Abstract
During Drosophila oogenesis, transposable element (TE) repression involves the Piwi-interacting RNA (piRNA) pathway which ensures genome integrity for the next generation. We developed a transgenic model to study repression of the Idefix retrotransposon in the germline. Using a candidate gene KD-approach, we identified differences in the spatio-temporal requirements of the piRNA pathway components for piRNA-mediated silencing. Some of them (Aub, Vasa, Spn-E) are necessary in very early stages of oogenesis within the germarium and appear to be less important for efficient TE silencing thereafter. Others (Piwi, Ago3, Mael) are required at all stages of oogenesis. Moreover, during early oogenesis, in the dividing cysts within the germarium, Idefix anti-sense transgenes escape host control, and this is associated with very low piwi expression. Silencing of P-element-based transgenes is also strongly weakened in these cysts. This region, termed the ‘Piwiless pocket’ or Pilp, may ensure that new TE insertions occur and are transmitted to the next generation, thereby contributing to genome dynamics. In contrast, piRNA-mediated silencing is strong in germline stem cells in which TE mobilization is tightly repressed ensuring the continued production of viable germline cysts.
INTRODUCTION
Transposable element (TE) activity represents a constant threat for the stability of eukaryotic genomes and as a result protection mechanisms have evolved that limit TE mobilization. Nevertheless, TEs have colonized genomes efficiently and are thus thought to provide evolutionary advantages through their effects on genome expression and dynamics. Therefore, TEs should be able to bypass host defense mechanisms and mobilize in cells that will ensure their propagation to the next generation. Oogenesis is thus an important stage during which an active host defense is required to protect germline integrity, while escape from this protection can be positive from an evolutionary point of view. Extensive studies performed in Drosophila and mice have identified the piRNA pathway (PIWI-interacting RNA pathway) that requires small RNAs associated with PIWI proteins, the piRNAs, as the major pathway for silencing TEs in the germline and thereby inhibiting their mobilization and transmission (1–3). TE silencing by the piRNA pathway occurs both transcriptionally (TGS for Transcriptional Gene Silencing) and post-transcriptionally (PTGS for Post-Transcriptional Gene Silencing) (2,4–7). TGS involves decreased RNA synthesis due to the formation of a compact chromatin structure at target promoters, and PTGS involves homology-dependent target RNA degradation by PIWI-RISCs (PIWI-RNA-Induced Silencing Complexes).
Much of our knowledge about the piRNA pathway comes from studies of Drosophila oogenesis. In Drosophila, an ovary consists of 15 to 18 ovarioles, each of which contain a series of egg chambers at progressively advanced stages of oogenesis (8). The germline and somatic stem cells (GSC and SSC, respectively) reside in a region called the germarium at the anterior tip of each ovariole (9). During GSC division, one daughter cell remains in a ‘niche’ and continues to divide as a GSC. The other daughter cell, called the cystoblast, undergoes four cycles of mitotic division to form interconnected cysts of successively 2, 4, 8 and 16 germ cells. When the mature egg chamber leaves the germarium, the germline cyst consists of the oocyte and 15 nurse cells, all surrounded by a monolayer of somatic follicle cells deriving from the SSC. Both germ cells and follicle cells in the Drosophila ovary possess a functional piRNA pathway, which differs, however, in piRNA biogenesis. In both types of cells, a pool of primary piRNAs is processed from putative long single-stranded transcripts containing sequences homologous to TEs. These long transcripts are produced from discrete genomic loci (piRNA clusters), which reside primarily in pericentric heterochromatin enriched in TEs or their relics. Only in the germline, do these primary piRNAs, which include transposon anti-sense transcripts, target transposon sense-transcripts resulting in the production of a secondary pool of piRNAs (2). Secondary sense piRNAs enhance cleavage of anti-sense piRNA precursors, which leads to amplification of piRNA production called the ping-pong cycle. The three Argonaute proteins—Piwi, Aub and Ago3—are major players in this pathway and have also been shown to play a crucial role in gonadal development. Other factors, in particular components of the germ cell perinuclear structure, called the nuage, such as Vasa (Vas), Maelstrom (Mael), Armitage (Armi) and Squash (Squ), have also been implicated in piRNA production and transposon repression (10–14).
Our current knowledge of the piRNA pathway has been mostly deduced from high-throughput sequencing and large-scale genetic screens. They clearly demonstrated the existence of different classes of TEs undergoing differential regulation (2,4,5,15). Thus, the challenge now is to perform functional analyses on specific TEs to uncover the underlying specificities in their silencing or alternatively to determine what allows them to escape from silencing. The silencing of the retrotransposon Idefix has been previously characterized in ovarian follicle cells where its promoter is active; it has been reported to be repressed by the flamenco piRNA cluster (also called the COM locus) in a Piwi-dependent manner (16). In other somatic tissues, Idefix was shown to be repressed in a Piwi-independent manner by a transcriptional silencing pathway involving Polycomb group proteins (Pc-G) (17). In the germline, Idefix transcripts were found to be significantly upregulated in piwi germline knockdown ovaries (5,15). Several studies have also reported that piRNAs with a ping-pong signature and homologous to Idefix are produced in the germline and that they are reduced in piwi knockdown ovaries (15). These results suggest that a repression capacity exists for this TE in the germline. Here, we report that Idefix-sensor transgenes, whose expression can be induced in the germline, are targets of the piRNA pathway. Our data show that two categories of piRNA pathway proteins with different temporal requirements are involved in the silencing of Idefix sequences. In addition, we identified a small developmental window, corresponding to dividing germline cysts in the germarium, during which piRNA-mediated silencing is strongly reduced. Spatio-temporal regulation of TEs may contribute to the balance between TE repression and mobilizaton in the germline.
MATERIALS AND METHODS
Drosophila strains and transgenic lines
All experiments were performed at 22°C except when indicated. The fly strains Act5C-Gal4 (3954 and 4414) and nos-Gal4 (4937) came from the Bloomington stock center as did the RNAi lines ago3 (35232), aub (35201), mael (35202), spn-E (35303), piwi (33724), w (35573, 33644), vasa (34950). αtub-Gal4 were kind gifts from V. Mirouse, piwiNT from J. Brennecke.
The Idefix-sensor transgenic constructions were generated by inserting 419 bp of the Idefix gag coding region (1003–1422) in either the sense (pGgIds) or anti-sense (pGgIdas) orientation with respect to gfp transcription within the UASp-gfp vector. Six independent transgenic lines were generated for pGgIds and pGgIdas. BC69 (kindly provided by JL Couderc) is a P-lacZ enhancer trap line (18) and contains an in-frame translational fusion of the Escherichia coli lacZ gene to the second exon of the P transposase gene and a rosy transformation marker (FBtp0000154). RS3 is a P-FRT-white transgene (FBms0003945). It is inserted in the Telomeric Associated Sequences (TAS) of the 3R chromosomal arm (site 100E3). It is homozygous viable and fertile (Bloomington #123282). Crosses involving BC69 were performed at 25°C.
The pTomato-piwi plasmid is the result of the LR clonase II reaction (Invitrogen) between on one hand pUASp Tomato N term Gateway (Kind gift from V. Mirouse), constructed by the initial insertion of tomato coding sequence (1431 bp; Genbank number AY678269.1) between KpnI and XhoI restriction sites of pUASp vector followed by the insertion of Gateway Cassette (Invitrogen) between BglII and SpeI restriction sites, and on the other hand pDONR piwi Gateway plasmid, obtained by BP clonase II reaction (Invitrogen) between pDONR221 (Invitrogen) and PCR fragment amplified from piwi cDNA-containing pBluscript_SK(−) vector (Genbank number BT011138) using primers attB1_Piwi_F: GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT GGC TGA TGA TCA GGG ACG T and attB2_Piwi_R GGG GAC CAC TTT GTA CAA GAA AGC TGG GTT TAT AGA TAA TAA AAC TTC TTT TC. UASp-tomato-piwi was introduced into the Drosophila genome using standard P-element-mediated transformation techniques at the Fly Facility platform (www.flyfacility.fr).
Fluorescent in situ hybridization
A gfp fragment was cloned into pGemt-easy (Promega) using the following primers gfp-probe_for: 5′-TAGATGGTGATGTTAATGGGC-3′ and gfp-probe_rev: 5′-GTTTGTATAGTTCATCCATGCC-3′. RNA-FISH was performed as described in (19). Briefly, ovaries were dissected in PBT (PBS-0.2% Tween) on ice, fixed with 4% formaldehyde/PBT at room temperature (RT) for 10 min and rinsed three times with PBT. After permeabilization (1 h in PBS-0.3% Triton) prehybridization was carried out as follows: 10 min in HYB-(Formamide 50%, SSC 5×, Tween 0.02%)/PBT 1:1, 10 min in HYB-, 1 h in HYB+ (HYB- with yeast tRNA 0.1 μg/μl, heparin 0.25 mg/ml) at 60°C. Hybridization was carried out overnight at 60°C with 1 μg of RNA UTP-Dig-labeled probe. Ovaries were rinsed in HYB- and HYB-/PBT at 60°C then four times in PBT at RT. Blocking was done for 1 h at RT with TNB 0.3% triton (Perkin-Elmer TSA kit) and immunodetection 1.5 h at RT with anti-Dig-HRP (Roche) in TNB 0.2% tween. Ovaries were rinsed three times in PBT, incubated for 10 min with TSA-Cy3 in 1/25 amplification diluent (Perkin-Elmer), then rinsed three times and stained with DAPI (1/10 000). For RNA visualization, RNaseH treatment for 30 min at 37°C was performed before TSA amplification to destroy RNA/DNA hybrids.
Immunostaining
Ovaries were dissected in PBS on ice, fixed with 4% formaldehyde/PBS at RT for 15 min and rinsed three times with PBT (PBS with 0.2%Triton X-100). After blocking in BBT (PBT with 0.1% BSA) for 2 h at RT, ovaries were incubated with primary antibodies in BBT overnight at 4°C. After three washes in PBT, ovaries were incubated with secondary antibodies for 2 h at RT. The primary antibodies used were α-Aub and α-Ago3 [1:500, rabbit; a kind gift from J. Brennecke (2)], α-Piwi P3G11 [directed against Piwi N-term; 1:1000, mouse; a kind gift from MC Siomi (20)], α-Piwi (ab5207Abcam rabbit polyclonal antibody obtained against the peptide corresponding to amino acids 350 to 450 in the Piwi protein), α-HP1 (1:100, mouse C1a9 from Developmental Studies Hybridoma Bank), α-Vasa (1:100, rat from Developmental Studies Hybridoma Bank, DSHB), Rhino (1:1000, guinea pig from a kind gift from P. Zamore), αGFP (1:1000, chicken from abcam ab13970), α-β-Galactosidase (1 :500, rabbit; Rockland immunochemicals Inc) and α-H3K9-tri-methylation (1:2000, #07-523 Millipore). The GFP recovery experiments were done as previously described in (17). lacZ expression assays were carried out using X-gal overnight staining as described in (18), except that ovaries were fixed for 6 min.
Microscope analysis and image treatment
Immunostaining analysis was performed on a LEICA SP5 confocal microscope. GFP was viewed in whole-mount ovaries using the LEICA SP5 confocal microscope and analyzed using ImageJ software. 3D reconstruction was carried out using the Imaris software.
qRT–PCR analysis
First strand cDNA was obtained by using random primers on Trizol-extracted total ovarian RNA from 2- to 3-day-old flies. Quantitative PCR was performed using Roche FastStart SYBR Green Master on the LC480 on two independent insertions for each transgene. Steady-state RNA levels were calculated from the threshold cycle for amplification using the 2−Δ Δ CT method (21). rp49 was used for the normalization. Average levels and standard deviations were calculated from at least four biological replicates according to (21). In RNAi experiments, fold enrichments correspond to the comparison with a pGgIds/RNAi-white control sample.
Primers for qRT–PCR analysis were
rp49_for: 5′-GACGCTTCAAGGGACAGTATCTG-3′
rp49_rev: 5′-AAACGCGGTTCTGCATGAG-3′
gfp_for : 5′-TACCTGTCCACACAATCTGC-3′
gfp_rev : 5′-ATCCATGCCATGTGTAATCC-3′
HeT-A_for : 5′-CGCGCGGAACCCATCTTCAGA-3′
HeT-A_rev : 5′-CGCCGCAGTCGTTTGGTGAGT-3′
TART_for : 5′-TTTTCCGGATCCAAGTGAAC-3′
TART_rev : 5′-TCTGGTCGTCGGAAGTTGTT-3′
I _for : 5′-CAAAAACAACAATACCGCTAAT-3′
I _rev : 5′-AGCAGGTTGCCGTCTCTTGTA-3′
roo_for: 5′-CGTCTGCAATGTACTGGCTCT-3′
roo_rev: 5′-CGGCACTCCACTAACTTCTCC-3′
stalker4_for: 5′-TTTGGAAGATTACCAAGGCAGTTCGC-3′
stalker4_rev: 5′-GGATCTAACTTATGACCCGATTCGTTCC-3′
RESULTS
Engineered transgenes functionally demonstrate the ability of the germline to silence Idefix
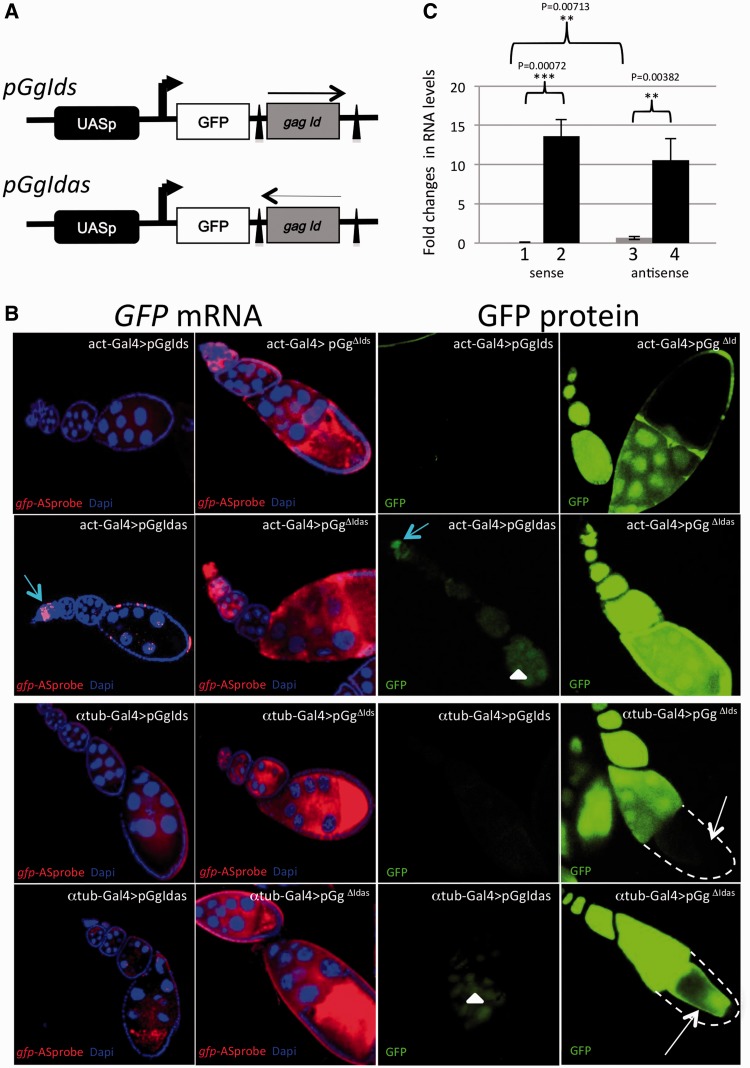
We developed a transgenic model that provides a convenient read-out to study piRNA-linked repression of the retrotransposon Idefix in the germline. The Idefix-sensor transgenes contain a gfp reporter gene (G) linked to a fragment of the Idefix gag gene in either a sense (gIds) or an anti-sense (gIdas) orientation (Figure 1A). The Idefix fragment is flanked by FRT sequences. Expression of the transgene is under the control of the germline UASp promoter (p). Six independent lines each carrying a single insertion were generated for each transgene (pGgIds and pGgIdas) using P-element transformation and the chromosomal mapping of each insertion was established. For each of the 12 transgenic lines, pGgIds and pGgIdas were expressed using two different Gal4 drivers, i.e. the ubiquitous actin-Gal4 (Figure 1B, rows 1 and 2) and the germline-specific α4tubuline-Gal4 (αtub-Gal4) (Figure 1B, rows 3 and 4). For the two drivers and the six pGgIds insertions tested, GFP was not detected, both at neither the RNA nor protein levels, in the germline throughout oogenesis (an example is presented Figure 1B, first and third columns, rows 1 and 3). For pGgIdas, faint GFP expression was generally observed in the nurse cells with both drivers (Figure 1B, first and third columns, rows 2 and 4), except for early stages of oogenesis within the germarium during which marked GFP protein and RNA signals were always detected (Figure 1B, row 2). When the Idefix sequence was excised upon recombination between the flanking FRTs, GFP RNA and protein signals were high for all the resulting pGgΔId transgenes (Figure 1B, second and fourth columns) indicating that the Idefix-sequence in the fusion transcript is responsible for GFP repression in the germline.
Figure 1.
Structure and silencing of pGgIds and pGgIdas transgenes. (A) Structure of pGgIds and pGgIdas: The minimal promoter and the Gal4 target sequences (UASp), the gfp reporter gene (GFP) and 419 bp of the Idefix gag coding region [from nt 1003 to 1422 (gag Id)] are indicated. Arrows indicate the orientation of the Idefix sequence, which was inserted in either the sense (pGgIds) or anti-sense orientation (pGgIdas). The two FRT sites flanking the Idefix fragment are indicated as black triangles. The transcription initiation sites between UASp and GFP are indicated by arrows. (B) Representative images of ovariole expression of transgenic constructs with the Idefix sequence in either the sense or anti-sense orientation (pGgIds and pGgIdas, respectively) or without the Idefix sequence (pGgΔIds and pGgΔIdas). Two drivers were used: the ubiquitous driver actin-Gal4 (rows 1 and 2) and the germline driver αtub-Gal4 (rows 3 and 4). gfp expression is presented at the mRNA (left, red) and protein (right, green) levels. Ovarioles are oriented with the germarium to the top or left. DNA was counterstained with DAPI (first and second columns, blue). Using both drivers, the transgene carrying the Idefix sequence in the sense orientation (pGgIds) exhibits undetectable levels of GFP mRNA and protein (columns 1 and 3). Using αtub-Gal4 and act-Gal4 drivers, the transgene carrying the Idefix sequence in the anti-sense orientation (pGgIdas) exhibits a low GFP expression in the nurse cells (white arrowheads) while a relatively strong GFP expression is detected only in the germarium (blue arrow) with the act-Gal4 driver. Upon excision of the Idefix fragment (pGgΔIds and pGgΔIdas), strong derepression of both gfp mRNA and protein is observed for both transgenes using both drivers. No gfp expression in somatic cells is observed when αtub-Gal4 is used (dashed line and white arrow in fourth column). (C) Quantitative RT–PCR analysis of steady-state levels of transcripts encoded by Idefix-sensors in ovaries. pGgIds insertion s1 (1), pGgΔIds insertion s1 (2), pGgIdas insertion as1 (3) and pGgΔIdas insertion as1 (4) driven by the αtub-Gal4. Experimental quadruplicates were carried out using gfp specific primers (see primer sets in ‘Materials and Methods’ section, mean ± SD and standard *P < 0.05, **P < 0.01, ***P < 0.001) and normalization, see ‘Materials and Methods’ section.
Quantitative RT–PCR with gfp-specific primers to measure the expression of the gfp-Idefix fusion transgenes confirmed that Idefix is a target of the germline repression (Figure 1C, histograms 1 and 3). In addition, pGgIdas displayed 2-fold higher expression than pGgIds (P-value = 0.0071). After Idefix was flipped out, the amount of RNA produced by pGgIds and pGgIdas increased >13-fold (P-value = 0.0007) and 10-fold (P-value = 0.0038), respectively (Figure 1C, histograms 2 and 4).
We conclude that the Idefix-based reporter constructs pGgIds and pGgIdas are targets of germline repression and that this repression is correlated with the presence of Idefix sequences within the transgenes.
Mutations in components of the piRNA pathway impact Idefix-sensor silencing with different temporal requirements
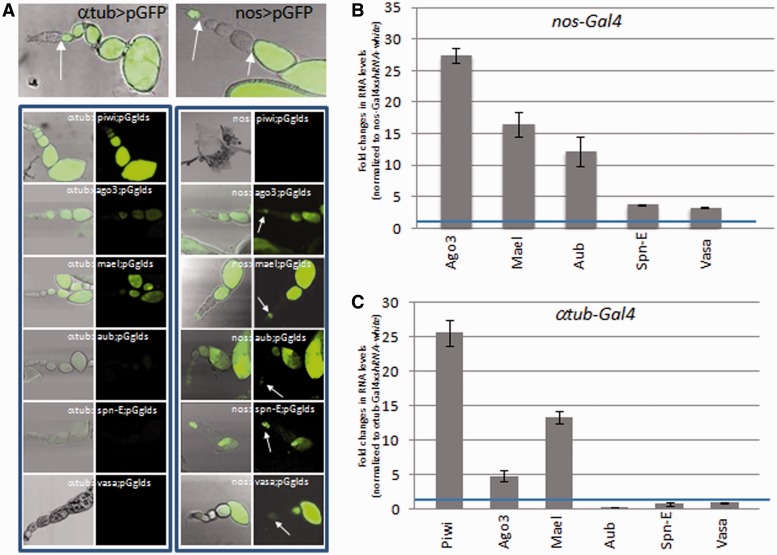
We then addressed the nature of the silencing exerted on Idefix-sensors in the germline in particular with respect to the piRNA pathway. We made use of TRIP RNAi lines (22,23), driven by αtub-Gal4, to target piwi, ago3, aub mael, spn-E and vasa (24–26). Since most of the genes tested have been shown to be key regulators of germline development, the fact that RNAi-mediated knockdown of all of these genes resulted in sterility with more or less severe ovarian phenotypes indicated the validity of the RNAi lines (data not shown). piwi, ago3 and mael knockdown released silencing exerted on pGgIds leading to GFP expression as soon as the αtub-Gal4driver was active from Stage 3 of oogenesis. Surprisingly this was not the case when aub, spn-E or vasa were targeted by RNAi since GFP expression was never recovered (Figure 2A, left). Results of quantitative RT–PCR with primers specific for aub, spn-E and vasa indicated that transcripts for these genes were indeed strongly reduced in ovaries compared with controls (Supplementary Figure S1A). At the protein level, immunofluorescent staining showed that αtub >aub- and >vasa-RNAi combinations resulted in strong depletion of Aub and Vasa proteins, respectively, as of Stage 3 of oogenesis when the driver is active, but these proteins were easily detected at earlier stages, in particular in the germarium, when this driver is inactive (Figure 2, top left and Supplementary Figure S1B).
Figure 2.
piRNA pathway components show different temporal requirements for Idefix silencing during Drosophila oogenesis. (A) GFP fluorescence is shown as a readout for pGgIds silencing release using two drivers, αtub-Gal4 (left column) and nos-Gal4 (right column) and RNAi constructs targeting piwi, ago3, mae1, aub, spnE and vasa transcripts. The expression profile of both drivers is presented at the top of each column using a GFP reporter construct (pGFP) and arrows delimit stages where these drivers are active. Ovarioles are oriented with the germarium to the top or left. nos > piwi RNAi results in atrophic ovaries. (B and C) gfp RNA was quantified by qRT–PCR using gfp-specific primers upon RNAi expression targeting the indicated genes and normalized to RNAi against white (the blue line represents no enrichment, value = 1). The transgene pGgIds was driven with nos-Gal4 (B) or α tub-Gal4 (C) (n = 4 biological replicates. Error bars represent SEM). Two transgenic lines pGgIds (s1 and s6) were tested in this experiment.
We next expressed the RNAi-constructs in the GSC present in the germarium up to Stage 2 using the nanos-Gal4 (nos-Gal4) driver. This driver is subsequently inactive between Stages 3 and 6 of oogenesis and re-activated at later stages as shown in Figure 2A (top right). When knockdown of ago3, mael, aub, spnE and vasa was performed using the nos-Gal4 driver, we found that the Idefix-sensors were derepressed and GFP expression recovered within the germarium and after Stage 6 but not between Stages 3 and 6 as expected according to nos-Gal4 activity (Figure 2A, right panel).
To confirm these results, we used quantitative RT–PCR with gfp-specific primers. Under RNAi conditions leading to derepression of GFP at the protein level (nos > ago3, mael, aub, spn-E and vasa; αtub > piwi, ago3 and mael), an increase in gfp RNA was also detected although in a lower amount in nos > spn-E and >vasa flies (Figures 2B and C). In contrast, the amount of gfp RNA was unchanged in αtub > aub, spn-E and vasa RNAi ovaries compared to controls, as was observed with GFP protein expression (Figure 2C).
Sensor de-repression in late egg chambers from nos > aub and not from αtub > aub flies could potentially be explained by a higher strength of the nos-Gal4 driver leading to a higher efficiency in knocking down target genes during late oogenesis. To compare the efficiency of the two drivers, we expressed an UASp-gfp transgene (pGgΔId) driven by nos-Gal4 or αtub-Gal4 driver, and quantified the gfp RNAs produced. We found almost twice as much gfp RNAs in (pGgΔId, αtub-Gal4) as in (pGgΔId, nos-Gal4) flies (Supplementary Figure S2) which indicates that nos-Gal4 is weaker than αtub-Gal4.
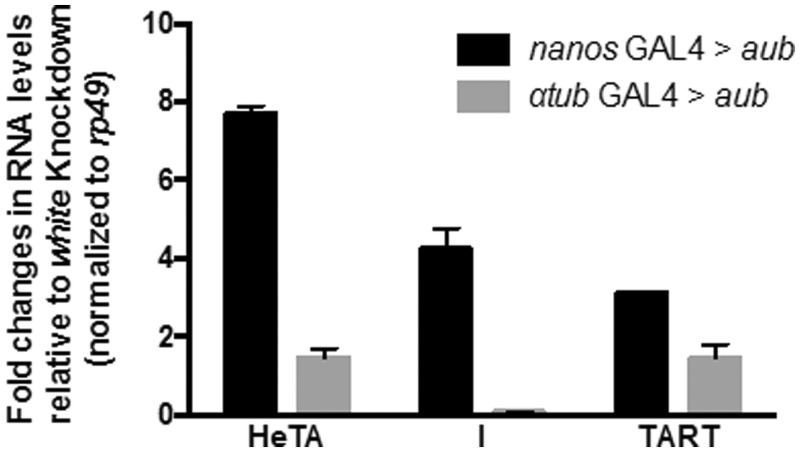
If the silencing exerted on pGgIds is released when knockdown of aub is driven by nos-Gal4 and not αtub-Gal4, then, the silencing exerted on endogenous TE should also be differently affected in these mutant backgrounds. Quantitative RT–PCR was performed with specific primers targeting germline TEs: HeT-A, I-element, and TART. Similarly to the results obtained with Idefix sensors, a higher increase of HeT-A, I and TART RNAs was observed in nos-Gal4 > aub than in αtub-Gal4 > aub ovaries (Figure 3). These findings also indicate that knock down of aub after Stage 3 of oogenesis fails to release HeT-A, I and TART silencing.
Figure 3.
Knockdown of aub driven by either nos-Gal4 or αtub-Gal4 driver has different impact on germline transposon de-silencing. Relative expression levels of the indicated TEs upon knockdown of aubergine using either nos-Gal4 or αtub-Gal4 driver. Each knockdown is normalized to white knockdown controls and to rp49. n = 3 biological replicates. Error bars represent SD.
Thus, our findings suggest that a spatio-temporal regulation of components of the piRNA pathway exists throughout oogenesis. Genes such as aub, spn-E and vasa are only necessary during the first stages of oogenesis up to Stage 3 to mediate silencing. Genes such as piwi, ago3 and mael are needed to silence piRNA targets both at early stages, within the germarium, and constantly thereafter.
Germline repression of Idefix-sensors is highly impaired in dividing germline cells in the germarium
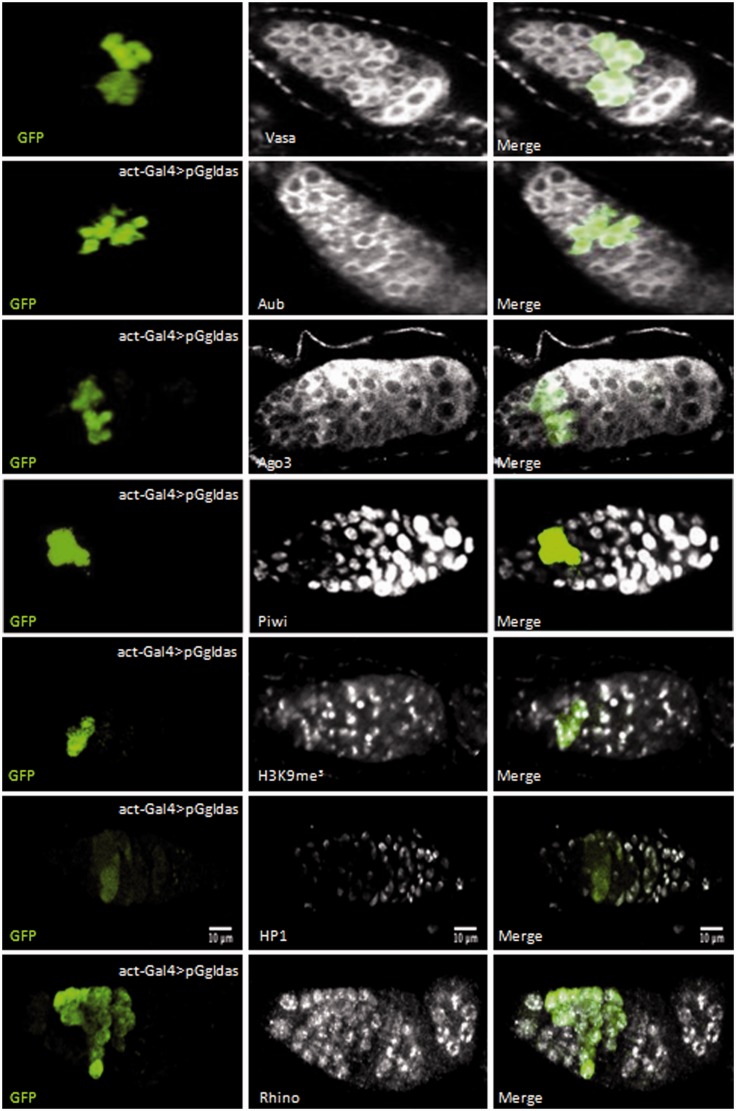
Although piRNA silencing of Idefix-sensor transgenes is efficient in the germline nurse cells, we observed that a small patch of cells located at the anterior tip of the germarium presented GFP fluorescence in ovaries in which the pGgIdas transgene was under control of the actin-Gal4 driver (Figure 1B, line 2). Co-immunofluorescence experiments revealed that GFP protein expressed from pGgIdas co-localized with the germline marker Vasa in the germarium (Figure 4, line 1). Upon examination of more than 100 germaria, co-localization of GFP and Vasa was only observed in dividing germline cysts of 4, 8 or 16 cells.
Figure 4.
Idefix silencing is reduced in the dividing germline cysts of the germarium. Immunodetection of Vasa, Aub, Ago3, Piwi, H3K9me3, HP1 and Rhino proteins (white) and GFP (green) in germaria of ovaries expressing the pGgIdas transgene under the control of the act-Gal4 driver. Germaria are oriented with anterior to the top or left. GFP signal is specifically observed in 4-, 8- and 16-cell germline cysts in which Piwi does not accumulate. Experiments were performed on pGgIdas lines as1 and as2.
A possible explanation for the defect in silencing of antisense Idefix-sensors in dividing germline cysts is that the piRNA pathway is less efficient in these cells. Thus, we performed immunostaining experiments to examine the expression of major actors of the piRNA pathway. Aub- and Ago3-antibodies revealed that both proteins were expressed throughout the germarium including in cysts expressing GFP from the Idefix-sensors (Figure 4, lines 2 and 3). We then examined Piwi whose expression has been reported to be weak in the germarium (27,28). In contrast to Aub and Ago3, although the Piwi antibody revealed the presence of Piwi protein in GSCs at the very anterior end of the germarium, as well as later in germarial region 2b, Piwi protein was barely detected in dividing germline cysts expressing GFP from the Idefix-sensors in more than 150 germaria examined (Figure 4, line 4 and Supplementary Movie S3).
To further characterize these cells, we also examined the presence of H3K9me3 and HP1 in early germline cells. Both have been recently reported to be present in Repressive Chromatin Centers (RCCs) which are required in the germline for piRNA production (29). We performed immunostaining experiments with an H3K9me3 antibody and observed, as Rangan et al. (29), a signal in prominent and discrete foci from the cystoblast to the late stages of germarial development (Figure 4, line 5). In contrast, HP1 signal was only detected at a low level at early stages in the germarium including in cells in which pGgIdas repression is inactive (Figure 4, line 6 and Supplementary Movie S4). Finally, we examined the expression of the Drosophila HP1 homolog, Rhino, required for piRNA cluster transcription and piRNAs production (30). Immunostaining experiments revealed that Rhino is uniformly expressed all along the germarium including the dividing cysts (Figure 4, line 7). Thus, although Aub, Ago3 and Rhino are expressed and H3K9me3 detected in RCCs, silencing of the Idefix-sensors is weak in dividing germline cysts that exhibit low levels of Piwi and HP1 proteins.
A post-transcriptional control represses Piwi in the dividing cysts
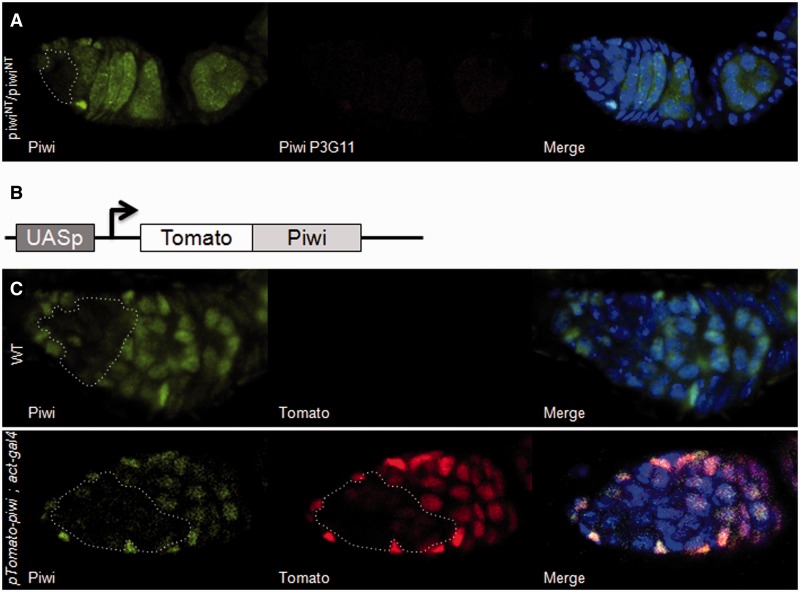
Piwi localization in germline and follicle cells is predominantly nuclear. We examined Piwi localization in mutant flies, piwiNT, in which Piwi nuclear accumulation is prevented (31). A polyclonal Piwi antibody revealed Piwi deleted from its N-terminus in the cytoplasm. The signal was intense in all the germline with the exception of the dividing cysts, where only a very faint signal similar to the wild-type Piwi signal could be detected (Figure 5A, left) (31). Thus, endogenous Piwi expression is highly reduced in dividing cysts.
Figure 5.
The weak expression of piwi in the dividing cysts is due to a post-transcriptional and/or translational repression. (A) Immunodetection of Piwi in germaria of piwiNT mutant ovaries. Polyclonal anti-Piwi (left, green) is directed against the central part of the Piwi protein. Monoclonal anti-Piwi Nter P3G11 (red) is directed against the amino-terminal part of Piwi which is deleted in the piwiNT mutant. As expected for a piwiNT mutant, no signal is revealed by the monoclonal antibody. In constrast, Piwi proteins are detected by the polyclonal antibody in the germ cells except within the dividing cysts. (B) Molecular structure of the pTomato-piwi transgene. (C) Tomato fluorescence is shown as a readout for pTomato-piwi expression. Endogenous Piwi proteins are detected using the polyclonal anti-Piwi (right). The expression profile of endogenous and transgenic Piwi is presented on the left and middle, respectively. When no driver is introduced in the line (upper panel), no Tomato fluorescence is detected in the germline. When pTomato-piwi is driven by actin-Gal4 in the germline (lower panel), Tomato fluorescence is revealed in the germline but remains undetected within the dividing cysts. DNA was counterstained with DAPI (blue).
We then speculated whether Piwi signal could be recovered in these cells if piwi was placed under the control of the heterologous promoter UASp. We established a transgenic line with a piwi cDNA fused to the Tomato fluorescent protein gene under the control of UASp (Figure 5B and ‘Materials and Methods’ section). When driven by actin-Gal4 to achieve germline expression, Tomato-Piwi fluorescence was detected, like endogenous wild-type Piwi, in the germ cells except the dividing cysts, in which the signal was faint (Figure 5C). Given that actin-Gal4 is active in these cells as illustrated Figure 4, we concluded that post-transcriptional and/or translational repression of Piwi exists during this short window of germline development.
Germline repression of a transgenic model mimicking the P-element repression is also impaired in dividing germline cells in the germarium
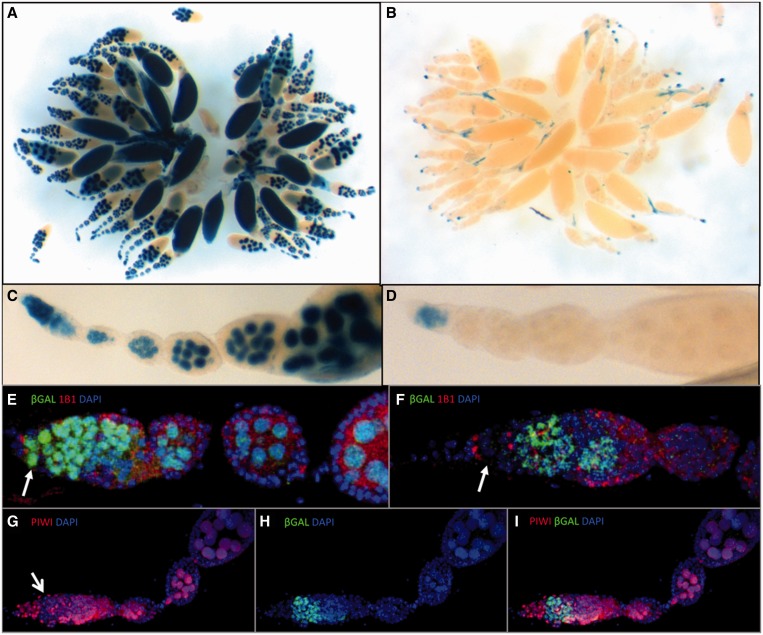
To determine whether the silencing defect in dividing germline cells is unique to Idefix-sensors, we investigated the silencing exerted on another TE, the P-element, a DNA transposon. P repression has been shown to be primarily established by P copies inserted in sub-telomeric heterochromatin loci (TAS) (32), which are strong piRNA-producing loci (2). Indeed, a single defective P-element inserted in the TAS of the X-chromosome can repress the activity of 80 P copies in trans (33). A transgenic model that mimics P repression was previously established using P-lacZ enhancer-trap transgenes (34). Telomeric P-lacZ inserted in TAS can repress a homologous P-lacZ in trans, irrespective of the genomic location of the latter (35,36). This trans-silencing effect (TSE) is restricted to the germline. Incomplete repression is frequently observed in the germarium and occasionally during later stages of oogenesis which results in an ‘on’ or ‘off’ target expression between egg chambers (variegation). However, we found that with some combinations of silencer-target transgenes, incomplete repression was strictly restricted to the germarium. The BC69 P-lacZ enhancer trap (18) for example, showed strong lacZ expression in all the germ cells of the ovarioles, including the GSCs (Figure 6A, C and E). When this transgene was submitted to trans-silencing mediated by a maternally-inherited telomeric P-FRT-white transgene (RS3) inserted in the TAS of the 3R chromosomal arm, complete repression was observed at all stages except for in the germarium (Figure 6B, D and F). Co-immunostaining experiments using 1B1, a marker for the GSC spectrosome and the branched fusomes of dividing germline cysts, and anti-β-galactosidase antibodies, showed that repression occurs in GSCs and at later stages outside of the germarium, but that dividing germline cysts in the middle of the germarium escape strong repression (Figure 6F). Finally, co-immunostaining experiments, using anti-Piwi and anti-β-galactosidase antibodies, revealed that impaired repression in germline cysts in the germarium correlated with low Piwi staining (Figure 6G–I).
Figure 6.
Trans-silencing in the female germline associated to P-element repression is strongly weakened in dividing cysts in the germarium. (A, C, E) Ovaries from BC69 females carrying one copy of a P-lacZ enhancer-trap inserted into the vasa gene. (B, D, F–I) Ovaries from females produced by the cross of BC69 males with RS3 females, which carry a P-FRT-white silencer transgene in the TAS of the 3R chromosomal arm. (A–D) X-gal staining. BC69 females carrying a P-lacZ transgene which contains the sequence for a NLS (Nuclear Localization Signal) fused to the lacZ coding sequence shows nuclear staining (A, C). βgalactosidase is strongly reduced in the presence of the TAS-associated silencer transgene except in germaria (B, D). (E, F) Double immunostaining of a germarium for 1B1 (red), which marks the spectrosomes of GSCs (arrows), and β-galactosidase (green). DNA was counterstained with DAPI (blue). In the presence of the TAS-associated silencer transgene (F), β-galactosidase expression is incompletely repressed in the region of the dividing germline cysts just next to the GSCs. (G–I) Double immunostaining of a germarium for Piwi (red) and β-galactosidase (green). DNA was counterstained with DAPI (blue). Incomplete repression correlates with weak Piwi staining in the germarium (marked by an arrow in G).
Overall, our results indicate that two different types of TEs (the Idefix retrotransposon and the DNA-based P-element) may escape piRNA-mediated repression during oogenesis in a window when germline cysts divide within germarial region 2a. This stage-specific release from piRNA-mediated repression is probably made possible, at least in part, through reduced Piwi expression at these stages. We propose to call this region the ‘Piwiless pocket’ or Pilp.
DISCUSSION
Drosophila female germline cells have the ability to repress Idefix via the piRNA pathway even though the Idefix promoter is not active in these cells
The Idefix promoter is active in follicular somatic cells and inactive in other somatic cells and in the germline (37), but Idefix repression mechanisms exist in all three tissue types. In follicular cells, the linear piRNA pathway is responsible for silencing Idefix, while in other somatic tissues we have shown that PTGS and TGS cooperate to silence Idefix reporter transgenes (17). We now show that the presence of an Idefix fragment allows silencing of engineered transgenes in the germline via the piRNA pathway. Since numerous Idefix copies are present in heterochromatic piRNA clusters (2,38), which produce piRNAs complementary to Idefix mRNAs, these are likely responsible for specific recognition of the transgene transcripts (37). Thus, the huge reservoir of multiple TEs constituting heterochromatin may protect the germline by anticipating sudden activation of a TE promoter that previously displayed somatic specificity, or from newly-incoming homologous TEs that have invaded the species by horizontal transfer (39).
Idefix piRNA-mediated silencing involves two categories of genes with different temporal requirement throughout oogenesis
Since the silencing that targets Idefix-reporters in the germline is removed once the targeted sequence is lost, we used these genetic tools to dissect spatio-temporal requirements for piRNA-mediated silencing in Drosophila germline.
In contrast to previous studies in which components of the piRNA pathway in the germline were disrupted by expressing RNAi using nos or MTD driver that are both active in the GSC (40–42), we decided to compare piRNA-mediated silencing when two drivers whose activity patterns differ during oogenesis, nos and αtub, are used (Figure 2A, upper panel). This comparison allowed the identification of two categories of piRNA components distinguished according to their temporal requirement. The activity of one of them includes Piwi, Ago3 and Mael and is needed in germline cells to repress Idefix-sensors continuously during oogenesis. Indeed depletion of these proteins within the germarium and after Stage 6 of oogenesis as a result of nos-Gal4 activity, or as of Stage 3 of oogenesis following αtub activity, leads to sensor-transgene desilencing (Figure 3, rows 1, 2 and 3). Another category of proteins, including Aub, Vasa and Spn-E, is required for silencing only in the earliest stages of germline development, before Stage 3, for efficient TE silencing all along the ovariole. If their depletion occurs after the germarium stage following αtub driver activity, Idefix-sensor silencing is never released. Idefix sensors are suitable tools to follow temporal requirements of piRNA pathway components because of their easy GFP read-out. However, we also reported that the silencing of endogenous TEs, HeT-A, I-element and TART, is differently affected by nos> and atub > aub knock down. As observed with Idefix-sensors, these TEs remain silenced when aub is depleted after the germarium stage whereas their silencing is released if aub depletion occurs as from the GSC. Nevertheless, the sensitivity of silencing to piRNA component knocked-down may differ between TEs. Several observations made by others indicated that loss of Spn-E has more severe effects on several endogenous TEs than loss of Aub (43,44). Here, we found that Idefix-sensor release is weaker when Spn-E is depleted than Ago3, Aub or Mael. Overall, our observations are further evidence of the complexity of TE silencing in the germline.
Interestingly, RNAi knockdown of aub, vasa and spn-E genes with both the early nos-Gal4 and the later αtub-Gal4 driver resulted in sterility. This is consistent with the axis patterning defects previously characterized for mutations in these genes (45). Hence, loss of function of this category of genes after Stage 3 of oogenesis impairs fecundity, but does not impair Idefix-sensor silencing nor the silencing of some TEs tested in this study such as HeT-A, I-element and TART repression. This suggests that defects in oogenesis due to aub, vasa and spn-E RNAi-depletion after Stage 3 might not be caused by transposition of TEs.
The perinuclear structure, the nuage, is involved in TE silencing (46,47). Many genes have been reported to be required for its assembly including aub, vasa and spn-E. The early requirement for these genes for Idefix-sensor silencing might be explained by their role at the initiation phase of nuage formation. Immunostaining experiments performed with an Ago3 antibody revealed that, if aub and vasa are knocked down after Stage 3 of oogenesis (αtub-Gal4 driving RNAi), the nuage is still observed around the germline nuclei of late-stage egg chambers (although fainter with vasa RNAi, see Supplementary Figure S5). Thus, it is conceivable that aub, vasa and spn-E function at the initiation phase of nuage formation. Once established, the nuage (or a sufficient structure) may persist throughout oogenesis independently of the factors that initiated its assembly, and thereby assure TE silencing. Alternatively, could an heritable silenced state be established on TEs before Stage 3 when aub, vasa or spn-E are active owing to the non-functional αtub driver? Why would this silenced state not be established when piwi, ago3 and mael are knocked down by the same driver? Further investigations are needed at this stage to understand the roles played by these components during oogenesis.
Our present data raise another important issue. In the ping-pong cycle of piRNA production, Ago3-bound-piRNAs have been shown to pair with Aub-bound piRNAs (2). Thus, the ping-pong cycle would be expected to collapse when either of the two PIWI proteins is depleted. However, Idefix-sensors are derepressed in αtub > Ago3-RNAi and not in αtub > aub-RNAi ovaries. In their study, Brennecke et al. (2007) showed that self-complementarity also occurs for both Aub- and Ago3-bound piRNA 5′-ends. Furthermore, Zhang et al. reported that both heterotypic and homotypic piRNA ping-pongs exist (48). The findings of our study support the existence of homotypic Ago3-Ago3 ping-pong for Idefix-sensor repression in ovaries depleted for Aub after Stage 3 of oogenesis. Taken together, our results suggest that aub, vasa and spn-E are necessary for an initial step of the piRNA pathway, possibly nuage formation, while piwi, ago3 and mael are required continuously for piRNA-mediated silencing.
TEs can escape host piRNA silencing in the ‘Piwiless pocket’ during early oogenesis
A detailed analysis of the repression exerted on Idefix-sensor transgenes throughout oogenesis revealed that its repression is partially released during a short window within the germarium corresponding to cystocytes undergoing mitotic divisions to form the interconnected 16-cell germline cysts. While pGgIdas silencing is largely absent in germline cysts of 4–16 cells, pGgIds repression is never released in these cells. Since silencing of pGgIdas is weaker than that of pGgIds as revealed by occasional expression along the ovariole, it is likely that pGgIdas expression in the dividing germline cysts corresponds to a decrease in the efficiency of the silencing pathway. Accordingly, we found that, although aub and ago3 are both constantly expressed in the germline, piwi expression is low in dividing germline cysts, but is clearly detected earlier in GSCs, and later, from region 2b of the germarium onwards. Our data further indicated that this decrease in Piwi is not due to a transcriptional regulation exerted on its own promoter but rather to a post-transcriptional repression exerted specifically in these cells. To elucidate at the molecular level why the Piwi protein level is lower even when it is expressed from the pTomato-piwi transgene, we are currently investigating other differences in piRNA-depending factors within and outside the dividing cysts.
If, as suggested above, piRNA silencing is weak in dividing germline cysts, Idefix would not be the only TE whose repression is affected in these cells. In fact, we found that germline cysts of 4–16 cells are also impaired for a homology-dependent silencing of the P-element, a transposon whose repression is established in the germline primarily by P copies inserted in subtelomeric piRNA-producing heterochromatin. P-element repression was found to be efficient earlier in GSCs and cystoblasts. Thus, although our experiments were performed on transgenes, our data suggest that repression of both the P-element and Idefix is impaired specifically in dividing germline cysts in which Piwi expression is low (the ‘Piwiless pocket’ or Pilp). It has to be noticed that, in a recent article, Shpiz et al. observed similar results with the retroelement HeT-A (44). Endogenous HeT-A copies as well as HeT-A-LacZ constructs are silenced in wild-type ovaries by the piRNA-mediated pathway. In homozygous piwi/piwi mutants, their expression is recovered along the whole ovariole. Interestingly, in heterozygotes piwi/+ females, their silencing is partially released giving rise to HeT-A or HeT-A-LacZ expression in germarial cysts only. The restricted expression of HeT-A in these cells could be due to an increased weakness of the piRNA pathway resulting from the heterozygous piwi/+ background in the Pilp, as seen with P and Idefix transgenes.
Since in the Pilp we observed RCCs, which present the H3K9me3 repressive mark and HP1a, but in a lower amount than in the rest of the ovarioles, it is possible that, in the dividing cysts, the absence of Piwi prevents H3K9me3 from recruiting HP1a. The activity of the piRNA clusters within RCCs would then be affected and TE silencing released or decreased. Further characterization of the Pilp will certainly help address more precisely how Piwi functions in germline transposon silencing, in particular in these few cells of the germarium.
Overall, our data strongly suggest that a window exists during early stages of oogenesis, the Pilp, during which TEs within germline cells can escape from host restraint and transpose. The fact that this window corresponds to dividing germline cysts may provide advantages for both TE regulation and genome dynamics. Since this phenomenon occurs in the germline, transmission of new, potentially beneficial, TE genomic insertions to the next generation is assured. Conversely, since TE derepression does not occur in GSCs, lethal or highly damaging mobilization events will not be produced in these cells and the potential to produce new viable germline cysts will be maintained. In addition, the existence of a Pilp in the germline may explain how sporadic reactivation of TEs occurs (49,50). Overall, these findings provide insight into the reasons for the highly successful invasion of genomes by TEs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-aid from Clermont University, the Centre National pour la Recherche Scientifique (CNRS), the Institut National pour la Santé et la Recherche Médicale (INSERM); This work was supported by the Ministry of Education (to J.D.); the Région Auvergne [Nouveau chercheur to E.B.]; and European Union (to P.P.); and Agence pour la Recherche contre le Cancer (ARC to J.D.); the Ligue régionale contre le Cancer (to C.V.). Funding for open access charge: INSERM U1103, CNRS 6293.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are very grateful to Anne Marie Pret and Aline Probst for critical review of the manuscript, members of our team for helpful discussions, M. Miller, F. Pellissier for technical assistance, Flyfacility for transgenic lines (www.Fly-facility.com), the Bloomington Drosophila Stock Center for flies, flybase and flyfacility. The authors thank J. Brennecke, W. Theurkauf and M. Siomi for antibodies.
REFERENCES
- 1.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 2.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 3.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 4.Sienski G, Donertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and Maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang SH, Elgin SC. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc. Natl Acad. Sci. USA. 2011;108:21164–21169. doi: 10.1073/pnas.1107892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donertas D, Sienski G, Brennecke J. Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex. Genes Dev. 2013;27:1693–1705. doi: 10.1101/gad.221150.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohtani H, Iwasaki YW, Shibuya A, Siomi H, Siomi MC, Saito K. DmGTSF1 is necessary for Piwi-piRISC-mediated transcriptional transposon silencing in the Drosophila ovary. Genes Dev. 2013;27:1656–1661. doi: 10.1101/gad.221515.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King RC. The meiotic behavior of the Drosophila oocyte. Int. Rev. Cytol. 1970;28:125–168. doi: 10.1016/s0074-7696(08)62542-5. [DOI] [PubMed] [Google Scholar]
- 9.Spradling AC. Germline cysts: communes that work. Cell. 1993;72:649–651. doi: 10.1016/0092-8674(93)90393-5. [DOI] [PubMed] [Google Scholar]
- 10.Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116:817–829. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- 11.Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pane A, Wehr K, Schupbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev. Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pek JW, Lim AK, Kai T. Drosophila maelstrom ensures proper germline stem cell lineage differentiation by repressing microRNA-7. Dev. Cell. 2009;17:417–424. doi: 10.1016/j.devcel.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 14.Vagin VV, Klenov MS, Kalmykova AI, Stolyarenko AD, Kotelnikov RN, Gvozdev VA. The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol. 2004;1:54–58. [PubMed] [Google Scholar]
- 15.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desset S, Meignin C, Dastugue B, Vaury C. COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics. 2003;164:501–509. doi: 10.1093/genetics/164.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dufourt J, Brasset E, Desset S, Pouchin P, Vaury C. Polycomb group-dependent, heterochromatin protein 1-independent, chromatin structures silence retrotransposons in somatic tissues outside ovaries. DNA Res. 2011;18:451–461. doi: 10.1093/dnares/dsr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemaitre B, Ronsseray S, Coen D. Maternal repression of the P element promoter in the germline of Drosophila melanogaster: a model for the P cytotype. Genetics. 1993;135:149–160. doi: 10.1093/genetics/135.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dennis C, Zanni V, Brasset E, Eymery A, Zhang L, Mteirek R, Jensen S, Rong YS, Vaury C. ‘Dot COM’, a nuclear transit center for the primary piRNA pathway in Drosophila. PLoS One. 2013;8:e72752. doi: 10.1371/journal.pone.0072752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Ni JQ, Liu LP, Binari R, Hardy R, Shim HS, Cavallaro A, Booker M, Pfeiffer BD, Markstein M, Wang H, et al. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics. 2009;182:1089–1100. doi: 10.1534/genetics.109.103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 25.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 26.Yang Z, Ebright YW, Yu B, Chen X. HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 2006;34:667–675. doi: 10.1093/nar/gkj474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- 28.Zamparini AL, Davis MY, Malone CD, Vieira E, Zavadil J, Sachidanandam R, Hannon GJ, Lehmann R. Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila. Development. 2011;138:4039–4050. doi: 10.1242/dev.069187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rangan P, Malone CD, Navarro C, Newbold SP, Hayes PS, Sachidanandam R, Hannon GJ, Lehmann R. piRNA production requires heterochromatin formation in Drosophila. Curr. Biol. 2011;21:1373–1379. doi: 10.1016/j.cub.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klenov MS, Sokolova OA, Yakushev EY, Stolyarenko AD, Mikhaleva EA, Lavrov SA, Gvozdev VA. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc. Natl Acad. Sci. USA. 2011;108:18760–18765. doi: 10.1073/pnas.1106676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ronsseray S, Lehmann M, Nouaud D, Anxolabehere D. The regulatory properties of autonomous subtelomeric P elements are sensitive to a Suppressor of variegation in Drosophila melanogaster. Genetics. 1996;143:1663–1674. doi: 10.1093/genetics/143.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marin L, Lehmann M, Nouaud D, Izaabel H, Anxolabehere D, Ronsseray S. P-Element repression in Drosophila melanogaster by a naturally occurring defective telomeric P copy. Genetics. 2000;155:1841–1854. doi: 10.1093/genetics/155.4.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roche SE, Rio DC. Trans-silencing by P elements inserted in subtelomeric heterochromatin involves the Drosophila Polycomb group gene, Enhancer of zeste. Genetics. 1998;149:1839–1855. doi: 10.1093/genetics/149.4.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Josse T, Maurel-Zaffran C, de Vanssay A, Teysset L, Todeschini AL, Delmarre V, Chaminade N, Anxolabehere D, Ronsseray S. Telomeric trans-silencing in Drosophila melanogaster: tissue specificity, development and functional interactions between non-homologous telomeres. PLoS One. 2008;3:e3249. doi: 10.1371/journal.pone.0003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Josse T, Teysset L, Todeschini AL, Sidor CM, Anxolabehere D, Ronsseray S. Telomeric trans-silencing: an epigenetic repression combining RNA silencing and heterochromatin formation. PLoS Genet. 2007;3:1633–1643. doi: 10.1371/journal.pgen.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tcheressiz S, Calco V, Arnaud F, Arthaud L, Dastugue B, Vaury C. Expression of the Idefix retrotransposon in early follicle cells in the germarium of Drosophila melanogaster is determined by its LTR sequences and a specific genomic context. Mol. Genet. Genomics. 2002;267:133–141. doi: 10.1007/s00438-002-0641-1. [DOI] [PubMed] [Google Scholar]
- 38.Desset S, Conte C, Dimitri P, Calco V, Dastugue B, Vaury C. Mobilization of two retroelements, ZAM and Idefix, in a novel unstable line of Drosophila melanogaster. Mol. Biol. Evol. 1999;16:54–66. doi: 10.1093/oxfordjournals.molbev.a026038. [DOI] [PubMed] [Google Scholar]
- 39.Werren JH. Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc. Natl Acad. Sci. USA. 2011;108(Suppl. 2):10863–10870. doi: 10.1073/pnas.1102343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Czech B, Preall JB, McGinn J, Hannon GJ. A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol. Cell. 2013;50:749–761. doi: 10.1016/j.molcel.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Handler D, Meixner K, Pizka M, Lauss K, Schmied C, Gruber FS, Brennecke J. The genetic makeup of the Drosophila piRNA pathway. Mol. Cell. 2013;50:762–777. doi: 10.1016/j.molcel.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petrella LN, Smith-Leiker T, Cooley L. The Ovhts polyprotein is cleaved to produce fusome and ring canal proteins required for Drosophila oogenesis. Development. 2007;134:703–712. doi: 10.1242/dev.02766. [DOI] [PubMed] [Google Scholar]
- 43.Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shpiz S, Olovnikov I, Sergeeva A, Lavrov S, Abramov Y, Savitsky M, Kalmykova A. Mechanism of the piRNA-mediated silencing of Drosophila telomeric retrotransposons. Nucleic Acids Res. 2011;39:8703–8711. doi: 10.1093/nar/gkr552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pek JW, Kai T. A role for vasa in regulating mitotic chromosome condensation in Drosophila. Curr. Biol. 2011;21:39–44. doi: 10.1016/j.cub.2010.11.051. [DOI] [PubMed] [Google Scholar]
- 46.Nagao A, Sato K, Nishida KM, Siomi H, Siomi MC. Gender-Specific Hierarchy in Nuage Localization of PIWI-Interacting RNA Factors in Drosophila. Front. Genet. 2011;2:55. doi: 10.3389/fgene.2011.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development. 2003;130:859–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Z, Xu J, Koppetsch BS, Wang J, Tipping C, Ma S, Weng Z, Theurkauf WE, Zamore PD. Heterotypic piRNA Ping-Pong requires qin, a protein with both E3 ligase and Tudor domains. Mol. Cell. 2011;44:572–584. doi: 10.1016/j.molcel.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vieira C, Biemont C. Transposable element dynamics in two sibling species: Drosophila melanogaster and Drosophila simulans. Genetica. 2004;120:115–123. doi: 10.1023/b:gene.0000017635.34955.b5. [DOI] [PubMed] [Google Scholar]
- 50.Yang HP, Hung TL, You TL, Yang TH. Genomewide comparative analysis of the highly abundant transposable element DINE-1 suggests a recent transpositional burst in Drosophila yakuba. Genetics. 2006;173:189–196. doi: 10.1534/genetics.105.051714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.