Abstract
The human immunodeficiency virus type 1 (HIV-1) nucleocapsid (NC) protein contains 15 basic residues located throughout its 55-amino acid sequence, as well as one aromatic residue in each of its two CCHC-type zinc finger motifs. NC facilitates nucleic acid (NA) rearrangements via its chaperone activity, but the structural basis for this activity and its consequences in vivo are not completely understood. Here, we investigate the role played by basic residues in the N-terminal domain, the N-terminal zinc finger and the linker region between the two zinc fingers. We use in vitro ensemble and single-molecule DNA stretching experiments to measure the characteristics of wild-type and mutant HIV-1 NC proteins, and correlate these results with cell-based HIV-1 replication assays. All of the cationic residue mutations lead to NA interaction defects, as well as reduced HIV-1 infectivity, and these effects are most pronounced on neutralizing all five N-terminal cationic residues. HIV-1 infectivity in cells is correlated most strongly with NC’s NA annealing capabilities as well as its ability to intercalate the DNA duplex. Although NC’s aromatic residues participate directly in DNA intercalation, our findings suggest that specific basic residues enhance these interactions, resulting in optimal NA chaperone activity.
INTRODUCTION
The retroviral nucleocapsid (NC) protein is the major nucleic acid (NA) binding domain of the Gag polyprotein, which is known to be necessary for virion assembly, as well as viral genome (gRNA) selection and packaging (1,2). The major domains of Gag include NC, capsid (CA), matrix (MA) and p6. After Gag processing by human immunodeficiency virus type 1 (HIV-1) protease, CA re-assembles to form a mature virus CA, NC binds to the gRNA inside the CA and MA binds to the cellular plasma membrane (3). Although the sequences of NC domains of Gag vary across different retroviruses, they are generally highly cationic in character (4,5). An exception is HTLV-1 NC, which overall is neutral but contains several highly charged regions (6). One or two CCHC-type zinc fingers, each containing one or two aromatic residues, are common structural elements present in all orthoretroviral NC proteins. The aromatic residues appear to confer some of the specificity of gRNA selection by NC (7,8). In contrast, NC’s basic residues are typically distributed over the entire protein, and are generally responsible for nonspecific electrostatic interactions with NAs (9–14).
HIV-1 NC is a NA chaperone protein that facilitates the rearrangement of NAs into their lowest energy configuration (2,15–23). These rearrangements are essential for many viral replication processes, such as reverse transcription and recombination (14,24–28). Three key steps of reverse transcription, tRNA primer annealing (29), minus-strand transfer and plus-strand transfer (30–34), require significant rearrangement of NA secondary structure. Previous work demonstrated that HIV-1 NC greatly facilitates these processes through its chaperone activity, which includes NA aggregation, destabilization and rapid protein–NA interaction kinetics (15,16,18,35–39). In vitro studies suggest that the ability of NC proteins to aggregate NA is due to their cationic character, and this activity is largely independent of their specific zinc-finger structures (9,36,40). In contrast, the other major component of the chaperone activity, NA destabilization, primarily requires properly folded zinc fingers (8,41,42). In the case of HIV-1 NC, this activity depends on the preferential binding of the zinc fingers to unpaired NA bases (43,44). The duplex destabilization activity differs significantly between different retroviral NC proteins, as well as related retrotransposons (6,45–47).
Although many aspects of NC’s chaperone activity have been extensively investigated, the detailed relationship between HIV-1 NC’s structure and its ability to aggregate and destabilize NAs is still incompletely understood. For example, although the effective cationic charge of HIV-1 NC, defined as a negative slope of the log-log dependence of the Kd versus salt concentration, is ∼+3.5 (48–52), the total number of positively charged residues on this protein is 15, with only four negatively charged residues. Thus, it is unclear how and to what extent specific HIV-1 NC charged residues participate in nonspecific versus specific NA binding. The role of specific basic residues in other chaperone activities, such as NA aggregation, is also unknown.
To clarify the interplay between specific basic residues and HIV-1 NC’s NA chaperone function, we use several complementary in vitro approaches. Ensemble assays are used to quantify NA binding, aggregation and annealing activities of wild-type (WT) and mutant HIV-1 NC proteins (6,8). These results are compared with the results obtained from single-molecule DNA stretching experiments. We find that cationic HIV-1 NC variants are defective in their overall NA binding affinity, aggregation and strand-annealing activities, but retain significant NA stacking capability at sufficiently high concentrations. These results are in contrast to previous work showing that aromatic residue variants are completely defective in stacking with NA bases, and have greatly reduced single-stranded DNA (ssDNA) binding affinity (8). Cell-based assays showed that all of the cationic residue mutations investigated lead to reduced virus infectivity, which correlated strongly with measurements of trans-activation response (TAR) RNA and DNA annealing as well as the capability to intercalate DNA at high force in single-molecule stretching experiments.
MATERIALS AND METHODS
Plasmids, mutagenesis and recombinant protein production
Site-directed mutagenesis was performed using the Agilent QuickChange kit, with verification by NA sequence analysis, for the generation of full-length proviral pNL4-3 plasmids [pNL4-3, GenBank accession no. AF324493 was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH from Dr Malcolm Martin (53)] or recombinant expression plasmids essentially as described (8). Mutations are as follows with numbering of nucleotide positions based on the pNL4-3 clone of HIV-1 (GenBank accession no. AF324493): K3A changes nucleotide 1927 to 1928 from aa to gc, R7A changes nucleotide 1939 to 1940 from ag to gc, R10A changes nucleotide 1948 to 1949 from ag to gc, K11A changes nucleotide 1951 to 1952 from aa to gc, K14A changes nucleotide 1960 to 1961 from aa to gc, K20A changes nucleotide 1978 to 1980 from aaa to gcc, K26A changes nucleotide 1996 to 1997 from aa to gc, R29A changes nucleotide 2005 to 2007 from agg to gca, R32A changes nucleotide 2014 to 2016 from agg to gcc, K33A changes nucleotide 2017 to 2019 from aaa to gcc, K34A changes nucleotide 2020 to 2022 from aag to gcc. All mutations were verified by NA sequence analysis. Recombinant NC proteins, 55 amino acids in length, were expressed and purified as described (33,45,54). The mutant NC amino acid sequences examined in either viruses or as purified NCp7 are presented in Figure 1.
Figure 1.
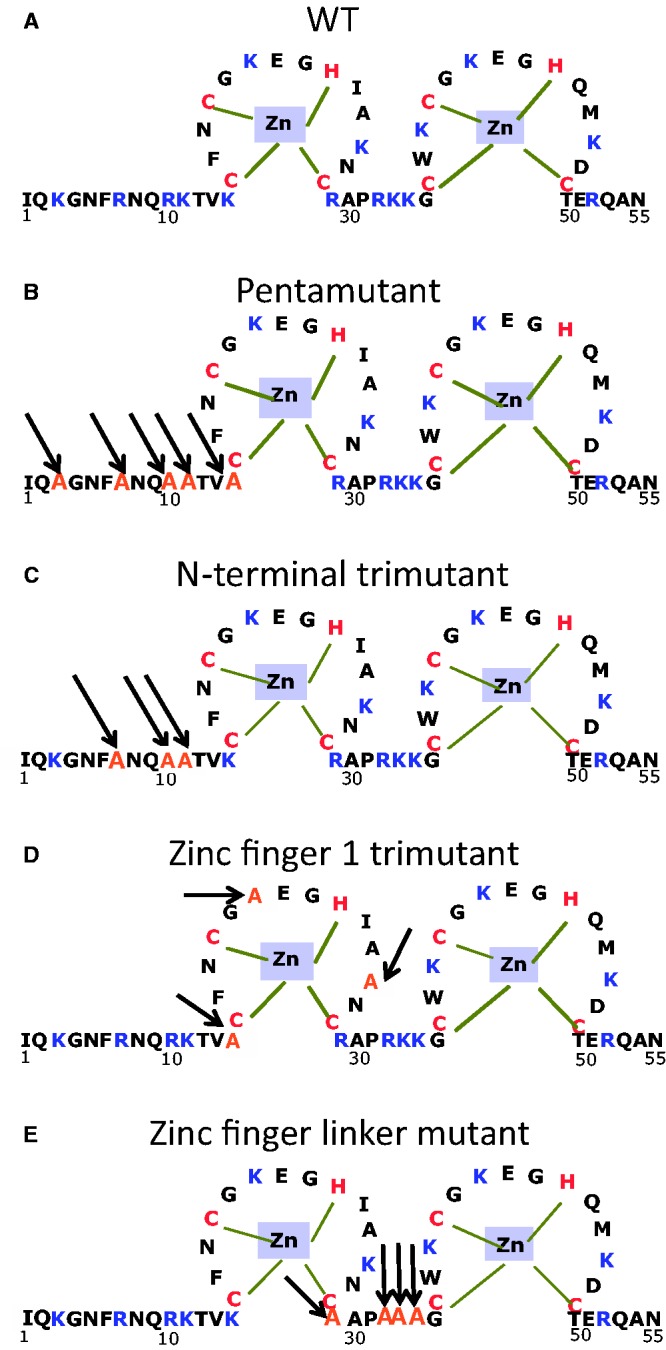
Sequence of WT HIV-1 NC (NL4-3 isolate) and variants investigated in this work. (A) WT; (B) K3A/R7A/R10A/K11A/K14A (pentamutant); (C) R7A/R10A/K11A (N-terminal trimutant); (D) K14A/K20A/K26A (Zinc finger 1 trimutant); (E) R29A/R32A/K33A/K34A (Zinc finger linker mutant).
Single-molecule DNA stretching studies
In the single-molecule stretching experiments performed essentially as described (55), two laser beams are overlapped to trap one streptavidin-coated bead. A second streptavidin-coated bead is attached to a glass micropipette. Bacteriophage λ DNA, which was labeled with biotin on both 5′ ends as described (56), is caught between the beads due to strong noncovalent bonds between biotin and streptavidin. All stretching experiments were performed at a constant pulling rate of ∼100 nm/s in 10 mM HEPES, 50 mM Na+, pH 7.5 buffer. After attachment of one DNA molecule, buffer was used to rinse out the other DNA molecules, and solutions containing specific protein concentrations were flowed around the DNA to investigate protein effects on DNA stretching curves.
Ensemble studies
Fluorescence anisotropy binding studies
Twenty nanomolar 5′ fluorescein-labeled 18-mer microTAR RNA (8) (Dharmacon, Inc.) or 10 nM 5′-carboxyfluorescein (FAM)-labeled-(TG)4 DNA (8) (Integrated DNA Technologies, Coralville, IA) was incubated in the absence or presence of varying concentrations of WT or mutant NC proteins at 20°C in 20 mM HEPES, pH 7.5, 50 mM NaCl, 10 µM tris (2-carboxyethyl)phosphine, 5 mM β-mercaptoethanol and 1 µM zinc acetate for 30 min. Fluorescence anisotropy (FA) measurements were carried out in duplicate for each protein concentration using 20 µl of solution per measurement. Assays were performed in Corning® 384-well low volume polystyrene NBS™ microplates (Corning, NY) using a SpectraMax® M5 multimode microplate reader (Molecular Devices, Sunnyvale, CA). Excitation and emission wavelengths were set at 485 nm (9 nm bandwidth) and 525 nm (15 nm bandwidth), respectively. Anisotropy (A) was calculated using SoftMax® Pro (Molecular Devices) software. The resulting plot of A versus NC concentration (N) was fit using Kaleidagraph (Synergy Software, Reading, PA) according to the following one-site binding model (56):
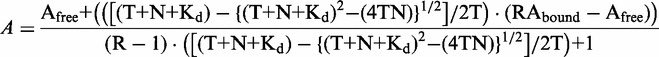 |
(1) |
where T, Abound, Afree, Kd and R represent the concentration of oligonucleotide, anisotropy of completely bound oligonucleotide, anisotropy of unbound oligonucleotide, equilibrium dissociation constant and the ratio of fluorescence intensity of completely bound oligonucleotide relative to unbound nucleotide, respectively. The anisotropy of completely bound oligonucleotide was determined as a fitting parameter representing saturated binding for each measurement.
TAR RNA/DNA annealing assays
Annealing assays were carried out with 59-nt TAR RNA and 59-nt complementary TAR (cTAR) DNA as described previously (8). Immediately prior to each experiment, 32P-labeled TAR RNA (1.5 µM) and cTAR DNA (6 µM) solutions were refolded in 25 mM HEPES, pH 7.5, and 20 mM NaCl by heating at 80°C for 2 min, then cooling to 60°C for 2 min, followed by addition of MgCl2 (to 10 mM final) and placement on ice. For annealing assays, 32P-labeled TAR RNA (15 nM) and cTAR DNA (45 nM) were combined in a solution containing 20 mM HEPES, pH 7.5, 20 mM NaCl, 0.2 mM MgCl2 and 5 mM dithiothreitol (DTT). The solution was incubated at 37°C for 5 min before addition of NC to a final concentration of 0.88 µM, corresponding to a 4:1 nt:NC ratio, unless otherwise indicated. At desired time points, an aliquot of the reaction mixture was quenched by addition of 1% sodium dodecyl sulphate, followed by incubation at room temperature for 5 min and placement on ice. The aliquots were phenol-chloroform extracted twice (to remove any NC bound to the NA) followed by addition of glycerol (5%) and separation on 12% polyacrylamide gel electrophoresis (19:1 acrylamide:bisacrylamide). The gels were visualized using a Bio-Rad (Hercules, CA) Molecular Imager FX and quantified with Bio-Rad Quantity One Software.
Aggregation assays
Sedimentation assays to monitor NC-induced NA aggregation activity were performed as previously described (36). Briefly, TAR RNA (unlabeled) was combined with 32P-labeled TAR RNA to a final concentration of 1.5 µM. Both 32P-labeled TAR RNA and cTAR DNA (6 µM) were folded as described for annealing assays (see below) and diluted to a final concentration of 15 and 45 nM, respectively, in a buffer containing 20 mM HEPES, pH 7.5, 20 mM NaCl, 0.2 mM MgCl2, and 5 mM DTT in the absence or presence of 0.88 µM NC, unless otherwise indicated. The samples (30 µl) were incubated at 37°C for 30 min and then spun in a Micromax RF (Thermo IEC) centrifuge rotor at a speed of 12 000 rpm for 10 min at 4°C. An aliquot (4 µl) of supernatant was taken and the amount of radioactivity measured using scintillation counting.
Cell culture-based studies
Full-length mutant or WT proviral plasmids were transfected into 293T cells and viruses were harvested as described previously (57). Viral genome packaging was assessed by measuring gRNA by quantitative reverse transcriptase-polymerase chain reaction and normalizing to exogenous-template reverse trancriptase (RT) activity as described (57). Single-round infectivity assays using HCLZ or TZM-bl cells as well as multiple-round replication assays using H9 cells were performed as described (57,58).
Correlation measurements
Correlation coefficients were calculated according to the following equation:
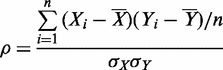 |
(2) |
where ρ is the correlation coefficient, 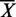 and
and 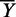 are the average values of
are the average values of 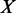 and
and 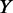 ,
, 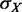 and
and 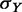 are the standard deviations of
are the standard deviations of  and
and  (59).
(59).
RESULTS
DNA stretching in the presence of WT HIV-1 NC
Figure 1 shows the sequences of WT HIV-1 NC and the basic residue variants studied in this work. Single-molecule λ-DNA stretching studies were carried out as previously described (8). As shown in Figure 2, in the absence of protein, little force is required to stretch the DNA to its B-form contour length, which is ∼0.34 nm/bp. As the contour length is approached, the force increases dramatically, reflecting the elasticity of the DNA helix. As the DNA is further stretched, it undergoes a force-induced overstretching transition from double-stranded DNA (dsDNA) to ssDNA. During this process, the DNA extension increases from 0.34 nm/bp to 0.6 nm/bp at an approximately constant force of 60 pN. The overstretched state of DNA can be either completely strand-separated with one or both strands under tension, or stretched to an unwound but still double-stranded structure, depending on solution conditions, DNA sequence and pulling rate (60–64). Under conditions of relatively low solution ionic strength as used here, or in the presence of ssDNA binding proteins, the DNA becomes single-stranded during the transition (62,63). In contrast, in the presence of proteins or ligands that preferentially bind to both DNA strands, the two strands may never separate. In the absence of protein, as the DNA is released back to lower extension, the force-extension curve is almost completely reversible, showing little hysteresis.
Figure 2.
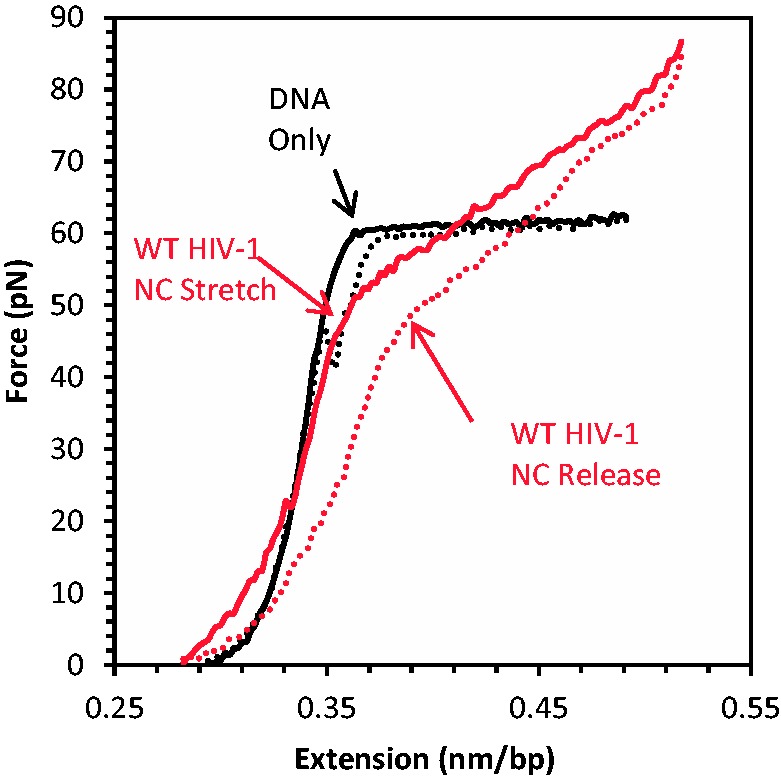
Force-extension (solid) and release (dashed) curves of DNA alone (black) and in the presence of 30 nM HIV-1 NC (red). Adapted from reference (8).
In the presence of WT HIV-1 NC, the overstretching transition becomes dramatically sloped, as the DNA starts to elongate beyond its B-form contour length from 20 to 30 pN (16,56) (Figure 2). The transition in this case is poorly defined, and extends over a broad range of forces between 20 and 90 pN, leading to an apparent force-extension slope up to 350 pN/nm/bp (Figure 2). A similar effect on DNA stretching was previously observed in the presence of intercalators such as ethidium (65), several ruthenium complexes (66,67) and threading- and bis-intercalators (68). In contrast to NC, these intercalators bind preferentially to dsDNA and increase the overall stability of duplex DNA, and this is usually reflected in an overall increase in the DNA stretching force. An example of a destabilizing intercalator is the anticancer drug Actinomycin D (Act D), which was recently characterized by DNA stretching (69). The stretching curves in the presence of Act D strongly resemble those observed in the presence of NC. This observation, when combined with our recent observation that the two DNA strands do not separate on overstretching in the presence of NC (8), suggests that NC acts as a weak intercalator, which on binding holds the two DNA strands together while simultaneously destabilizing the duplex form. Thus, the significant DNA elongation observed in the presence of NC at forces >20 pN results from the stacking of some NC residues between dsDNA bases, resulting in an intercalation process, similar to that observed for Act D (69), although it occurs at a much faster rate. In the case of HIV-1 NC, this duplex elongation likely occurs on intercalation of the aromatic residues (Phe16 and Trp37) (70,71), similar to the stacking with bases that was observed in the nuclear magnetic resonance (NMR) structures of HIV-1 NC bound to HIV-1 stem loops SL2 and SL3 (43,44,51,72–79). Here, we will use the value of the slope as a primary quantitative characteristic of NC-DNA binding, which reflects the ability of these aromatic residues to optimally stack with the DNA bases.
DNA stretching with HIV-1 NC basic residue mutants
Presented in Figure 3 are the stretch and release curves obtained in the presence of HIV-1 NC basic residue mutants. All of these curves differ significantly, showing high sensitivity to even a few amino acid substitutions. To summarize the protein–DNA interaction information contained in these data, we use two quantitative parameters describing the stretch–release cycle for the protein–DNA complex: the slope of the overstretching transition, S, and the scaled hysteresis in the stretch–release cycle, H, which is derived from the area between the stretch and release curves (8). S is measured as the slope of the tangent line to the force-extension curve at the transition midpoint of the extension (∼0.48 nm/bp). The protein-free slope, S0, is ∼20 pN/nm/bp, and the protein-saturated maximum slope, Smax, reflects the protein’s ability to intercalate into dsDNA on saturated binding at high forces. A small transition slope close to S0 indicates a force-induced DNA strand separation. In contrast, Smax>>S0 reflects strong intercalation by the protein at high force. The theoretical maximum amount of hysteresis would be achieved if DNA could be stretched as dsDNA and released as ssDNA. We define the scaled hysteresis, H, as the actual area of the protein–DNA stretch–release cycle divided by this maximum possible value. If the protein intercalates DNA, this may result in small H because it prevents the two strands from separating when being stretched. If the protein does not intercalate DNA and the strands are separated by force during overstretching, then small H indicates that the strands rapidly anneal on the time scale of stretching. Larger values of H may occur if the DNA intercalates but the intercalation is slow, as is the case for Act D. If there is no intercalation and the strands separate during stretching, large hysteresis may result from protein binding to ssDNA and subsequent slow dissociation. We will use these observed features of DNA stretching curves below to probe the effects of cationic mutations on NC–DNA interactions.
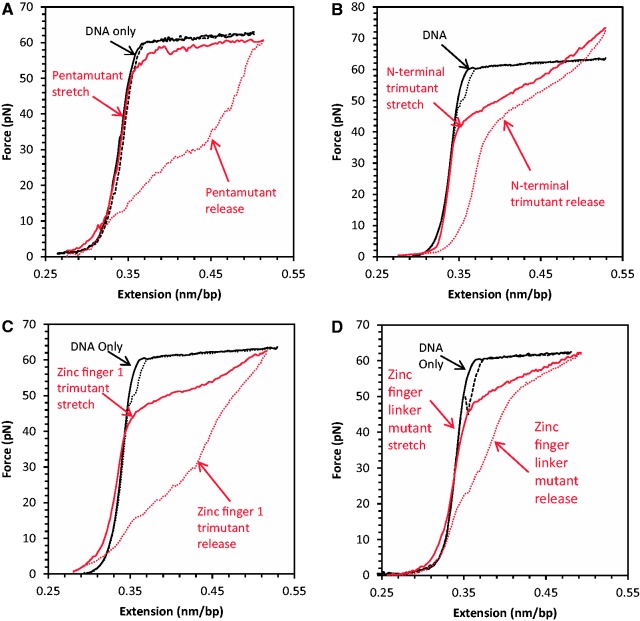
Figure 3.
DNA stretching (solid line) and release (dotted line) in the presence (red) and absence (black) of HIV-1 NC cationic mutants: (A) 200 nM N-terminal pentamutant (K3A/R7A/R10A/K11A/K14A); (B) 200 nM N-terminal trimutant (R7A/R10A/K11A); (C) ZF-1 cationic mutant (K14A/K20A/K26A); (D) Zinc finger linker mutant (R29A/R32A/K33A/K34A).
Effect of HIV-1 NC basic residue mutations
Presented in Figure 3A is a typical DNA stretch–release cycle in the presence of an HIV-1 NC variant with 5 N-terminal basic residues changed to alanine (K3A/R7A/R10/K11A/K14; pentamutant) (see Figure1B). Surprisingly, the dsDNA stretching curve in the presence of 200 nM pentamutant resembles the curve in the absence of protein. The fact that the transition slope remains close to the protein-free value (Figure 3A) suggests that the protein does not intercalate DNA and the DNA is melted by force in the presence of the pentamutant NC, consistent with the large hysteresis observed on DNA release. The large hysteresis also suggests strong and relatively slow binding of the pentamutant to ssDNA. It is initially surprising that the pentamutant NC is unable to intercalate DNA, as there is still an aromatic residue on each of the two zinc fingers. However, it has been shown that the strength of DNA intercalation by proteins depends strongly on interactions with amino acid side chains that are outside the intercalative wedge of the protein, and this must be the case for NC as well (80). In addition, it is clear that the intercalative ability of the pentamutant is weakened but not eliminated, as the DNA stretching slope increases with concentration.
Figure 3B shows the result of the DNA stretch–release curves in the presence of an N-terminal NC variant with only three of the basic residues mutated to alanine (R7A/R10A/K11A; N-terminal trimutant). This variant displayed a greatly increased S relative to the pentamutant, but a reduced H, suggesting efficient DNA intercalation by the N-terminal trimutant, similar to that observed for WT NC. Two additional basic residue mutants containing three (K14A/K20A/K26A; zinc finger 1 trimutant) or four (R29A/R32A/K33A/K34A; zinc finger linker mutant) changes displayed features suggesting weakened intercalative binding by these proteins as well as partial DNA melting by force, as determined by the smaller slopes and larger hysteresis observed compared with WT or N-terminal trimutant NC (Figure 3C and D). A summary of the S and H parameters calculated for all the cationic NC mutants investigated as a function of protein concentration is presented in Figures 4 and 5, respectively. For all mutants, the S parameter increases with concentration, suggesting that the ability to intercalate is strongly enhanced by binding that is facilitated by basic residues, and this enhancement is different depending on the location and number of the residues. The H parameter also increases with concentration for all basic residue mutants studied, in contrast to WT NC (Figure 5). This result reflects the loss of the ability of all of these mutants to intercalate DNA, along with defects in their abilities to aggregate ssDNA and promote strand annealing after separation by force. Based on these results, we can conclude that changes to any cluster of basic residues in the N-terminus, N-terminal zinc finger or linker region of HIV-1 NC significantly impacts at least one of these characteristics, which are associated with NA chaperone activity.
Figure 4.
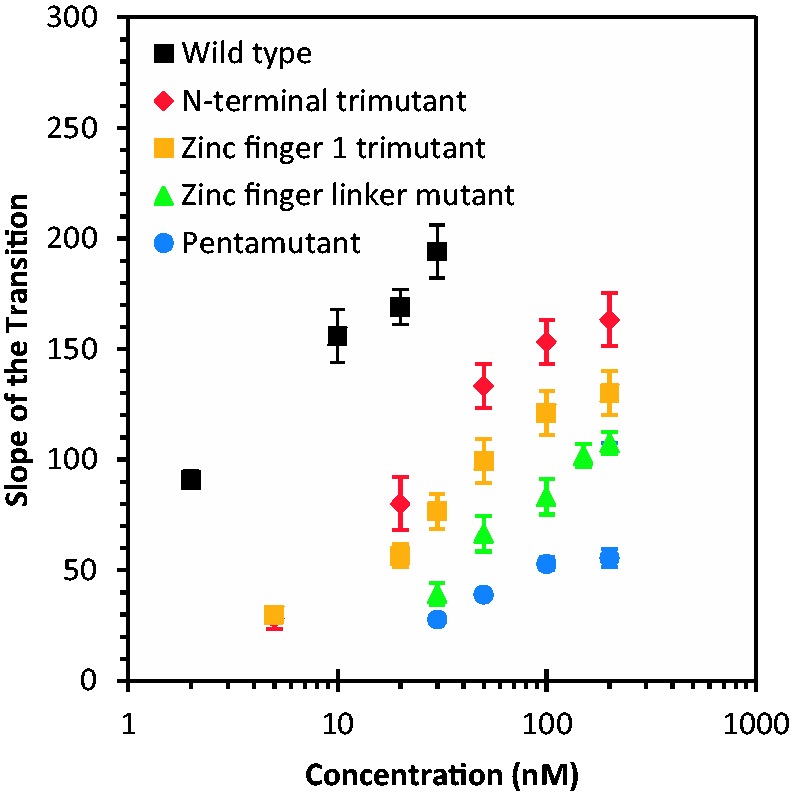
Slope of the transition versus concentration for WT HIV-1 NC and cationic mutants. Note that the 5 nM data points for the zinc finger 1 linker mutant and the N-terminal tri-mutant are coincident.
Figure 5.
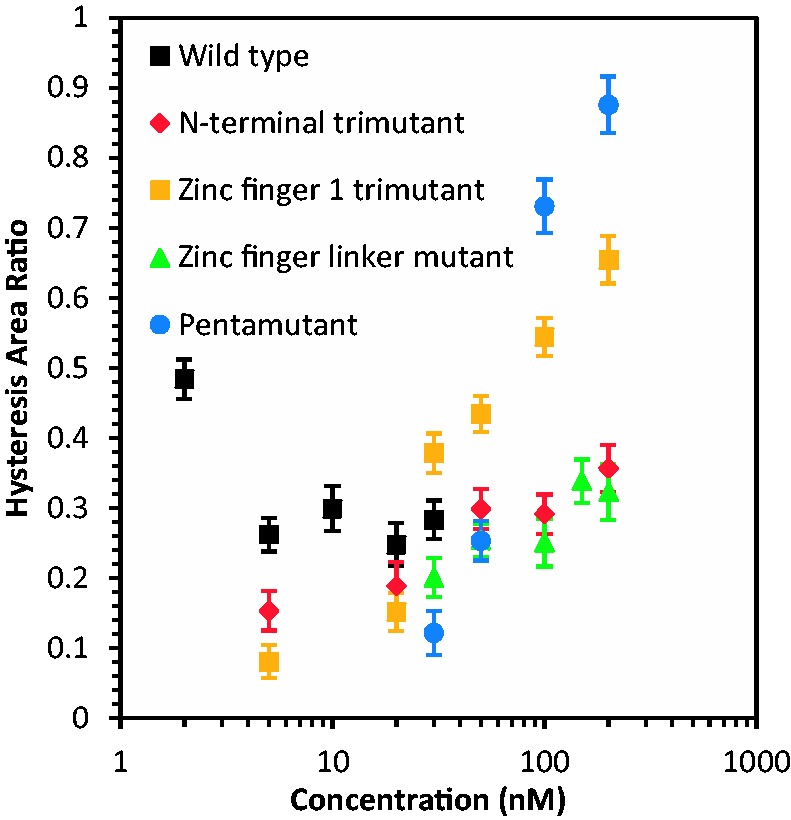
Hysteresis ratio versus concentration curves for WT HIV-1 NC and cationic mutants.
The fact that all of these mutations alter DNA interactions is notable, as the net effective charge of WT HIV-1 NC binding to NA is only ∼+3.5, which is much smaller than the total number of cationic residues (+15) (48–52) distributed throughout the protein. The effective charge implies that ∼3.5 Na+ ions are released from the NA on HIV-1 NC binding. The fact that mutation of several different subsets of cationic residues along the protein sequence affects the binding implies that a majority of the cationic residues participate in direct interactions with the NA phosphates, but to different extents. Thus, the extent to which mutations contribute to HIV-1 NC-DNA interaction defects does not completely correlate with the total number of altered basic residues (3, 4 or 5). The quantity most reflective of the ability of NC to intercalate DNA, S, is greatest for the least number of residues changed (N-terminal trimutant) and lowest for the greatest number of residues changed (pentamutant). The N-terminal trimutant and the zinc finger 1 trimutant exhibit different slopes within uncertainty, despite having the same number of basic residues mutated. These observations illustrate the differential contributions of the cationic mutations to the ability of NC to intercalate. These results are consistent with the effects observed for cationic mutations on high mobility group proteins and other intercalating proteins (80).
Ensemble studies of HIV-1 NC basic residue variants
FA binding studies
We next examined the effect of basic residue mutations on binding to two oligonucleotides using FA. We chose an 18-nt RNA derived from TAR (micro-TAR RNA) and a ssDNA sequence known to be a preferred NC-binding substrate, the 8-nt (TG)4 oligomer (82). Relative to WT HIV-1 NC, which bound to micro-TAR and (TG)4 with relatively high affinity (315 and 5 nM Kd, respectively), the pentamutant bound with 8- and 12-fold reduced affinity to these oligonucleotides, respectively (Table 1). The N-terminal trimutant was less defective in NA binding than the pentamutant, displaying 5-fold lower affinity binding to both oligonucleotides (Table 1). The zinc finger 1 mutant only displayed 2- to 4-fold lower affinity binding to micro-TAR RNA and (TG)4, respectively. The linker mutant resulted in the largest binding defect to (TG)4 (16-fold) but only a <2-fold decreased affinity to micro-TAR. Overall, the cationic mutations in NC significantly alter binding to both nucleotide sequences. Interestingly, mutations of NC’s aromatic residues resulted in poor binding to the ssDNA sequence (TG)4, while the binding of aromatic residue mutants to micro-TAR RNA was only marginally affected (8). These results are consistent with the hypothesis that the cationic residues of NC are responsible primarily for the nonspecific electrostatic binding to NAs. This nonspecific binding is strongest for double-stranded NA molecules, while the aromatic residues in the zinc fingers of NC are responsible for the specific stacking interactions involved in binding to TG-rich single-stranded sequences (51,82,83).
Table 1.
Binding, aggregation and annealing parameters measured for WT HIV-1 NC and basic residue mutants
| HIV-1 NC variant | Micro-TAR RNAa Kd (nM) | (TG)4 DNAa Kd (nM) | %TAR RNA aggregatedb | TAR RNA/DNA annealing ratec min−1 |
|---|---|---|---|---|
| WT | 315 ± 48 | 5 ± 1 | 94.2 ± 1.2 | >6 |
| N-terminal trimutant | 1720 ± 129 | 27 ± 2 | 84.7 ± 3.1 | 1.19 ± 0.56 |
| ZF1 trimutant | 591 ± 138 | 19 ± 10 | 93.8 ± 1.5 | 0.51 ± 0.38 |
| ZF linker mutant | 552 ± 20 | 82 ± 54 | 79.3 ± 9.5 | 0.60 ± 0.41 |
| Pentamutant | 2440 ± 370 | 62 ± 20 | 85.4 ± 2.5 | 0.09 ± 0.07 |
aApparent equilibrium dissociation constants, Kd, for binding to NA oligomers measured at room temperature in 50 mM NaCl by FA as described in ‘Materials and Methods’ section.
bPercent TAR RNA aggregated by 10 µM protein in a solution containing 15 nM radiolabeled TAR RNA and 45 nM TAR DNA at 37°C in 50 mM NaCl.
cTAR RNA-DNA annealing rate in the presence of 10 µM protein under the same solution conditions used in the aggregation assay.
NA annealing
To assess the cationic HIV-1 NC mutants in chaperone function, we performed gel-shift TAR RNA/DNA annealing assays as previously described (8). This assay requires both aggregation and destabilization activity and is therefore a good indicator of overall chaperone function. When low amounts of NC were used [0.88 µM or 1 NC per 4 nt (36)], the WT annealing reaction proceeds with a rate of >∼6 s−1, showing >104-fold annealing rate enhancement over the background rate of annealing in the absence of protein. None of the four cationic HIV-1 NC mutants studied in this work were able to facilitate the annealing kinetics to any measurable degree when added at this concentration (Figure 6A). However, at high concentrations (10 µM), all of the NC basic residue variants facilitated annealing, but to varying extents (Figure 6B). The fitted values of the TAR RNA/DNA annealing rates under these conditions are summarized in Table 1. As 10 µM significantly exceeds the measured Kd values for any of these cationic HIV-1 NC mutants binding to either double-stranded or single-stranded NA, the differences in their ability to facilitate TAR annealing reflects differences in chaperone function.
Figure 6.
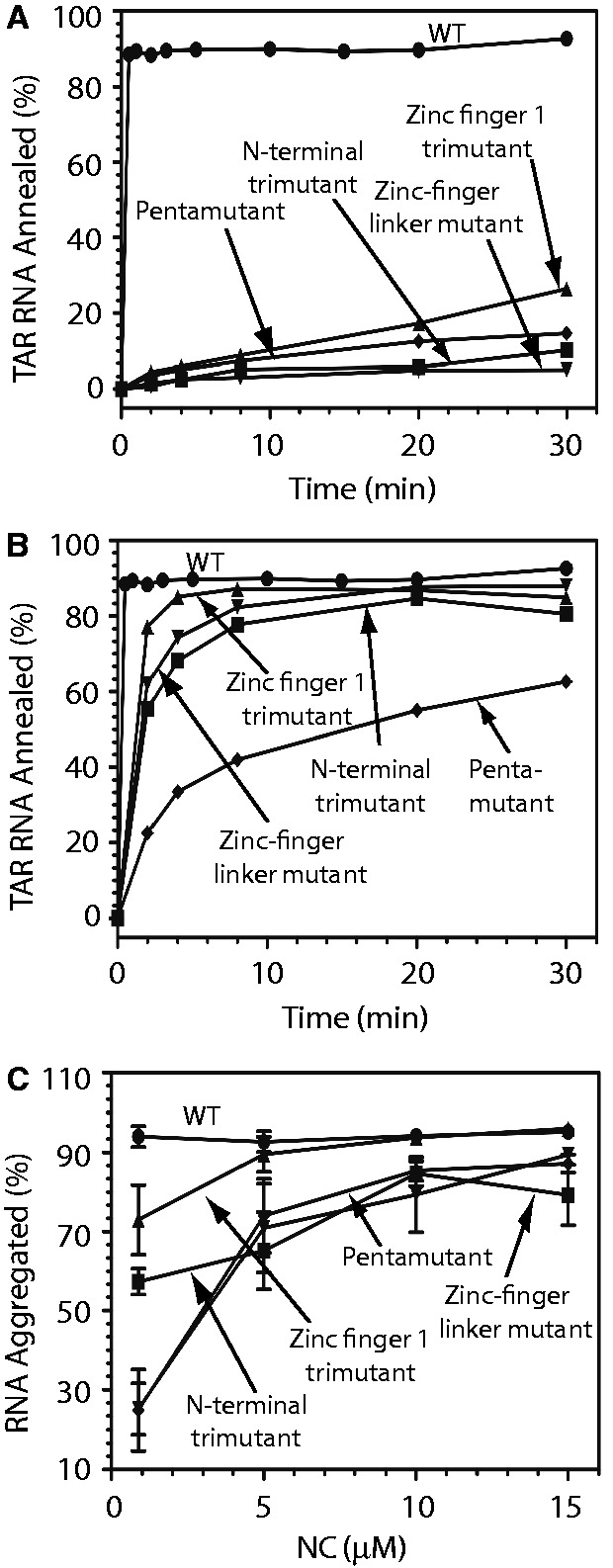
Percent TAR RNA annealed to TAR DNA (A, B) or aggregated (C) by WT or mutant HIV-1 NC. Annealing assays were conducted using 0.88 µM WT or mutant NC (panel A) or 0.88 µM WT and 10 µM mutant NC (panel B). Aggregation assays were performed as a function of protein concentration as indicated.
NA aggregation
The effect of basic residue mutations on NA aggregation is summarized in Figure 6C. All cationic HIV-1 NC mutants displayed reduced NA aggregation activity relative to WT HIV-1 NC, as expected from their reduced NA binding affinity. The effect generally increased with increasing numbers of basic residues mutated. Substitution of the three basic residues in the N-terminus (trimutant) had a greater effect on RNA aggregation than neutralizing the three basic residues in zinc finger 1. The linker mutant and pentamutant were the most defective at low NC concentrations but were almost as effective at NA aggregation as WT NC at 10 µM concentration (Table 1). NC aggregates all NA nonspecifically through an electrostatic mechanism that is due to its overall high charge density (6,12,13,19,45). This electrostatic attraction mechanism is similar to that observed for multivalent cations of charge ≥+3 (84–87). The fact that NC mutants with 3 to 5 positive charges neutralized still retain the ability to aggregate NA at saturated binding illustrates the adjustable nature of NC–NA interactions. Here it appears that the role of the neutralized residues can be replaced by the remaining positive charges distributed through the rest of the molecule.
Cell culture-based studies
Table 2 summarizes the effects of the NC basic residue mutations expressed in HIV-1 virions on gRNA packaging, and single- and multiple-round infectivity. Single-round infectivity levels reflect the combined effect of the mutations on the ability of virions to ultimately produce the Tat protein, resulting in positive events in the HCLZ or TZM-bl cells used (58). The fractional gRNA packaging levels reported in the second column of Table 2 show relatively minor defects for all but the pentamutant NC, which only packages ∼3% WT-levels of gRNA. Packaging levels for the zinc finger 1 trimutant are in agreement with the previous study by Poon and Aldovini (88). However, in all cases, single-round infectivities (column 3) are more defective than genome packaging, with the pentamutant being considerably more defective in single-round replication. These defects are compounded in subsequent replication cycles, leading to exponential defectiveness reflected in the multiple-round infectivity assay data in the far right column of Table 2. Although the uncertainties for multiple-round infectivity are close to the mean, the overall measurements vary by orders of magnitude for different mutants. Thus, reduction of the basic character of NC affects genome packaging but more significantly disrupts reverse transcription processes (discussed below) as observed by (i) a reduction in Tat-mediated focus forming activity in single-round assays and (ii) severe reductions in multiple-round replication.
Table 2.
Properties of WT HIV-1 NC and basic residue mutants measured in cells
| NC protein | RNA packaginga | Single-round infectivityb | Relative multiple-round H9 infectivityc |
|---|---|---|---|
| WT | 100 | 100 | 1.0 |
| N-terminal trimutant | 51 ± 19 | 7.4 ± 1.6 | (3.8 ± 3.3)×10−3 |
| Zinc finger 1 trimutant | 31 ± 14 | 8.8 ± 2.4 | (5.0 ± 4.4)×10−5 |
| Zinc finger linker mutant | 61 ± 11 | 3.2 ± 0.7 | (7.2 ± 4.5)×10−5 |
| Pentamutant | 3.0 ± 0.3 | 0.016 ± 0.007 | ≤(7.1)×10−6d |
aDetermined by normalizing genome quantities to equivalent RT activities and reported as % of WT. Results from at least two separate experiments with standard deviations reported.
bEither HCLZ or TZM-bl cells were used for these analyses. Titers (reported as % of WT) were determined by taking the BCFU/ml of the mutant and dividing by WT titer, corrected for input virus (based on exogenous template RT activity). Results are from at least four separate experiments. Errors are standard deviations.
cDetermined by taking the minimum dilution that gives rise to a spreading infection over 8 weeks (average of at least three infection experiments), normalized for equivalent exogenous template RT activities. Titers of mutants reported relative to a WT infection. Errors represent the standard error of the mean. The uncertainties are close to the mean due to the choice of dilutions used.
dThree independent analyses were performed with the undiluted sample of the pentamutant being negative in each assay.
To understand how the observed biophysical effects contribute to viral infectivity and RNA packaging, we calculated the correlation coefficient between several of the in vitro measurements and measurements in cells. The results can be found in Table 3. While all of the in vitro measurements show some correlation with RNA packaging and replication in cells, only the TAR RNA/DNA annealing and single-molecule measurements of the DNA overstretching slope show overall strong correlations.
Table 3.
Correlation of in vitro WT and mutant HIV-1 NC measurements with cellular replication and packaging measurements for the same mutants
| Correlation | RNA packaging | Single-round infectivity | Log (multiple-round infectivity) |
|---|---|---|---|
| Kd, microTAR RNA | −0.74 | −0.53 | −0.48 |
| Kd,(TG)4 DNA | −0.42 | −0.65 | −0.69 |
| RNA/DNA annealing rate | 0.85 | 0.99 | 0.94 |
| DNA stretching hysteresis | −0.91 | −0.52 | −0.76 |
| Transition slope | 0.89 | 0.81 | 0.96 |
Correlation coefficients are determined from Equation (2), using values from Table 1 for Kd or annealing rate and Table 2 for measurements in cells. Single molecule transition slope and hysteresis were evaluated at 200 nM NC for the mutants and 20 nM for WT NC. To take into account the exponential effect of multiple rounds of replication, we used the logarithm of the infectivity for correlation calculations. Positive results mean that the two quantities are correlated and negative results mean inverse correlation. Assuming a one-tailed distribution, for five measurements the correlation must be greater than 0.805 for 95% confidence in the correlation, and 0.687 for 90% confidence (59). Therefore, correlations >0.69 should be considered strong for this number of measurements.
DISCUSSION
In this work, we study the effect of HIV-1 NC cationic mutants on protein–NA interactions and retroviral replication. All of the basic residue mutations that were tested reduced micro-TAR RNA binding affinity, with the pentamutant showing the greatest reduction in affinity, as expected. However, the N-terminal trimutant exhibits the strongest reduction in binding relative to the number of residues changed. In contrast, the strongest reduction in binding affinity to (TG)4 DNA comes from the zinc finger linker mutant and the pentamutant, with the linker mutant having the strongest effect per residue mutated. Therefore, the ability of NC to bind short NA sequences decreases with the number of neutralized basic residues, but the positions of these residues are also important. These results suggest that the cationic residue mutations on the N-terminal domain most strongly alter the binding affinity to nonspecific NA sequences, represented by the micro-TAR RNA, while mutations on the zinc finger linker have a greater effect on the protein’s ability to stack with NAs, as required for optimal binding to (TG)4. Although all of the mutants aggregated NA at high concentration, they were less effective aggregating agents at the lowest concentration tested (0.88 µM) and the percent aggregated even at 10 µM was somewhat less for all of the mutants except the zinc finger 1 mutant.
The single-molecule studies also revealed defects in DNA interactions due to basic residue mutations. For all of the mutants, the ability of NC to alter the slope of the DNA stretching curve was significantly compromised, but the slope change was recovered to some extent by increasing the concentration of protein used. The ability to alter the stretching slope was weakest for the pentamutant, followed by the zinc finger linker mutant, the zinc finger 1 trimutant and finally the N-terminal trimutant, which was closest to WT NC activity. However, while these effects increase with the number of residues mutated, the zinc finger 1 and N-terminal trimutants exhibit significantly different effects, supporting the conclusion that the positions of the residues that are mutated are critical. Similarly, the amount of hysteresis measured for the mutants also increases with the number of residues mutated, with the hysteresis area ratio at 200 nM being greatest for the pentamutant. However, in this case the zinc finger 1 trimutant is almost as defective as the pentamutant, and the other mutants are closer to WT NC. Overall, the single molecule studies show that each set of mutated residues decreases NC’s ability to optimally interact with DNA to varying extents, depending on the location of the mutated residues.
To determine the extent to which the results of these in vitro measurements reflect the ability of NC to facilitate viral replication in cells, we calculated the correlation coefficient between the in vitro single molecule, binding and annealing measurements and measurements of RNA packaging, single-round infectivity and multiple-round infectivity in cells (Table 3). We have also plotted the measurements in cells as a function of the in vitro measurements, along with their linear fits, and these results are shown in Supplementary Figures S1 and S2. Because cationic residue mutations are expected to primarily reduce electrostatic binding interactions, it would be reasonable to expect the cell-based measurements to result in a negative correlation coefficient with Kd. Surprisingly, however, the measurements in cells correlate only moderately with binding affinity to microTAR or (TG)4 DNA, with the absolute value of these correlation coefficients less than the 90% confidence interval for two of the three measurements, and less than 95% confidence for correlation with all three. Visual examination of the plots in Supplementary Figure S1 (a-f) confirms that there is not a strong correlation between in vitro binding affinity and measurements in cells, although removal of the WT data point from Supplementary Figure S1 (e) results in a strong negative correlation between the binding affinity of mutant NC to (TG)4 DNA and single-round infectivity. The lack of strong correlation between the binding of the cationic NC mutants and the extent of RNA packaging and replication defects observed in cells is consistent with the hypothesis that the aromatic zinc finger residues play the primary role in gRNA selection and chaperone function, while the cationic residues play only a secondary role in these functions. This result is also consistent with the hypothesis that the chaperone activity of NC occurs in vivo when NC concentrations are in excess of the Kd values observed here even for the most binding-defective cationic NC mutants (Table 1). For example, cumulative evidence suggests that HIV-1 reverse transcription is mechanistically linked to CA uncoating (89–98) and the early steps of reverse transcription are likely to occur within an intact CA core.
The ability of NC to facilitate TAR RNA/DNA annealing appears to correlate strongly with RNA packaging and replication measurements based on Table 3, consistent with the importance of overall NA chaperone activity for viral replication (8). However, because the results of the TAR RNA/DNA annealing measurements for WT protein are significantly different than those of the mutants, this leads to an anomalously high correlation between annealing and measurements in cells, as shown in Supplementary Figure S1 (g–i). When the WT NC data point is removed from these graphs, the correlation coefficients become 0.75, 0.66 and 0.99 for correlation of annealing rate with RNA packaging, single-round infectivity and the logarithm of multiple-round infectivity, respectively. This suggests a moderate to strong correlation between TAR RNA/DNA annealing and replication measurements in cells. Similarly, the ability of NC to prevent DNA strand separation by force, as reflected in small hysteresis, also correlates strongly with RNA packaging, and moderately with multiple-round infectivity (Table 3, DNA stretching hysteresis). The hysteresis measurements do not change significantly when influential data points are removed. In addition to RNA/DNA annealing, the other strong correlation between in vitro measurements and measurements in cells is in the slope of the single-molecule DNA stretching curve. All three measurements in cells correlate strongly with the slope S, even when influential data points are removed. As discussed above, this change in slope reflects the ability of the protein to elongate dsDNA without allowing the strands to separate. This elongation at high forces represents a measurement of DNA intercalation by NC, which only occurs at high forces. However, the correlation with replication measurements suggests that this optimal ability to intercalate is a critical component of NC’s NA chaperone activity. Similar stacking interactions between the F16 and W37 residues of NC and single-stranded bases were recently characterized by NMR (79). These interactions resemble the hemi-intercalation observed between the aromatic rings of the dsDNA intercalator Act D and ssDNA bases (99). While the observed intercalation is too weak to occur frequently at zero force when NAs are fully double-stranded, the intercalation measured in these single-molecule experiments likely becomes important in cases of locally unstable elements of NA secondary structure, such as duplexes containing mismatches, loops or bulges. Thus, these combined experiments demonstrate a strong correlation between in vitro NA chaperone activity and cellular replication measurements, illustrating the importance of specific HIV-1 NC basic residues for these processes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E with Leidos Biomedical Research, Inc. (RJG); National Institutes of Health [GM065056 to K.M.-F. and GM072462 to M.C.W.]; National Science Foundation [MCB-1243883 to M.C.W]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. We wish to thank Donald Johnson and Catherine Hixson for the assistance in preparing the recombinant NC proteins used in this study.
REFERENCES
- 1.Berkowitz RD, Ohagen A, Höglund S, Goff SP. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J. Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 3.Freed EO. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 4.Covey SN. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 1986;14:623–633. doi: 10.1093/nar/14.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg JM. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986;232:485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- 6.Qualley DF, Stewart-Maynard KM, Wang F, Mitra M, Gorelick RJ, Rouzina I, Williams MC, Musier-Forsyth K. C-terminal domain modulates the nucleic acid chaperone activity of human T-cell leukemia virus type 1 nucleocapsid protein via an electrostatic mechanism. J. Biol. Chem. 2010;285:295–307. doi: 10.1074/jbc.M109.051334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorfman T, Luban J, Goff SP, Haseltine WA, Göttlinger HG. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu H, Mitra M, McCauley MJ, Thomas JA, Rouzina I, Musier-Forsyth K, Williams MC, Gorelick RJ. Aromatic residue mutations reveal direct correlation between HIV-1 nucleocapsid protein's nucleic acid chaperone activity and retroviral replication. Virus Res. 2013;171:263–277. doi: 10.1016/j.virusres.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darlix JL, Lapadat-Tapolsky M, de Rocquigny H, Roques BP. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 10.Lapadat-Tapolsky M, de Rocquigny H, Van Gent D, Roques B, Plasterk R, Darlix J-L. Interaction between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21:831–839. doi: 10.1093/nar/21.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lapadat-Tapolsky M, Pernelle C, Borie C, Darlix JL. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res. 1995;23:2434–2441. doi: 10.1093/nar/23.13.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Cam E, Coulaud D, Delain E, Petitjean P, Roques BP, Gérard D, Stoylova E, Vuilleumier C, Stoylov SP, Mély Y. Properties and growth mechanism of the ordered aggregation of a model RNA by the HIV-1 nucleocapsid protein: an electron microscopy investigation. Biopolymers. 1998;45:217–229. doi: 10.1002/(SICI)1097-0282(199803)45:3<217::AID-BIP4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 13.Stoylov SP, Vuilleumier C, Stoylova E, De Rocquigny H, Roques BP, Gérard D, Mély Y. Ordered aggregation of ribonucleic acids by the human immunodeficiency virus type 1 nucleocapsid protein. Biopolymers. 1997;41:301–312. doi: 10.1002/(SICI)1097-0282(199703)41:3<301::AID-BIP5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 14.Onafuwa-Nuga A, Telesnitsky A. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol. Mol. Biol. Rev. 2009;73:451–480. doi: 10.1128/MMBR.00012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 16.Williams MC, Gorelick RJ, Musier-Forsyth K. Specific zinc finger architecture required for HIV-1 nucleocapsid protein’s nucleic acid chaperone function. Proc. Natl Acad. Sci. USA. 2002;99:8614–8619. doi: 10.1073/pnas.132128999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuchihashi Z, Brown PO. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams MC, Rouzina I, Wenner JR, Gorelick RJ, Musier-Forsyth K, Bloomfield VA. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc. Natl Acad. Sci. USA. 2001;98:6121–6126. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu H, Rouzina I, Williams MC. Single-molecule stretching studies of RNA chaperones. RNA Biol. 2010;7:712–723. doi: 10.4161/rna.7.6.13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 2010;7:754–774. doi: 10.4161/rna.7.6.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darlix JL, Godet J, Ivanyi-Nagy R, Fosse P, Mauffret O, Mely Y. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J. Mol. Biol. 2011;410:565–581. doi: 10.1016/j.jmb.2011.03.037. [DOI] [PubMed] [Google Scholar]
- 22.Godet J, Mely Y. Biophysical studies of the nucleic acid chaperone properties of the HIV-1 nucleocapsid protein. RNA Biol. 2010;7:687–699. doi: 10.4161/rna.7.6.13616. [DOI] [PubMed] [Google Scholar]
- 23.Liu HW, Cosa G, Landes CF, Zeng Y, Kovaleski BJ, Mullen DG, Barany G, Musier-Forsyth K, Barbara PF. Single-molecule FRET studies of important intermediates in the nucleocapsid-protein-chaperoned minus-strand transfer step in HIV-1 reverse transcription. Biophys. J. 2005;89:3470–3479. doi: 10.1529/biophysj.105.065326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson JA, Teufel RJ, II, Yin PD, Hu WS. Correlated Template-Switching Events during Minus-Strand DNA Synthesis: a Mechanism for High Negative Interference during Retroviral Recombination. J. Virol. 1998;72:1186–1194. doi: 10.1128/jvi.72.2.1186-1194.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bampi C, Jacquenet S, Lener D, Décimo D, Darlix JL. The chaperoning and assistance roles of the HIV-1 nucleocapsid protein in proviral DNA synthesis and maintenance. Curr. HIV Res. 2004;2:79–92. doi: 10.2174/1570162043485022. [DOI] [PubMed] [Google Scholar]
- 26.Mark-Danieli M, Laham N, Kenan-Eichler M, Castiel A, Melamed D, Landau M, Bouvier NM, Evans MJ, Bacharach E. Single point mutations in the zinc finger motifs of the human immunodeficiency virus type 1 nucleocapsid alter RNA binding specificities of the gag protein and enhance packaging and infectivity. J. Virol. 2005;79:7756–7767. doi: 10.1128/JVI.79.12.7756-7767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Negroni M, Buc H. Recombination during reverse transcription: an evaluation of the role of the nucleocapsid protein. J. Mol. Biol. 1999;286:15–31. doi: 10.1006/jmbi.1998.2460. [DOI] [PubMed] [Google Scholar]
- 28.Negroni M, Buc H. Mechanisms of retroviral recombination. Ann. Rev. Genet. 2001;35:275–302. doi: 10.1146/annurev.genet.35.102401.090551. [DOI] [PubMed] [Google Scholar]
- 29.Hargittai MRS, Gorelick RJ, Rouzina I, Musier-Forsyth K. Mechanistic insights into the kinetics of HIV-1 Nucleocapsid Protein-facilitated tRNA annealing to the primer binding site. J. Mol. Biol. 2004;337:951–968. doi: 10.1016/j.jmb.2004.01.054. [DOI] [PubMed] [Google Scholar]
- 30.You JC, McHenry CS. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J. Biol. Chem. 1994;269:31491–31495. [PubMed] [Google Scholar]
- 31.Peliska JA, Balasubramanian S, Giedroc DP, Benkovic SJ. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry. 1994;33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- 32.Rodríguez-Rodríguez L, Tsuchihashi Z, Fuentes GM, Bambara RA, Fay PJ. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J. Biol. Chem. 1995;270:15005–15011. doi: 10.1074/jbc.270.25.15005. [DOI] [PubMed] [Google Scholar]
- 33.Guo J, Wu T, Anderson J, Kane BF, Johnson DG, Gorelick RJ, Henderson LE, Levin JG. Zinc Finger Structures in the Human Immunodeficiency Virus Type 1 Nucleocapsid Protein Facilitate Efficient Minus- and Plus-Strand Transfer. J. Virol. 2000;74:8980–8988. doi: 10.1128/jvi.74.19.8980-8988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson PE, Turner RB, Wu ZR, Hairston L, Guo J, Levin JG, Summers MF. a mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription. Biochemistry. 2000;39:9084–9091. doi: 10.1021/bi000841i. [DOI] [PubMed] [Google Scholar]
- 35.Cruceanu M, Gorelick RJ, Musier-Forsyth K, Rouzina I, Williams MC. Rapid kinetics of protein-nucleic acid interaction is a major component of HIV-1 nucleocapsid protein's nucleic acid chaperone function. J. Mol. Biol. 2006;363:867–877. doi: 10.1016/j.jmb.2006.08.070. [DOI] [PubMed] [Google Scholar]
- 36.Vo MN, Barany G, Rouzina I, Musier-Forsyth K. Mechanistic studies of mini-TAR RNA/DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. J. Mol. Biol. 2006;363:244–261. doi: 10.1016/j.jmb.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 37.Williams MC, Rouzina I, Bloomfield VA. Thermodynamics of DNA interactions from single molecule stretching experiments. Acc. Chem. Res. 2002;35:159–166. doi: 10.1021/ar010045k. [DOI] [PubMed] [Google Scholar]
- 38.Cruceanu M, Stephen AG, Beuning PJ, Gorelick RJ, Fisher RJ, Williams MC. Single DNA molecule stretching measures the activity of chemicals that target the HIV-1 nucleocapsid protein. Anal. Biochem. 2006;358:159–170. doi: 10.1016/j.ab.2006.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hergott CB, Mitra M, Guo J, Wu T, Miller JT, Iwatani Y, Gorelick RJ, Levin JG. Zinc finger function of HIV-1 nucleocapsid protein is required for removal of 5'-terminal genomic RNA fragments: a paradigm for RNA removal reactions in HIV-1 reverse transcription. Virus Res. 2013;171:346–355. doi: 10.1016/j.virusres.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mirambeau G, Lyonnais S, Coulaud D, Hameau L, Lafosse S, Jeusset J, Justome A, Delain E, Gorelick RJ, Le Cam E. Transmission electron microscopy reveals an optimal HIV-1 nucleocapsid aggregation with single-stranded nucleic acids and the mature HIV-1 nucleocapsid protein. J. Mol. Biol. 2006;364:496–511. doi: 10.1016/j.jmb.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 41.Beltz H, Clauss C, Piemont E, Ficheux D, Gorelick RJ, Roques B, Gabus C, Darlix JL, de Rocquigny H, Mély Y. Structural determinants of HIV-1 nucleocapsid protein for cTAR DNA binding and destabilization, and correlation with inhibition of self-primed DNA synthesis. J. Mol. Biol. 2005;348:1113–1126. doi: 10.1016/j.jmb.2005.02.042. [DOI] [PubMed] [Google Scholar]
- 42.Godet J, Ramalanjaona N, Sharma KK, Richert L, de Rocquigny H, Darlix JL, Duportail G, Mely Y. Specific implications of the HIV-1 nucleocapsid zinc fingers in the annealing of the primer binding site complementary sequences during the obligatory plus strand transfer. Nucleic Acids Res. 2011;39:6633–6645. doi: 10.1093/nar/gkr274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the Psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 44.de Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 45.Stewart-Maynard KM, Cruceanu M, Wang F, Vo MN, Gorelick RJ, Williams MC, Rouzina I, Musier-Forsyth K. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J. Virol. 2008;82:10129–10142. doi: 10.1128/JVI.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaurasiya KR, Geertsema H, Cristofari G, Darlix JL, Williams MC. A single zinc finger optimizes the DNA interactions of the nucleocapsid protein of the yeast retrotransposon Ty3. Nucleic Acids Res. 2012;40:751–760. doi: 10.1093/nar/gkr726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin SL, Bushman D, Wang F, Li PW, Walker A, Cummiskey J, Branciforte D, Williams MC. A single amino acid substitution in ORF1 dramatically decreases L1 retrotransposition and provides insight into nucleic acid chaperone activity. Nucleic Acids Res. 2008;36:5845–5854. doi: 10.1093/nar/gkn554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urbaneja MA, Kane BP, Johnson DG, Gorelick RJ, Henderson LE, Casas-Finet JR. Binding Properties of the Human Immunodeficiency Virus Type 1 Nucleocapsid Protein p7 to a Model RNA: Elucidation of the Structural Determinants for Function. J. Mol. Biol. 1999;287:59–75. doi: 10.1006/jmbi.1998.2521. [DOI] [PubMed] [Google Scholar]
- 49.Urbaneja MA, Wu M, Casas-Finet JR, Karpel RL. HIV-1 nucleocapsid protein as a nucleic acid chaperone: spectroscopic study of its helix-destabilizing properties, structural binding specificity, and annealing activity. J. Mol. Biol. 2002;318:749–764. doi: 10.1016/S0022-2836(02)00043-8. [DOI] [PubMed] [Google Scholar]
- 50.Vo MN, Barany G, Rouzina I, Musier-Forsyth K. Effect of Mg(2+) and Na(+) on the nucleic acid chaperone activity of HIV-1 nucleocapsid protein: implications for reverse transcription. J. Mol. Biol. 2009;386:773–788. doi: 10.1016/j.jmb.2008.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vuilleumier C, Bombarda E, Morellet N, Gerard D, Roques BP, Mély Y. Nucleic acid sequence discrimination by the HIV-1 nucleocapsid protein NCp7: a fluorescence study. Biochemistry. 1999;38:16816–16825. doi: 10.1021/bi991145p. [DOI] [PubMed] [Google Scholar]
- 52.Athavale SS, Ouyang W, McPike MP, Hudson BS, Borer PN. Effects of the nature and concentration of salt on the interaction of the HIV-1 nucleocapsid protein with SL3 RNA. Biochemistry. 2010;49:3525–3533. doi: 10.1021/bi901279e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carteau S, Gorelick RJ, Bushman FD. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 1999;73:6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCauley MJ, Williams MC. Optical tweezers experiments resolve distinct modes of DNA-protein binding. Biopolymers. 2009;91:265–282. doi: 10.1002/bip.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, Fivash MJ, Stephen AG, Fisher RJ, Gorelick RJ, Casas-Finet JR, et al. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 2006;34:593–605. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gorelick R, Gagliardi T, Bosche W, Wiltrout T, Coren L, Chabot D, Lifson J, Henderson L, Arthur A. Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: Characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology. 1999;256:92–104. doi: 10.1006/viro.1999.9629. [DOI] [PubMed] [Google Scholar]
- 58.Morcock DR, Thomas JA, Sowder RC, II, Henderson LE, Crise BJ, Gorelick RJ. HIV-1 inactivation by 4-vinylpyridine is enhanced by dissociating Zn(2+) from nucleocapsid protein. Virology. 2008;375:148–158. doi: 10.1016/j.virol.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bevington PR, Robinson DK. Data Reduction and Error Analysis for the Physical Sciences. Boston: McGraw-Hill; 1992. [Google Scholar]
- 60.Shokri L, McCauley MJ, Rouzina I, Williams MC. DNA overstretching in the presence of glyoxal: structural evidence of force-induced DNA melting. Biophys. J. 2008;95:1248–1255. doi: 10.1529/biophysj.108.132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaurasiya KR, Paramanathan T, McCauley MJ, Williams MC. Biophysical characterization of DNA binding from single molecule force measurements. Phys. Life Rev. 2010;7:299–341. doi: 10.1016/j.plrev.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Mameren J, Gross P, Farge G, Hooijman P, Modesti M, Falkenberg M, Wuite GJ, Peterman EJ. Unraveling the structure of DNA during overstretching by using multicolor, single-molecule fluorescence imaging. Proc. Natl Acad. Sci. USA. 2009;106:18231–18236. doi: 10.1073/pnas.0904322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.King GA, Gross P, Bockelmann U, Modesti M, Wuite GJ, Peterman EJ. Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching using fluorescence microscopy. Proc. Natl Acad. Sci. USA. 2013;110:3859–3864. doi: 10.1073/pnas.1213676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang X, Chen H, Le S, Rouzina I, Doyle PS, Yan J. Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching by single-molecule calorimetry. Proc. Natl Acad. Sci. USA. 2013;110:3865–3870. doi: 10.1073/pnas.1213740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vladescu ID, McCauley MJ, Rouzina I, Williams MC. Mapping the phase diagram of single DNA molecule force-induced melting in the presence of ethidium. Phys. Rev. Lett. 2005;95:158102. doi: 10.1103/PhysRevLett.95.158102. [DOI] [PubMed] [Google Scholar]
- 66.Mihailovic A, Vladescu L, McCauley M, Ly E, Williams MC, Spain EM, Nunez ME. Exploring the interaction of ruthenium(II) polypyridyl complexes with DNA using single-molecule techniques. Langmuir. 2006;22:4699–4709. doi: 10.1021/la053242r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vladescu ID, McCauley MJ, Nunez ME, Rouzina I, Williams MC. Quantifying force-dependent and zero-force DNA intercalation by single-molecule stretching. Nat. Methods. 2007;4:517–522. doi: 10.1038/nmeth1044. [DOI] [PubMed] [Google Scholar]
- 68.Kleimann C, Sischka A, Spiering A, Tonsing K, Sewald N, Diederichsen U, Anselmetti D. Binding kinetics of bisintercalator Triostin a with optical tweezers force mechanics. Biophys. J. 2009;97:2780–2784. doi: 10.1016/j.bpj.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paramanathan T, Vladescu I, McCauley MJ, Rouzina I, Williams MC. Force spectroscopy reveals the DNA structural dynamics that govern the slow binding of Actinomycin D. Nucleic Acids Res. 2012;40:4925–4932. doi: 10.1093/nar/gks069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Rocquigny H, Shvadchak V, Avilov S, Dong CZ, Dietrich U, Darlix JL, Mely Y. Targeting the viral nucleocapsid protein in anti-HIV-1 therapy. Mini Rev. Med. Chem. 2008;8:24–35. doi: 10.2174/138955708783331603. [DOI] [PubMed] [Google Scholar]
- 71.Mely Y, Piemont E, Sorinas-Jimeno M, De Rocquigny H, Jullian N, Morellet N, Roques BP, Gerard D. Structural and dynamic characterization of the aromatic amino acids of the human immunodeficiency virus type I nucleocapsid protein zinc fingers and their involvement in heterologous tRNA(Phe) binding: a steady-state and time-resolved fluorescence study. Biophys. J. 1993;65:1513–1522. doi: 10.1016/S0006-3495(93)81222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paoletti AC, Shubsda MF, Hudson BS, Borer PN. Affinities of the nucleocapsid protein for variants of SL3 RNA in HIV-1. Biochemistry. 2002;41:15423–15428. doi: 10.1021/bi026307n. [DOI] [PubMed] [Google Scholar]
- 73.Morellet N, Demene H, Teilleux V, Huynh-Dinh T, de Rocquigny H, Fournie-Zaluski MC, Roques BP. Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACGCC) J. Mol. Biol. 1998;283:419–434. doi: 10.1006/jmbi.1998.2098. [DOI] [PubMed] [Google Scholar]
- 74.Amarasinghe GK, Zhou J, Miskimon M, Chancellor KJ, McDonald JA, Matthews AG, Miller RR, Rouse MD, Summers MF. Stem-loop SL4 of the HIV-1 psi RNA packaging signal exhibits weak affinity for the nucleocapsid protein. structural studies and implications for genome recognition. J. Mol. Biol. 2001;314:961–970. doi: 10.1006/jmbi.2000.5182. [DOI] [PubMed] [Google Scholar]
- 75.Morellet N, de Rocquigny H, Mély Y, Jullian N, Déméné H, Ottmann M, Gérard D, Darlix JL, Fournie-Zaluski MC, Roques BP. Conformational behaviour of the active and inactive forms of the nucleocapsid NCp7 of HIV-1 studied by 1H NMR. J. Mol. Biol. 1994;235:287–301. doi: 10.1016/s0022-2836(05)80033-6. [DOI] [PubMed] [Google Scholar]
- 76.Morellet N, Jullian N, De Rocquigny H, Maigret B, Darlix JL, Roques BP. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by1H NMR. EMBO J. 1992;11:3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Summers MF, South TL, Kim B, Hare DR. High resolution structure of an HIV zinc finger via a new NMR-based distance geometry aproach. Biochemistry. 1990;29:329–340. doi: 10.1021/bi00454a005. [DOI] [PubMed] [Google Scholar]
- 78.Tisné C, Roques B, Dardel F. Heteronuclear NMR studies of the interaction of tRNA(Lys)3 with HIV-1 nucleocapsid protein. J. Mol. Biol. 2001;306:443–454. doi: 10.1006/jmbi.2000.4391. [DOI] [PubMed] [Google Scholar]
- 79.Bazzi A, Zargarian L, Chaminade F, Boudier C, De Rocquigny H, Rene B, Mely Y, Fosse P, Mauffret O. Structural insights into the cTAR DNA recognition by the HIV-1 nucleocapsid protein: role of sugar deoxyriboses in the binding polarity of NC. Nucleic Acids Res. 2011;39:3903–3916. doi: 10.1093/nar/gkq1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Werner MH, Gronenborn AM, Clore GM. Intercalation, DNA kinking, and the control of transcription. Science. 1996;271:778–784. doi: 10.1126/science.271.5250.778. [DOI] [PubMed] [Google Scholar]
- 81.Shubsda MF, Paoletti AC, Hudson BS, Borer PN. Affinities of packaging domain loops in HIV-1 RNA for the nucleocapsid protein. Biochemistry. 2002;41:5276–5282. doi: 10.1021/bi016045+. [DOI] [PubMed] [Google Scholar]
- 82.Fisher RJ, Rein A, Fivash M, Urbaneja MA, Casas-Finet JR, Medaglia M, Henderson LE. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J. Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fisher RJ, Fivash MJ, Stephen AG, Hagan NA, Shenoy SR, Medaglia MV, Smith LR, Worthy KM, Simpson JT, Shoemaker R, et al. Complex interactions of HIV-1 nucleocapsid protein with oligonucleotides. Nucleic Acids Res. 2006;34:472–484. doi: 10.1093/nar/gkj442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bloomfield VA. DNA condensation by multivalent cations. Biopolymers. 1997;44:269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 85.Pelta J, Livolant F, Sikorav JL. DNA condensation by polyamines and cobalthexamine. J. Biol. Chem. 1996;271:13644–13651. doi: 10.1074/jbc.271.10.5656. [DOI] [PubMed] [Google Scholar]
- 86.Rouzina I, Bloomfield VA. Macroion attraction due to electrostatic correlation between screening counterions.1. Mobile surface-adsorbed ions and diffuse ion cloud. J. Phys. Chem. 1996;100:9977–9989. [Google Scholar]
- 87.Nguyen TT, Rouzina I, Shklovskii BI. Reentrant Condensation of DNA induced by Multivalent Counterions. J. Chem. Phys. 2000;112:2562–2568. [Google Scholar]
- 88.Poon DT, Wu J, Aldovini A. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J. Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang Y, Fricke T, Diaz-Griffero F. Inhibition of reverse transcriptase activity increases stability of the HIV-1 core. J. Virol. 2013;87:683–687. doi: 10.1128/JVI.01228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Diaz-Griffero F, Kar A, Lee M, Stremlau M, Poeschla E, Sodroski J. Comparative requirements for the restriction of retrovirus infection by TRIM5alpha and TRIMCyp. Virology. 2007;369:400–410. doi: 10.1016/j.virol.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Perron MJ, Stremlau M, Lee M, Javanbakht H, Song B, Sodroski J. The human TRIM5alpha restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J. Virol. 2007;81:2138–2148. doi: 10.1128/JVI.02318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl Acad. Sci. USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ohagen A, Gabuzda D. Role of Vif in stability of the human immunodeficiency virus type 1 core. J. Virol. 2000;74:11055–11066. doi: 10.1128/jvi.74.23.11055-11066.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Roa A, Hayashi F, Yang Y, Lienlaf M, Zhou J, Shi J, Watanabe S, Kigawa T, Yokoyama S, Aiken C, et al. RING domain mutations uncouple TRIM5alpha restriction of HIV-1 from inhibition of reverse transcription and acceleration of uncoating. J. Virol. 2012;86:1717–1727. doi: 10.1128/JVI.05811-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Diaz-Griffero F, Perron M, McGee-Estrada K, Hanna R, Maillard PV, Trono D, Sodroski J. A human TRIM5alpha B30.2/SPRY domain mutant gains the ability to restrict and prematurely uncoat B-tropic murine leukemia virus. Virology. 2008;378:233–242. doi: 10.1016/j.virol.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Arfi V, Lienard J, Nguyen XN, Berger G, Rigal D, Darlix JL, Cimarelli A. Characterization of the behavior of functional viral genomes during the early steps of human immunodeficiency virus type 1 infection. J. Virol. 2009;83:7524–7535. doi: 10.1128/JVI.00429-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Berger G, Durand S, Fargier G, Nguyen XN, Cordeil S, Bouaziz S, Muriaux D, Darlix JL, Cimarelli A. APOBEC3A is a specific inhibitor of the early phases of HIV-1 infection in myeloid cells. PLoS Pathog. 2011;7:e1002221. doi: 10.1371/journal.ppat.1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hulme AE, Perez O, Hope TJ. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc. Natl Acad. Sci. USA. 2011;108:9975–9980. doi: 10.1073/pnas.1014522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wadkins RM, Jares-Erijman EA, Klement R, Rudiger A, Jovin TM. Actinomycin D binding to single-stranded DNA: sequence specificity and hemi-intercalation model from fluorescence and 1H NMR spectroscopy. J. Mol. Biol. 1996;262:53–68. doi: 10.1006/jmbi.1996.0498. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.