Preface
Biofilms are ubiquitous communities of tightly associated bacteria encased in an extracellular matrix. Bacillus subtilis has long-served as a robust model organism to examine the molecular mechanisms of biofilm formation and a number of studies have revealed that this process is subject to a number of integrated regulatory pathways. In this Review, we focus on the molecular mechanisms controlling biofilm assembly and briefly summarize the current state of knowledge regarding their disassembly. We also discuss recent progress that has expanded our understanding of biofilm formation on plant roots, which are a natural habitat for this soil bacterium.
Introduction
“At the surface of the liquid… …The rods adhere together by their sides after the manner of the elements of columnar epithelium, but there is, I think, strong reason to believe that this adhesion is not direct, i.e that they are not in actual contact but glued together by a viscous intermediary substance.”
The “viscous intermediary substance” described here by Burton-Sanderson in 1870 is a hallmark feature of biofilms and the image he portrays is in all likelihood a Bacillus subtilis biofilm. From the dawn of microbiology this Gram-positive bacterium has been the subject of thorough investigation. Its capacity to sporulate and form biofilms was already beautifully described in the classic work of Ferdinand Cohn in 18771.
Biofilms are communities of surface-associated microorganisms encased in a self-produced extracellular matrix. Biofilm formation is a nearly universal bacterial trait and biofilms are found on almost all natural and artificial surfaces2,3. They are widely studied because they represent a fascinating example of microbial development and also because they can be problematic in many man-made settings4-6. In clinical settings they form on virtually any indwelling device and in industrial settings they often clog pipes and tubing7. But there is also interest in exploiting the beneficial aspects of biofilms; they play a major role in wastewater treatment and are potential sources of energy in the form of microbial fuel cells8-10. While most natural biofilms are polymicrobial communities, much has been learned about the basic biology of biofilms through the study of single species biofilms using model bacteria. Because of the clinical relevance of biofilms, most of the model systems that were initially studied involved pathogenic bacteria that were predominantly Gram-negative. For example, Pseudomonas aeruginosa is arguably the most studied bacterium in the biofilm field7,11.
Over the past decade, B. subtilis, which is a non-pathogenic Gram-positive bacterium, has emerged as an alternative model organism for studying the molecular basis of biofilm formation. A general schematic depicting the different stages of B. subtilis biofilm formation is shown in Figure 1. Within the biofilm, genetically identical cells express different genes and produce subpopulations of functionally distinct co-existing cell types. The process begins with the expression of matrix genes in response to some external signal (such as surfactin). Initially, cells are short, motile rods but as the biofilm develops, they form long chains of non-motile cells that adhere to each other and the surface by secreting an extracellular matrix12-14. This substance is essential to the integrity of the biofilm as it holds the community together15-17. As the biofilm matures the cell clusters enlarge and the community is protected and organized by the extracellular matrix. In addition to matrix producers, motile cells and spores are also present and are spatially organized within the maturing biofilm (reviewed in18,19). The presence and localization of the different cell types is dynamic and there appears to be an ordered sequence of differentiation in which motile cells become matrix-producing cells, which go on to become spores20. Importantly, this is not terminal differentiation; as conditions change it is possible for cells to alter gene expression (in the case of motile or matrix-producing cells) or germinate (in the case of the spores). Phenotypic heterogeneity in B. subtilis is not limited to these three cell types. The topic of heterogeneity and the processes that regulate this heterogeneity have been extensively covered in several reviews19,21-24. In laboratory conditions, biofilms have a limited life span and they eventually disassemble in response to self-generated signals25,26. As biofilms disassemble, spores are released from the matrix giving them the potential to disperse and encounter environmental conditions that are propitious for germination.
Figure 1.
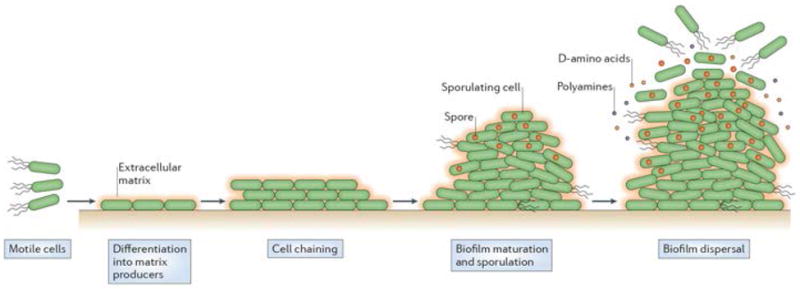
The life-cycle of a Bacillus subtilis biofilm. This process occurs in stages which comprise the development, maturation and disassembly of the community. Motile cells with flagella differentiate into non-motile matrix-producing cells that become organized in chains and are surrounded by extracellular matrix (orange). In mature biofilms matrix-producing cells sporulate (dark brown spores). In aged biofilms, some cells secrete small molecules such as D-amino acids and polyamines that break down the extracellular matrix and the cells disperse in the environment.
Much is known about the molecular mechanisms that regulate entry into biofilm formation, the structural components that comprise the extracellular matrix and how the biofilm eventually disassembles. In this review we describe the major features of B. subtilis biofilms with a focus on the various signals and mechanisms that regulate expression of the matrix genes, which induce biofilm formation. We also discuss recent findings relating to the secreted molecules produced by cells within the biofilm that target the extracellular matrix to disassemble the community. This research has shed light on the potential to control biofilm growth of other pathogenic bacteria. Finally, we discuss the use of plant roots as a natural habitat for the study of B. subtilis biofilms.
Biofilm morphology and structural components
Study conditions and biofilm morphology
Several laboratory conditions have been used to study B. subtilis biofilm formation including colonies at the air-agar interface, floating biofilms that form at the air-liquid interface (also termed pellicles) and, in the case of certain domesticated strains, submerged, surface-adhered biofilms that form at the liquid-solid interface (Fig. 2). Colony biofilms are produced when cells are placed on a solid agar surface containing a medium that promotes the expression of genes required for extracellular matrix production. Subsequent growth of the cells leads to the appearance of complex wrinkled colonies within a few days13 (supplementary information S1 (movie)). Wrinkles form as a consequence of localized cell death coupled with the stiffness provided by the extracellular matrix27. The B. subtilis matrix is primarily composed of exopolysaccharide (EPS) and proteins (Table 1)15,16. This matrix, in conjunction with the rough surface topography, provides the biofilm with a remarkably hydrophobic surface that is largely impermeable to aqueous liquids and organic solvents28. In liquid conditions cells will either float to the surface of the liquid where they produce extracellular matrix and form a pellicle at the air-liquid interface, or remain under the surface of the liquid where they adhere to the side of the container and form a submerged biofilm. The particular type of biofilm and its robustness varies depending on the strain of B. subtilis and the experimental conditions used (Box 1). In addition to the artificial methods described above, B. subtilis also forms biofilms on natural surfaces such as plant roots where the bacteria provide the plant with many benefits (Box 2).
Figure 2.
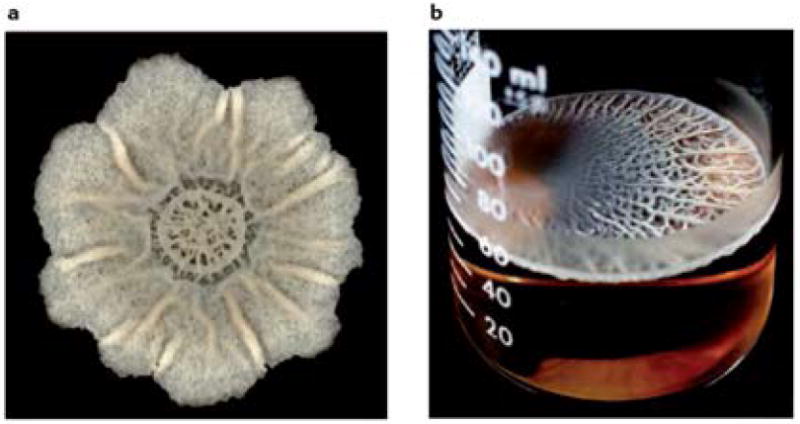
A) Top-down view of a colony grown at room temperature on biofilm-inducing medium (MSgg) for 7 days. Scale bar is 2 mm. A time-lapse movie of the growth of this colony can be viewed in the supplemental material (H. Vlamakis, unpublished movie). B) Top-down view of a pellicle grown at room temperature for 5 days (image from 13).
Table 1.
Genes involved in extracellular matrix production.
| Gene | Role in matrix production | Mutant phenotype |
|---|---|---|
| epsA – O operon | Enzymes that produce exopolysaccharide (EPS). | Flat colonies (Δ operon, epsE 41, epsG 13, epsH 13; other genes not tested) |
| Thin pellicles (Δ operon, epsE 41, epsG 13, epsH 13; other genes not tested) | ||
| Defective submerged biofilm (epsG 33; other genes not tested) | ||
|
| ||
| tapA (formerly yqxM) | TasA accessory/anchoring protein. Minor component of TasA fibers. | Flat colonies 12 |
| Thin pellicles 12 | ||
| Wild type submerged biofilm 50 | ||
|
| ||
| sipW | Bifunctional signal peptidase required for TapA and TasA processing and secretion and for eps gene expression. | Flat colonies 12 |
| Thin pellicles 12 | ||
| Defective submerged biofilm 50 | ||
|
| ||
| tasA | Major protein component of TasA fibers. | Flat colonies 12 |
| Thin pellicles 12 | ||
| Wild type submerged biofilm 50 | ||
|
| ||
| pgsB (formerly ywsC) | First gene in operon that encodes enzymes that produce γ-poly-DL-glutamic acid (PGA). | Wild type colonies 12 |
| Wild type pellicles 12 | ||
| Over expression enhances submerged biofilms 44 | ||
|
| ||
| bslA (formerly yuaB) | Surface hydrophobic layer protein, BslA. | Flat colonies 55 |
| Thin pellicles 52,55 | ||
| Untested in submerged assay | ||
Box 1. Domestication of lab strains.
As cells are passaged in liquid culture in the laboratory, mutations can arise that decrease the ability of these cells to form biofilms. For example, Branda et al. analyzed a common laboratory strain (PY79) and a “less domesticated” strain (NCIB 3610), which is a close relative of PY79109. NCIB 3610, forms robust wrinkled colonies (a hallmark of biofilm formation) and floating pellicle biofilms in a defined medium, but adheres poorly to glass surfaces (See Fig. 2 and movie of colony growing)12,13. At the same time, Hamon and Lazzazzera developed a submerged surface-adhered biofilm assay with the commonly used lab strain JH64231. This strain formed submerged surface-adhered biofilms on polyvinylchloride (PVC) and glass surfaces, but this strain and its close relatives (strains 168 and PY79) do not produce robust wrinkled colonies13,110. All of these strains are extremely similar at the nucleotide level, thus the specific genotypic differences that conferred these phenotypes were investigated. Using gross morphology of complex colony wrinkling (which is easily observed by visual screening of colonies on agar plates) as a read-out for matrix production, five specific genes were identified as being central to this process. Point mutations in four genes sfp, epsC, degQ, and swrA and the lack of rapP, a regulatory gene found on the plasmid of strain NCIB 3610 were responsible for the diminished matrix gene expression in the domesticated strain 168 relative to NCIB 3610110. The sfp gene encodes a phosphopantetheinyl transferase that is required for surfactin production and the point mutation in strain 168 impairs surfactin production. The epsC gene is in the epsA – O operon and the point mutation results in a decrease in EPS production. Both surfactin and EPS are required for biofilm formation. DegQ is a small protein that stimulates phosphotransfer from DegS to DegU and has previously been shown to be involved in biofilm formation44,52. SwrA is a regulatory protein that is important in swarming motility and PGA synthesis44,52. The mechanism by which RapP regulates biofilm formation has yet to be determined, although this protein is similar at the amino acid level to a family of regulatory proteins that antagonize response regulators, including Spo0F, a member of the Spo0A phosphorelay111. It is possible that the effect of RapP is mediated by interactions with a regulatory protein. More specific details regarding the genome differences of different B. subtilis strains have been discussed in several papers109,112.
Box 2. Plant roots as a natural habitat for B. subtilis biofilm formation.
In order to proliferate in the soil, B. subtilis requires a nutrient source such as decaying organic material or plant roots113. The rhizosphere, or region of soil directly surrounding plant roots, is rich in plant secretions that can provide bacteria with nutrients114-116. Bacteria in the rhizosphere can benefit the plant, and Bacillus species - including B. subtilis - are sold commercially as biological control agents for agriculture115,117,118. Bacillus species can promote growth and protect plants from infections by pathogenic bacteria, fungi and even nematodes. This protection is due to the secretion of antimicrobial compounds by B. subtilis, coupled with induced systemic resistance in response to B. subtilis that enhances the capacity of the plant to resist various pathogens119-123.
B. subtilis is readily isolated from the rhizosphere of plants and the majority of root-associated strains are capable of forming robust biofilms in laboratory conditions124,125. In addition, several other Bacillus species form biofilms on plant roots126-128. As shown in the figure, biofilm formation on plant roots parallels in vitro biofilm formation in that the matrix exopolysaccharide is required (94,125 and P. Beauregard unpublished results). Similarly, the master regulator Spo0A and the antirepressor SinI are also required for root colonization (125 and P. Beauregard unpublished results). In many wild isolates, the presence of these genes, and thus the capacity to form a biofilm on the root, is also required for the strain to exert its maximal biocontrol effect125.
B. subtilis colonization of Arabidopsis thaliana roots also requires the production of surfactin, a lipopeptide antimicrobial that is also important for biofilm formation in vitro126. The production of surfactin and other lipopeptides by Bacillus cells is one of main mechanisms for plant biocontrol since these molecules can induce systemic resistance as well as strongly inhibit the growth of common plant pathogens such as Pseudomonas syringae125,126,129.
To recruit B. subtilis, plants secrete small molecules. For example, when A. thaliana is infected with P. syringae, malic acid is secreted and this enhances B. subtilis biofilm formation on the root127. Furthermore, root exudates from P. syringae-infected plants or purified malic acid induce matrix gene expression in B. subtilis127. This phenomenon is not specific to A. thaliana, malic acid is also found in tomato root exudates and, at high concentrations, can stimulate matrix gene expression and biofilm formation in vitro94. Tomato root exudates stimulate matrix gene expression in a manner that is dependent on the Spo0A kinase KinD. Mutants specifically lacking the extracellular CACHE domain of KinD are less efficient colonizers of tomato roots94.
Figure.
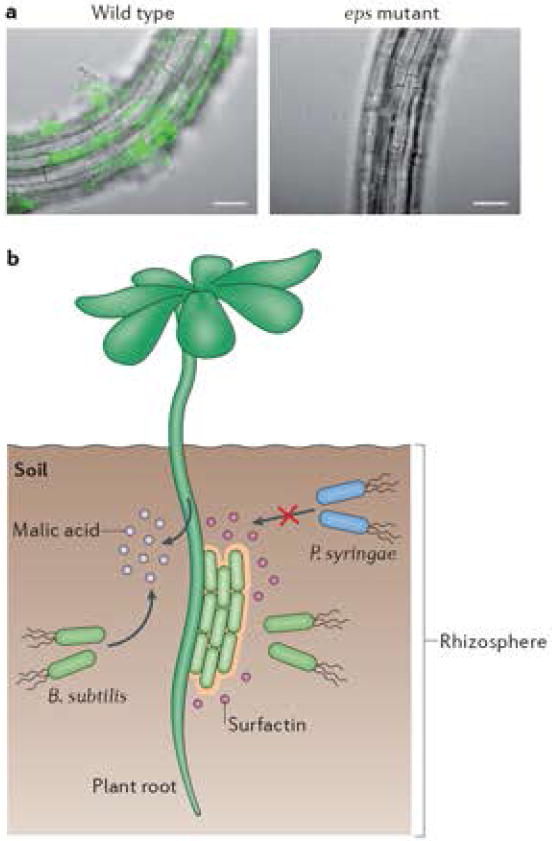
A) Wild-type B. subtlis cells or an eps mutant constitutively expressing YFP were inoculated with 6 day-old seedlings of A. thaliana. Colonization of the root was observed after 24h. Overlays of fluorescence (false-coloured green) and transmitted light images (gray) are shown. Bars are 50 μm. Images in A) by P. Beauregard (unpublished). B) Schematic illustration of B. subtilis colonizing a plant root. B. subtilis secretes the lipopetide surfactin, which is required for B. subtilis (green rods) biofilm formation on the root. A second trigger for B. subtilis matrix gene expression is malic acid, which is constitutively secreted in the rhizosphere by tomato plants but secreted by A. thaliana only when the plant is infected with the pathogen P. syringae. B. subtilis exerts beneficial effects on the plant by promoting its growth and helping to fight of pathogens (such as P. syringae), directly via the secretion of surfactin and other antimicrobials and indirectly by eliciting induced systemic resistance in the plant.
In all biofilms, a series of morphological changes occurs in cells during biofilm development (Fig. 1). Although the number of motile cells decreases as the biofilm develops, a small subpopulation of motile cells remains, even in mature biofilms20. The role of these motile cells in B. subtilis biofilms varies depending on growth conditions. Motility-defective mutants that do not have flagella are delayed in forming pellicle biofilms14 and are defective in the formation of submerged surface-adhered biofilms29, however, their colony morphology is akin to that of the wild type20,30. Similarly, although some cells in the biofilm eventually sporulate, sporulation per se is not a requirement for biofilm formation13,31.
Exopolysaccharide and polymer components
The major exopolysaccharide component required for each biofilm type is synthesized by the products of the epsA-epsO operon 13,32,33. Mutations in the eps genes result in defective biofilm formation, as do mutations in pgcA (formerly yhxB - encoding α-phosphoglucomutase) and gtaB (encoding UTP-glucose-1-phosphate uridylyltransferase), which are involved in the production of nucleotide sugars that likely feed into the eps pathway34,35. Indeed, mutants defective in the synthesis of uridine diphosphate-galactose (UDP-Gal), which is a precursor metabolite required for EPS biosynthesis36, are defective in biofilm formation. UDP-Gal is a toxic intermediate product in galactose metabolism that is normally converted to the non-toxic UDP-glucose by the UDP-glucose 4-epimerase GalE. When the galE gene is mutated, growth on galactose is toxic because UDP-Gal accumulates. Interestingly, galE mutants grown in biofilm-inducing conditions or galE mutants that overexpress the eps genes can survive even in the presence of galactose because the UDP-Gal is shunted into the EPS pathway36.
Of the 15 genes in the epsA-O operon, only a subset has been studied individually13,37-40. The best-studied gene product of this operon, EpsE, is a bifunctional protein that coordinates the production of EPS with the cessation of motility40. In addition to displaying glycosyltransferase activity required for EPS synthesis, EpsE also functions as a molecular clutch that inhibits flagella rotation by interacting with the flagella rotor protein, FliG40,41. Motility inhibition occurs independently of the glycosyltransferase activity of EpsE. This remarkable mechanism of regulation ensures that cells shut off motility when matrix production occurs for biofilm formation. Interestingly, in colony biofilms, it is EPS and not motility that is important for colony spreading: EPS is thought to generate osmotic pressure gradients that allow the colony to spread outwards and thus acquire nutrients30. This could explain the growth defect observed in colonies of mutants that are unable to produce EPS42.
Another extracellular polymer, γ-poly-DL-glutamic acid (PGA), is produced in copious amounts by some B. subtilis strains, and can enhance submerged biofilm formation43,44. However, PGA is not required for wrinkled colony morphology or pellicle formation12,14.
Protein components
In addition to several uncharacterized proteins that are present within the matrix, two structural protein components have been described for B. subtilis biofilms: TasA and BslA. TasA was the first described protein component of the extracellular matrix of colony and pellicle biofilms12. TasA assembles into long amyloid-like fibres that are attached to the cell wall by the protein TapA (TasA anchoring/assembly protein; formerly YqxM)45,46. TapA is found in the cell wall fraction of cells grown as pellicles or colonies and it plays a role not only in anchoring TasA fibres to the cell, but also in assembly of the amyloid fibres46. In addition to its cell wall localization, TapA can be purified as a minor component of the amyloid fibres46. These proteins are encoded by the tapA-sipW-tasA operon, which also encodes SipW, the signal peptidase that processes both TapA and TasA47-49. SipW processes TapA and TasA by recognizing an N-terminal signal sequence and cleaving the proteins as they are secreted so that they can be released from the membrane and become cell-wall associated fibers.
While they are essential in colony and pellicle biofilms, TasA and TapA are not required for submerged biofilm formation50. However, mutation of sipW in certain domesticated strains results in defective attachment to glass or polyvinyl chloride surfaces12,50. This is because SipW is a bifunctional protein whose signal peptidase activity can process TasA and TapA, but whose C-terminal domain actually functions to activate eps gene expression. This activation is essential for attachment and occurs only when cells are growing in a submerged surface-adhered mode. Consistent with this, overexpression of the eps operon is sufficient to restore submerged biofilm formation in a sipW mutant33.
In addition to TasA, another secreted protein, BslA (formerly YuaB) is important for surface hydrophobicity, complex colony morphology and pellicle formation51-54. The biofilm-defective phenotype of a bslA mutant can be extracellularly complemented by mixing this mutant with a mutant lacking eps and tasA55. This observation suggests that EPS and TasA are provided by the bslA mutant and BslA is provided by the eps tasA mutant55. BslA forms a hydrophobic layer on the surface of biofilms and is termed BslA for biofilm surface layer protein51. It has amphiphilic properties and when purified, BslA forms polymers in solution when the air – surface interface is increased by the addition of bubbles51. However, it is currently unclear exactly how BslA functions to confer hydrophobicity to the biofilm surface.
Regulatory pathways that control biofilm formation
Given all of the components that are necessary to assemble the matrix, how does this bacterium regulate their production and assembly? Indeed, B. subtilis has a complex regulatory network to coordinate expression of matrix genes in response to the shifting environmental conditions that it encounters in its natural environment. Figure 3 is a simplified schematic of this network and the four numbered sub-networks represent four pathways that regulate the expression of matrix genes (see Table 1).
Figure 3.
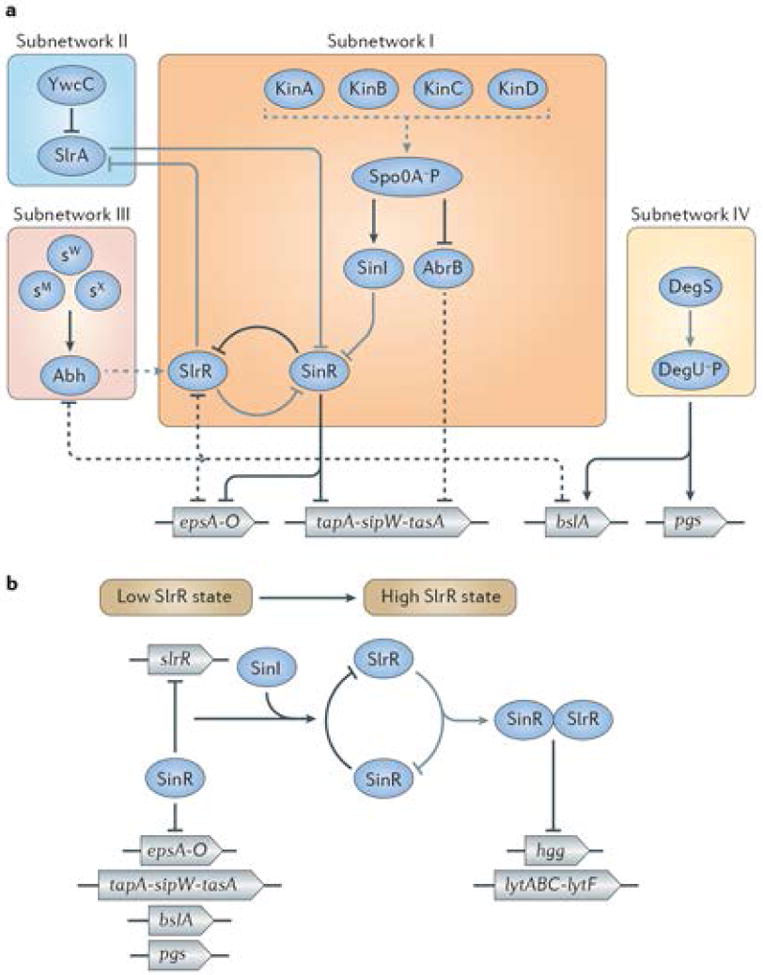
Simplified schematic of the regulatory network that controls biofilm formation in B. subtilis. A) Several sub-networks (I-IV) are integrated to activate (arrows) or repress (T-bars) matrix gene expression depending on the environmental condition. Details are discussed in the text. The genes encoding components of the extracellular matrix are coloured in blue and encode: EPS (epsA-O), TasA (tapA-sipW-tasA), BslA (bslA), and PGA (pgs). Red and pink lines indicate gene regulation whereas yellow lines indicate protein-protein interactions. Dashed lines indicate indirect activity. B) The double negative feedback loop (involving the slrR gene, the SlrR protein and the SinR protein) exists in SlrR low (left side of figure) and SlrR high (right side of figure) states. The SinR–SlrR switch regulates matrix genes (epsA-O and tapA-sipW-tasA), autolysin (lytABC and lytF) and motility genes (hag), as well as the gene (slrR) for SlrR itself. In the SlrR low state (left) the SinR protein represses the slrR gene, keeping the levels of the SlrR protein low. In the SlrR high state (right), SlrR binds to SinR, trapping it in the heteromeric SinR•SlrR complex. This titrates SinR, resulting in derepression of matrix genes and slrR, setting up a self-reinforcing switch leading to high SlrR levels. At the same time, SlrR repurposes SinR in that the SinR•SlrR complex represses autolysin and motility genes (right). Image adapted from72. The double negative loop is epigenetic in that both the SlrR low and SlrR high states self-reinforcing and are stable for many generations. During biofilm formation, SinI produced under the control of Spo0A~P (see text) drives the switch into the high state by binding to and inhibiting SinR.
The Spo0A pathway
Spo0A is a central transcriptional regulator that controls the expression of over one hundred genes, including those necessary for biofilm matrix gene expression and sporulation56,57. The activity of this protein is regulated by phosphorylation at a single aspartate residue and both phosphorylated and unphosphorylated forms of Spo0A are always found in the cell. The concentration of phosphorylated Spo0A (Spo0A~P) in a given cell determines the gene expression profile and changes in its concentration facilitate differential gene regulation56. For example, intermediate levels of Spo0A~P result in matrix gene expression and higher levels induce sporulation genes. In this way, when Spo0A is initially phosphorylated biofilm formation is induced as a result of matrix gene expression. As the biofilm matures, Spo0A~P accumulates in certain cells and activates sporulation.
The concentration of Spo0A~P is determined by the activity of at least four kinases (KinA, KinB, KinC and KinD) that either act directly on Spo0A or indirectly via a phosphorelay58. The phosphorelay consists of Spo0F, which is phosphorylated by either KinA, KinB, KinC or KinD and then passes its phosphoryl group to Spo0B, which goes on to phosphorylate Spo0A. There is a fifth kinase, KinE, which can also feed into this pathway, but it does not appear to play a role in matrix gene expression59. There are many levels of regulation within the phosphorelay and this topic has been reviewed previously60,61. No single kinase is solely responsible for matrix gene expression, but rather the contribution of different kinases changes depending on the signals present in the growth conditions being analyzed31,59,62. Specific signalling molecules that trigger phosphorylation of Spo0A by these kinases are discussed in more detail later in this review.
Spo0A~P governs the regulatory pathway for matrix gene expression by influencing the activity of the master regulator SinR, a repressor of the epsA-O and the tapA operons. Derepression of the matrix genes is accomplished by the action of the SinR antirepressor SinI, which is under the control of Spo0A~P (see below). In addition to the matrix genes, SinR also represses the regulatory gene slrR (Fig. 3A, subnetwork I)32,63,64. However, when SinI is expressed, it blocks SinR-mediated repression through the formation of a SinI-SinR protein-protein complex that renders SinR incapable of DNA binding65. SinR is produced in all cells but is only inactivated by SinI in a fraction of cells, thus only a subpopulation of cells expresses the tapA and eps operons20,66.
In addition to determining which cells express matrix genes, Spo0A~P levels also determine the duration of matrix gene expression. The promoter of sinI contains both a high-affinity activator and multiple low affinity operators for Spo0A~P66. When Spo0A~P levels are relatively low, the high affinity activator is bound and sinI is expressed. As the levels of Spo0A~P increase, the lower affinity operators are also occupied and further sinI expression is curtailed66. Meanwhile, sporulation genes become activated by the high levels of Spo0A~P56. A second and embedded mechanism exists to turn off matrix genes once sporulation commences. The function of SinI and SinR are remarkably sensitive to gene dose: a mere doubling of the sinI and sinR genes completely blocks matrix production67. While actively dividing cells do not maintain two chromosomes for very long, in early sporulation there is a prolonged presence of two copies of the chromosome in the mother cell that results in two copies of sinI and sinR which is sufficient to inhibit matrix gene expression. Together, Spo0A~P affinity for the sinI promoter and gene copy number of sinI and sinR ensures that matrix gene expression is transient and that sporulating cells do not expend energy producing the extracellular matrix.
Spo0A~P also represses a second matrix gene repressor, AbrB68. Like SinR, AbrB represses both the tapA and the epsA-O operons32,50,63,64,69. Furthermore, AbrB represses expression of the matrix protein BslA54 and the regulatory proteins SlrR64 and Abh69. The presence of two Spo0A-regulated repressors, SinR and AbrB, with highly overlapping targets is likely a means to fine-tune the regulation of biofilm formation and to ensure the coordinated expression of all of the matrix genes.
The SlrR – SinR epigenetic switch
As mentioned above, SinR and AbrB inhibit expression of the regulatory protein SlrR64,70,71. SlrR is essential for the control of biofilm formation in two ways. First, SlrR binds to SinR (to form a SinR•SlrR complex), titrating SinR away and thus preventing it from repressing matrix gene promoters (epsA-O, tapA-sipW-tasA and slrR itself). Because SinR represses the slrR gene and the SlrR protein inhibits SinR, this results in a selfreinforcing double-negative feedback loop whereby the slrR gene remains de-repressed because SlrR prevents repression by SinR. Thus, once SlrR levels are high enough, they tend to stay high for many generations. In this state, the matrix genes are de-repressed because free SinR levels are low. Conversely, when SlrR levels are low, SinR is not inhibited and the matrix operons are switched off (Fig. 3B). Second, the complex of SinR•SlrR represses the promoters of the hag gene (which encodes flagellin and is required for motility), as well as genes involved in cell separation (lytABC and lytF, which encode autolysins) (Fig. 3B) 72,73. Thus, SlrR acts on SinR in two ways: it blocks it from repressing matrix gene and also re-purposes SinR in the form of the SinR•SlrR complex to become a repressor of autolysin and motility genes.
As mentioned earlier, cell chaining is essential at the onset of biofilm formation in B. subtilis (see Fig. 1). This cell chaining is achieved by a SinR•SlrR-mediated repression of genes encoding autolysins, which are required to separate chains of cells. SlrR is a member of the LexA family of autopeptidases and is proteolytically unstable and undergoes self-cleavage. In addition, cleavage of SlrR is dependent on the ClpCP protease. This instability results in the eventual degradation of SlrR and the derepression of genes encoding the autolysins, which allows the chains of cells to separate72. Inhibiting the separation of chains by using a non-cleavable mutant of SlrR72 or by deleting three of the autolysins required for cell separation14 does not alter pellicle formation. However, expression of the autolysin lytC prevents chaining and results in featureless colonies and pellicles containing cells that are delayed in sporulation72.
As we have seen the SlrR-SinR switch can exist in two states: a state in which SlrR levels are low (corresponding to single, motile cells) and a state in which SlrR levels are high (corresponding to chains of matrix producing cells). How does the protein switch from the low state to the high state? This is accomplished by SinI, which is produced under the control of Spo0A~P in response to environmental signals that activate the histidine kinases that govern biofilm formation. SinI, in turn, is like SlrR, an antagonist of SinR that binds to and inhibits the SinR repressor. Thus, the production of SinI inhibits SinR, leading to the depression of the gene for SlrR. This results in the accumulation of SlrR to high levels and further inhibition of SinR by SlrR, driving the switch into an SlrR high state. Because the switch in self-reinforcing, it persists in the high state for many generations and can be said to be epigenetic.
The components of the epigenetic switch are subject to additional regulation from another pathway comprised of YwcC and SlrA (Fig. 3A, subnetwork II). SlrA is a paralogous antirepressor for SinR and functions similarly to SinI70. The slrA gene is repressed by YwcC, a TetR-type transcription repressor70,71. When YwcC receives an as-yet-unknown signal, slrA is derepressed and the matrix genes are induced by SlrA-mediated inactivation of SinR70. Unlike SinI, SlrA is produced in almost all cells, which transiently boosts matrix production in the entire population. In this sense, the YwcC-SlrA pathway might constitute a stress-response pathway to ensure that cells respond quickly to changing environmental conditions by forming a biofilm to protect the bacterial community70.
Transcription of slrR is indirectly activated by the regulatory protein Abh (Fig. 3A, subnetwork III)74. The abh gene is itself repressed by AbrB69 and its transcription is controlled by several extracytoplasmic function (ECF) sigma factors including σM, σX, and σW 14,74-78. ECF sigma factors are activated by a variety of external stimuli including cell wall stresses and specific antibiotics79, thereby providing a Spo0A-independent mechanism for responding to changes in external conditions.
Expression of slrR is positively regulated by several other proteins aside from Abh. For example, YmdB, a putative phosphoesterase, is necessary for slrR expression80. In addition, slrR gene expression also requires two small proteins RemA and RemB81. Genetic analyses show that RemA and RemB activate expression of the eps and tapA matrix operons via SlrR and in a manner independent of SlrR. The exact mechanism by which these small proteins function remains to be determined, however it appears that they act in parallel with SinR, AbrB and DegU (see below), the other known matrix gene regulatory proteins81.
Other regulation
A fourth pathway that only regulates the expression of bslA and the pgs operon involves the DegS-DegU two-component system (Fig. 3A, subnetwork IV)44,52,54,82. In this system, DegS is a sensor histidine kinase that phosphorylates DegU, the response regulator. DegU is a global regulator in B. subtilis that is involved in the control of a variety of cellular processes such as competence, motility and secretion of degradative enzymes83. In addition, a degU mutant is defective in submerged biofilm formation, which requires the polymer PGA, the product of the enzymes encoded by the pgs operon 44. Furthermore, colony biofilm formation is defective in a DegU mutant due to the loss of the surface hydrophobicity protein BslA51,52,54,82.
Finally, in addition to the various transcription factors that are described above, the epsA-O operon is under the control of a cis-acting RNA element that is located between the second and third genes of the operon. This element is termed the “EAR” (eps-associated RNA element) and it is conserved among a subset of the Bacillales order. This element is thought to act as an anti-terminator and increases eps gene expression by interacting with RNA polymerase84.
The foregoing list of regulators underscores the remarkably complex and multi-layered regulatory mechanisms controlling biofilm formation in B. subtilis. The particular regulators are specific for this organism and it is highly unlikely that we will find homologs with similar roles in other bacterial species. However, the take-home message is that such complex regulatory mechanisms are likely to have evolved to adequately respond to environmental changes by forming biofilms at the right time and under the right conditions.
Triggers of biofilm formation
As is clear from the previous section, much is known about the molecular mechanisms that regulate matrix gene expression. But what are the signals that trigger these pathways? Because of the numerous inputs to the system (the four histidine kinases, the ECF sigma factors, YwcC, and the DegS-DegU system) it is likely that many conditions exist that could trigger biofilm formation. As is described below, several signals and mechanisms that result in increased expression of extracellular matrix genes have been identified. The known mechanisms occur via two of the sensor kinases that phosphorylate Spo0A: KinC and KinD. Currently, the mechanisms by which the other kinases affect biofilm formation have not been elucidated. As we explore additional conditions that B. subtilis encounters in its natural environment, many more triggers for biofilm formation are likely to be identified.
KinC-mediated matrix gene expression
The first molecule identified as an inducer of matrix gene expression was surfactin, a molecule produced by constituent cells of the biofilm62. Surfactin has been studied for its potent surfactant and anti-microbial activities as well as its role in surface motility in B. subtilis85-87. Surprisingly, surfactin also acts as a signal that triggers phosphorylation of Spo0A via the sensor kinase KinC and positively regulates matrix gene expression62. Interestingly, surfactin is only produced by a sub-population of cells, and the cells that produce surfactin are not the same cells that respond to the molecule (i.e. those cells that express matrix genes or sinI, which are also only produced by a subset of biofilm cells)88. This concept represents a new way of thinking about self-generated quorum sensing signals in bacteria and has been referred to as paracrine signalling, whereby the signal is unidirectional and the signal producer does not respond to the signal it makes. This is in contrast to previously described quorum sensing systems where every cell in a population is thought to produce and respond to the signalling molecule (reviewed in 18,89).
Induction of gene expression in response to surfactin does not occur by the canonical mechanism involving a sensor protein binding to a ligand. Instead of responding to the structure of surfactin, KinC is activated by the function of the molecule. Surfactin is a lipopeptide that inserts into the membrane and results in potassium leakage. This potassium leakage activates KinC by an unknown mechanism and the matrix genes are expressed62. Other compounds with different structures to surfactin that cause potassium leakage, such as the fungicide nystatin and the antibiotic valinomycin62, also induce matrix gene expression via KinC. Importantly, molecules such as surfactin, nystatin and valinomycin are natural products produced by other organisms that reside in the soil, thus it is likely that B. subtilis encounters these molecules in nature. The fact that the antifungal agent nystatin, which functions by binding and displacing ergosterol in the membrane, affected signalling in B. subtilis led to the finding that B. subtilis harbors membrane microdomains analogous to lipid rafts in eukaryotes90 (see Box 3). Chlorine dioxide, which is a potent biocide at high concentrations, induces matrix gene expression in a KinC dependent manner at sublethal concentrations91. Unlike surfactin, chlorine dioxide is thought to trigger KinC activation by collapsing the membrane potential of the cell. Thus it appears that KinC can be activated by at least two distinct membrane-disrupting mechanisms: potassium leakage and a decreased membrane potential. Increasing matrix production in the presence of membrane disruptors may be beneficial for B. subtilis survival; at least in the case of chlorine dioxide, wherein the presence of exopolysaccharide provides protection against the lethal effects of the molecule91.
Box 3. Lipid rafts coordinate signalling molecules in bacterial membranes.
Membrane microdomains, which are analogous to the cholesterol-rich lipid rafts found in eukaryotic cells, also exist in bacteria. B. subtilis membranes do not contain sterols, however, lipids that are synthesized by the same precursor (isoprenyl pyrophosphate) are present. Different lipid components of the membrane can be separated by their ability to withstand detergent treatment and proteins found in detergent-resistant microdomains can be purified from the membrane of B. subtilis using techniques similar to those used for the purification of detergent-resistant lipids in eukaryotes88,124. KinC localizes in these microdomains along with a variety of other proteins involved in signalling90. In addition, FloT (formerly YuaG) and YqfA, two homologs of the eukaryotic lipid raft protein Flotillin-1, are associated with the detergent-resistant microdomains and localize in a punctate pattern throughout the membrane90,130,131. The figure shows a FloT-YFP protein fusion localized in puncta throughout the B. subtilis membrane. A mutant in the gene yisP, which encodes a squalene synthase that is required for the production of the detergent-resistant lipids is defective in biofilm formation. Furthermore, known inhibitors of squalene synthase, such as zaragozic acid, disrupt these domains and inhibit biofilm formation. The series of images show dissipation of FloT-YFP puncta over time after treatment with zaragozic acid. By eight hours, all of the FloT-YFP puncta have dispersed90. As is discussed at the end of this review, this knowledge led to studies in other bacteria where the disruption of membrane microdomains using inhibitory molecules of lipid biosynthesis also inhibited biofilm formation and blocked the production of several virulence factors.
Figure.
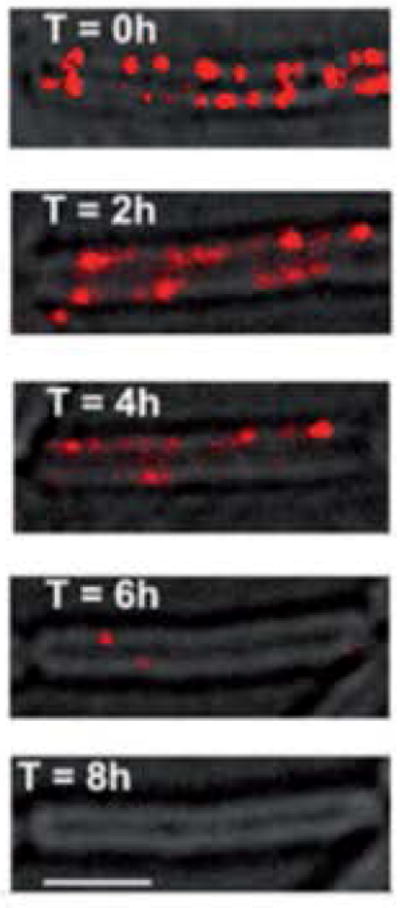
FloT-YFP localizes in a punctate pattern in the cell membrane and is disrupted by inhibitors of squalene synthase. Fluorescence image (false-coloured red) is overlayed on the transmitted light image. An untreated cell is shown (top image) and a series of images taken over a period of 8 h after treatment with zaragozic acid. Scale bar is 2 μm. Images from 90.
KinD-mediated matrix gene expression
Matrix itself appears to regulate matrix gene expression and mutants that are unable to produce EPS and TasA show prolonged expression of the promoters of the eps and tasA operons (as observed using transcriptional reporters) and delayed sporulation in biofilm conditions20,42. This is at least in part due to the activity of KinD. Like many other two component sensor kinases92, KinD displays both kinase and phosphatase activity. KinD appears to function as a phosphatase to maintain low levels of Spo0A~P until matrix (or a component thereof) is sensed, at which point KinD functions as a kinase to promote sporulation42. Therefore, a checkpoint exists in which the differentiation into spores during biofilm conditions relies on the production of an extracellular matrix. KinD appears to be important in the response to extracellular matrix and it also activates matrix gene expression in response to compounds produced by soil microbes93 and (as discussed in Box 2) in tomato root exudate94.
Cannibalism to increase the matrix-producing sub-population
Matrix gene expression can also be increased by a non-signalling mechanism. In addition to activating the genes for extracellular matrix production, low levels of Spo0A~P also induce the expression of two “cannibalism” operons that produce secreted toxin peptides: SKF (for sporulation killing factor) and SDP (for sporulation delaying protein). Furthermore, these cells also express the resistance machinery for the toxins95,96. In the case of SKF, the exact mechanism of resistance is unclear, but requires an ABC transporter encoded within the operon that pumps the toxin out of the cell96. SDP resistance requires the membrane-associated immunity protein SdpI which is transcribed divergently relative to the gene encoding the toxin. SdpI is induced in the presence of the toxin but only in those cells that have high enough levels of Spo0A~P97. Thus the cannibal cells secrete toxins that kill siblings that are not expressing these genes. Because the cannibalism genes and matrix-producing genes are activated by low levels of Spo0A~P, the population of matrix-producing cells and cannibalism toxin-producing cells is highly overlapping56,98. Thus the same cells that have initiated matrix production also secrete toxins that effectively decrease the population of non-matrix producers. This ultimately results in a population consisting of an amplified number of matrix-producing cells98.
The cannibalism toxins do not only kill B. subtilis siblings. In fact, they preferentially kill different species when B. subtilis is grown in mixed cultures99,100. Consistent with this, cannibalism-like toxins secreted by close relatives of B. subtilis likely play a role in increasing the B. subtilis matrix-producing sub-population in mixed-species soil communities93. This was discovered in a screen for natural inducers of matrix gene expression in which a B. subtilis strain harboring a fluorescent reporter for matrix gene expression was co-cultured with soil organisms. Surprisingly, despite the diversity of bacteria in the soil samples, the vast majority of inducing organisms were other Bacillus species. In some instances, the inducing organisms produced a secreted molecule (cannibalism-like toxin) that preferentially killed non-matrix producing cells resulting in an increase in the matrix producing population93. The other inducing molecules required a functional KinD protein in the responding cells in order to be sensed.
In summary, diverse signalling molecules ranging from self-produced surfactin and cannibalism toxins to small molecules produced by other soil bacteria can trigger an increase in the number of matrix-producing cells in a population to stimulate biofilm formation. This can occur either via signalling, which results in differential gene expression, or by the selective killing of non-matrix producing B. subtilis cells.
Biofilm dispersal
Escaping the extracellular matrix
This review has focused on the process of building a biofilm. However, as the biofilm matures, it may be beneficial for the constituent cells to disperse owing to resource limitation and waste product accumulation101,102. One mechanism that B. subtilis has exploited to escape biofilms is the release of D-amino acids, a stationary phase phenomenon that naturally occurs in a number of organisms26,103. Cells in mature B. subtilis biofilms release a mixture of D-amino acids (D-tyrosine, D-leucine, D-tryptophan and D-methionine), which results in dissolution of the mature biofilm or inhibition of biofilm formation26. Furthermore, the accumulation of D-amino acids appears to be regulated by racemase enzymes (which catalyze the stereochemical conversion of L- to D-amino acids), as mutations in the encoding genes result in significantly delayed biofilm disassembly26. D-amino acids disrupt biofilm formation by becoming incorporated into peptidoglycan and thereby altering the association of certain proteins to the cell wall, including TapA, the protein that anchors TasA fibres to the cell wall. Thus, D-amino acids result in the release of the TasA amyloid fibres from the cell26,46.
In addition to D-amino acids, the supernatant of aging B. subtilis biofilms also harbors the polyamine norspermidine, which efficiently disperses biofilms25. The inhibitory activity of norspermidine is synergistic with that of D-amino acids suggesting that it acts by a different mechanism. Indeed, norspermidine does not affect association of TasA to the cell wall; instead, it interacts directly and specifically with the exopolysaccharide component of the matrix25. This interaction results in a collapse of the EPS as visualized by microscopy and a change in cell size using light scattering25. This biofilm disrupting activity is also a feature of other polyamines such as norspermine, which harbor three methylene groups flanked by two amino groups. Similar molecules such as spermidine and spermine (which have an additional methylene group) do not exhibit inhibitory activity and it was proposed that the additional methelyne group alters the interaction with EPS25. Disrupting genes that interfere with norspermidine production result in more robust biofilms25. Interestingly, spermidine, which is also produced by B. subtilis, has the opposite effect; disrupting spermidine biosynthesis results in less-robust biofilms and this phenotype can be rescued by the addition of exogenous spermidine104. Although spermidine and norspermidine are similar in a chemical sense, different genes are required for the biosynthesis of each and it is currently unknown how exactly spermidine functions to enhance biofilm formation or how the production of these two molecules is regulated. However, it is clear that B. subtilis is able to modulate biofilm formation using a number of secreted compounds.
Controlling biofilms in other species with small molecules
Efforts to decipher the molecular mechanisms regulating biofilm formation in B. subtilis have led to the discovery of a number of compounds that could function as general biofilm inhibitors. For example, D-amino acids are also able to disrupt biofilm formation in the pathogens Staphylococcus aureus and Pseudomonas aeruginosa26. Similar to what was observed in B. subtilis, where D-amino acids disrupt TapA association with the cell wall, D-amino acids also prevent surface localization of proteins in S. aureus105. The molecular basis for the inhibitory effect of D-amino acids on biofilm formation in Gram-negative bacteria is currently unknown. However, a recent study that analyzed the effect of D-amino acids on P. aeruginosa strain PAO1 flow cell biofilms showed a lethal effect on cells as well as an increase in extracellular matrix production and no biofilm inhibition. This suggests that there is condition or strain specificity in the response, at least in P. aeruginosa 106.
The polyamines norspermine and norspermidine are also potent inhibitors of biofilm formation in S. aureus and Escherichia coli 25. Studies have yet to be performed showing exactly what the targets of the polyamines are in these systems, however it is likely that the EPS component of the matrix is altered in the presence of the polyamines.
In a similar vein, inhibitors of key lipid synthesis enzymes (such as the squalene synthase inhibitor zaragozic acid (see box 3) and cholesterol-lowering statins) are effective against biofilm formation in S. aureus. At the same time, these molecules also disrupt the production of virulence factors such as proteases and the carotenoid staphyloxanthin90. This might contribute to the ability of cholesterol biosynthesis inhibitors to block S. aureus virulence107.
Importantly, zaragozic acid, D-amino acids and polyamines do not inhibit growth of the target Gram-positive organisms. This is an appealing feature of these compounds as their use should reduce the selection pressure for the emergence of resistant mutants, which is associated with traditionally used antibiotics. Since many nosocomial infections are associated with biofilm formation, these molecules might represent a promising alternative to antibiotics101,108. Alternatively, using these compounds to disrupt biofilms prior to treatment with other antimicrobials could provide a more effective means of eliminating harmful bacteria.
Concluding remarks
B. subtilis, a bacterium for which a multitude of genetic and cell-biology tools have been developed, has proven to be an ideal model organism for characterizing the molecular mechanisms underpinning biofilm formation and disassembly. B. subtilis has evolved a number of regulatory pathways that trigger and control biofilm formation. Among these, activation of Spo0A is central for the induction of matrix gene expression in response to a wide variety of extracellular signals. Matrix producers also use cannibalism to amplify the population of matrixproducing cells and this appears to be a more general feature observed when B. subtilis is in contact with close relatives that produce similar cannibalism-like molecules. This leads to enhanced biofilm formation in the presence of potential competition. It is very likely that as B. subtilis is studied in more complex environments, such as on plant roots or in the presence of other bacteria, even more mechanisms to induce matrix gene expression will be discovered. Understanding how cells are able to form biofilms at the appropriate time in the presence of diverse inputs is the next step in enhancing our understanding of biofilm formation in this organism. Another major challenge will be to decipher how the many regulatory pathways, often with overlapping outputs, converge to control matrix gene expression. Perhaps in different conditions only subsets of matrix genes are important (such as the dispensability of TasA in submerged biofilms) and this is why such complexity in regulation has evolved.
Unlike the abundance of information that has been amassed on the regulation of biofilm assembly, studies on biofilm disassembly in B. subtilis are still in their infancy. There are many questions that remain to be addressed. How is the production of small molecules such as norspermidine and D-amino acids regulated to ensure that biofilms do not disassemble at inappropriate times? Is their production regulated by even more external stimuli? What are the exact mechanisms by which these molecules exert their inhibitory effects?
Many features, such as the requirement for extracellular matrix components comprising exopolysaccharides and proteins, are general for the formation of all bacterial biofilms. This has prompted the successful use of small molecules that disrupt B. subtilis biofilms for the targeting of biofilms produced by pathogenic organisms including S. aureus. Much remains to be done as far as understanding how the biofilm disassembly factors identified in B. subtilis are able to inhibit biofilm formation in other diverse organisms. Despite the many outstanding mechanistic questions, it is exciting to imagine how combinatorial therapy, such as the coupling of these molecules with improved antibiotics, has the potential for more successful eradication of detrimental biofilms.
Supplementary Material
Movie of B. subtilis cells forming a colony biofilm at room temperature over a period of approximately 9 days on MSgg medium. Images were taken every 4h. Each second of the movie is equivalent to 16 h real time.
Online summary.
B. subtilis is a model non-pathogenic bacterium that has been used to study biofilms. These ubiquitous communities of tightly associated bacteria are encased in a selfproduced extracellular matrix that holds cells together.
Within a biofilm, genetically identical B. subtilis cells differentiate and cells expressing different sets of genes serve distinct functions.
B. subtilis cells are able to sense and respond to diverse extracellular cues using a complex regulatory network to ultimately express matrix genes and form a biofilm. These cues range from self-produced signals, such as surfactin, to natural products produced by other organisms found in the soil.
Several self-produced molecules are secreted late during the life cycle of a biofilm to trigger disassembly of the community. These molecules appear to have properties that are active against biofilms formed by other bacteria.
B. subtilis naturally colonizes plant roots and is commonly used as a biocontrol agent. Biofilm formation is important for plant root colonization and protection.
Acknowledgments
We are grateful to Elizabeth Shank for her invaluable feedback on the preparation of this review. Work in our labs related to Bacillus subtilis biofilms was funded by National Institutes of Health grants GM58213 to R.K. and GM18546 to R.L.
Definitions
- Biofilms
Surface-associated communities of microbial cells that are encased in a self-produced extracellular matrix composed of exopolysaccharide, protein and occasionally DNA.
- Bimodality
In statistics, bimodality represents a continuous probability distribution with two different modes, appearing often as two distinct peaks in the probability density function.
- Population heterogeneity
A population of genetically identical cells that demonstrate heterogeneous phenotypes.
- Epigenetic switch
A regulatory switch that can impose heritable changes in gene expression or phenotype caused by mechanisms other than genetic changes.
- Chaining
Occurs when daughter cells divide but are remain connected by polar cell wall peptidoglycans.
- Biocontrol
A method to protect against disease using biological control agents such as beneficial bacteria.
- Rhizosphere
A narrow region of soil that surrounds the root of a plant and is directly influenced by plant root secretions and associated soil microorganisms.
- TetR-type transcriptional repressor
Two-domain proteins with a DNA-binding domain and a small ligand-binding domain. Normally, repression is relieved upon binding of a small molecule to the ligand-binding domain.
- Cannibalism
In the context of B. subtilis, this term is used to indicate that in a genetically identical population some cells secrete toxins that kill siblings that are not expressing the genes encoding resistance to these toxins.
Biographies
Hera Vlamakis received her B.S. from the University of IL at Urbana-Champaign and her Ph.D. from the University of CA at Berkeley. She currently holds an instructor position in Roberto Kolter’s lab at Harvard Medical School and she is interested in how individual bacteria coordinate their behaviors in complex communities.
Yunrong Chai received his Ph.D in microbiology from Cornell University, studying bacterial quorum sensing mechanisms. He then carried out postdoctoral training with Dr. Richard Losick at Harvard University, working on biofilm formation in Bacillus subtilis. Yunrong Chai is now an assistant professor in the Biology Department at Northeastern University.
Pascale B. Beauregard received her Ph.D. under the supervision of Luis Rokeach (Université de Montréal, Quebec, Canada) where she worked on a prion-like phenomenon in yeast. She is now a post-doctoral fellow in Roberto Kolter’s laboratory (Harvard Medical School, Massachusetts), where she studies Bacillus subtilis biofilm formation on plant roots.
Richard Losick received his A.B. in Chemistry at Princeton University and his Ph.D. from MIT. He then moved to Harvard University where he is a Professor. He studies spore and biofilm formation by Bacillus subtilis and the role of RNA polymerase sigma factors in the control of gene transcription.
Professor Roberto Kolter studied at Carnegie-Mellon University (B.S.), UCSD (Ph.D.), and Stanford (post-doc). He has been a faculty member at Harvard Medical School since 1983. There he has worked on antibiotic synthesis, bacterial starvation physiology, experimental evolution, bacterial biofilms, and chemical communication in the microbial world.
References
- 1.Cohn F. Untersuchungen über bacterien. IV. Beiträge zur biologie der Bacillen. Beiträge zur biologie der Pflanzen. 1877;7:249–276. [Google Scholar]
- 2.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 3.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 4.Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 5.Mah TF, O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 6.Stewart PS. Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol. 2002;292:107–113. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- 7.Hall-Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cell Microbiol. 2009;11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 8.Erable B, Duteanu NM, Ghangrekar MM, Dumas C, Scott K. Application of electro-active biofilms. Biofouling. 2010;26:57–71. doi: 10.1080/08927010903161281. [DOI] [PubMed] [Google Scholar]
- 9.Logan BE. Exoelectrogenic bacteria that power microbial fuel cells. Nat Rev Microbiol. 2009;7:375–381. doi: 10.1038/nrmicro2113. [DOI] [PubMed] [Google Scholar]
- 10.Singh R, Paul D, Jain RK. Biofilms: implications in bioremediation. Trends Microbiol. 2006;14:389–397. doi: 10.1016/j.tim.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Monds RD, O’Toole GA. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 2009;17:73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006;59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. This study shows that TasA and exopolysaccharide are components of the extracellular matrix and essential for biofilm formation. [DOI] [PubMed] [Google Scholar]
- 13.Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. This was the first paper describing B. subtilis colony and pellicle biofilms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi K. Bacillus subtilis pellicle formation proceeds through genetically defined morphological changes. J Bacteriol. 2007;189:4920–4931. doi: 10.1128/JB.00157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Branda SS, Vik S, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Marvasi M, Visscher PT, Casillas Martinez L. Exopolymeric substances (EPS) from Bacillus subtilis : polymers and genes encoding their synthesis. FEMS Microbiol Lett. 2010 doi: 10.1111/j.1574-6968.2010.02085.x. [DOI] [PubMed] [Google Scholar]
- 17.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 18.Lopez D, Vlamakis H, Kolter R. Biofilms. Cold Spring Harb Perspect Biol. 2010;2 doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vlamakis H, Kolter R. In: Bacterial Stress Responses. 2. Storz G, Hengge R, editors. ASM Press; 2011. pp. 365–373. [Google Scholar]
- 20.Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008;22:945–953. doi: 10.1101/gad.1645008. This work describes spatio-temporal regulation of gene expression within biofilms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubnau D, Losick R. Bistability in bacteria. Mol Microbiol. 2006;61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- 22.Smits WK, Kuipers OP, Veening JW. Phenotypic variation in bacteria: the role of feedback regulation. Nat Rev Microbiol. 2006;4:259–271. doi: 10.1038/nrmicro1381. [DOI] [PubMed] [Google Scholar]
- 23.Veening JW, Smits WK, Kuipers OP. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol. 2008;62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. Excellent review discussing phenotypic variability in clonal bacterial populations. [DOI] [PubMed] [Google Scholar]
- 24.Lopez D, Kolter R. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol Rev. 2010;34:134–149. doi: 10.1111/j.1574-6976.2009.00199.x. [DOI] [PubMed] [Google Scholar]
- 25.Kolodkin-Gal I, et al. A Self-Produced Trigger for Biofilm Disassembly that Targets Exopolysaccharide. Cell. 2012;149:1–9. doi: 10.1016/j.cell.2012.02.055. The self-produced compound norspermidine is described as an inhibitor of EPS in B. subtilis biofilms. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Kolodkin-Gal I, et al. D-amino acids trigger biofilm disassembly. Science. 2010;328:627–629. doi: 10.1126/science.1188628. Identified that D-amino acids that were secreted late in biofilm formation inhibit B. subtilis biofilms as well as those of other species. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asally M, et al. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proc Natl Acad Sci U S A. 2012;109:18891–18896. doi: 10.1073/pnas.1212429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Epstein AK, Pokroy B, Seminara A, Aizenberg J. Bacterial biofilm shows persistent resistance to liquid wetting and gas penetration. Proc Natl Acad Sci U S A. 2011;108 doi: 10.1073/pnas.1011033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chagneau C, Saier MH., Jr Biofilm-defective mutants of Bacillus subtilis. Journal of molecular microbiology and biotechnology. 2004;8:177–188. doi: 10.1159/000085790. [DOI] [PubMed] [Google Scholar]
- 30.Seminara A, et al. Osmotic spreading of Bacillus subtilis biofilms driven by an extracellular matrix. Proc Natl Acad Sci U S A. 2012;109:1116–1121. doi: 10.1073/pnas.1109261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamon MA, Lazazzera BA. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol Microbiol. 2001;42:1199–1209. doi: 10.1046/j.1365-2958.2001.02709.x. This was the first paper describing B. subtilis submerged surface-adhered biofilms. [DOI] [PubMed] [Google Scholar]
- 32.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 33.Terra R, Stanley-Wall NR, Cao G, Lazazzera BA. Identification of Bacillus subtilis SipW as a Bifunctional Signal Peptidase that Controls Surface-Adhered Biofilm Formation. J Bacteriol. 2012;194:2781–2790. doi: 10.1128/JB.06780-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Branda SS, et al. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol. 2004;186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazarevic V, et al. Bacillus subtilis alpha-phosphoglucomutase is required for normal cell morphology and biofilm formation. Appl Environ Microbiol. 2005;71:39–45. doi: 10.1128/AEM.71.1.39-45.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chai Y, Beauregard PB, Vlamakis H, Losick R, Kolter R. Galactose Metabolism Plays a Crucial Role in Biofilm Formation by Bacillus subtilis. MBio. 2012;3(4):e00184–12. doi: 10.1128/mBio.00184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren D, et al. Gene expression in Bacillus subtilis surface biofilms with and without sporulation and the importance of yveR for biofilm maintenance. Biotechnol Bioeng. 2004;86:344–364. doi: 10.1002/bit.20053. [DOI] [PubMed] [Google Scholar]
- 38.Nagorska K, Ostrowski A, Hinc K, Holland IB, Obuchowski M. Importance of eps genes from Bacillus subtilis in biofilm formation and swarming. Journal of applied genetics. 2010;51:369–381. doi: 10.1007/BF03208867. [DOI] [PubMed] [Google Scholar]
- 39.Nagorska K, Hinc K, Strauch MA, Obuchowski M. Influence of the sigmaB stress factor and yxaB, the gene for a putative exopolysaccharide synthase under sigmaB Control, on biofilm formation. J Bacteriol. 2008;190:3546–3556. doi: 10.1128/JB.01665-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science. 2008;320:1636–1638. doi: 10.1126/science.1157877. This is a beautiful example of the multiple levels of regulation that ensure that matrix-producing cells are not motile. [DOI] [PubMed] [Google Scholar]
- 41.Guttenplan SB, Blair KM, Kearns DB. The EpsE flagellar clutch is bifunctional and synergizes with EPS biosynthesis to promote Bacillus subtilis biofilm formation. PLoS Genet. 2010;6(12):e1001243. doi: 10.1371/journal.pgen.1001243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aguilar C, Vlamakis H, Guzman A, Losick R, Kolter R. KinD is a checkpoint protein linking spore formation to extracellular-matrix production in Bacillus subtilis biofilms. MBio. 2010;1(1):e00035–10. doi: 10.1128/mBio.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morikawa M, et al. Biofilm formation by a Bacillus subtilis strain that produces gamma-polyglutamate. Microbiology. 2006;152:2801–2807. doi: 10.1099/mic.0.29060-0. [DOI] [PubMed] [Google Scholar]
- 44.Stanley NR, Lazazzera BA. Defining the genetic differences between wild and domestic strains of Bacillus subtilis that affect poly-gamma-dl-glutamic acid production and biofilm formation. Mol Microbiol. 2005;57:1143–1158. doi: 10.1111/j.1365-2958.2005.04746.x. [DOI] [PubMed] [Google Scholar]
- 45.Romero D, Aguilar C, Losick R, Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. In this work, TasA was shown to form amyloid-like fibers in the extracellular matrix. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romero D, Vlamakis H, Losick R, Kolter R. An accessory protein required for anchoring and assembly of amyloid fibres in B. subtilis biofilms. Mol Microbiol. 2011;80:1155–1168. doi: 10.1111/j.1365-2958.2011.07653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stover AG, Driks A. Control of synthesis and secretion of the Bacillus subtilis protein YqxM. J Bacteriol. 1999;181:7065–7069. doi: 10.1128/jb.181.22.7065-7069.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stover AG, Driks A. Regulation of synthesis of the Bacillus subtilis transition-phase, spore-associated antibacterial protein TasA. J Bacteriol. 1999;181:5476–5481. doi: 10.1128/jb.181.17.5476-5481.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tjalsma H, et al. Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases. Genes Dev. 1998;12:2318–2331. doi: 10.1101/gad.12.15.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamon MA, Stanley NR, Britton RA, Grossman AD, Lazazzera BA. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol Microbiol. 2004;52:847–860. doi: 10.1111/j.1365-2958.2004.04023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kobayashi K, Iwano M. BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol Microbiol. 2012;85:51–66. doi: 10.1111/j.1365-2958.2012.08094.x. Recent study showing that BslA (formerly YuaB) plays a role in the hydrophobicity of B. subtilis biofilms. [DOI] [PubMed] [Google Scholar]
- 52.Kobayashi K. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol Microbiol. 2007;66:395–409. doi: 10.1111/j.1365-2958.2007.05923.x. [DOI] [PubMed] [Google Scholar]
- 53.Kovacs AT, Kuipers OP. Rok regulates yuaB expression during architecturally complex colony development of Bacillus subtilis 168. J Bacteriol. 2011;193:998–1002. doi: 10.1128/JB.01170-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verhamme DT, Murray EJ, Stanley-Wall NR. DegU and Spo0A jointly control transcription of two loci required for complex colony development by Bacillus subtilis. J Bacteriol. 2009;191:100–108. doi: 10.1128/JB.01236-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ostrowski A, Mehert A, Prescott A, Kiley TB, Stanley-Wall NR. YuaB functions synergistically with the exopolysaccharide and TasA amyloid fibers to allow biofilm formation by Bacillus subtilis. J Bacteriol. 2011;193:4821–4831. doi: 10.1128/JB.00223-11. In this work, YauB was first described as a matrix protein that is required for biofilm formation in addition to TasA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fujita M, Gonzalez-Pastor JE, Losick R. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol. 2005;187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Molle V, et al. The Spo0A regulon of Bacillus subtilis. Mol Microbiol. 2003;50:1683–1701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- 58.Jiang M, Shao W, Perego M, Hoch JA. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol Microbiol. 2000;38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x. [DOI] [PubMed] [Google Scholar]
- 59.McLoon AL, Kolodkin-Gal I, Rubinstein SM, Kolter R, Losick R. Spatial regulation of histidine kinases governing biofilm formation in Bacillus subtilis. J Bacteriol. 2011;193:679–685. doi: 10.1128/JB.01186-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Piggot PJ, Hilbert DW. Sporulation of Bacillus subtilis. Curr Opin Microbiol. 2004;7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 61.Perego M, Hoch JA. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc Natl Acad Sci U S A. 1996;93:1549–1553. doi: 10.1073/pnas.93.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lopez D, Fischbach MA, Chu F, Losick R, Kolter R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci U S A. 2009;106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chu F, Kearns DB, Branda SS, Kolter R, Losick R. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol. 2006;59:1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x. [DOI] [PubMed] [Google Scholar]
- 64.Chu F, et al. A novel regulatory protein governing biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;68:1117–1127. doi: 10.1111/j.1365-2958.2008.06201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lewis RJ, Brannigan JA, Smith I, Wilkinson AJ. Crystallisation of the Bacillus subtilis sporulation inhibitor SinR, complexed with its antagonist, SinI. FEBS Lett. 1996;378:98–100. doi: 10.1016/0014-5793(95)01432-2. [DOI] [PubMed] [Google Scholar]
- 66.Chai Y, Chu F, Kolter R, Losick R. Bistability and biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;67:254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chai Y, Norman T, Kolter R, Losick R. Evidence that metabolism and chromosome copy number control mutually exclusive cell fates in Bacillus subtilis. EMBO J. 2011;30:1402–1413. doi: 10.1038/emboj.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strauch M, Webb V, Spiegelman G, Hoch JA. The SpoOA protein of Bacillus subtilis is a repressor of the abrB gene. Proc Natl Acad Sci U S A. 1990;87:1801–1805. doi: 10.1073/pnas.87.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Strauch MA, et al. Abh and AbrB control of Bacillus subtilis antimicrobial gene expression. J Bacteriol. 2007;189:7720–7732. doi: 10.1128/JB.01081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chai Y, Kolter R, Losick R. Paralogous Antirepressors Acting on the Master Regulator for Biofilm Formation in Bacillus subtilis. Mol Microbiol. 2009;74:876–887. doi: 10.1111/j.1365-2958.2009.06900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kobayashi K. SlrR/SlrA control the initiation of biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;69:1399–1410. doi: 10.1111/j.1365-2958.2008.06369.x. [DOI] [PubMed] [Google Scholar]
- 72.Chai Y, Kolter R, Losick R. Reversal of an epigenetic switch governing cell chaining in Bacillus subtilis by protein instability. Mol Microbiol. 2010;78:218–229. doi: 10.1111/j.1365-2958.2010.07335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chai Y, Norman T, Kolter R, Losick R. An epigenetic switch governing daughter cell separation in Bacillus subtilis. Genes Dev. 2010;24:754–765. doi: 10.1101/gad.1915010. Here SlrR is shown to bind to and re-purpose the SinR repressor in order to regulate a different set of genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murray EJ, Strauch MA, Stanley-Wall NR. SigmaX is involved in controlling Bacillus subtilis biofilm architecture through the AbrB homologue Abh. J Bacteriol. 2009;191:6822–6832. doi: 10.1128/JB.00618-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang X, Fredrick KL, Helmann JD. Promoter recognition by Bacillus subtilis sigmaW: autoregulation and partial overlap with the sigmaX regulon. J Bacteriol. 1998;180:3765–3770. doi: 10.1128/jb.180.15.3765-3770.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang X, Helmann JD. Identification of target promoters for the Bacillus subtilis sigma X factor using a consensus-directed search. J Mol Biol. 1998;279:165–173. doi: 10.1006/jmbi.1998.1765. [DOI] [PubMed] [Google Scholar]
- 77.Luo Y, Asai K, Sadaie Y, Helmann JD. Transcriptomic and phenotypic characterization of a Bacillus subtilis strain without extracytoplasmic function sigma factors. J Bacteriol. 2010;192:5736–5745. doi: 10.1128/JB.00826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mascher T, Hachmann AB, Helmann JD. Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function sigma factors. J Bacteriol. 2007;189:6919–6927. doi: 10.1128/JB.00904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Helmann JD. The extracytoplasmic function (ECF) sigma factors. Advances in microbial physiology. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- 80.Diethmaier C, et al. A novel factor controlling bistability in Bacillus subtilis: the YmdB protein affects flagellin expression and biofilm formation. J Bacteriol. 2011;193:5997–6007. doi: 10.1128/JB.05360-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Winkelman JT, Blair KM, Kearns DB. RemA (YlzA) and RemB (YaaB) regulate extracellular matrix operon expression and biofilm formation in Bacillus subtilis. J Bacteriol. 2009;191:3981–3991. doi: 10.1128/JB.00278-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Verhamme DT, Kiley TB, Stanley-Wall NR. DegU co-ordinates multicellular behaviour exhibited by Bacillus subtilis. Mol Microbiol. 2007;65:554–568. doi: 10.1111/j.1365-2958.2007.05810.x. [DOI] [PubMed] [Google Scholar]
- 83.Murray EJ, Kiley TB, Stanley-Wall NR. A pivotal role for the response regulator DegU in controlling multicellular behaviour. Microbiology. 2009;155:1–8. doi: 10.1099/mic.0.023903-0. [DOI] [PubMed] [Google Scholar]
- 84.Irnov I, Winkler WC. A regulatory RNA required for antitermination of biofilm and capsular polysaccharide operons in Bacillales. Mol Microbiol. 2010;76:559–575. doi: 10.1111/j.1365-2958.2010.07131.x. [DOI] [PubMed] [Google Scholar]
- 85.Kearns DB. A field guide to bacterial swarming motility. Nat Rev Microbiol. 2010;8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.James BL, Kret J, Patrick JE, Kearns DB, Fall R. Growing Bacillus subtilis tendrils sense and avoid each other. FEMS Microbiol Lett. 2009;298:12–19. doi: 10.1111/j.1574-6968.2009.01665.x. [DOI] [PubMed] [Google Scholar]
- 87.Das P, Mukherjee S, Sen R. Genetic regulations of the biosynthesis of microbial surfactants: an overview. Biotechnology & genetic engineering reviews. 2008;25:165–185. doi: 10.5661/bger-25-165. [DOI] [PubMed] [Google Scholar]
- 88.Lopez D, Vlamakis H, Losick R, Kolter R. Paracrine signaling in a bacterium. Genes Dev. 2009;23:1631–1638. doi: 10.1101/gad.1813709. This work showed that not all of the genes in a clonal population produce quorum sensing signals and that the cells that produce the signal are not the same cells that respond to it. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shank EA, Kolter R. Extracellular signaling and multicellularity in Bacillus subtilis. Curr Opin Microbiol. 2011;14:741–747. doi: 10.1016/j.mib.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lopez D, Kolter R. Functional microdomains in bacterial membranes. Genes Dev. 2010 doi: 10.1101/gad.1945010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shemesh M, Kolter R, Losick R. The biocide chlorine dioxide stimulates biofilm formation in Bacillus subtilis by activation of the histidine kinase KinC. J Bacteriol. 2010;192:6352–6356. doi: 10.1128/JB.01025-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 93.Shank EA, et al. Interspecies interactions that result in Bacillus subtilis forming biofilms are mediated mainly by members of its own genus. Proc Natl Acad Sci U S A. 2011;108:E1236–1243. doi: 10.1073/pnas.1103630108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen Y, et al. A Bacillus subtilis sensor kinase involved in triggering biofilm formation on the roots of tomato plants. Mol Microbiol. 2012;85:418–430. doi: 10.1111/j.1365-2958.2012.08109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gonzalez-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- 96.Gonzalez-Pastor JE. Cannibalism: a social behavior in sporulating Bacillus subtilis. FEMS Microbiol Rev. 2011;35:415–424. doi: 10.1111/j.1574-6976.2010.00253.x. [DOI] [PubMed] [Google Scholar]
- 97.Ellermeier CD, Hobbs EC, Gonzalez-Pastor JE, Losick R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell. 2006;124:549–559. doi: 10.1016/j.cell.2005.11.041. [DOI] [PubMed] [Google Scholar]
- 98.Lopez D, Vlamakis H, Losick R, Kolter R. Cannibalism enhances biofilm development in Bacillus subtilis. Mol Microbiol. 2009;74:609–618. doi: 10.1111/j.1365-2958.2009.06882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nandy SK, Bapat PM, Venkatesh KV. Sporulating bacteria prefers predation to cannibalism in mixed cultures. FEBS Lett. 2007;581:151–156. doi: 10.1016/j.febslet.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 100.Liu WT, et al. Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc Natl Acad Sci U S A. 2010;107:16286–16290. doi: 10.1073/pnas.1008368107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kaplan JB. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J Dent Res. 2010;89:205–218. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol. 2012;10:39–50. doi: 10.1038/nrmicro2695. [DOI] [PubMed] [Google Scholar]
- 103.Lam H, et al. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;325:1552–1555. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Burrell M, Hanfrey CC, Murray EJ, Stanley-Wall NR, Michael AJ. Evolution and multiplicity of arginine decarboxylases in polyamine biosynthesis and essential role in Bacillus subtilis biofilm formation. J Biol Chem. 2010;285:39224–39238. doi: 10.1074/jbc.M110.163154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hochbaum AI, et al. Inhibitory effects of D-amino acids on Staphylococcus aureus biofilm development. J Bacteriol. 2011;193:5616–5622. doi: 10.1128/JB.05534-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sanchez Z, Tani A, Kimbara K. Extensive reduction in cell viability and enhanced matrix production in Pseudomonas aeruginosa PAO1 flow biofilms treated with D-amino acid mixture. Appl Environ Microbiol. 2012 doi: 10.1128/AEM.02911-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu CI, et al. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science. 2008;319:1391–1394. doi: 10.1126/science.1153018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Richards JJ, Melander C. Controlling bacterial biofilms. Chembiochem : a European journal of chemical biology. 2009;10:2287–2294. doi: 10.1002/cbic.200900317. [DOI] [PubMed] [Google Scholar]
- 109.Zeigler DR, et al. The origins of 168, W23, and other Bacillus subtilis legacy strains. J Bacteriol. 2008;190:6983–6995. doi: 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McLoon AL, Guttenplan SB, Kearns DB, Kolter R, Losick R. Tracing the domestication of a biofilm-forming bacterium. J Bacteriol. 2011;193:2027–2034. doi: 10.1128/JB.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lazazzera BA. The intracellular function of extracellular signaling peptides. Peptides. 2001;22:1519–1527. doi: 10.1016/s0196-9781(01)00488-0. [DOI] [PubMed] [Google Scholar]
- 112.Srivatsan A, et al. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 2008;4:e1000139. doi: 10.1371/journal.pgen.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Earl AM, Losick R, Kolter R. Ecology and genomics of Bacillus subtilis. Trends Microbiol. 2008;16:269–275. doi: 10.1016/j.tim.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Danhorn T, Fuqua C. Biofilm formation by plant-associated bacteria. Annu Rev Microbiol. 2007;61:401–422. doi: 10.1146/annurev.micro.61.080706.093316. [DOI] [PubMed] [Google Scholar]
- 115.Lugtenberg B, Kamilova F. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 116.Ramey BE, Koutsoudis M, von Bodman SB, Fuqua C. Biofilm formation in plant-microbe associations. Curr Opin Microbiol. 2004;7:602–609. doi: 10.1016/j.mib.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 117.Morikawa M. Beneficial biofilm formation by industrial bacteria Bacillus subtilis and related species. J Biosci Bioeng. 2006;101:1–8. doi: 10.1263/jbb.101.1. [DOI] [PubMed] [Google Scholar]
- 118.Emmert EA, Handelsman J. Biocontrol of plant disease: a (gram-) positive perspective. FEMS Microbiol Lett. 1999;171:1–9. doi: 10.1111/j.1574-6968.1999.tb13405.x. [DOI] [PubMed] [Google Scholar]
- 119.Kloepper JW, Ryu CM, Zhang S. Induced Systemic Resistance and Promotion of Plant Growth by Bacillus spp. Phytopathology. 2004;94:1259–1266. doi: 10.1094/PHYTO.2004.94.11.1259. [DOI] [PubMed] [Google Scholar]
- 120.Ryu CM, Murphy JF, Mysore KS, Kloepper JW. Plant growth-promoting rhizobacteria systemically protect Arabidopsis thaliana against Cucumber mosaic virus by a salicylic acid and NPR1-independent and jasmonic acid-dependent signaling pathway. Plant J. 2004;39:381–392. doi: 10.1111/j.1365-313X.2004.02142.x. [DOI] [PubMed] [Google Scholar]
- 121.Choudhary DK, Johri BN. Interactions of Bacillus spp. and plants--with special reference to induced systemic resistance (ISR) Microbiol Res. 2009;164:493–513. doi: 10.1016/j.micres.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 122.Perez-Garcia A, Romero D, de Vicente A. Plant protection and growth stimulation by microorganisms: biotechnological applications of Bacilli in agriculture. Curr Opin Biotechnol. 2011;22:187–193. doi: 10.1016/j.copbio.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 123.Vessey JK. Plant growth promoting rhizobacteria as biofertilizers. Plant and Soil. 2003;255:571–586. [Google Scholar]
- 124.Fall R, Kinsinger RF, Wheeler KA. A simple method to isolate biofilm-forming Bacillus subtilis and related species from plant roots. Systematic and applied microbiology. 2004;27:372–379. doi: 10.1078/0723-2020-00267. [DOI] [PubMed] [Google Scholar]
- 125.Chen Y, et al. Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ Microbiol. 2012 doi: 10.1111/j.1462-2920.2012.02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bais HP, Fall R, Vivanco JM. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 2004;134:307–319. doi: 10.1104/pp.103.028712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rudrappa T, Czymmek KJ, Pare PW, Bais HP. Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 2008;148:1547–1556. doi: 10.1104/pp.108.127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rudrappa T, Quinn WJ, Stanley-Wall NR, Bais HP. A degradation product of the salicylic acid pathway triggers oxidative stress resulting in down-regulation of Bacillus subtilis biofilm formation on Arabidopsis thaliana roots. Planta. 2007;226:283–297. doi: 10.1007/s00425-007-0480-8. [DOI] [PubMed] [Google Scholar]
- 129.Ongena M, et al. Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ Microbiol. 2007;9:1084–1090. doi: 10.1111/j.1462-2920.2006.01202.x. [DOI] [PubMed] [Google Scholar]
- 130.Donovan C, Bramkamp M. Characterization and subcellular localization of a bacterial flotillin homologue. Microbiology. 2009;155:1786–1799. doi: 10.1099/mic.0.025312-0. [DOI] [PubMed] [Google Scholar]
- 131.Huang X, Gaballa A, Cao M, Helmann JD. Identification of target promoters for the Bacillus subtilis extracytoplasmic function sigma factor, sigma W. Mol Microbiol. 1999;31:361–371. doi: 10.1046/j.1365-2958.1999.01180.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie of B. subtilis cells forming a colony biofilm at room temperature over a period of approximately 9 days on MSgg medium. Images were taken every 4h. Each second of the movie is equivalent to 16 h real time.


