Abstract
Many bacterial, viral and parasitic pathogens undergo antigenic variation to counter host immune defense mechanisms. In Plasmodium falciparum, the most lethal of human malaria parasites, switching of var gene expression results in alternating expression of the adhesion proteins of the Plasmodium falciparum-erythrocyte membrane protein 1 class on the infected erythrocyte surface. Recombination clearly generates var diversity, but the nature and control of the genetic exchanges involved remain unclear. By experimental and bioinformatic identification of recombination events and genome-wide recombination hotspots in var genes, we show that during the parasite’s sexual stages, ectopic recombination between isogenous var paralogs occurs near low folding free energy DNA 50-mers and that these sequences are heavily concentrated at the boundaries of regions encoding individual Plasmodium falciparum-erythrocyte membrane protein 1 structural domains. The recombinogenic potential of these 50-mers is not parasite-specific because these sequences also induce recombination when transferred to the yeast Saccharomyces cerevisiae. Genetic cross data suggest that DNA secondary structures (DSS) act as inducers of recombination during DNA replication in P. falciparum sexual stages, and that these DSS-regulated genetic exchanges generate functional and diverse P. falciparum adhesion antigens. DSS-induced recombination may represent a common mechanism for optimizing the evolvability of virulence gene families in pathogens.
INTRODUCTION
The pathogenicity of Plasmodium falciparum malaria is linked to the parasite’s capacity to modify infected erythrocyte surfaces to cause sequestration in various host organs, thus allowing the parasites to avoid clearance by the spleen (1). Sequestration of infected erythrocytes is mediated by binding of parasite-encoded Plasmodium falciparum-erythrocyte membrane protein 1 (PfEMP1) adhesion antigens to host endothelial receptors (2). PfEMP1 are major, immunodominant targets of protective antibody mediated immunity (3). To escape antibody mediated immunity, which will tend to be antigen variant-specific, parasites have evolved to contain a repertoire of ∼60 var genes encoding different PfEMP1 variants (1). Inter-genome comparisons have revealed an immense variation in the global pool of var gene sequences (4,5). This opposing evolutionary pressure on PfEMP1 molecules to generate immune-evading antigenic diversity, while maintaining receptor binding capacity, has resulted in highly organized genetic structures harbouring alternating loci with relatively conserved and extremely diverse sequences. Var genes can be classified into three major groups (A–C) on the basis of their 5′ upstream sequences (upsA, B and C) and chromosomal location and orientation (6). The encoded PfEMP1 consist of restricted compositions of different subtypes of Duffy Binding Like (DBL) and cysteine-rich interdomain region (CIDR) domains, which are associated with the ups type of the genes (7), specific cytoadhesion properties (i.e. human receptor specificities (2) and, ultimately, disease outcome (8,9).
This genetic organization and functional compartmentalization is thought to be maintained not only through selection, but also as a consequence of a restricted recombination imposed by e.g. the different chromosomal positioning and orientation of the centromeric upsC var genes and the inversely oriented subtelomeric upsA and upsB genes (6,10). There is general consensus from experimental and sequence analysis studies that var genes are subject to ectopic recombination causing gene conversion (11–15). Studies on experimental genetic crosses have presented evidence that recombination between var genes on heterologous chromosomes occurs more frequently than expected from the overall estimated rate of meiotic crossing-over (12,14,16,17), and data from these and other studies indicate that chimeric var genes are products of recombination between isogenous var paralogs (i.e. var genes originating from the same haploid P. falciparum genome) (12,14,18,19). These data suggest that var recombination is preferentially initiated in the sexual stages, although var genes have also been documented to recombine during the mitotic divisions of asexual blood-stage parasites (18,20,21). However, as noted by Bopp et al. in 2013, it appears unlikely that var chimeras generated during the sexual reproduction and by mitotic recombination occurring in asexual blood-stages are products of the same recombination mechanism. It has been proposed that clustering of chromosome-ends into bouquet-like configurations, observed in the sexual parasite forms, facilitates genetic exchanges between subtelomeric var genes by securing the necessary proximity (12). The identification of inducing and regulating factors that can explain how and when var recombination is initiated is essential to understand how the parasite is able to employ a strategy of immune evasion through var gene diversification.
Here we document a tight association between predicted DNA secondary structures (DSS) and var gene recombination sites in four chimeric var genes, known recombination hotspots and boundaries of protein domains. Altogether these data indicate that DSS serve as var recombination inducers and suggest a hitherto unseen arrangement to facilitate ordered recombination.
MATERIALS AND METHODS
P. falciparum culture
P. falciparum blood-stage progeny clones (X2, X4, X6, X8, X10, X11, X12, X30, X44, X47, X50, X56 and X67) from the HB3x3D7 cross (16) were cultured as described (22).
Pulsed field gel electrophoresis and quantitative real-time polymerase chain reaction
P. falciparum chromosomal DNA blocks were prepared as described (23). Pulsed field gel electrophoresis (PFGE) was performed as described (24). Separated chromosomes were excised from the PFGE gels and DNA purified using spin columns. Primers for 60 3D7 (25,26) and 49 HB3 (27) var genes, and 14 syntenic chromosome marker genes (Supplementary Table S1a), were used in quantitative real-time polymerase chain reaction (QPCR) to determine the chromosomes and var genes contained in each excision (28). QPCR primers (Supplementary Table S1b) targeting 5′and 3′ends of 49 3D7 and 29 HB3 var genes were applied to progeny genomic DNA to identify chimeric genes lacking either the 5′ or 3′ end of the original parental genes. Primer amplification efficiencies were validated by QPCR on serial dilutions of genomic DNA. QPCR was performed using the Rotorgene 6000, version 1.7 system (Corbett Research). Reactions were prepared in 20-µl aliquots using the QuantiTect SYBER Green PCR kit (Qiagen) and 1 µM primer concentrations. PCR cycling was 95°C for 15 min, followed by 40 cycles of 95°C for 30 s, 54°C for 20 s and 65°C for 40 s, with final extension at 68°C for 40 s. The cycle threshold was set at 0.025 and all products were authenticated by melting point analysis.
Amplification of var exon 1 for sequencing and restriction fragment length polymorphism analysis
PCR amplification of var exon 1
var exon 1 sequences were amplified using primers listed in Supplementary Table S1c. PCR reactions were done using TaKaRa LA Taq™ polymerase (Fisher) following the manufacturer’s recommendations. Two-step PCR conditions were one cycle of 94°C for 1 min, followed by 33 cycles of 98°C for 10 s and an annealing/extension at 60°C for 5 min. The identity of individually amplified var exon 1 sequences was tested by QPCR using primers against all 3D7 var genes.
RFLP analysis
Seventy-one amplified var exon 1 from 3D7 and seven progeny clones were digested with five different restriction enzymes (Taq I, Eco RI, Hpa I, Hha I and Cla I) and visualized on agarose gels.
Sequencing
Fifty-one amplified var exon 1 sequences (size 5–10 Kb), together with chromosomes 2–4 of progeny X5, were sequenced on a Roche FLX platform using Titanium sequencing chemistry. Amplified var gene sequences from the same progeny clone were pooled in equimolar concentrations (20–50 ng/µl), and then fragmented using Fragmentase (New England Biolabs) to an average size of <1000 bp. DNA fragments were purified using a Minelute column (Qiagen), then build into MID labelled sequencing libraries according to the manufacturer’s guidelines and the Rapid FLX library build kit (Roche). Each library was subsequently subjected to emulsion PCR and sequenced. Post-sequencing data were firstly split by the multiplex identifier (MID) of each library, then de novo assembled by Newbler v2.3 (454 Life Sciences Corp., A Roche Company, Brandford, CT 06405, USA), and finally analysed using BioEdit.
Identification and validation of X4 PFB0010w/PFA0765c chimera
Using QPCR and primer pairs specific for 5′ and 3′ ends of 78 parental var genes, the 5′ but not the 3′end of 3D7 chromosome 2 gene PFB0010w and the 3′ but not the 5′ end of 3D7 chromosome 1 gene PFA0765c could be amplified from genomic DNA of progeny clone X4. To test if the two parental genes, PFB0010w and PFA0765c, had recombined to create a chimeric gene in the X4 progeny clone, the PFB0010w 5′-end forward primer and the PFA0765c 3′-end reverse primer were subsequently used to amplify the possible chimeric sequence. These primers successfully amplified a 5500-bp sequence from genomic DNA of progeny X4, but failed to amplify any product from 3D7 genomic DNA. Sequencing of the 5500-bp PCR product showed a chimeric sequence containing the 5′-end of PFB0010w and the 3′-end of PFA0765c. The gene-specific QPCR primer pairs specific for the 5′ and 3′-end of the X4 PFB0010w/PFA0765c chimera were then tested on PFGE-separated chromosomal DNA from progeny clone X4 together with chromosome marker primers for each of 14 chromosomes. This analysis showed that both ends of the chimeric X4 PFB0010w/PFA0765c gene were contained within the chromosome block containing chromosomes 2–4, and thus confirmed that ectopic recombination had resulted in translocation of the PFA0765 3′-end from chromosome 1 to chromosomes 2–4. To exclude the possibility that PCR amplification had produced an artefact chimeric sequence, we designed PCR primer pairs based on the X4 PFB0010w/PFA0765c chimeric sequence that specifically should amplify regions containing the observed recombination breakpoints. PCR amplification using these primer pairs failed to amplify any products from 3D7 genomic DNA, but successfully amplified sequences of the right sizes from genomic DNA of the X4 progeny clone. Sequencing of these PCR products confirmed the presence of each of the sequences containing the observed recombination breakpoints in the X4 PFB0010w/PFA0765c chimeric sequence. Finally, we were able to extract single sequence reads from whole-genome sequencing data of 3D7 × HB3 progeny (available from Sanger Institute Malaria Program).
Identification and validation of X5 PFA0005w/PFB1055c chimera
Gene mapping of inherited var genes in recombinant progeny clones showed that a central region of the PFA0005w gene located on chromosome 1 in 3D7 mapped to chromosomes 2–4 in progeny clone X5. This indicated that ectopic recombination had resulted in translocation of this PFA0005w-specific sequence from chromosome 1 to chromosomes 2–4. A primer pair specific for the 3′-end of the PFA0005w gene failed to amplify any sequence from genomic DNA of the X5 progeny clone. A forward primer specific for the 5′-end of the PFA0005 gene was then used together with a reverse primer targeting a conserved 3′ end sequence contained by most var genes to amplify the possible chimeric sequence in the X5 progeny. PCR amplification resulted in a ∼5500-bp product, which then was sequenced. The sequenced PCR product showed a chimeric sequence containing the 5′-end of the PFA0005w gene and the 3′-end of the PFB1055c gene located on chromosome 2 in 3D7. To ensure that this sequence did not result from a PCR artefact, we designed primers specific for each of the regions covering the observed recombination breakpoints. PCR amplification using these primer pairs failed to amplify any products from 3D7 genomic DNA, but successfully amplified sequences of the right sizes from genomic DNA of the X5 progeny clone. Sequencing of these PCR products confirmed the presence of each of the sequences containing the observed recombination breakpoints in the X5 PFA0005w/PFB1055c chimeric sequence. Finally, we were able to extract single sequence reads from whole-genome sequencing data of 3D7 × HB3 progeny (available from Sanger Institute Malaria Program and from shotgun sequencing of chromosome 2–4 of X5) covering each of the breakpoints.
Identification and validation of X96 DD2var18/DD2var23 and X98 HB3var10/HB3var14 chimeras
Using BWA mapping software (version 1.2.2) (29) available on the public Galaxy server (usegalaxy.org) (30) Illumina GA II, 75-bp paired-end sequencing reads with at least one read matching one of the published DD2 and HB3 (31) var gene sequences were extracted from whole-genome sequencing data from 18 DD2 × HB3 progeny available from Sanger Institute Malaria Program (ENA accession number ERP000199). Paired-end reads from each progeny were de novo assembled separately using SOAPdenovo (32) (settings: k-mer size of 57 bp and median insert size as determined from the mapping) and compared with parental genes by alignment. This resulted in discovery of two contigs representing chimeric genes X96 DD2var18/DD2var23 (ERS009996) and X98 HB3var10/HB3var14 (ERS009998). Each breakpoint was validated by identification of paired reads, where one read contained the breakpoint and the mate read matched one of the parental genes.
Bioinformatics
DNA 50-mer folding free energy calculations
All gene groups (var genes from six genomes as previously described (31), and all 3D7 CDS divided into var, rif, stevor and other genes) were subjected to a moving window analysis, using a window size of 50 nt and step size of 1 nt. The minimum free energy structure was calculated for all 50-mers using RNAfold v2.1.2 with thermodynamic parameters specifically for folding single-stranded DNA sequences (parameter file dna_mathews1999.par from the ViennaRNA package v2.1.2) (33). GU and lonely pairs were disallowed in the secondary structures. Graphical models of shown 50-mer DSS were drawn using Context fold (34).
Shuffled versions of the gene groups were also analysed as described earlier. The shuffled gene groups were generated by randomizing the nucleotide sequence of each gene, thus producing sequence sets with same gene number, length and nucleotide composition. Density and frequency plots were produced using scripts of Python v2.5 and R v2.8. Plots and statistical tests were generated for the 1%, 2%, 3%, 4% and 5% of the lowest 50-mer folding free energies calculated in the shuffled var genes, to define an energy threshold of 50-mer sequences predicted to form DSS. Calculations based on all five thresholds gave similar results and conclusions, but only the results of the 3rd percentile data (corresponding to an energy threshold of −6.27 kcal/mol) are presented.
Statistical analysis of correlation between recombination sites and predicted DSS
Statistical significance of the correlation between recombination sites and predicted DSS in the four chimeric genes was tested using re-sampling based analysis. The null hypothesis was that recombination sites occurred at random distances from DSS. First the average distance between recombination sites and the nearest DSS in either of the two parental donor sequences was calculated (average distance was 44.46 nucleotides). This value was compared with the corresponding distribution of average distances between randomly chosen locations in the four sets of parental genes and the nearest DSS. Specifically, the procedure was as follows: (i) One random location was selected in each of the two parental genes for a given chimera. (ii) The nearest DSS for this pair of locations was found (i.e. the DSS closest to either of the two random locations in the two parental genes). (iii) Steps 1 + 2 were repeated 13 times over the four parental gene pairs, such that the pairs were sampled the same number of times as for the actual observed chimeric genes (i.e. for each random average, we sampled three random distances from PFB0010w + PFA0765c, four from HB3var10 + HB3var14, one from DD2var23 + DD2var18 and five from PFB1055c + PFA0005w).
(iv) The average of these 13 random DSS-distances was computed. (v) Steps 1–4 were repeated 1 000 000 times. (vi) Finally, the real average distance (44.46) was compared with the resulting distribution of 1 000 000 random average distances. Among these values, 9032 were found to be less than or equal to the real value. The probability of observing a value less than or equal to the real average distance if the null hypothesis is true (the P-value) is therefore 9032/1 000 000 = 0.009032.
Statistical analysis of correlation between predicted DSS and previously defined recombination hotspots
Re-sampling based statistical analysis was performed on each PfEMP1 DBL domain type to test whether the number of DBL sequences containing a DSS very near to the previously defined recombination hotspot, located between the structural sub-domains S2 and S3, was significantly higher than expected for random reasons. Specifically, for each DBL domain type, the number of genes that contained one or more DSS within a window of ±50 nucleotides, surrounding the defined recombination hotspot, was counted. The same analysis was subsequently repeated on 1 000 000 shuffled datasets, in which the position of DSS found in each gene was randomly placed. The number of DSS in the real dataset was then finally compared with the number of DSS in the shuffled dataset to calculate the P-value. A similar procedure was performed within a window of ±50 nucleotides around the other previously defined recombination hotspot (31,35) at the ‘mid-var region’ of PfEMP1 type 1 genes (36) (specifically defined as the point halfway between the 3′-end of the NTS-DBLα-CIDR1 domains and the 5′-end of DBLδ-CIDR2 domains).
Construction of yeast recombination strains
To insert a DSS sequence into a previously described direct-repeat recombination assay strain ML144-8C (37), the KANMX4 selection marker was amplified from yeast genomic DNA (38) using homology and DSS-adapted primers LEU2-U2-KanMX-F (5′-GGATATCGTCCATTCCGACAGCATCGCCAGTCACTATGGCGTGCTGCTAG CGTACGCTGCAGGTCGAC) and PFB1055c-D2-KanMX-R (5′-ACTTTTGCCAGTGGCACCAATCGATGAATTCGAGCTCG), PFB1055cS-D2-KanMX-R (5′-CCTAGCCAGCAATTTCAGGATCGATGAATTCGAGCTCG), PFB1055bc-D2-KanMX-R (5′-TGCCGTTTTCGTCCTTACATCGATGAATTCGAGCTCG), PFB1055bcS-D2-KanMX-R (5′-TGTTGGTTTCCAGGCTTATACATCGATGAATTCGAGCTCG), PFA0765c-D2-KanMX-R (5′-GGTTTTCTCACCAGGTGTTTGATCGATGAATTCGAGCTCG) or PFA0765cS-D2-KanMX-R (5′-GGCTTCCTTGCATATCAGTGCATCGATGAATTCGAGCTCG). KANMX4 marker homology sequence is shown in bold and DSS in italic. The two PCR products were subsequently extended with the full DSS sequence and 50 bp of homology (underscored) to the yeast genomic region of the direct-repeat recombination assay using primers PFB1055c-LEU2-R (5′-ACTATTTCTCATCATTTGCGTCATCTTCTAACACCGTATATGATAATATATGGGTGGCACACAAATGGCACCCTTATCACCACTTTTGCCAGTGGCACCA), PFB1055cS-LEU2-R (5′-ACTATTTCTCATCATTTGCGTCATCTTCTAACACCGTATATGATAATATATTATAGGTCTACCCCGGGCACATCTAGGACCCCTAGCCAGCAATTTCAGG), PFB1055bc-LEU2-R (5′-ACTATTTCTCATCATTTGCGTCATCTTCTAACACCGTATATGATAATATAGGGGACTTGGTCGGCATTTGAGCCGGGCTTTTTGCCGTTTTCGTCCTTAC), PFB1055bcS-LEU2-R (5′-ACTATTTCTCATCATTTGCGTCATCTTCTAACACCGTATATGATAATATACGCTTGGTAGGCGGGTTTCGGCCTTTCGCTGTTGGTTTCCAGGCTTATAC), PFA0765c-LEU2-R (5′-ACTATTTCTCATCATTTGCGTCATCTTCTAACACCGTATATGATAATATAGCACCCTGGTTAGTACCACTAGGTGGGGTGGTTTTCTCACCAGGTGTTTG) or PFA0765cS-LEU2-R (5′-ACTATTTCTCATCATTTGCGTCATCTTCTAACACCGTATATGATAATATAAGAGGCTTCGCGTCTTTGAGGATTGCTGGGGCTTCCTTGCATATCAGTGC) together with primer LEU2-U2-KanMX-F. The two fusion PCR products were separately transformed into yeast strain ML144-8C using the lithium acetate method (39). Transformants were selected on YPD containing 200 µg/ml G418 (Sigma-Aldrich). Correct integration of the PCR products was confirmed by sequencing.
Recombination assay
Direct-repeat mitotic recombination was measured in haploid yeast strains. The procedure for determining mitotic recombination frequencies and their standard deviation was essentially as described before (40), with the following exception: All cultures were grown in liquid synthetic complete medium supplemented with 100 µg/ml adenine (41). Single-strand annealing was distinguished from gene conversion by replica-plating the Leu+ recombinants to synthetic complete medium lacking leucine and uracil after 2 days to score for loss of the URA3 marker. The median frequency for 15 trials was used to determine the recombination rate by the method of the median (41).
RESULTS
Identification of chimeric var genes
To study var gene recombination events, progeny clones of a genetic cross (16) between two P. falciparum clones, 3D7 and HB3, were searched for novel chimeric var genes using three complementary approaches. Var genes consist of a large 3–10-kb polymorphic exon 1 encoding the variable extracellular domains and a conserved 1.3-kb exon 2 encoding the intracellular domain. Fourteen of the cross progeny were analysed using QPCR and primer pairs specific for the 5′ and 3′ ends of exon 1 of 78 parental var genes (this constitutes 72% of all the parental vars). To detect ectopic recombination events between non-allelic var genes, the chromosomal localization of all parental var genes was also mapped in both parents and seven of the progeny clones. This was carried out using var gene and chromosome-specific marker primers in QPCR screens of PFGE-separated chromosomes (28), and the experimental results were subsequently compared against the complete 3D7 genome data available from the PlasmoDB database (http://plasmodb.org/plasmo/) (Supplementary Figure S1). Finally, 120 full-length randomly chosen var exon 1 sequences were PCR amplified and either sequenced (n = 49) or compared for RFLPs (n = 71). In addition to these experiments, whole-genome sequencing data from 18 progeny clones of a second cross (clone HB3 × DD2), available from Sanger Institute, were also searched in an effort to identify other recombinant var chimeras.
Four recombinant var genes were identified in these screens. In two of the 3D7 × HB3 progeny clones, the QPCR analysis indicated that ectopic recombination had occurred between the 3D7 chromosome 1 and 2 var genes. The novel chimeric genes were located on chromosome 1 in progeny clone X4 and chromosome 2 in progeny clone X5. Additional PCR analyses and shotgun sequencing of clone X5 chromosomes 2–4 (not shown) confirmed that both recombinant genes (X5PFA0005w/PFB1055c and X4PFA0765c/PFB0010w) were chimeras resulting from an ectopic recombination between two upsB var genes that had the same orientation and telomeric position, although they were situated on different chromosomes of the 3D7 parent.
PCR experiments confirmed both presence of the two donor genes and the absence of the chimeric genes found in the progeny, in the parental 3D7 genome. Chromosome mapping of var genes in progeny clone X5 showed that this parasite genome only contains HB3 chromosome 1 var genes on its chromosome 1 (Supplementary Figure S1). This indicates that the process of meiotic crossing-over has replaced 3D7 chromosome 1 loci, originally containing the recombining PFA0005w gene, with HB3 DNA after the recombination event. Together with PCR experiments, confirming both the presence of donor genes and the absence of chimeric progeny genes in the parental 3D7 genome, this indicates that the isogenous var recombination creating X5PFA0005w/PFB1055c occurred sometime between the onset of gametogenesis and the meiotic reduction divisions in the ookinetes in the mosquito midgut.
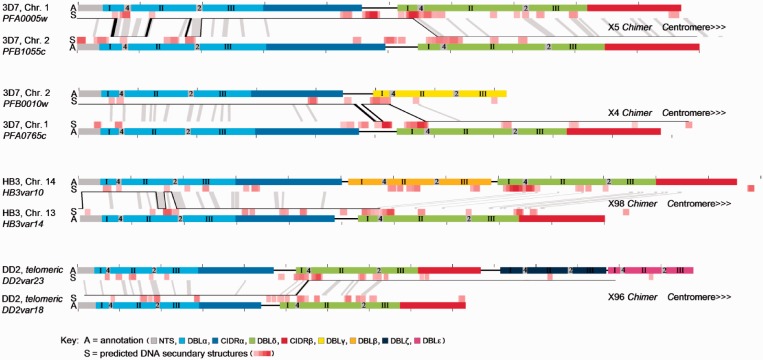
Two additional chimeric var sequences (X96DD2var18/DD2var23 and X98HB3var10/HB3var14) also generated by isogenous recombination between upsB var genes were identified in the analysis of the whole-genome sequencing data resulting from the other genetic cross between parental clones HB3 and DD2. A schematic diagram of the sequenced chimeric var genes showing the recombination breakpoints relative to the parent donor genes is shown in Figure 1 and in high detail in Supplementary Figure S2.
Figure 1.
Schematic diagram of the origin of four novel var chimeric genes. The verified sequences from four chimeric var genes (exon 1) from two 3D7 × HB3 progeny (X5 and X4) and two HB3 × DD2 progeny (X98 and X96) are shown as continuous black lines crossing between domain-annotated parental genes. These are all upsB type var genes, i.e. telomeric genes being transcribed towards the centromere. PfEMP1 are composed of the N-terminal segment (NTS) and different sub-classes of DBL and CIDR domains (coloured). DBL domains contain three structural elements, sub-domains 1–3 (marked I–III) (42,43). Both DBL and CIDR domains can be described as being composites selected from a repertoire of 628 different short semi-conserved homology blocks (HB) (31). Six breakpoints occurred at the boundaries of the structural DBL sub-domains 1–3 marked by HB4 and HB2 (grey boxes ‘2’ and ‘4’) and two breakpoints occurred in low-complexity sequence inter-domain regions. The quasi-palindromes with highest potential to form DSS shown in red (S) are frequently found near recombination breakpoints. Predicted DSS are identified by calculations of folding free energy in 50-mer windows (Supplementary Figure S2) and coloured increasingly intense red with decreasing folding free energy levels from −6.27 kcal/mol. Regions of donor sequence identity of at least 90% over 20 bp are shown in grey shades between genes. 500-bp intervals are marked.
Recombination breakpoint analysis
The recombination patterns of the chimeric genes are similar and give a novel insight into the mechanism of var recombination. All four sequences have between one and five recombination breakpoints and retain open reading frames (Supplementary Figure S2), indicating that they could be expressed as functional proteins. Interestingly, 8 of the 13 breakpoints are located at or close to the boundaries of sequences encoding distinct PfEMP1 tertiary structures (Figure 1) i.e. six recombination breakpoints are located at the boundaries of the three structural sub-domains that make up the conserved fold of DBL domains (42,43), whereas two breakpoints are located in the low-complexity inter-domain region of X4PFA0765c/PFB0010w. Both the region separating DBL sub-domains 2 and 3 and the low-complexity inter-domain region have previously been associated with high frequencies of recombination (31). This indicates that these regions are recombination hotspots per se (i.e. recombination occurs more frequently within these defined regions than elsewhere in the genes) and that recombination is being actively directed to these regions.
The sequences around each breakpoint were closely examined for features possibly relating to their recombinogenic propensity. All breakpoints occurred within highly similar regions between the parental donor sequences, with a minimal required sequence identity of around 20 base pairs with 10% mismatch. Interestingly, numerous short quasi-palindromes (i.e. imperfect inverted repeats separated by very few base pairs) were also observed located near the recombination breakpoints (Supplementary Figure S2). The recombinogenic potential of secondary hairpin or cruciform DNA structures formed by quasi-palindromes has recently attracted considerable interest, with several studies presenting examples of individual recombination sites being marked by the presence of a palindromic DNA sequence [reviewed in (44,45)]. Mitotic recombination analysis in Saccharomyces cerevisiae indicates that recombination breakpoints occur at distances up to several kilobases from an initiating DSS. The highest probability of recombination being in homologous regions closest to the DNA secondary structure (46). This is comparable with the recombination events observed here. If DSS are a primary initiator of var recombination, during the course of evolution, these structures should become concentrated at or near recombination hotspots. To test this hypothesis, we investigated the location of predicted DSS in the donor sequences of the var chimeras as well as within a large sample of sequenced var genes.
DSS locations are associated with var recombination hotspots and PfEMP1 domain borders
The folding free energy of single-stranded DNA was calculated in a sliding 50-mer window across all P. falciparum genes. The var genes were found to harbour a high proportion of particularly low folding free energy 50-mers with the highest likelihood of forming DSS (Supplementary Figure S3 and examples of 50-mer DSS given in Supplementary Figure S4). To visualize the positioning of these sites with the potential to form DSS, the lowest folding free energy DSS were plotted onto the parental donor sequences of the identified var chimeras (Figure 1) and onto 366 full-length domain-aligned PfEMP1 sequences, following a recently published general PfEMP1 domain annotation scheme (31) (Supplementary Figure S5). Plots were generated using energy thresholds corresponding to 1st–5th percentiles of 50-mers in randomized var gene sequences. All plots exhibited similar DSS localization patterns, but only the 3rd percentile data are shown here (folding free energies below −6.27 kcal/mol).
Visual inspection of the four var chimeras indicated a correlation between the identified recombination sites and locations of low folding free energy 50-mers (predicted DSS) in the corresponding parental donor sequences. To investigate this correlation statistically, we performed a re-sampling based analysis (see ‘Materials and Methods’ section), which showed that the average distance between a recombination break site and the nearest corresponding DSS (in either of the donor sequences) was significantly lower than that expected for random associations (P = 0.009032).
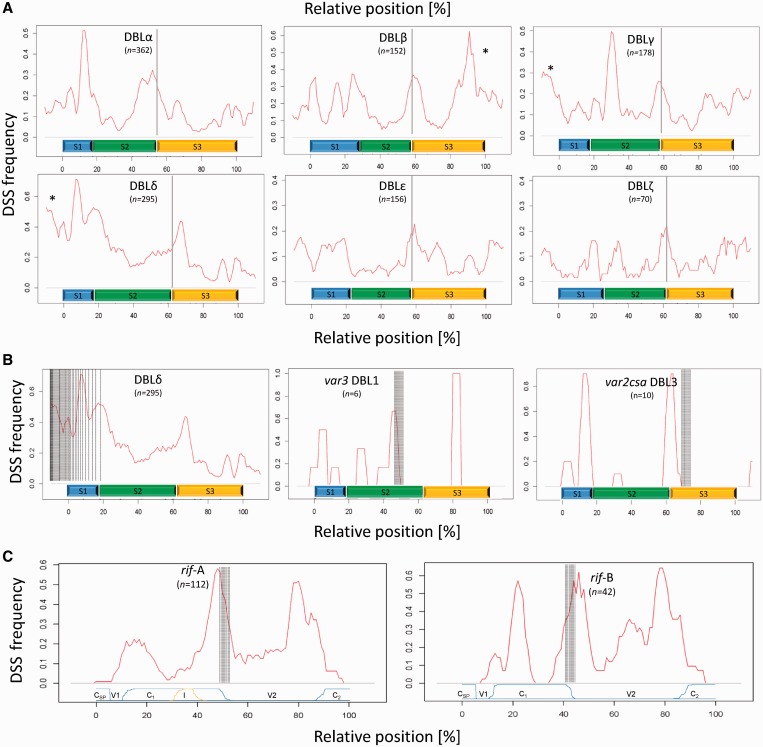
Mapping of low folding free energy 50-mers in 366 domain-aligned PfEMP1 sequences (Supplementary Figure S5) showed that predicted DSS appeared to be non-randomly localized, with a striking tendency to occur at recombination hotspots previously proposed by studies of sequence homology disequilibria within the PfEMP1 family. To investigate this in detail, the frequency of DSS occurrence was plotted along the sequence of each of the main DBL and CIDR domain types (Figure 2 and Supplementary Figure S6). The high frequency of predicted DSS in and around the 5′ end of DBLδ domains is particularly evident. This ‘mid-var gene region’ has been reported to be a site of frequent recombination between the so-called type 1 var paralogs (i.e. DBLα-CIDR1- DBLδ -CIDR2; Supplementary Figure S5) (31,35). A re-sampling based statistical analysis confirmed that the number of var genes containing a DSS within a window of ±50 nucleotides around this ‘mid-var gene region’ (defined as the point halfway between the 3′ end of the CIDR1 domain and the 5′ end of the DBLδ domain of type 1 var genes) was significantly higher than the random expectation (Table 1). Sequence analysis of DBL sub-domains 1–3 has also identified the border between DBL sub-domains 2 and 3 as a hotspot for recombination (31). Figure 2 shows a high frequency of predicted DSS at the borders of DBL sub-domains 2–3, which also were tested to be significantly higher than expected by chance (Table 1). The DSS frequencies at previously in silico defined recombination hotspots in the conserved var2csa (47) and var3 genes (31) were also investigated (Figure 2b) and again found to be associated with high frequencies of DSS.
Figure 2.
Relative location of DSS in selected var gene domains and rif genes. (A) The frequency of predicted DSS (red graph) is shown relative to the position in the PfEMP1 domain types DBLα–ζ. The relative position of DBL sub-domains S1–S3 (blue, green and orange bars) and the previously defined recombination hotspot in DBL domains (vertical grey lines) (31) is shown. In general, peaks in DSS frequency are observed at the recombination hotspot located at the boundary of DBL S2 and S3 and at the end of the domains (indicated by asterisk). The frequency of DSS at the DBL S2–S3 recombination hotspot was found to be significantly higher than expected by chance (Table 1). (B) The association between DSS localization and the ‘mid var’ recombination region (marked by vertical punctuated lines) is particularly evident in the DSS frequency plot of DBLδ domains (left plot). The recombination hotspots (vertical grey lines) defined in var3 DBL1 sub-domain 2 (31) and var2csa DBL3 (47) also co-localize with peaks in DSS frequency (middle and right plots). (C) The frequency of predicted DSS (red graphs) is shown relative to their position in annotated rif-A and B genes. Blue line indicates the relative positions of conserved (Csp = conserved signal peptide, C1 and C2) and variable (V1 and V2) regions. Yellow line indicates position of the 75-bp insert (I) unique to rif-A. In both rif-A and B genes, the highest DSS frequency peaks are found at the border between major conserved (C1) and variable (V2) regions (grey shadow), previously defined as a hotspot for recombination (48).
Table 1.
Test results of correlation between predicted DSS and previously defined recombination hotspots
| Hotspot | Number of sequences | Number of sequences with DSS within ±50 bp of hotspot |
||
|---|---|---|---|---|
| Real | Random [CI] | P | ||
| ‘Mid var’ | 146 | 112 | 36 [15, 66] | <0.000001 |
| DBLα S2–S3 | 362 | 136 | 85 [48, 124] | <0.000001 |
| DBLβ S2–S3 | 152 | 85 | 34 [12, 63] | <0.000001 |
| DBLγ S2–S3 | 178 | 59 | 37 [15, 64] | 0.000109 |
| DBLδ S2–S3 | 295 | 189 | 71 [35, 109] | <0.000001 |
| DBLε S2–S3 | 156 | 45 | 28 [9, 54] | 0.000690 |
| DBLζ S2–S3 | 70 | 21 | 13 [0, 31] | 0.014570 |
DSS locations are associated with rif gene recombination hotspots and RIFIN domain borders
The rif genes constitute P. falciparum’s largest variant surface antigen family with ∼150 gene copies/genome (36). These genes also contain a high density of low folding free energy 50-mers, relative to other P. falciparum genes (Supplementary Figure S3). The fact that rif and var genes share both hyper-variation and frequent telomeric location suggests that similar recombination mechanisms may operate on both gene families (11,48). RIFINs can be divided into A and B types, based on a 75-bp insert specific to RIFIN-A types. Recombination is expected to occur most frequently at the border between the major conserved (C1) and variable (V2) domain (48), coinciding with the recombination breakpoint in the only known rif chimera (11,48). Figure 2c shows that predicted DSS are concentrated around the expected recombination hotspot, supporting a hypothesis that rif and var genes evolve via similar DSS-induced recombination.
Var 50-mers predicted to form DSS induce recombination in yeast
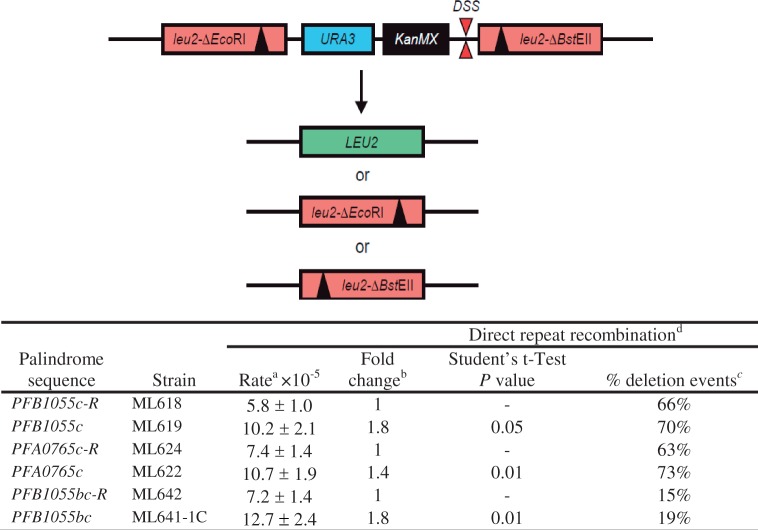
To assess the recombinogenic potential of specific 50-mers associated with actual recombination breakpoints in the X5PFA0005w/PFB1055c and X4 PFA0005c/PFB0010w chimeras, the PFB1055c2886–2935, PFB1055884-933 and PFA07652863–2912 50-mers and their randomized counterparts (of higher folding free energy) were cloned into the yeast S. cerevisiae and tested in a recombination assay commonly used to study homologous recombination (49). All the var 50-mers exhibited a significantly higher recombination rate (1.4–1.8-fold) compared with the randomized versions of these sequences (Figure 3).
Figure 3.
Effects of predicted DNA secondary structure sequences on mitotic leu2 direct-repeat recombination. The schematic illustrates the assay for spontaneous direct-repeat recombination between two non-functional leu2-ΔEcoRI and leu2-ΔBstEII alleles showing the position of the inserted DSS sequences (DSS). Recombination between the leu2 alleles to produce a functional LEU2 allele leads to prototrophy for leucine (Leu+). The assay scores for Leu+ recombinants generated by single-strand annealing, replication slippage or gene conversion between the leu2 alleles. Single-strand annealing leads to loss of the URA3 gene and uracil autotrophy. The fraction of URA3 deletion events were the same for the pair-wise combinations of strains and their scrambled counterpart, indicating that the 50-mer with the lowest folding free energy stimulated the different types of recombination equally well.
aRecombination rate is presented as events per cell per generation ± standard deviation, as described in ‘Materials and Methods’ section.
bRelative to a randomized sequence (R).
cPercentage of deletion events among Leu+ recombinants.
dStrains harbouring the palindrome sequences PFB1055c (ML619: 5′-TGGTGCCACTGGCAAAAGTGGTGATAAGGGTGCCATTTGTGTGCCACCCA), PFA0765c (ML622: 5′-CAAACACCTGGTGAGAAAACCACCCCACCTAGTGGTACTAACCAGGGTGC) and PFB1055bc (ML641-1C: 5′-GTAAGGACGAAAACGGCAAAAAGCCCGGCTCAAATGCCGACCAAGTCCCC) or their randomized counterparts PFB1055c-R (ML618: 5′-CCTGAAATTGCTGGCTAGGGGTCCTAGATGTGCCCGGGGTAGACCTATAA), PFA0765c-R (ML624: 5′-GCACTGATATGCAAGGAAGCCCCAGCAATCCTCAAAGACGCGAAGCCTCT) and PFB1055bc-R (ML642: 5′-GTATAAGCCTGGAAACCAACAGCGAAAGGCCGAAACCCGCCTACCAAGCG), respectively, were analysed for mitotic direct-repeat recombination at 30°C, as described (49).
DISCUSSION
Genetic recombination requires that participating donor sequences have sufficient sequence identity, that the chromosomes involved in the event are in proximity and that some distinct factor initiates the process (50). While physical clustering of heterologous chromosome-ends may result in the necessary proximity (12), quasi-palindromic sequences prone to forming base-paired DSS appear to act as the inducing factors initiating recombination in var genes. DSS are known to induce recombination in other organisms by making the DNA more accessible to structure-specific nucleases (51), by replication fork stalling and collapse followed by micro-homology-mediated strand invasion (52,53) or by strand invasion-independent faulty template switching of the DNA polymerase during DNA replication (54). The proximity of multiple recombination breakpoints within the var chimeras identified here, which are the result of recombination between unlinked var paralogs, is most consistent with a template switching mechanism. Such template switching has been previously observed when the DNA replication fork has stalled (55).
Our screens to identify novel chimeric var genes in genetic cross-progeny clones yielded four chimeras all created by ectopic recombination between var genes of the same genome. This result proves the previously raised hypothesis that var recombination mainly occurs between isogenous var paralogs during sexual reproduction (12) and is also suggestive of a replication-dependent mechanism. Replication-dependent recombination may not be restricted to the sexual stages but could, in theory, also operate during the mitotic divisions of blood-stage parasites. However, the differences between the recombination events creating the chimeric var sequences identified here and the var chimeras created from mitotic recombination during culturing of asexual blood-stages (20) suggest that different responsible mechanisms have been in play. Specifically, whereas the chimeras created in asexual blood-stage parasites are generated by single crossing-over events with no association to DSS or PfEMP1 domain structures, the chimeras generated during sexual reproduction contain multiple closely located recombination breaks resulting in a mosaic gene composition characteristic of var genes (13,15).
The observation that subtelomeric regions of heterologous chromosomes associate differently in asexual and sexual parasite forms (in clusters near the nuclear periphery and in bouquet-like configurations near one pole of the elongated nuclei, respectively) (12) may have influence on the ability of var genes to recombine at the different time points. In addition, given the likely association with DNA replication, a stage-specific var recombination mechanism could be associated with the DNA replication phase when the chromosome complement doubles from 2N to 4N in the short-lived zygotic stage known as the ookinete (56) or with the three rapid genome doublings, which create the ‘male’ microgametes, although previous studies have reported that the Dd2 clone is unable to produce male gametocytes, making the latter notion less likely (57,58).
In consensus, our evidence supports the view that virulence gene diversification in P. falciparum results from ectopic recombination between isogenous paralogs caused by a DSS-induced mechanism during DNA replication and explains previous observations that the P. falciparum parasite is able to generate new var genes during self-mating of male and female gametes derived from a single clone (18). This ability has probably been advantageous to the parasite, as it enables diversification of progeny antigen repertoires despite high inbreeding rates, which have been measured at 0.34 under high-intensity transmission in Tanzania (59) and at 0.90 under lower-intensity transmission in Papua New Guinea (60).
PfEMP1 proteins can be understood as composites of partially conserved homology blocks, resulting from shuffling var gene segments under the constraint of maintaining functional cytoadhesive structures while modulating antigenicity in the attempt to evade variant-specific immunity. The finding that the var DNA with the highest likelihood of forming DSS co-localize with recombination hotspots at, or close to, breaks of homology and boundaries of structural elements in the conserved superstructure of PfEMP1/var genes is significant. It indicates that DNA structural features, and not just the protein phenotype, have been selected to increase the frequency of recombination at positions that optimize the chances that an antigenically novel PfEMP1 structure retains essential functional domains. These data are the first evidence of a DSS-dependent recombination mechanism regulating and directing the evolution of a gene family. The structural boundary ‘rules’ being followed may aid the definition and re-engineering of minimal binding regions of complex PfEMP1 adhesins, in the ongoing effort to develop PfEMP1-based vaccines and cytoadhesion-blocking anti-malaria therapy.
The DSS-directed mechanism of P. falciparum virulence gene recombination seems to have evolved under host selection, to optimize conservation of essential protein functions while generating sufficient antigenic diversity to escape preexisting immunity. The var DSS 50-mers were shown to induce recombination when tested in a S. cerevisiae recombination assay, indicating that the var DSS have an intrinsic recombinogenic potential, but it does not exclude that protein factors specific to P. falciparum may also contribute to promoting recombination at DSS sequences. The relatively low but nonetheless significant recombinogenic potential of individual var DSS 50-mers may reflect the constraint of allowing DNA replication to occur with reasonable fidelity while at the same time stimulating above background levels of recombination.
A recent study of African trypanosome species, the first identified and best understood model of protozoan antigenic variation, has shown that DNA helix-destabilizing TAA:TTA repeats within the VSG antigen genes induce antigenic variation and VSG sequence diversification through a recombination pathway that shares some resemblance to the mechanism of var gene diversification outlined here (61). Both the VSG and var multi-gene families evolve by ectopic recombination events through a mechanism of break-induced replication (62,63), but the inducing DNA structures appear to be different in the two parasites. Where specific TAA:TTA repeats destabilize the DNA helix of trypanosome VSG genes (64), Plasmodium var gene DSS are formed from diverse sequences and are predicted to form stable secondary structures. These differences may reflect the fundamentally different functional constraints on PfEMP1 (receptor binding) versus VSG (antigenic variation, ‘smoke screen’), as well as the fact that VSG gene expression frequently is switched by recombination (often of gene fragments from a large repertoire of pseuodogenes) into an active expression site (65), whereas switching of var gene expression appears to be independent of recombination (66) and involves in situ activation of intact genes, thus reducing the importance of pseudogenes. Various DNA structure-induced recombination pathways may thus have evolved in pathogens, each balancing immune selection pressure to create novel antigenic variants with disease-specific functional constraints.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Danish National Research Foundation (http://www.dg.dk/) through a Niels Bohr Visiting Professorship award [Project: 312000-50-64920 to A.F.S. and D.E.A.]; Funded by Carlsbergfondet [2012_01_0393 to A.F.S.]; The Danish Council for Independent Research (to T.L.); The [FP7/2007-2013] under grant agreement [#200889 (STOPPAM) to A.S.]; the European Research Council (to M.L.); The Danish National Research Foundation and Lundbeck Foundation (to S.L.F.); a grant from the Danish Center for Scientific Computing (DCSC) (to T.S.R. and A.G.P.). Funding for open-access charge: Centre for Medical Parasitology, ISIM, University of Copenhagen.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Hashim El Hussein for excellent technical assistance; the NIAID Microbial Sequencing Centre at the Broad Institute and the Pathogen Genomics group at the Wellcome Trust Sanger Institute for sequencing and making available the HB3, 3D7 and progeny genome sequences.
REFERENCES
- 1.Su X, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, Ravetch JA, Wellems TE. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 2.Rowe JA, Claessens A, Corrigan RA, Arman M. Adhesion of Plasmodium falciparum-infected erythrocytes to human cells: molecular mechanisms and therapeutic implications. Exp. Rev. Mol. Med. 2009;11:e16. doi: 10.1017/S1462399409001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bull PC, Lowe BS, Kortok M, Molyneux CS, Newbold CI, Marsh K. Parasite antigens on the infected red cell are targets for naturally acquired immunity to malaria. Nat. Med. 1998;4:358–360. doi: 10.1038/nm0398-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barry AE, Leliwa-Sytek A, Tavul L, Imrie H, Migot-Nabias F, Brown SM, McVean GA, Day KP. Population genomics of the immune evasion (var) genes of Plasmodium falciparum. PLoS. Pathog. 2007;3:e34. doi: 10.1371/journal.ppat.0030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kyes S, Taylor H, Craig A, Marsh K, Newbold C. Genomic representation of var gene sequences in Plasmodium falciparum field isolates from different geographic regions. Mol. Biochem. Parasitol. 1997;87:235–238. doi: 10.1016/s0166-6851(97)00071-6. [DOI] [PubMed] [Google Scholar]
- 6.Lavstsen T, Salanti A, Jensen A, Arnot D, Theander T. Sub-grouping of Plasmodium falciparum 3D7 var genes based on sequence analysis of coding and non-coding regions. Malar. J. 2003;2:27. doi: 10.1186/1475-2875-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rask TS, Hansen DA, Theander TG, Gorm PA, Lavstsen T. Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes–divide and conquer. PLoS Comput. Biol. 2010;6 doi: 10.1371/journal.pcbi.1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lavstsen T, Turner L, Saguti F, Magistrado P, Rask TS, Jespersen JS, Wang CW, Berger SS, Baraka V, Marquard AM, et al. Plasmodium falciparum erythrocyte membrane protein 1 domain cassettes 8 and 13 are associated with severe malaria in children. Proc. Natl Acad. Sci. USA. 2012;109:E1791–E1800. doi: 10.1073/pnas.1120455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner L, Lavstsen T, Berger SS, Wang CW, Petersen JE, Avril M, Brazier AJ, Freeth J, Jespersen JS, Nielsen MA, et al. Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature. 2013;498:502–505. doi: 10.1038/nature12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kraemer SM, Smith JD. A family affair: var genes, PfEMP1 binding, and malaria disease. Curr. Opin. Microbiol. 2006;9:374–380. doi: 10.1016/j.mib.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Frank M, Kirkman L, Costantini D, Sanyal S, Lavazec C, Templeton TJ, Deitsch KW. Frequent recombination events generate diversity within the multi-copy variant antigen gene families of Plasmodium falciparum. Int. J. Parasitol. 2008;38:1099–1109. doi: 10.1016/j.ijpara.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freitas-Junior LH, Bottius E, Pirrit LA, Deitsch KW, Scheidig C, Guinet F, Nehrbass U, Wellems TE, Scherf A. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- 13.Kraemer SM, Kyes SA, Aggarwal G, Springer AL, Nelson SO, Christodoulou Z, Smith LM, Wang W, Levin E, Newbold CI, et al. Patterns of gene recombination shape var gene repertoires in Plasmodium falciparum: comparisons of geographically diverse isolates. BMC Genomics. 2007;8:45. doi: 10.1186/1471-2164-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor HM, Kyes SA, Newbold CI. Var gene diversity in Plasmodium falciparum is generated by frequent recombination events. Mol. Biochem. Parasitol. 2000;110:391–397. doi: 10.1016/s0166-6851(00)00286-3. [DOI] [PubMed] [Google Scholar]
- 15.Ward CP, Clottey GT, Dorris M, Ji DD, Arnot DE. Analysis of Plasmodium falciparum PfEMP-1/var genes indicates that recombination rearranges constrained sequences. Mol. Biochem. Parasitol. 1999;102:167–177. doi: 10.1016/s0166-6851(99)00106-1. [DOI] [PubMed] [Google Scholar]
- 16.Walliker D, Quakyi IA, Wellems TE, McCutchan TF, Szarfman A, London WT, Corcoran LM, Burkot TR, Carter R. Genetic analysis of the human malaria parasite Plasmodium falciparum. Science. 1987;236:1661–1666. doi: 10.1126/science.3299700. [DOI] [PubMed] [Google Scholar]
- 17.Jiang H, Li N, Gopalan V, Zilversmit MM, Varma S, Nagarajan V, Li J, Mu J, Hayton K, Henschen B, et al. High recombination rates and hotspots in a Plasmodium falciparum genetic cross. Genome Biol. 2011;12:R33. doi: 10.1186/gb-2011-12-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dharia NV, Plouffe D, Bopp SE, Gonzalez-Paez GE, Lucas C, Salas C, Soberon V, Bursulaya B, Kochel TJ, Bacon DJ, et al. Genome scanning of Amazonian Plasmodium falciparum shows subtelomeric instability and clindamycin-resistant parasites. Genome Res. 2010;20:1534–1544. doi: 10.1101/gr.105163.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flick K, Chen Q. var genes, PfEMP1 and the human host. Mol. Biochem. Parasitol. 2004;134:3–9. doi: 10.1016/j.molbiopara.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 20.Bopp SE, Manary MJ, Bright AT, Johnston GL, Dharia NV, Luna FL, McCormack S, Plouffe D, McNamara CW, Walker JR, et al. Mitotic evolution of Plasmodium falciparum shows a stable core genome but recombination in antigen families. PLoS Genet. 2013;9:e1003293. doi: 10.1371/journal.pgen.1003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duffy MF, Byrne TJ, Carret C, Ivens A, Brown GV. Ectopic recombination of a malaria var gene during mitosis associated with an altered var switch rate. J. Mol. Biol. 2009;389:453–469. doi: 10.1016/j.jmb.2009.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trager W, Jensen JB. Human malaria parasites in continuous culture. 1976. J. Parasitol. 2005;91:484–486. doi: 10.1645/0022-3395(2005)091[0484:HMPICC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez-Rivas R, Scherf A. Separation and mapping of chromosomes of parasitic protozoa. Mem. Inst. Oswaldo Cruz. 1997;92:815–819. doi: 10.1590/s0074-02761997000600017. [DOI] [PubMed] [Google Scholar]
- 24.Sander AF, Salanti A, Lavstsen T, Nielsen MA, Magistrado P, Lusingu J, Ndam NT, Arnot DE. Multiple var2csa-type PfEMP1 genes located at different chromosomal loci occur in many Plasmodium falciparum isolates. PLoS One. 2009;4:e6667. doi: 10.1371/journal.pone.0006667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dahlback M, Lavstsen T, Salanti A, Hviid L, Arnot DE, Theander TG, Nielsen MA. Changes in var gene mRNA levels during erythrocytic development in two phenotypically distinct Plasmodium falciparum parasites. Malar. J. 2007;6:78. doi: 10.1186/1475-2875-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salanti A, Staalsoe T, Lavstsen T, Jensen A, Sowa M, Arnot D, Hviid L, Theander TG. Selective up-regulation of a single distinctly structured var gene in CSA-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol. Microbiol. 2003;49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- 27.Soerli J, Barfod L, Lavstsen T, Bernasconi NL, Lanzavecchia A, Hviid L. Human monoclonal IgG selection of Plasmodium falciparum for the expression of placental malaria-specific variant surface antigens. Parasite Immunol. 2009;31:341–346. doi: 10.1111/j.1365-3024.2009.01097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su XZ, Wellems TE. Plasmodium falciparum: assignment of microsatellite markers to chromosomes by PFG-PCR. Exp. Parasitol. 1999;91:367–369. doi: 10.1006/expr.1998.4390. [DOI] [PubMed] [Google Scholar]
- 29.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rask TS, Hansen DA, Theander TG, Gorm Pedersen A, Lavstsen T. Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes– divide and conquer. PLoS Comput. Biol. 2010;6:e1000933. doi: 10.1371/journal.pcbi.1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lorenz R, Bernhart SH, Honer Zu SC, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA Package 2.0. Algorithms. Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zakov S, Goldberg Y, Elhadad M, Ziv-Ukelson M. Rich parameterization improves RNA structure prediction. J. Comput. Biol. 2011;18:1525–1542. doi: 10.1089/cmb.2011.0184. [DOI] [PubMed] [Google Scholar]
- 35.DePristo MA, Zilversmit MM, Hartl DL. On the abundance, amino acid composition, and evolutionary dynamics of low-complexity regions in proteins. Gene. 2006;378:19–30. doi: 10.1016/j.gene.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 36.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Germann SM, Oestergaard VH, Haas C, Salis P, Motegi A, Lisby M. Dpb11/TopBP1 plays distinct roles in DNA replication, checkpoint response and homologous recombination. DNA Repair (Amst) 2011;10:210–224. doi: 10.1016/j.dnarep.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 38.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 39.Gietz D, St JA, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith J, Rothstein R. An allele of RFA1 suppresses RAD52-dependent double-strand break repair in Saccharomyces cerevisiae. Genetics. 1999;151:447–458. doi: 10.1093/genetics/151.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eckert-Boulet N, Rothstein R, Lisby M. Cell biology of homologous recombination in yeast. Methods Mol. Biol. 2011;745:523–536. doi: 10.1007/978-1-61779-129-1_30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Higgins MK. The structure of a chondroitin sulfate-binding domain important in placental malaria. J. Biol. Chem. 2008;283:21842–21846. doi: 10.1074/jbc.C800086200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh SK, Hora R, Belrhali H, Chitnis CE, Sharma A. Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature. 2006;439:741–744. doi: 10.1038/nature04443. [DOI] [PubMed] [Google Scholar]
- 44.Bikard D, Loot C, Baharoglu Z, Mazel D. Folded DNA in action: hairpin formation and biological functions in prokaryotes. Microbiol. Mol. Biol. Rev. 2010;74:570–588. doi: 10.1128/MMBR.00026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith GR. Meeting DNA palindromes head-to-head. Genes Dev. 2008;22:2612–2620. doi: 10.1101/gad.1724708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nickoloff JA, Sweetser DB, Clikeman JA, Khalsa GJ, Wheeler SL. Multiple heterologies increase mitotic double-strand break-induced allelic gene conversion tract lengths in yeast. Genetics. 1999;153:665–679. doi: 10.1093/genetics/153.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dahlback M, Rask TS, Andersen PH, Nielsen MA, Ndam NT, Resende M, Turner L, Deloron P, Hviid L, Lund O, et al. Epitope mapping and topographic analysis of VAR2CSA DBL3X involved in P. falciparum placental sequestration. PLoS Pathog. 2006;2:e124. doi: 10.1371/journal.ppat.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joannin N, Abhiman S, Sonnhammer EL, Wahlgren M. Sub-grouping and sub-functionalization of the RIFIN multi-copy protein family. BMC Genomics. 2008;9:19. doi: 10.1186/1471-2164-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith J, Rothstein R. An allele of RFA1 suppresses RAD52-dependent double-strand break repair in Saccharomyces cerevisiae. Genetics. 1999;151:447–458. doi: 10.1093/genetics/151.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lettier G, Feng Q, de Mayolo AA, Erdeniz N, Reid RJ, Lisby M, Mortensen UH, Rothstein R. The role of DNA double-strand breaks in spontaneous homologous recombination in S. cerevisiae. PLoS Genet. 2006;2:e194. doi: 10.1371/journal.pgen.0020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leach DR, Stahl FW. Viability of lambda phages carrying a perfect palindrome in the absence of recombination nucleases. Nature. 1983;305:448–451. doi: 10.1038/305448a0. [DOI] [PubMed] [Google Scholar]
- 52.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Payen C, Koszul R, Dujon B, Fischer G. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 2008;4:e1000175. doi: 10.1371/journal.pgen.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paek AL, Kaochar S, Jones H, Elezaby A, Shanks L, Weinert T. Fusion of nearby inverted repeats by a replication-based mechanism leads to formation of dicentric and acentric chromosomes that cause genome instability in budding yeast. Genes Dev. 2009;23:2861–2875. doi: 10.1101/gad.1862709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 56.Sinden RE, Hartley RH. Identification of the meiotic division of malarial parasites. J. Protozool. 1985;32:742–744. doi: 10.1111/j.1550-7408.1985.tb03113.x. [DOI] [PubMed] [Google Scholar]
- 57.Guinet F, Dvorak JA, Fujioka H, Keister DB, Muratova O, Kaslow DC, Aikawa M, Vaidya AB, Wellems TE. A developmental defect in Plasmodium falciparum male gametogenesis. J. Cell Biol. 1996;135:269–278. doi: 10.1083/jcb.135.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaidya AB, Muratova O, Guinet F, Keister D, Wellems TE, Kaslow DC. A genetic locus on Plasmodium falciparum chromosome 12 linked to a defect in mosquito-infectivity and male gametogenesis. Mol. Biochem. Parasitol. 1995;69:65–71. doi: 10.1016/0166-6851(94)00199-w. [DOI] [PubMed] [Google Scholar]
- 59.Babiker HA, Ranford-Cartwright LC, Currie D, Charlwood JD, Billingsley P, Teuscher T, Walliker D. Random mating in a natural population of the malaria parasite Plasmodium falciparum. Parasitology. 1994;109(Pt. 4):413–421. doi: 10.1017/s0031182000080665. [DOI] [PubMed] [Google Scholar]
- 60.Paul RE, Packer MJ, Walmsley M, Lagog M, Ranford-Cartwright LC, Paru R, Day KP. Mating patterns in malaria parasite populations of Papua New Guinea. Science. 1995;269:1709–1711. doi: 10.1126/science.7569897. [DOI] [PubMed] [Google Scholar]
- 61.Jackson AP, Berry A, Aslett M, Allison HC, Burton P, Vavrova-Anderson J, Brown R, Browne H, Corton N, Hauser H, et al. Antigenic diversity is generated by distinct evolutionary mechanisms in African trypanosome species. Proc. Natl Acad. Sci. USA. 2012;109:3416–3421. doi: 10.1073/pnas.1117313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boothroyd CE, Dreesen O, Leonova T, Ly KI, Figueiredo LM, Cross GA, Papavasiliou FN. A yeast-endonuclease-generated DNA break induces antigenic switching in Trypanosoma brucei. Nature. 2009;459:278–281. doi: 10.1038/nature07982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim HS, Cross GA. TOPO3alpha influences antigenic variation by monitoring expression-site-associated VSG switching in Trypanosoma brucei. PLoS Pathog. 2010;6:e1000992. doi: 10.1371/journal.ppat.1000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ohshima K, Kang S, Larson JE, Wells RD. TTA.TAA triplet repeats in plasmids form a non-H bonded structure. J. Biol. Chem. 1996;271:16784–16791. doi: 10.1074/jbc.271.28.16784. [DOI] [PubMed] [Google Scholar]
- 65.Horn D, McCulloch R. Molecular mechanisms underlying the control of antigenic variation in African trypanosomes. Curr. Opin. Microbiol. 2010;13:700–705. doi: 10.1016/j.mib.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scherf A, Hernandez-Rivas R, Buffet P, Bottius E, Benatar C, Pouvelle B, Gysin J, Lanzer M. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 1998;17:5418–5426. doi: 10.1093/emboj/17.18.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.