [DOI] [PubMed] [Google Scholar]
- 16.Joyce CM, Benkovic SJ. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 17.Doublie S, Sawaya MR, Ellenberger T. An open and closed case for all polymerases. Structure (London, England:1993) 1999;7:R31–R35. doi: 10.1016/S0969-2126(99)80017-3. [DOI] [PubMed] [Google Scholar]
- 18.Tsai YC, Johnson KA. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markiewicz RP, Vrtis KB, Rueda D, Romano LJ. Single-molecule microscopy reveals new insights into nucleotide selection by DNA polymerase I. Nucleic Acids Res. 2012;40:7975–7984. doi: 10.1093/nar/gks523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang W. Damage repair DNA polymerases Y. Curr. Opin. Struct. Biol. 2003;13:23–30. doi: 10.1016/s0959-440x(02)00003-9. [DOI] [PubMed] [Google Scholar]
- 21.Wong JH, Fiala KA, Suo Z, Ling H. Snapshots of a Y-family DNA polymerase in replication: substrate-induced conformational transitions and implications for fidelity of Dpo4. J. Mol. Biol. 2008;379:317–330. doi: 10.1016/j.jmb.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 22.Zhou BL, Pata JD, Steitz TA. Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol. Cell. 2001;8:427–437. doi: 10.1016/s1097-2765(01)00310-0. [DOI] [PubMed] [Google Scholar]
- 23.Rechkoblit O, Malinina L, Cheng Y, Kuryavyi V, Broyde S, Geacintov NE, Patel DJ. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 2006;4:e11. doi: 10.1371/journal.pbio.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beckman JW, Wang Q, Guengerich FP. Kinetic analysis of correct nucleotide insertion by a Y-family DNA polymerase reveals conformational changes both prior to and following phosphodiester bond formation as detected by tryptophan fluorescence. J. Biol. Chem. 2008;283:36711–36723. doi: 10.1074/jbc.M806785200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu C, Maxwell BA, Brown JA, Zhang L, Suo Z. Global conformational dynamics of a Y-family DNA polymerase during catalysis. PLoS Biol. 2009;7:e1000225. doi: 10.1371/journal.pbio.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunkel TA, Soni A. Mutagenesis by transient misalignment. J. Biol Chem. 1988;263:14784–14789. [PubMed] [Google Scholar]
- 27.Kokoska RJ, Bebenek K, Boudsocq F, Woodgate R, Kunkel TA. Low fidelity DNA synthesis by a y family DNA polymerase due to misalignment in the active site. J. Biol. Chem. 2002;277:19633–19638. doi: 10.1074/jbc.M202021200. [DOI] [PubMed] [Google Scholar]
- 28.Boudsocq F, Iwai S, Hanaoka F, Woodgate R. Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): an archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic poleta. Nucleic Acids Res. 2001;29:4607–4616. doi: 10.1093/nar/29.22.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamichhane R, Solem A, Black W, Rueda D. Single-molecule FRET of protein-nucleic acid and protein-protein complexes: surface passivation and immobilization. Methods (San Diego, Calif.) 2010;52:192–200. doi: 10.1016/j.ymeth.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, et al. Real-time DNA sequencing from single polymerase molecules. Science (New York, N.Y.) 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 31.DeLucia AM, Grindley ND, Joyce CM. Conformational changes during normal and error-prone incorporation of nucleotides by a Y-family DNA polymerase detected by 2-aminopurine fluorescence. Biochemistry. 2007;46:10790–10803. doi: 10.1021/bi7006756. [DOI] [PubMed] [Google Scholar]
- 32.Vaisman A, Ling H, Woodgate R, Yang W. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 2005;24:2957–2967. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irimia A, Zang H, Loukachevitch LV, Eoff RL, Guengerich FP, Egli M. Calcium is a cofactor of polymerization but inhibits pyrophosphorolysis by the Sulfolobus solfataricus DNA polymerase Dpo4. Biochemistry. 2006;45:5949–5956. doi: 10.1021/bi052511+. [DOI] [PubMed] [Google Scholar]
- 34.Vipond IB, Halford SE. Specific DNA recognition by EcoRV restriction endonuclease induced by calcium ions. Biochemistry. 1995;34:1113–1119. doi: 10.1021/bi00004a002. [DOI] [PubMed] [Google Scholar]
- 35.Joyce CM, Potapova O, Delucia AM, Huang X, Basu VP, Grindley ND. Fingers-closing and other rapid conformational changes in DNA polymerase I (Klenow fragment) and their role in nucleotide selectivity. Biochemistry. 2008;47:6103–6116. doi: 10.1021/bi7021848. [DOI] [PubMed] [Google Scholar]
- 36.Tsai YC, Jin Z, Johnson KA. Site-specific labeling of T7 DNA polymerase with a conformationally sensitive fluorophore and its use in detecting single-nucleotide polymorphisms. Anal. Biochem. 2009;384:136–144. doi: 10.1016/j.ab.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joyce CM. Techniques used to study the DNA polymerase reaction pathway. Biochim. Biophys. Acta. 2010;1804:1032–1040. doi: 10.1016/j.bbapap.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christian TD, Romano LJ. Monitoring the conformation of benzo[a]pyrene adducts in the polymerase active site using fluorescence resonance energy transfer. Biochemistry. 2009;48:5382–5388. doi: 10.1021/bi900148t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bronson JE, Fei J, Hofman JM, Gonzalez RL, Jr, Wiggins CH. Learning rates and states from biophysical time series: a Bayesian approach to model selection and single-molecule FRET data. Biophys. J. 2009;97:3196–3205. doi: 10.1016/j.bpj.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fiala KA, Hypes CD, Suo Z. Mechanism of abasic lesion bypass catalyzed by a Y-family DNA polymerase. J. Biol. Chem. 2007;282:8188–8198. doi: 10.1074/jbc.M610718200. [DOI] [PubMed] [Google Scholar]
- 41.Sabanayagam CR, Eid JS, Meller A. Using fluorescence resonance energy transfer to measure distances along individual DNA molecules: corrections due to nonideal transfer. J. Chem. Phys. 2005;122:061103. doi: 10.1063/1.1854120. [DOI] [PubMed] [Google Scholar]
- 42.Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv. Protein Chem. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- 43.Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc. Natl Acad. Sci.USA. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fiala KA, Suo Z. Mechanism of DNA polymerization catalyzed by Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry. 2004;43:2116–2125. doi: 10.1021/bi035746z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.