Abstract
MicroRNAs, the non-coding single-stranded RNA of 19–25 nucleotides are emerging as robust players of gene regulation. Plethora of evidences support that the ability of microRNAs to regulate several genes of a pathway or even multiple cross talking pathways have significant impact on a complex regulatory network and ultimately the physiological processes and diseases. Brain being a complex organ with several cell types, expresses more distinct miRNAs than any other tissues. This review aims to discuss about the microRNAs in brain development, function and their dysfunction in brain tumors. We also provide a comprehensive summary of targets of brain specific and brain enriched miRNAs that contribute to the diversity and plasticity of the brain. In particular, we uncover recent findings on miRNA-128, a brain-enriched microRNA that is induced during neuronal differentiation and whose aberrant expression has been reported in several cancers. This review describes the wide spectrum of targets of miRNA-128 that have been identified till date with potential roles in apoptosis, angiogenesis, proliferation, cholesterol metabolism, self renewal, invasion and cancer progression and how this knowledge might be exploited for the development of future miRNA-128 based therapies for the treatment of cancer as well as metabolic diseases.
Keywords: miRNA, miRNA-128, Brain, Cancer, Apoptosis, Cholesterol
Introduction
MicroRNAs are small non-coding RNAs of 19–25 nucleotides in length and are known to regulate several protein-coding genes both in plants and animals. The first miRNA, lin-4 that controlled developmental timing in Caenorhabditis elegans was identified by two different groups in 1993 [1,2]. Later, let-7 miRNAs were found to control the timing of fate specification of neuronal and hypodermal cells during larval development [3-5]. Subsequently, numerous miRNAs have been implicated in a variety of cellular processes including differentiation, apoptosis, cell proliferation, embryonic development, stem cell renewal, stress response and metabolism [6-11]. Their profound impact on the regulation of numerous cellular processes clearly suggests that any aberration in miRNA biogenesis pathway or its regulation contributes to several human diseases such as cancer [12-14], cardiovascular diseases [15], schizophrenia [16], psoriasis [17], diabetes [18], chronic hepatitis [19], AIDS [20], and obesity [21].
MicroRNAs (miRNAs) interfere with target gene expression by binding to the 3′ UTRs of their target mRNAs and act primarily at the level of translation. Complete complementarity between miRNA and 3′UTR of its target leads to the degradation of mRNA targets as shown in plants whereas partial complementarity leads to inhibition of translation as seen in mammals [22-24]. Literature reveals that a single miRNA can target several mRNAs together, and a single mRNA can be targeted by different miRNAs in a concerted manner. Large number of microRNAs and the capacity of each miRNA to target several transcripts suggest a complex regulatory network to fine tune the gene expression and a mechanism by which they are thought to regulate various processes during health and disease [25].
The advancement of high-throughput sequencing techniques has led to the rapid growth in the number of annotated miRNA. The most recent miRBase Sequence Database, Release 20 ( http://www.mirbase.org/), harbours 24521 entries representing hairpin precursor miRNAs and expressing 30424 mature miRNA products in 193 species [26]. The sequences of most miRNAs are conserved across large evolutionary distances, suggesting a conserved role in regulation of various physiological processes [27].
The diversity and gene-regulatory capacity of miRNAs is particularly valuable in the brain, where persistent flow of information and functional specialization of neurons requires constant neuronal adaptation to environmental cues [28]. The brain expresses more distinct and largest number of miRNAs than any other tissue in vertebrates as it has wide variety of cell types, neuronal and nonneuronal (for e.g. astrocytes) [29]. In this review, we summarize present knowledge on the microRNA expression and functions in the brain and their potential involvement in relation to brain tumors. Herein, we also give an overview of the functions and targets of brain-enriched and brain-specific miRNAs before delving into specific example of miRNA-128, the most abundant brain-enriched miRNA. We believe that the understanding of the impact of microRNA-128 on regulation of proliferation, apoptosis and metabolic processes is still at its dawn and needs further research for the development of future miRNA-based therapies for the treatment of metabolic diseases and cancer.
MiRNA biogenesis and mechanism of action
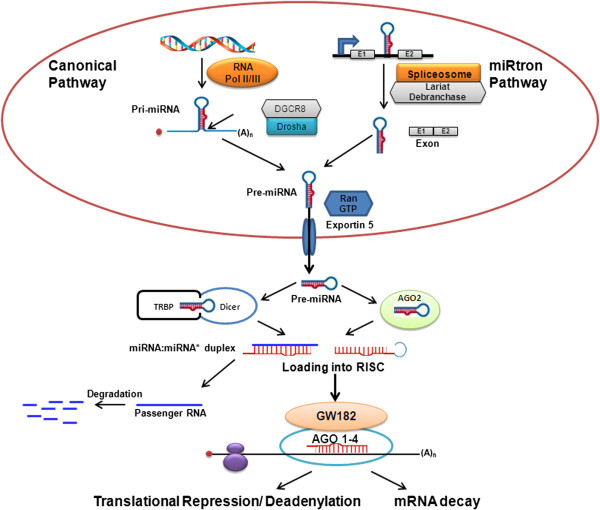
Approximately, 50% of the mammalian miRNAs have found their location in introns or exons of protein-coding genes or introns of long non-coding RNAs [30,31]. Their expression is either derived by independent transcriptional units or by protein-coding gene transcriptional units [32]. As shown in Figure 1, miRNAs are transcribed from genomic DNA by RNA polymerase II or III into long, primary transcripts (pri-miRNAs) just like other protein coding genes. These pri-miRNAs are several kilobases in length and usually possess a 5′ CAP and a 3′ poly (A) tail. These pri-miRNAs are processed by a microprocessor complex which consists of a ribonuclease III (RNase III) named Drosha, a RNA-binding protein DiGeorge syndrome critical region 8 (DGCR8/Pasha) and a variety of co-factors [DEAD box helicases p68 and p72 and the heterogeneous nuclear ribonucleoproteins (hnRNPs)] which are thought to promote the specificity and/or activity of Drosha cleavage [32-35]. Drosha processing occurs co-transcriptionally in most mammalian miRNAs i.e. before splicing of host RNA (canonical pathway). However, Drosha pathway can be evaded by miRtrons (a subset of intronic miRNAs) and are made by splicing and debranching of short hairpin introns [36,37]. The product of Drosha cleavage is a 70–100 nucleotide hairpin-shaped precursor referred to as pre-miRNA. These pre-miRNAs are exported to the cytoplasm by Ran-GTP and Exportin-5 dependent mechanisms [38]. In cytoplasm, these pre-miRNAs are excised by the RNase III enzyme Dicer into a double-stranded RNA of ~22 nucleotides in length, referred to as the miRNA:miRNA* duplex or by Ago2, an Argonaute protein that is part of the RISC complex and aligns the miRNA and messenger RNA [39,40]. The criteria for binding and cleavage by Ago2 after the 30th nucleotide are short stem and spanning of the loop by miRNA sequence. The duplex produced by either Dicer or Ago2 is loaded onto an Argonaute protein where one strand, i.e. guide strand, complementary to the target mRNA, is selected and subsequently forms the miRNA effector as part of a miRISC (miRNA-induced silencing complex), while the remaining strand (the "passenger strand") is released and degraded [41]. Similar to Drosha and Dicer assisting proteins, the formation of the miRISC and the execution of its activity involve many additional factors [42]. The two key factors involved in the assembly and function of miRISCs are Argonaute (AGO) proteins, which directly interact with miRNAs, and glycine-tryptophan protein of 182 kDa (GW182), which act as downstream effectors in the repression. miRNA then guides the miRISC to recognize the partially complementary binding sites located in the 3′UTR of their target mRNAs.
Figure 1.
miRNA biogenesis pathway and function: miRNAs are transcribed in the nucleus either from introns or exons of protein-coding genes or introns of long non-coding RNAs into primary transcripts (pri-miRNAs). Pri-miRNAs are then processed in two steps in the nucleus and cytoplasm, catalyzed by the RNase III type endonucleases Drosha and Dicer, in complexes with dsRNA-binding domain proteins, DGCR8 and TRBP respectively. In the canonical pathway, Drosha-DGCR8 processes the transcript to a stem loop-hairpin precursor (pre-miRNA). Intron derived miRNAs, called miRtrons, evade canonical pathway and processed by the spliceosome and the debranching enzyme into pre-miRNAs. Both canonical miRNAs and miRtrons are exported to the cytoplasm via Exportin 5, where they are further processed by Dicer-TRBP or by Ago2 to yield 20-25-bp miRNA duplexes. Dicer processing adds 5′ phosphate groups and two-nucleotide overhangs at the 3′ ends of the mature strands. The duplex produced by either Dicer or Ago2 is loaded onto an Argonaute protein of RISC where one strand is selected to function as mature miRNA while the partner miRNA* strand is preferentially degraded. The mature miRNA produced by these two mechanisms leads to translational repression or mRNA degradation.
The perfect binding between seed region (5′ 2–8 nucleotides 3′) of mature miRNA and 3′UTR of their target by Watson-Crick base-pairing is considered to be the major determinant in blocking the target mRNA either by translational repression or mRNA degradation [43]. However other 3′- supplementary and 3′- compensatory binding sites in miRNA sequence also play a significant role during interactions [24]. Although miRNA binding sites are most common in 3′UTRs of mRNAs, yet there are some reports of miRNA interaction within the 5′UTR, mRNA coding region and intron-exon junctions [44,45]. The detailed mechanisms underlying the inhibition of protein synthesis by miRNAs are not well understood, but literature suggests sequestration of mRNA into P bodies from ribosomes, blockage of translational initiation, translational repression or target deadenylation coupled to transcript degradation [23,46]. However, it is now believed that miRNA regulate gene expression in majority of cases by mRNA decay rather than translational repression [47]. Epigenetic modifications and transcription factors also play a decent role in the regulation of miRNA function. Recent reports also depict the role of pseudogenes as miRNA sequestering sponges or decoys in the regulation of miRNA function [48,49].
MicroRNAs in brain development and function
The brain is a complex organ, with various types of cells (neurons and non-neurons) that form an intricate communication network. Literature reveals that 70% of known miRNAs are expressed in the brain [50]. Surprisingly, only a handful of microRNAs are expressed in a brain specific or brain-enriched manner [51]. Since these miRNAs are dynamically regulated during brain development, have different targets and perform different functions in brain, herein we provide a comprehensive list of the recent validated roles of brain-enriched and brain-specific microRNAs along with their targets in brain in Tables 1 and 2.
Table 1.
Comprehensive list of brain enriched microRNAs and their targets and functions related to brain
| Brain enriched miRNAs | Target | Function | Ref |
|---|---|---|---|
| miR-9* |
SOX2 |
Induces neuronal differentiation, affects both proliferation and differentiation |
[52-54] |
| miRNA-128 |
Reelin, DCX, SUZ12, neurofibromin 1, BMI1, RTK, EGFR,PDGFRαUPF1, MLN51, NTRK3, WEE1, Bax, E2F3a, SNAP25 |
Synaptogenesis; reduces neuroblastoma cell motility and invasiveness; suppressor of PRC activity; renders glioma stemlike cells less radioresistant; suppressor of the colony formation ability and invasiveness of pituitary tumor cells; suppressor of growth and mediates differentiation; regulates Nonsense-mediated decay; regulates apoptosis, inhibits proliferation and self-renewal, |
[55-68] |
| miR-7 |
KLF4, α-synuclein, Sepp1b, EGFR, IRS-2 |
Suppresses brain metastasis, control neurite outgrowth, protects against oxidative stress, potential tumor suppressor, decreases viability and invasiveness of primary glioblastoma |
[65,69-74] |
| miR-125 a-b |
NR2A, SMG1, SMAD4 |
regulates synaptic plasticity; regulates Nonsense-mediated decay. |
[52,75-77] |
| miR-23 |
laminB1,X-linked inhibitor of apoptosis (XIAP) |
regulates oligodendrocyte development and myelination, regulates cerebral ischemia and neural specification |
[78,79] |
| miR-132 |
PTBP2, AChE, FoxP2, Sirt1, MeCP2, ATA2, DPYSL3, STAT4; p250RhoGAP, Mecp2, Ep300, Jarid1a, Btg2, Paip2a, For more targets view
[80] |
Regulates progressive supranuclear palsy, regulator of the brain-to-body resolution of inflammation, contribute to neurodevelopmental and neuromorphological pathologies, neuronal cell development, regulate synaptic plasticity, neuronal maturation, regulates basal and activity-induced neurite outgrowth, regulates recognition memory and synaptic plasticity, regulates Circadian Clock. |
[52,80-93] |
| miR-137 |
CDK6, MindBomb-1, CSMD1, C10orf26, CACNA1C, TCF4, ZNF804A, neurofibromin 1, CSE1L, Cox-2, LSD1, MITF, EZH2, KLF4, SPTLC1, For more targets view
[94] |
Inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells, neuronal maturation, regulates differentiation of neural stem cells, suppress growth and invasion of oligodendroglioma and glioma cells, |
[52,67,94-102] |
| miR-139 | Mcl-1, C-X-C chemokine receptor type 4 (CXCR4), FoxO1, CPG1, Bcl2 | Suppressor of the proliferation and enhances drug induced apoptosis, Reduced invasion and metastasis, Regulates Transcriptional activity. | [52,88,103-105] |
Table 2.
Comprehensive list of brain specific microRNAs and their targets and functions related to brain
| Brain specific miRNAs | Target | Function | Ref |
|---|---|---|---|
| miR-9 |
KCNMA1, cyclicAMP response element-binding protein (CREB), neurofibromin 1 (NF1), Hes1, FoxP2, prelamin A |
Promotes Neuronal differentiation, Inhibits proliferation, Promotes migration, Control neural stem cell differentiation |
[65,87,106-112] |
| miR-124 a-b |
SNAI2, NR3C2, SOS1, CDK4, Usp14, inhibitory member of the apoptosis-stimulating proteins of p53 family (iASPP), AMPA2 and AMPA3, SCP-1, PTBP1, Sox9, Ephrin-B1, JAG1, BAF53a, CDK6, p38α mitogen-activated protein kinase, CEBPa, RhoG, anachronism (ana), SNAI2, Lhx2, Ctdsp1, BACE1, NeuroD1 |
Promotes neuronal transcriptome/neurogenesis; inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells, regulation of renin-angiotensin-aldosterone system, radiosensitize Glioblastoma multiforme cells, promotes neuronal survival under ischemic conditions, induce differentiation into neurons, regulates the migration of glioma cells and the self-renewal of GSCs, inhibits growth of medulloblastoma xenograft tumors, regulates neuroblast proliferation, alleviates cell death. |
[53,65,95,113-134] |
| miR-134 |
Nanog, LRH1, Forkhead Box M1 (FOXM1), μ-opioid receptor (MOR), DPD gene (DPYD), Xenopus LIM kinase 1 (Xlimk1), cMYC, Pum2, Dcx and Chrdl-1, CREB, splicing factor SC35, Limk1 |
Controls dendritic spine development, control synaptic protein synthesis and plasticity, inhibits cell proliferation, invasion and migration capability and promotes apoptosis, inhibits epithelial to mesenchymal transition, guidance of nerve growth cones, growth-promoting effect on dendritogenesis; inducer of pluripotent stem cell differentiation; stage-specific modulation of cortical development, regulates memory, modify both alternative splicing and cholinergic neurotransmission |
[135-147] |
| miR-135 |
Focal Adhesion Kinase (FAK), EB1, NR3C2, Smad5, APC |
Decreased cell invasion and increased drug sensitivity, regulation of immunity, regulation of renin-angiotensin-aldosterone system, inhibit differentiation of osteoprogenitors, regulates Wnt signaling pathway. |
[52,118,148-151] |
| miR-153 |
SNCA, BSN, PCLO, amyloid-β (Aβ) precursor protein (APP), APLP2, alpha-synuclein, Bcl-2 and Mcl-1, SNAI1 and ZEB2 |
Promote neuronal differentiation, impairs self-renewal ability and induces differentiation, repress growth and induce apoptosis of GBM-stem cells, decreases cell proliferation and increases apoptosis in GBM cell line, regulates epithelial-mesenchymal transition and tumor metastasis, regulate gliomagenesis. |
[52,152-159] |
| miR-219 | EGFR, PLK2, Sox6, FoxJ3, PDGFRα, ZFP238, ELOVL7, CaMKIIgamma | Inhibits the proliferation, anchorage independent growth and migration of glioma cells, promote oligodendrocyte differentiation and myelination, modulates NMDA receptor-mediated neurobehavioral dysfunction, maintenance of lipids and redox homeostasis in mature Olligodendrocytes, regulates circadian rhythms of expression. | [52,160-166] |
The increasing variety of miRNAs being identified in the brain suggests a sheer connection between the biogenesis, dynamics of action and regulatory potential of miRNAs and the complexity of the brain. Numerous studies on depletion of the Dicer gene in the nervous system of animal models further demonstrate that microRNAs play essential roles in controlling neuronal proliferation, migration and precursor fates [167-171] and serve important roles in development and function in the brain [172,173]. Stark et al. in their study, have illustrated the contribution of defects in miRNA biogenesis to brain abnormalities in Dicer deficient mice and mouse model of schizophrenia [174].
An expression profiling study by Sempere et al. showed a group of 17 miRNAs were expressed in mouse and human brain (miR-7, -9, -9*, -124a, -124b, -125a, -125b, -128, -132, -135, -137, -139, -153, -149, -183, -190, -219). All these miRNAs have been found to regulate neuronal differentiation, maturation, and/or survival in mouse and human. Conservation of these miRNAs between mouse and human suggests that they may play a conserved role in the establishment and/or maintenance of a cell or tissue type of brain [52,55,106,175]. Specific expression of miR-9 and miR-132 is restricted to hippocampus and medal frontal gyrus [176] whereas miR-124 and miR-128 are unique for neurons and miR-23, miR-26 and miR-29 are specifically expressed in astrocytes [177]. In addition miR-195 displays a moderate to low expression level in the mammalian embryonic brain, with the highest level at the preadult brain developmental stage [178].
Studies have further shown that miR-9 expression is necessary for neurogenesis in cultured stem cells and miR-132 plays a role in neurite extension and neurogenesis [52,179]. MiR-132 has also been linked to BDNF (a member of the nerve growth factor family that is necessary for survival of striatal neurons in the brain) and MeCP2 (methyl-CpG DNA binding protein that plays an essential role in mammalian development) by a negative feedback loop [81]. Interestingly, miR-124 when overexpressed in non-neuronal HeLa cells shifts the gene expression profile from an immature cervix cell into a neuronal phenotype, suggesting that miR-124 downregulates mRNAs directing cells into a non-neuronal phenotype [180]. Recently, it has been observed that MiR-124 targets REST, BTBP and Sox9, all proteins have been known to antagonize the formation of neuronal cells during development [106,113,114]. Similar to miR-124, miRNA-128 is induced during brain development and in differentiating neuronal cells; leading to repressed NMD (Nonsense-mediated decay) and the consequent upregulation of batteries of mRNAs encoding proteins important for neuron differentiation and function [56]. MiR-23 is implicated in neural specification while miR-26 is required during neuronal cell differentiation. Report by Kole et al. showed that miR-29b is markedly induced during neuronal maturation and functions as a inhibitor of neuronal apoptosis [181]. Apart from development, aberrant microRNA expression has been discovered in human CNS (central nervous system) diseases including brain tumors in the past decade.
Dysfunction of microRNAs in brain tumors
Gliomas are the primary brain tumors that are made up of glial cells which provide important structural support for the nerve cells in the brain. Malignant gliomas are the most common and lethal tumors arising in the central nervous system and are classified by the World Health Organization (WHO) into four different grades based on malignancy (I, II, III, IV) [182,183]. Grade IV glioblastoma multiforme (GBM) is the most common lethal primary brain tumor in adults that is characterized by aggressive vascular proliferation, invasiveness, stem cell-like behaviour and chemoresistance to new and traditional therapies [183]. Accumulating evidences indicate the presence of different miRNAs with pro-oncogenic and anti-oncogenic properties in glioblastomas. Koshkin et al. recently observed gradual increase in miR-21 and miR-23a levels in all tumor grades and significant decrease of miR-7 and miR-137 depending on the glioma grade [184]. Further, miR-210 has been found to be highly expressed in human gliomas and confers a poor prognosis in glioma patients [185]. MicroRNA-206 has been found to be a tumor suppressor in human malignant cancers. Low miR-206 expression is associated with poor overall survival in patients with malignant astrocytomas, hence it could become a valuable marker of astrocytoma progression [186]. Apart from these, several independent studies observed that the expressions of miR-16, miR-503, miR-203, miR-34c-3p, miR-34c-5p, miR-106a, Let-7a, miR-218, miR-223, miR-34a, miR-329, miR-145, miR-124, miR-137, miR-138, miR-219-5p, miR-495, miR-383, miR-200b, miR-134, miR-153, miR-195, miR-143, miR-107, miR-326, miR-204 and miR-214 were significantly reduced in high WHO grade glioma tissues relative to low WHO grade glioma tissues and normal brain tissues [115,135,152,160,187-208]. The expression of most of these miRNAs decreases with increasing degrees of malignancy. The low expression of Let-7a was correlated with poor prognosis of primary glioblastoma patients [192]. Further, miR-708 and miR-17 ~ 92 cluster were downregulated in GBM tumor cell lines whereas three miR-17 ~ 92 cluster miRNAs (miR-2, -19a and -20) were upregulated in human medulloblastoma with aberrantly activated sonic hedgehog (SHH) signaling pathway [209-211]. Another group by Skalsky and Ciafre et al. in their cluster analysis further revealed that miR-139, miR-95 and miR-873 were down-regulated specifically in glioblastomas and miR-137 and miR-181a/b were down-regulated in gliomas whereas miR-7, miR-124 and miRNA-128 were down-regulated in both [212,213]. Interestingly, miRNA-128 and miR-124 are not only down regulated in gliomas but also other brain cancers including medulloblastomas and neuroblastomas [212,214]. Li et al. reported that decreased miR-146b-5p expression was strongly correlated with chromosome 10q loss in gliomas, especially glioblastomas [215]. The significance of the sequence of miRNA can be illustrated by the example of miR-23 in brain. The expression of miRNA-23b was gradually downregulated with the malignancy of glioma whereas miR-23a was upregulated in malignant glioma tissues [216,217]. Further, miR-328, miR-106b-5p, miR-155, miR-650, miR-92b, miR-30a-5p, miR-10b, miR-372, miR-183, miR-486 and miR-17 were found to be upregulated in invading glioma cells in vivo and glioma tissues respectively as revealed by miRNA expression profiling of microdissected human tumor biopsy specimens [218-228]. MiR-650 expression can be used as a significant prognostic indicator in glioma. MiR-19a, -19b and miR-9 have been found to be overexpressed in glioma cell lines and astrocytic glioma tissues, and their expression level is positively correlated with tumor grades [107,229]. Several studies showed that miR-21 and miR-10b are upregulated in glioblastomas and has recently been shown to be a significant contributor for tumor growth in vivo [230,231]. Wu et al. have recently documented that overall patient survival for those with low miR-21 expression was significantly longer than those patients with high miR-21 expression [232]. Further, miRNA-21, 221, 222, 181b, 181c, and 128a were found to be significantly deregulated in GBM tissues by Slaby et al. and Zhou et al. It was also observed that miRNA-181b and 181c were the most down regulated miRNAs in patients who responded to radiation therapy (RT) and temozolomide (TMZ) and hence could serve as predictors for RT/TMZ response. Several differentially expressed miRNAs such as miR-124, miR-21, -128, -181, -221 and -222 could serve as potential biomarkers in GBM in general since they play common role in the etiology of malignant brain tumors [233,234].
Role of pro-neural miRNA-128 in brain related disorders
MiRNA-128 is transcribed by two distinct genes, miRNA-128-1 and miRNA-128-2 in two primary transcripts, which are processed into an identical mature miRNA sequence. MiRNA-128-1 and miRNA-128-2 are both present in the intronic regions of two genes on two different chromosomes. MiRNA-128-1 is embedded in the R3HDM1 (R3H domain containing 1) gene on chromosome 2q21.3 and miRNA-128-2 is in the ARPP21 (cyclic AMP-regulated phosphoprotein, 21 kDa) on chromosome 3p22.3 [57]. Evidences in the literature reveal that miRNA-128 has tissue specific and developmental specific expression patterns. Apart from brain, miRNA-128 has also been found in the skeletal muscle and thymus and is highly expressed during neuronal differentiation. Down regulation of miRNA-128 has been reported in several brain cancers for example- glioblastoma [213] and medulloblastoma [235]. Allelic loss in chromosome 3p, where miRNA-128-2 is present, has also been associated with the most aggressive forms of neuroblastoma [213].
Cui et al. demonstrated that the down-regulation of miRNA-128 inversely correlates with tumor grade. They also observed that the decrease of miRNA-128 is coupled with significant increase in the expression of Bmi-1, the transcription factor E2F-3a and angiopoietin-related growth factor protein 5 (ARP5; ANGPTL6). Increased expression of these factors may explain the undifferentiated, self-renewing state of brain cells and de-regulated cell-cycle signaling pathways that support cellular proliferation in glioma and GBM [236]. Zhang et al. in his study showed that brain-enriched miRNA-128 was also down regulated in glioma tissues and cell-lines and overexpression of miRNA-128 inhibited cellular proliferation through negatively regulating E2F3a, which is highly expressed in glioma and important for cell cycle progression (Figure 2) [58]. Papagiannakopoulos et al. recently showed that miRNA-128 represses growth and enhances neuronal differentiation of glioma-initiating neural stem cells (giNSCs) by downregulating oncogenic receptor tyrosine kinases (RTKs), epithelial growth factor receptor (EGFR) and platelet-derived growth factor receptor-α (PDGFRα) (Figure 2) [59]. In an independent study, Godlewski et al., reported that overexpression of miRNA-128 reduces glioma cell proliferation by downregulating Bmi-1 (B lymphoma mouse Moloney leukemia virus insertion region 1), decrease in histone methylation (H3K27me3) and Akt phosphorylation and up-regulation of p21CIP1 levels [60]. As Bmi-1 is also known to promote the stem cell renewal, a process that is important in glioma, hence miRNA-128 may be used against the "stem cell-like" characteristics of glioma cells [237]. These data suggest that miRNA-128 may suppress cancer pathogenesis by inducing differentiation out of a stem cell-like state. Roth et al. recently reported upregulation of miRNA-128 in the blood samples of glioblastoma patients compared to healthy controls and speculated that this miRNA fingerprint may be used as suitable biomarker for glioblastoma [238].
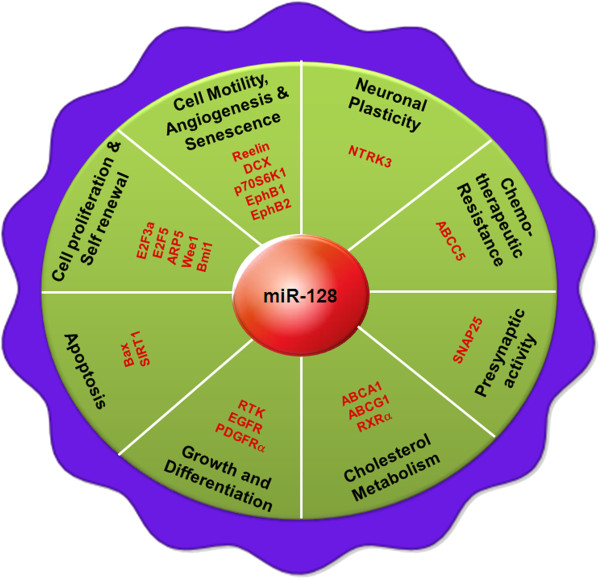
Figure 2.
Roles of miRNA-128 in different cellular processes: The role of miRNA-128 in the different biological processes and multistep events that lead to cancer are shown. The experimentally validated target genes of miRNA-128 are depicted along with the respective biological processes.
Besides these, levels of miRNA-128 have been reported to be deregulated in autism, prion-induced neurodegeneration, Huntington disease, Parkinson disease and Alzheimer disease [239-242]. Several evidences in the literature show that under different biological conditions, expression patterns of miRNA-128 varies i.e. in some instances, it is up regulated whereas in some, it is down regulated (Table 3). Furthermore, Eletto et al. in his study showed that miRNA-128a inhibits expression of the pre-synaptic protein SNAP25 by binding to its 3′UTR (Figure 2). They observed Tat mediated deregulation of miRNA-128, in primary cortical neurons during the infection of neurons by HIV-1. However, the role of miRNA-128a in regulating synaptic activity in normal and in neurodegenerative disorders including HIV-1 Encephalopathy (HIVE) needs to be determined [61].
Table 3.
The pathological conditions in which miRNA-128 is implicated[243](u - up, d – down; hsa-miRNA-128- refers to both hsa-miRNA-128a and b)
| miRNA | Disease | Status | Reference | Year |
|---|---|---|---|---|
| hsa-miRNA-128a |
Acute lymphoblastic leukemia (ALL) |
u |
[244] |
2007 |
| hsa-miRNA-128a |
Acute myeloid leukemia (AML) |
d |
[244] |
2007 |
| hsa-miRNA-128a |
Alzheimer’s disease |
u |
[242] |
2007 |
| hsa-miRNA-128a |
Autism spectrum disorder (ASD) |
u |
[239] |
2008 |
| hsa-miRNA-128a |
Glioblastoma |
d |
[213] |
2005 |
| hsa-miRNA-128a |
Glioblastoma multiforme (GBM) |
d |
[95] |
2008 |
| hsa-miRNA-128a |
Malignant melanoma |
d |
[245] |
2008 |
| hsa-miRNA-128a |
Oral Squamous Cell Carcinoma (OSCC) |
d |
[246] |
2008 |
| hsa-miRNA-128a |
Pituitary adenoma |
d |
[247] |
2007 |
| hsa-miRNA-128a |
Breast cancer |
u |
[248] |
2008 |
| hsa-miRNA-128b |
Lung cancer |
d |
[249] |
2008 |
| hsa-miRNA-128b |
Acute lymphoblastic leukemia (ALL) |
u |
[244] |
2007 |
| hsa-miRNA-128b |
Acute myeloid leukemia (AML) |
d |
[244] |
2007 |
| hsa-miRNA-128b |
Breast cancer |
u |
[250] |
2005 |
| hsa-miRNA-128b |
Chronic pancreatitis |
u |
[251] |
2007 |
| hsa-miRNA-128b |
Colorectal cancer |
u |
[252] |
2006 |
| hsa-miRNA-128b |
Lung cancer |
u |
[252] |
2006 |
| hsa-miRNA-128b |
Malignant melanoma |
d |
[245] |
2008 |
| hsa-miRNA-128b |
Pancreatic cancer |
u |
[252] |
2006 |
| hsa-miRNA-128b |
Hepatocellular carcinoma (HCC) |
u |
[253] |
2009 |
| hsa-miRNA-128b |
Acute promyelocytic leukemia (APL) |
d |
[254] |
2009 |
| hsa-miRNA-128 |
Glioma |
d |
[58] |
2008 |
| hsa-miRNA-128 |
Neurodegeneration |
u |
[240] |
2008 |
| hsa-miRNA-128 |
Neuroblastoma |
d |
[55] |
2009 |
| hsa-miRNA-128 | Huntington’s disease | d | [241] | 2010 |
Expression of pro-neural miRNA-128 in cancers other than brain
Cancer occurs due to accumulation of several genomic alterations and is characterized by unrestricted proliferation, invasion, and metastasis. miRNAs normally negatively regulate their transcript targets and recent evidence indicates that miRNAs may function as tumor suppressors (by binding to oncogenes and suppressing them) or oncogenes (by binding to tumor suppressor genes and suppressing them) and alterations in miRNA expression may play a critical role in the cancer initiation and progression [255,256]. Within the past few years, profiling of the miRNome (global miRNA expression levels) is common and abundant miRNome data are currently available for various cancers. MiRNA expression can be correlated with cancer type, stage, and other clinical variables which may be useful for the classification, diagnosis, or prognosis of some human malignancies [257]. With respect to miRNA-128, it is known that miRNA-128 may act as a tumor-suppressor. Kotani et al. in his study reported down regulation of miRNA-128 in MLL-AF4 Acute Lymphocytic Leukemia and Khan et al. revealed down regulation of miRNA-128 in invasive prostate cancer cells as compared to benign prostate epithelial cell lines, where its levels are elevated [254,258]. In addition, miRNA-128 was significantly reduced in chemoresistant breast tumor-initiating cells (BT-ICs) enriched from breast cancer cell lines and primary breast tumors (P < 0.01), accompanied by an overexpression of Bmi-1 and ABCC5, which were identified as targets of miRNA-128 [259]. In contrast to these studies, strong induction of miRNA-128 has been observed in endometrial cancer [260] as well as in acute lymphoblastic leukemia (ALL) [244]. Up regulation of miRNA-128 not only suppressed the colony formation ability and invasiveness of pituitary tumor cells but also suppressed pituitary GH3 tumor growth in xenografts via, Bmi-1. MiRNA-128 found to regulate its direct target Bmi-1 and PTEN-AKT pathway in pituitary tumors [62]. Allelic loss in chromosome 3p (where miRNA-128-2 is present) has been shown to be associated with the most aggressive form of lung carcinogenesis. Furthermore, it was observed that loss of heterozygosity (LOH) of MicroRNA-128b in tumor samples correlated significantly with clinical response and survival following Gefitinib via EGFR [249]. Although the increase/decrease of miRNA-128 has been reported in a number of the studies related to cancer but it is not known whether it is a cause or effect of the disease.
Pro-neural miRNA-128 as regulator of apoptosis
Alterations in susceptibility to apoptosis is a key factor for the survival of a malignant cell [261] and it enhances resistance to conventional anticancer therapies [262]. As the altered expression of pro-neural miRNA-128 was found in several cancers, numerous studies were undertaken to delineate the mechanism for the inhibition of cell proliferation and induction of apoptosis by miRNA-128. Sean Lawler’s laboratory demonstrated that ectopic expression of miRNA-128 in human glioma neurosphere cultures (having stem-like properties) led to reduction in glioma neurosphere number and volume by down regulating Bmi-1 (Figure 2) [60]. In addition to this, several independent studies have illustrated the anti-proliferative role of miRNA-128 in glioma cells and glioblastoma cell lines [58,236]. Infact, ginsenoside Rh2, a triterpene saponin has also been found to inhibit glioma cell proliferation by upregulating microRNA-128 [263]. Further, over expression of miRNA-128 leads to an alteration in the expression of genes implicated in cytoskeletal organization (via truncated isoform of NTRK3) as well as genes involved in apoptosis, cell survival and proliferation, including the anti-apoptotic factor BCL2 in SH-SY5Y neuroblastoma cells [63]. In our laboratory also, we recently observed that miRNA-128 overexpression induced apoptosis by down regulation of Bax and up regulation of p53 and Bak [57]. Furthermore, transcriptome analysis of miRNA-128 overexpressed cells revealed that miRNA-128 inhibits SIRT1 expression directly through a miRNA-128 binding site within the 3′UTR of SIRT1 (Figure 2). Finally, we found that miRNA-128 induces apoptosis in wild type (WT) p53 as well as in mutant p53-expressing cells in a p53-dependent and -independent manner via induction of PUMA in MCF7, MDA-MB-231, HCT116 p53 +/+ and HCT116 p53 -/- cells respectively [264]. In our study, we also demonstrated that miRNA-128 augments the antitumor effects of compounds (Etoposide and Cisplatin). Contrary to our findings, Yolanda’s group has shown that ectopic expression of miRNA-128 downregulated genes that induce apoptosis and upregulated genes implicated in cell survival [63]. In an another recent study, miRNA-128 was found to target Bax in breast cancer cell line MDA-MB-231 and downregulation of miRNA-128 sensitised MDA-MB-231 cells to chemodrugs [265]. Furthermore, Donzelli et al. observed that miRNA-128-2 expression in lung cancer cells inhibits apoptosis and confers increased resistance to cisplatin, doxorubicin and 5-Fluorouracyl treatment via E2F5 (Figure 2) [266]. Based on the above information, we can say that depending upon the cell type; miRNA-128 can have anti-apoptotic as well as pro-apoptotic functions. It seems that miRNA-128 can be targeted to facilitate cancer cell death and/or inhibit cancer cell growth; however, this aspect warrants further investigation.
Role of miRNA-128 in cell motility, angiogenesis and senescence
Ability to migrate and eventually disseminate to distal sites is one of the key characteristic of tumor cells, which is also responsible for the relative aggressiveness of the tumor. Data from several independent studies showed that overexpression of miRNA-128 inhibits cell motility and invasiveness. Evangelisti et al. proved that overexpression of miRNA-128 reduces neuroblastoma cell motility and invasiveness by targeting Reelin and DCX (Figure 2) [55]. DCX is a microtubule-associated protein required for neuroblastic migration during cerebral cortex development [267] while Reelin is a high-molecular-weight secreted glycoprotein, which is thought to play its role as a guide for migratory neurons [268]. Messi et al. described DCX as a marker of SK-N-SH neuroblastoma cells that show high motility and invasiveness [269]. DCX expression is also detectable in some tumors of the nervous system, such as GBM and neuroblastoma. Reelin has been shown to be a positive marker for prostate carcinoma aggressiveness [270] and is overexpressed in retinoblastoma and esophageal carcinoma and its expression is directly correlated with tumor aggressiveness [271,272]. Khan et al. investigated the proteomic alterations in a cohort of 15 prostate-derived tissues from adjacent benign prostate (Benign), clinically localized prostate cancer (PCA) and metastatic disease from distant sites (Mets). By coupling multidimensional protein fractionation and quantitative mass spectrometry with bioinformatics-based enrichment analysis, they demonstrated the involvement of miRNA-128 in the stages of prostate cancer progression. They have shown using qRT-PCR that miRNA-128 levels were reduced in invasive prostate cancer cells as compared to benign prostate epithelial cell lines. Further, over expression of miRNA-128 attenuated invasion in prostate cancer cells while its knockdown induced invasion in benign prostate epithelial cells as revealed by matrigel invasion assay [258]. These findings suggest that miRNA-128 reduces cell motility and invasiveness of tumor cells and thus prevents angiogenesis. In an independent study, Shi et al. observed that miRNA-128 overexpression inhibited tumorigenesis and angiogenesis through targeting p70S6K1 and suppressing downstream molecules of p70S6K1 such as HIF-1 and VEGF [273]. Their study identified a link between miRNA-128 and p70S6K1 axis, which plays a vital role in glioma angiogenesis (Figure 2).
Over expression of Bmi-1 oncogene has been found to promote NSC self-renewal by repressing the p16Ink4a and p19Arf senescence pathways [274]. As miRNA-128 directly targets Bmi-1 oncogene, the role of miRNA-128 in senescence is also evident. Observation by Venkatraman’s group of increased methylation of histone 3 lysine 9 (H3K9me2) (a mark of repressed gene expression mediated by the Bmi-1 polycomb repressor complex) after overexpression of miRNA-128 further confirms the role of miRNA-128 in promoting cellular senescence. They also observed that overexpression of miRNA-128a in medulloblastoma alters the intracellular redox state of the tumor cells. In our study also, over expression of miRNA-128 in HEK293T cells led to an increase in reactive oxygen species [57]. This is quite interesting for therapeutic scenario where miRNA-128 can be used as therapeutic modality for treating cancer, as cancer stem cells are more resistant to therapy due to a lower overall redox state, where it can induce ROS [235].
Insights into regulation of cholesterol metabolism by miRNA-128: a new key player in cholesterol related disorders
Aberrant regulation of cholesterol homeostasis is associated with obesity as well as multiple types of cancer. The regulation of cholesterol homeostasis pathways is complex with transcriptional regulation by sterol-regulatory element-binding protein (SREBP) and liver X receptor/retinoid X receptor (RXR) transcription factors but poorly understood at the post-transcriptional levels [275,276]. While investigating the mechanism of miRNA-128 induced apoptosis, we observed that besides regulating the genes of apoptosis, miRNA-128 also regulates cholesterol metabolism and fatty acid biosynthesis pathways. In our study, we discovered that miRNA-128 up-regulated cholesterol synthesis genes and down regulated fatty acid biosynthesis genes. miRNA-128 further affected cholesterol efflux pathway by direct targeting ABCA1, ABCG1 and RXRα (Figure 2). We provided the first evidence of miRNA-128-2 to be a new regulator of cholesterol homeostasis [277]. Our invitro results present a novel opportunity to investigate microRNA related interactions invivo and their role in cholesterol regulation. We believe validation using invivo model should not only provide novel insights into understanding of cholesterol regulation by miRNAs but should also help us to combat a variety of cholesterol related pathologies.
Biological relevance of miRNA-128 as revealed by bioinformatic analysis
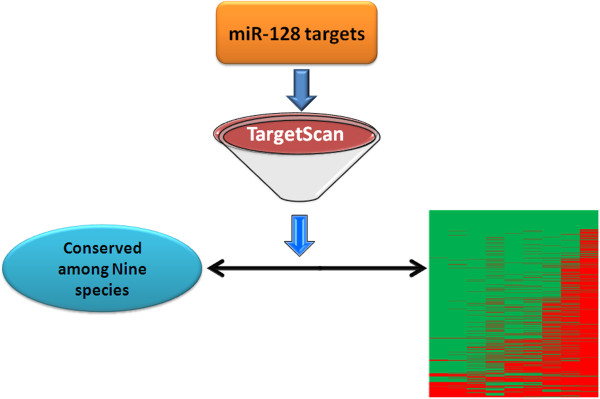
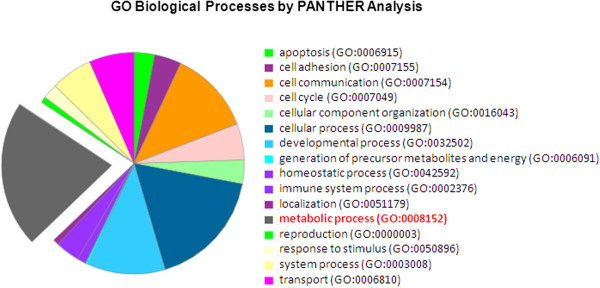
The overall cellular functions and pathways affected by this miRNA remains still undiscovered due to lack of high throughput target validation methods. To reveal biological significance of miRNA-128, a list of predicted targets of miRNA-128 was made using the miRNA target prediction software, TargetScan 5 program (Table 4) [278]. From this list, we discovered that 90 targets of miRNA-128 were conserved among 9 species (Human, Mouse, Chimpanzee, Rhesus, Cow, Chicken, Frog, Rat, Opossum); thereby indicating possible mechanistically conserved functions of this miRNA (Figure 3). To evaluate the specific pathways or processes that are targeted by miRNA-128, we used the list of these ninety conserved targets to find enriched pathways by the PANTHER and GeneCodis [279,280] analysis. Insulin/IGF pathway-mitogen activated protein kinase kinase/MAP kinase cascade, TGF-beta signaling pathway, Angiogenesis, Insulin/IGF pathway-protein kinase B signaling cascade, PI3 kinase pathway, Wnt signaling pathway were found to be the most enriched biological pathways as revealed by the PANTHER analysis (Figure 4). Interestingly, we observed that the Insulin signaling pathway and chemokine signaling pathway were the enriched categories (p-value < 0.05) in both PANTHER and GeneCodis analysis. Till now, only one study by Motohashi et al. describes about the regulation of Insulin signaling pathways by miRNA-128a via the regulation of INSR (insulin receptor), IRS1 (insulin receptor substrate 1) and PIK3R1 (phosphatidylinositol 3-kinases regulatory 1) [281]. There are a few reports which have talked about the involvement of miRNA-128 in TGF-β signaling [282] and PI3 kinase pathway [60]. The reports which have revealed the association between miRNA-128 and angiogenesis, have been described by us in the above section. However, regulation of Wnt signaling and chemokine signaling pathways by miRNA-128 needs further validation. Surprisingly, metabolic process came out to be the highest rated biological process with maximum number of genes during PANTHER Analysis (Figure 5). Our recent work on the regulation of cholesterol metabolism by miRNA-128 point towards a possible link between miRNA-128 and metabolic processes which is just beginning to be revealed and certainly merits further studies. Such discoveries not only provide new insights into mode of action of miRNA-128, but also raise hopes for translating miRNA-128 for therapy.
Table 4.
List of ninety conserved targets of miRNA-128 among nine species
| C1orf144 | FOXP2 | RELN | ARFGEF1 |
|---|---|---|---|
| AFF4 |
CORO1C |
ENAH |
C5orf41 |
| EYA4 |
WSB1 |
hCG_1757335 |
JMJD1C |
| PLK2 |
PLAG1 |
IRS1 |
NDUFS4 |
| ONECUT2 |
NRP2 |
RNF38 |
UPF1 |
| RYBP |
HAPLN1 |
PDE7B |
SPRY2 |
| SOCS6 |
CDH11 |
MAPK14 |
ELL2 |
| UBR5 |
ZHX1 |
UBE2N |
DLL4 |
| LBH |
STK39 |
MED13 |
MLL3 |
| C6orf60 |
PDS5B |
GRIA3 |
MEIS2 |
| SYT1 |
SEMA6A |
RAP1B |
SPOPL |
| BAZ2B |
ZNF827 |
UBE2W |
RAPGEF2 |
| APBA2 |
FLRT3 |
ZNF618 |
MARCKS |
| ISL1 |
KLF4 |
TMEFF1 |
ARID2 |
| UNC13C |
DNAJC13 |
tcag7.1228 |
ZFHX4 |
| FRYL |
SERTAD2 |
AFF3 |
CPEB3 |
| C5orf13 |
INSM1 |
CITED2 |
TMEM189-UBE2V1 |
| WNK1 |
SATB2 |
NARG1 |
UBE2V1 |
| FBXO33 |
HOXA10 |
TSC1 |
CPEB4 |
| TNPO1 |
OTX2 |
MED14 |
EN2 |
| ABL2 |
APPBP2 |
FUBP3 |
PDE3B |
| PPP1CC |
PELI2 |
NIPBL |
MAN2A1 |
| ARID1B | ATP2B1 |
Figure 3.
Strategy for filtering common genes among nine species: Total targets of miRNA-128 have been extracted using TargetScan 5 program for nine species (Human, Mouse, Chimpanzee, Rhesus, Cow, Chicken, Frog, Rat, Opossum). Data was arranged in a tabular format where the union of all genes from the mentioned species were represented as first column in each row (row head). The subsequent columns in first row had species names in them (column head). For every gene, 1 was written under the species where it was found to be present and zero otherwise. This way, a matrix of 1 and zeroes was populated for every gene where 1 means presence and zero means absence. In the last column, sum across the row was taken to count the number of species in which a particular gene was present. We chose only those genes with presence in all nine species. This led to a list of ninety genes which we called high confidence set and were conserved among these species. The total green area specifies ninety common targets whereas red specifies the absence of a particular target in a particular species out of nine species.
Figure 4.
Biological relevance of miRNA-128 as revealed by bioinformatic analysis: The biological pathways affected by miRNA-128 were revealed by the PANTHER and Gene Codis analysis using the list of common ninet y targets as input. Insulin signaling pathway and chemokine signaling pathway were the enriched categories in both PANTHER and GeneCodis analysis (p-value < 0.05).
Figure 5.
GO biological processes by PANTHER analysis: The highest rated biological process being affected by miRNA-128 came out to be metabolic process with maximum number of genes during PANTHER analysis.
Future directions/conclusions
MiRNA-128 is encoded by two distinct genes, viz., R3HDM1 and ARPP21. Interestingly, insilico analysis of transcription factor binding sites of these two genes reveals almost similar pattern of transcription factors (unpublised data). This suggests that these two genes may presumably resulted from a gene duplication event. Until now, neither transcription factors binding proteins of hsa-miRNA-128 gene nor epigenetic factors, have been identified that interact with the regulatory region of this miRNA. However, Monteys et al. have recently suggested dual regulation of miRNA-128-2 by both intronic (pol III) and host gene (Pol II) promoters in acute lymphoblastic leukemia [283]. The fact that miRNA-128 plays multiple roles - a pro-apoptotic molecule, a anti-apoptotic molecule as well as a regulator of cholesterol homeostasis, raise the possibility of exploiting miRNA-128 for therapeutic intervention and development of novel therapies. Further, therapeutic modalities either using replacement strategy by miRNA-128 mimetics (for upregulation of miRNA-128) or using antisense miRNA oligonucleotides (AMOs or antagomirs), LNAs (Locked nucleic Acid) (for downregulation) may now be pursued in an effort to target a particular disease. We believe that there are several fundamental questions that still need to be answered and are open for investigation which will help in the development of miRNA-128 as therapeutics.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YKA and NS conceived the study. The survey of the literature and the inferences were made by YKA and NS. The bioinformatics analysis was carried out by YKA. The manuscript was drafted by YKA and NS. Both authors have read and approved the final manuscript.
Contributor Information
Yogita K Adlakha, Email: yogita@igib.in.
Neeru Saini, Email: nsaini@igib.in.
Acknowledgements
The authors acknowledge the Council of Scientific and Industrial Research (CSIR) funded project ‘Genome dynamics in cellular organization, differentiation and enantiostasis’ (GENCODE-C, BSC-0123). YKA was supported with RA Fellowship from BSC-0123. We also acknowledge Dr. Amit Kumar Yadav for their help.
References
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. doi: 10.1016/S1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. doi: 10.1016/S1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev Cell. 2005;8:321–330. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Karp X, Ambros V. Developmental biology. Encountering microRNAs in cell fate signaling. Science. 2005;310:1288–1289. doi: 10.1126/science.1121566. [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Stadler BM, Ruohola-Baker H. Small RNAs: keeping stem cells in line. Cell. 2008;132:563–566. doi: 10.1016/j.cell.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Sun YH, Shi R, Clark C, Li L, Chiang VL. Novel and mechanical stress-responsive MicroRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell. 2005;17:2186–2203. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynam-Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos Soc. 2009;84:55–71. doi: 10.1111/j.1469-185X.2008.00061.x. [DOI] [PubMed] [Google Scholar]
- Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, Kittler EL, Zapp ML, Klattenhoff C, Schulz N, Theurkauf WE, Weng Z, Zamore PD. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latronico MV, Catalucci D, Condorelli G. Emerging role of microRNAs in cardiovascular biology. Circ Res. 2007;101:1225–1236. doi: 10.1161/CIRCRESAHA.107.163147. [DOI] [PubMed] [Google Scholar]
- Hansen T, Olsen L, Lindow M, Jakobsen KD, Ullum H, Jonsson E, Andreassen OA, Djurovic S, Melle I, Agartz I, Hall H, Timm S, Wang AG, Werge T. Brain expressed microRNAs implicated in schizophrenia etiology. PLoS One. 2007;2:e873. doi: 10.1371/journal.pone.0000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonkoly E, Wei T, Janson PC, Saaf A, Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B, Scheynius A, Ståhle M, Pivarcsi A. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One. 2007;2:e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MD, Mitchell GM. MicroRNAs in insulin resistance and obesity. Exp Diabetes Res. 2012;2012:484696. doi: 10.1155/2012/484696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- Hariharan M, Scaria V, Pillai B, Brahmachari SK. Targets for human encoded microRNAs in HIV genes. Biochem Biophys Res Commun. 2005;337:1214–1218. doi: 10.1016/j.bbrc.2005.09.183. [DOI] [PubMed] [Google Scholar]
- Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13:496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant MicroRNAs. Plant Cell. 2013;25:2383–2399. doi: 10.1105/tpc.113.113159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Sun M, Zou GM, Chen J. MicroRNA and cancer: current status and prospective. Int J Cancer. 2007;120:953–960. doi: 10.1002/ijc.22454. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KJ, Dietrich MR, McPeek MA. MicroRNAs and metazoan macroevolution: insights into canalization, complexity, and the Cambrian explosion. Bioessays. 2009;31:736–747. doi: 10.1002/bies.200900033. [DOI] [PubMed] [Google Scholar]
- O’Carroll D, Schaefer A. General principals of miRNA biogenesis and regulation in the brain. Neuropsychopharmacology. 2013;38:39–54. doi: 10.1038/npp.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motti D, Bixby JL, Lemmon VP. MicroRNAs and neuronal development. Semin Fetal Neonatal Med. 2012;17:347–352. doi: 10.1016/j.siny.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. Annotating noncoding RNA genes. Annu Rev Genomics Hum Genet. 2007;8:279–298. doi: 10.1146/annurev.genom.8.080706.092419. [DOI] [PubMed] [Google Scholar]
- Ul Hussain M. Micro-RNAs (miRNAs): genomic organisation, biogenesis and mode of action. Cell Tissue Res. 2012;349:405–413. doi: 10.1007/s00441-012-1438-0. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, Wolfe SA, Giraldez AJ. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol. 2008;20:214–221. doi: 10.1016/j.ceb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, Lieberman J, Rigoutsos I, Pandolfi PP. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64:303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Nowak JS, Michlewski G. miRNAs in development and pathogenesis of the nervous system. Biochem Soc Trans. 2013;41:815–820. doi: 10.1042/BST20130044. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roese-Koerner B, Stappert L, Koch P, Brustle O, Borghese L. Pluripotent stem cell-derived somatic stem cells as tool to study the role of microRNAs in early human neural development. Curr Mol Med. 2013;13:707–722. doi: 10.2174/1566524011313050003. [DOI] [PubMed] [Google Scholar]
- Jeon HM, Sohn YW, Oh SY, Kim SH, Beck S, Kim S, Kim H. ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9*-mediated suppression of SOX2. Cancer Res. 2011;71:3410–3421. doi: 10.1158/0008-5472.CAN-10-3340. [DOI] [PubMed] [Google Scholar]
- Evangelisti C, Florian MC, Massimi I, Dominici C, Giannini G, Galardi S, Bue MC, Massalini S, McDowell HP, Messi E, Gulino A, Farace MG, Ciafrè SA. MiR-128 up-regulation inhibits Reelin and DCX expression and reduces neuroblastoma cell motility and invasiveness. FASEB J. 2009;23:4276–4287. doi: 10.1096/fj.09-134965. [DOI] [PubMed] [Google Scholar]
- Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY, Song HW, Corbett MA, Gifford WD, Gecz J, Pfaff SL, Wilkinson MF. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell. 2011;42:500–510. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlakha YK, Saini N. MicroRNA-128 downregulates Bax and induces apoptosis in human embryonic kidney cells. Cell Mol Life Sci. 2011;68:1415–1428. doi: 10.1007/s00018-010-0528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chao T, Li R, Liu W, Chen Y, Yan X, Gong Y, Yin B, Qiang B, Zhao J, Yuan J, Peng X. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J Mol Med (Berl) 2009;87:43–51. doi: 10.1007/s00109-008-0403-6. [DOI] [PubMed] [Google Scholar]
- Papagiannakopoulos T, Friedmann-Morvinski D, Neveu P, Dugas JC, Gill RM, Huillard E, Liu C, Zong H, Rowitch DH, Barres BA, Verma IM, Kosik KS. Pro-neural miR-128 is a glioma tumor suppressor that targets mitogenic kinases. Oncogene. 2012;31:1884–1895. doi: 10.1038/onc.2011.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca EA, Lawler S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- Eletto D, Russo G, Passiatore G, Del Valle L, Giordano A, Khalili K, Gualco E, Peruzzi F. Inhibition of SNAP25 expression by HIV-1 Tat involves the activity of mir-128a. J Cell Physiol. 2008;216:764–770. doi: 10.1002/jcp.21452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo T, Faucz FR, Azevedo M, Xekouki P, Iliopoulos D, Stratakis CA. Functional screen analysis reveals miR-26b and miR-128 as central regulators of pituitary somatomammotrophic tumor growth through activation of the PTEN-AKT pathway. Oncogene. 2013;32:1651–1659. doi: 10.1038/onc.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi M, Muinos-Gimeno M, Kagerbauer B, Marti E, Estivill X, Espinosa-Parrilla Y. Overexpression of miR-128 specifically inhibits the truncated isoform of NTRK3 and upregulates BCL2 in SH-SY5Y neuroblastoma cells. BMC Mol Biol. 2010;11:95. doi: 10.1186/1471-2199-11-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peruzzi P, Bronisz A, Nowicki MO, Wang Y, Ogawa D, Price R, Nakano I, Kwon CH, Hayes J, Lawler SE, Ostrowski MC, Chiocca EA, Godlewski J. MicroRNA-128 coordinately targets polycomb repressor complexes in glioma stem cells. Neuro Oncol. 2013;15:1212–1224. doi: 10.1093/neuonc/not055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschou M, Doxakis E. Neurofibromin 1 is a miRNA target in neurons. PLoS One. 2012;7:e46773. doi: 10.1371/journal.pone.0046773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuchty S, Arjona D, Li A, Kotliarov Y, Walling J, Ahn S, Zhang A, Maric D, Anolik R, Zenklusen JC, Fine HA. Prediction of Associations between microRNAs and Gene Expression in Glioma Biology. PLoS One. 2011;6:e14681. doi: 10.1371/journal.pone.0014681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury NR, de Lima AF, de Andres-Aguayo L, Graf T, Caceres JF, Rappsilber J, Michlewski G. Tissue-specific control of brain-enriched miR-7 biogenesis. Genes Dev. 2013;27:24–38. doi: 10.1101/gad.199190.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda H, Xing F, Pandey PR, Sharma S, Watabe M, Pai SK, Mo YY, Iiizumi-Gairani M, Hirota S, Liu Y, Wu K, Pochampally R, Watabe K. miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. 2013;73:1434–1444. doi: 10.1158/0008-5472.CAN-12-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A. 2009;106:13052–13057. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewing AS, Rueli RH, Robles MJ, Nguyen-Wu ED, Zeyda T, Berry MJ, Bellinger FP. Expression and regulation of mouse selenoprotein P transcript variants differing in non-coding RNA. RNA Biol. 2012;9:1361–1369. doi: 10.4161/rna.22290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Shalom-Feuerstein R, Riley J, Zhang SD, Tucci P, Agostini M, Aberdam D, Knight RA, Genchi G, Nicotera P, Melino G, Vasa-Nicotera M. miR-7 and miR-214 are specifically expressed during neuroblastoma differentiation, cortical development and embryonic stem cells differentiation, and control neurite outgrowth in vitro. Biochem Biophys Res Commun. 2010;394:921–927. doi: 10.1016/j.bbrc.2010.03.076. [DOI] [PubMed] [Google Scholar]
- Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S, Purow B. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68:3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Mao W, Zheng S, Ye J. Epidermal growth factor receptor-regulated miR-125a-5p–a metastatic inhibitor of lung cancer. FEBS J. 2009;276:5571–5578. doi: 10.1111/j.1742-4658.2009.07238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissart C, Nissan X, Giraud-Triboult K, Peschanski M, Benchoua A. miR-125 potentiates early neural specification of human embryonic stem cells. Development. 2012;139:1247–1257. doi: 10.1242/dev.073627. [DOI] [PubMed] [Google Scholar]
- Lin ST, Fu YH. miR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. Dis Model Mech. 2009;2:178–188. doi: 10.1242/dmm.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel C, Li J, Liu F, Benashski SE, McCullough LD. miR-23a regulation of X-linked inhibitor of apoptosis (XIAP) contributes to sex differences in the response to cerebral ischemia. Proc Natl Acad Sci U S A. 2011;108:11662–11667. doi: 10.1073/pnas.1102635108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanet A, Tacheny A, Arnould T, Renard P. miR-212/132 expression and functions: within and beyond the neuronal compartment. Nucleic Acids Res. 2012;40:4742–4753. doi: 10.1093/nar/gks151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- Smith PY, Delay C, Girard J, Papon MA, Planel E, Sergeant N, Buee L, Hebert SS. MicroRNA-132 loss is associated with tau exon 10 inclusion in progressive supranuclear palsy. Hum Mol Genet. 2011;20:4016–4024. doi: 10.1093/hmg/ddr330. [DOI] [PubMed] [Google Scholar]
- Shaked I, Meerson A, Wolf Y, Avni R, Greenberg D, Gilboa-Geffen A, Soreq H. MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity. 2009;31:965–973. doi: 10.1016/j.immuni.2009.09.019. [DOI] [PubMed] [Google Scholar]
- Miller BH, Zeier Z, Xi L, Lanz TA, Deng S, Strathmann J, Willoughby D, Kenny PJ, Elsworth JD, Lawrence MS, Roth RH, Edbauer D, Kleiman RJ, Wahlestedt C. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc Natl Acad Sci U S A. 2012;109:3125–3130. doi: 10.1073/pnas.1113793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognini P, Pizzorusso T. MicroRNA212/132 family: molecular transducer of neuronal function and plasticity. Int J Biochem Cell Biol. 2012;44:6–10. doi: 10.1016/j.biocel.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clovis YM, Enard W, Marinaro F, Huttner WB, De Pietri Tonelli D. Convergent repression of Foxp2 3′UTR by miR-9 and miR-132 in embryonic mouse neocortex: implications for radial migration of neurons. Development. 2012;139:3332–3342. doi: 10.1242/dev.078063. [DOI] [PubMed] [Google Scholar]
- Lages E, Guttin A, El Atifi M, Ramus C, Ipas H, Dupre I, Rolland D, Salon C, Godfraind C, de Fraipont F. MicroRNA and target protein patterns reveal physiopathological features of glioma subtypes. PLoS One. 2011;6:e20600. doi: 10.1371/journal.pone.0020600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott HL, Tamagnini F, Narduzzo KE, Howarth JL, Lee YB, Wong LF, Brown MW, Warburton EC, Bashir ZI, Uney JB. MicroRNA-132 regulates recognition memory and synaptic plasticity in the perirhinal cortex. Eur J Neurosci. 2012;36:2941–2948. doi: 10.1111/j.1460-9568.2012.08220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strum JC, Johnson JH, Ward J, Xie H, Feild J, Hester A, Alford A, Waters KM. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol Endocrinol. 2009;23:1876–1884. doi: 10.1210/me.2009-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Lei Y, Zhang H, Hou L, Zhang M, Dayton AI. MicroRNA regulation of STAT4 protein expression: rapid and sensitive modulation of IL-12 signaling in human natural killer cells. Blood. 2011;118:6793–6802. doi: 10.1182/blood-2011-05-356162. [DOI] [PubMed] [Google Scholar]
- Godoy J, Nishimura M, Webster NJ. Gonadotropin-releasing hormone induces miR-132 and miR-212 to regulate cellular morphology and migration in immortalized LbetaT2 pituitary gonadotrope cells. Mol Endocrinol. 2011;25:810–820. doi: 10.1210/me.2010-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Saavedra M, Antoun G, Yanagiya A, Oliva-Hernandez R, Cornejo-Palma D, Perez-Iratxeta C, Sonenberg N, Cheng HY. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum Mol Genet. 2011;20:731–751. doi: 10.1093/hmg/ddq519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C, Turner JA, Calhoun VD, Perrone-Bizzozero N. Potential Impact of miR-137 and Its Targets in Schizophrenia. Front Genet. 2013;4:58. doi: 10.3389/fgene.2013.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello JF, Bergers G, Weiss WA, Alvarez-Buylla A, Hodgson JG. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrt RD, Szulwach KE, Pfeiffer RL, Li X, Guo W, Pathania M, Teng ZQ, Luo Y, Peng J, Bordey A, Jin P, Zhao X. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28:1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon E, Wang W, Tsai LH. Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets. Mol Psychiatry. 2013;18:11–12. doi: 10.1038/mp.2011.170. [DOI] [PubMed] [Google Scholar]
- Li KK, Yang L, Pang JC, Chan AK, Zhou L, Mao Y, Wang Y, Lau KM, Poon WS, Shi Z, Ng HK. MIR-137 suppresses growth and invasion, is downregulated in oligodendroglial tumors and targets CSE1L. Brain Pathol. 2013;23:426–439. doi: 10.1111/bpa.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang X, Wang H, Li Y, Yan W, Han L, Zhang K, Zhang J, Wang Y, Feng Y, Pu P, Jiang T, Kang C, Jiang C. miR-137 is frequently down-regulated in glioblastoma and is a negative regulator of Cox-2. Eur J Cancer. 2012;48:3104–3111. doi: 10.1016/j.ejca.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Sun G, Ye P, Murai K, Lang MF, Li S, Zhang H, Li W, Fu C, Yin J, Wang A, Ma X, Shi Y. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat Commun. 2011;2:529. doi: 10.1038/ncomms1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen MH, Valles A, Kirkels LA, Mastebroek M, Olde Loohuis N, Kos A, Wissink-Lindhout WM, de Brouwer AP, Nillesen WM, Pfundt R, Holder-Espinasse M, Vallée L, Andrieux J, Coppens-Hofman MC, Rensen H, Hamel BC, van Bokhoven H, Aschrafi A, Kleefstra T. Chromosome 1p21.3 microdeletions comprising DPYD and MIR137 are associated with intellectual disability. J Med Genet. 2011;48:810–818. doi: 10.1136/jmedgenet-2011-100294. [DOI] [PubMed] [Google Scholar]
- Geekiyanage H, Chan C. MicroRNA-137/181c regulates serine palmitoyltransferase and in turn amyloid beta, novel targets in sporadic Alzheimer’s disease. J Neurosci. 2011;31:14820–14830. doi: 10.1523/JNEUROSCI.3883-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li RY, Chen LC, Zhang HY, Du WZ, Feng Y, Wang HB, Wen JQ, Liu X, Li XF, Sun Y, Yang DB, Jiang T, Li YL, Jiang CL. MiR-139 inhibits Mcl-1 expression and potentiates TMZ-induced apoptosis in glioma. CNS Neurosci Ther. 2013;19:477–483. doi: 10.1111/cns.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Fu HJ, Xie QS, Wang L, Zhang R, Guo ZY, Zhao J, Meng YL, Ren XL, Wang T, Li Q, Jin BQ, Yao LB, Wang RA, Fan DM, Chen SY, Jia LT, Yang AG. HER2 interacts with CD44 to up-regulate CXCR4 via epigenetic silencing of microRNA-139 in gastric cancer cells. Gastroenterology. 2011;141:2076–2087. doi: 10.1053/j.gastro.2011.08.050. [DOI] [PubMed] [Google Scholar]
- Hasseine LK, Hinault C, Lebrun P, Gautier N, Paul-Bellon R, Van Obberghen E. miR-139 impacts FoxO1 action by decreasing FoxO1 protein in mouse hepatocytes. Biochem Biophys Res Commun. 2009;390:1278–1282. doi: 10.1016/j.bbrc.2009.10.135. [DOI] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Wang S, Yang B, Zhu L, Yin B, Chao T, Zhao J, Yuan J, Qiang B, Peng X. The CREB-miR-9 negative feedback minicircuitry coordinates the migration and proliferation of glioma cells. PLoS One. 2012;7:e49570. doi: 10.1371/journal.pone.0049570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatro ET, Hefler S, Shumaker-Armstrong S, Soontornniyomkij B, Yang M, Yermanos A, Wren N, Moore DJ, Achim CL. Modulation of BK Channel by MicroRNA-9 in Neurons After Exposure to HIV and Methamphetamine. J Neuroimmune Pharmacol. 2013;8(5):1210–1223. doi: 10.1007/s11481-013-9446-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SL, Ohtsuka T, Gonzalez A, Kageyama R. MicroRNA9 regulates neural stem cell differentiation by controlling Hes1 expression dynamics in the developing brain. Genes Cells. 2012;17:952–961. doi: 10.1111/gtc.12009. [DOI] [PubMed] [Google Scholar]
- Jung HJ, Coffinier C, Choe Y, Beigneux AP, Davies BS, Yang SH, Barnes RH 2nd, Hong J, Sun T, Pleasure SJ, Young SG, Fong LG. Regulation of prelamin A but not lamin C by miR-9, a brain-specific microRNA. Proc Natl Acad Sci U S A. 2012;109:E423–E431. doi: 10.1073/pnas.1111780109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Z, Yang L. MiR-124 inhibits the growth of glioblastoma through the downregulation of SOS1. Mol Med Rep. 2013;8:345–349. doi: 10.3892/mmr.2013.1561. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, Cheung WK, Ng SS, Jiang X, Jiang S, Sze J, Leung GK, Lu G, Chan DT, Bian XW, Kung HF, Poon WS, Lin MC. Loss of brain-enriched miR-124 microRNA enhances stem-like traits and invasiveness of glioma cells. J Biol Chem. 2012;287:9962–9971. doi: 10.1074/jbc.M111.332627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sober S, Laan M, Annilo T. MicroRNAs miR-124 and miR-135a are potential regulators of the mineralocorticoid receptor gene (NR3C2) expression. Biochem Biophys Res Commun. 2010;391:727–732. doi: 10.1016/j.bbrc.2009.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Ma L, Wu M, Zhang G, Jin C, Guo Y, Liu R. miR-124 radiosensitizes human glioma cells by targeting CDK4. J Neurooncol. 2013;114:263–274. doi: 10.1007/s11060-013-1179-2. [DOI] [PubMed] [Google Scholar]
- Doeppner TR, Doehring M, Bretschneider E, Zechariah A, Kaltwasser B, Muller B, Koch JC, Bahr M, Hermann DM, Michel U. MicroRNA-124 protects against focal cerebral ischemia via mechanisms involving Usp14-dependent REST degradation. Acta Neuropathol. 2013;126:251–265. doi: 10.1007/s00401-013-1142-5. [DOI] [PubMed] [Google Scholar]
- Liu X, Li F, Zhao S, Luo Y, Kang J, Zhao H, Yan F, Li S, Ji X. MicroRNA-124-mediated regulation of inhibitory member of apoptosis-stimulating protein of p53 family in experimental stroke. Stroke. 2013;44:1973–1980. doi: 10.1161/STROKEAHA.111.000613. [DOI] [PubMed] [Google Scholar]
- Dutta R, Chomyk AM, Chang A, Ribaudo MV, Deckard SA, Doud MK, Edberg DD, Bai B, Li M, Baranzini SE, Fox RJ, Staugaitis SM, Macklin WB, Trapp BD. Hippocampal demyelination and memory dysfunction are associated with increased levels of the neuronal microRNA miR-124 and reduced AMPA receptors. Ann Neurol. 2013;73:637–645. doi: 10.1002/ana.23860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvanitis DN, Jungas T, Behar A, Davy A. Ephrin-B1 reverse signaling controls a posttranscriptional feedback mechanism via miR-124. Mol Cell Biol. 2010;30:2508–2517. doi: 10.1128/MCB.01620-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev ED, Veremeyko T, Barteneva NS. Visualization and quantitation of the expression of microRNAs and their target genes in neuroblastoma single cells using imaging cytome. BMC Res Notes. 2011;4:517. doi: 10.1186/1756-0500-4-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber J, Hashizume R, Felix T, Hariono S, Yu M, Berger MS, Huse JT, VandenBerg SR, James CD, Hodgson JG, Gupta N. Expression of miR-124 inhibits growth of medulloblastoma cells. Neuro Oncol. 2013;15:83–90. doi: 10.1093/neuonc/nos281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson SK, Dobrikova EY, Shveygert M, Gromeier M. p38alpha mitogen-activated protein kinase depletion and repression of signal transduction to translation machinery by miR-124 and -128 in neurons. Mol Cell Biol. 2013;33:127–135. doi: 10.1128/MCB.00695-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat Med. 2011;17:64–70. doi: 10.1038/nm.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke K, Otto W, Johannes S, Baumgart J, Nitsch R, Schumacher S. miR-124-regulated RhoG reduces neuronal process complexity via ELMO/Dock180/Rac1 and Cdc42 signalling. EMBO J. 2012;31:2908–2921. doi: 10.1038/emboj.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng R, Cohen SM. Drosophila miR-124 regulates neuroblast proliferation through its target anachronism. Development. 2012;139:1427–1434. doi: 10.1242/dev.075143. [DOI] [PubMed] [Google Scholar]
- Sanuki R, Onishi A, Koike C, Muramatsu R, Watanabe S, Muranishi Y, Irie S, Uneo S, Koyasu T, Matsui R, Chérasse Y, Urade Y, Watanabe D, Kondo M, Yamashita T, Furukawa T. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat Neurosci. 2011;14:1125–1134. doi: 10.1038/nn.2897. [DOI] [PubMed] [Google Scholar]
- Fang M, Wang J, Zhang X, Geng Y, Hu Z, Rudd JA, Ling S, Chen W, Han S. The miR-124 regulates the expression of BACE1/beta-secretase correlated with cell death in Alzheimer’s disease. Toxicol Lett. 2012;209:94–105. doi: 10.1016/j.toxlet.2011.11.032. [DOI] [PubMed] [Google Scholar]
- Liu K, Liu Y, Mo W, Qiu R, Wang X, Wu JY, He R. MiR-124 regulates early neurogenesis in the optic vesicle and forebrain, targeting NeuroD1. Nucleic Acids Res. 2011;39:2869–2879. doi: 10.1093/nar/gkq904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu CS, Yang Y, Cheng CD. MiR-134 regulates the proliferation and invasion of glioblastoma cells by reducing Nanog expression. Int J Oncol. 2013;42:1533–1540. doi: 10.3892/ijo.2013.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Bicker S, Khudayberdiev S, Weiss K, Zocher K, Baumeister S, Schratt G. The DEAH-box helicase DHX36 mediates dendritic localization of the neuronal precursor-microRNA-134. Genes Dev. 2013;27:991–996. doi: 10.1101/gad.211243.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay YM, Tam WL, Ang YS, Gaughwin PM, Yang H, Wang W, Liu R, George J, Ng HH, Perera RJ, Lufkin T, Rigoutsos I, Thomson AM, Lim B. MicroRNA-134 modulates the differentiation of mouse embryonic stem cells, where it causes post-transcriptional attenuation of Nanog and LRH1. Stem Cells. 2008;26:17–29. doi: 10.1634/stemcells.2007-0295. [DOI] [PubMed] [Google Scholar]
- Li J, Wang Y, Luo J, Fu Z, Ying J, Yu Y, Yu W. miR-134 inhibits epithelial to mesenchymal transition by targeting FOXM1 in non-small cell lung cancer cells. FEBS Lett. 2012;586:3761–3765. doi: 10.1016/j.febslet.2012.09.016. [DOI] [PubMed] [Google Scholar]
- Ni J, Gao Y, Gong S, Guo S, Hisamitsu T, Jiang X. Regulation of mu-opioid type 1 receptors by microRNA134 in dorsal root ganglion neurons following peripheral inflammation. Eur J Pain. 2013;17:313–323. doi: 10.1002/j.1532-2149.2012.00197.x. [DOI] [PubMed] [Google Scholar]
- Hirota T, Date Y, Nishibatake Y, Takane H, Fukuoka Y, Taniguchi Y, Burioka N, Shimizu E, Nakamura H, Otsubo K, Ieiri I. Dihydropyrimidine dehydrogenase (DPD) expression is negatively regulated by certain microRNAs in human lung tissues. Lung Cancer. 2012;77:16–23. doi: 10.1016/j.lungcan.2011.12.018. [DOI] [PubMed] [Google Scholar]
- Han L, Wen Z, Lynn RC, Baudet ML, Holt CE, Sasaki Y, Bassell GJ, Zheng JQ. Regulation of chemotropic guidance of nerve growth cones by microRNA. Mol Brain. 2011;4:40. doi: 10.1186/1756-6606-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayanithy V, Sarver AL, Kartha RV, Li L, Angstadt AY, Breen M, Steer CJ, Modiano JF, Subramanian S. Perturbation of 14q32 miRNAs-cMYC gene network in osteosarcoma. Bone. 2012;50:171–181. doi: 10.1016/j.bone.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, Kim TK, Greenberg ME, Schratt G. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaughwin P, Ciesla M, Yang H, Lim B, Brundin P. Stage-specific modulation of cortical neuronal development by Mmu-miR-134. Cereb Cortex. 2011;21:1857–1869. doi: 10.1093/cercor/bhq262. [DOI] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerson A, Cacheaux L, Goosens KA, Sapolsky RM, Soreq H, Kaufer D. Changes in brain MicroRNAs contribute to cholinergic stress reactions. J Mol Neurosci. 2010;40:47–55. doi: 10.1007/s12031-009-9252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovskaya VM, Sumbler B, Ho B, Yemma M, Cance WG. miR-138 and MiR-135 directly target focal adhesion kinase, inhibit cell invasion, and increase sensitivity to chemotherapy in cancer cells. Anticancer Agents Med Chem. 2014;14(1):18–28. doi: 10.2174/187152061401140108113435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Qian JC, Deng ZL, Cai Z, Tang T, Wang P, Zhang KH, Cai JP. Expression of miR-21, miR-31, miR-96 and miR-135b is correlated with the clinical parameters of colorectal cancer. Oncol Lett. 2012;4:339–345. doi: 10.3892/ol.2012.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hassan MQ, Volinia S, van Wijnen AJ, Stein JL, Croce CM, Lian JB, Stein GS. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci U S A. 2008;105:13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, Meijer GA, Agami R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795–5802. doi: 10.1158/0008-5472.CAN-08-0951. [DOI] [PubMed] [Google Scholar]
- Zhao S, Deng Y, Liu Y, Chen X, Yang G, Mu Y, Zhang D, Kang J, Wu Z. MicroRNA-153 is tumor suppressive in glioblastoma stem cells. Mol Biol Rep. 2013;40:2789–2798. doi: 10.1007/s11033-012-2278-4. [DOI] [PubMed] [Google Scholar]
- Stappert L, Borghese L, Roese-Koerner B, Weinhold S, Koch P, Terstegge S, Uhrberg M, Wernet P, Brustle O. MicroRNA-based promotion of human neuronal differentiation and subtype specification. PLoS One. 2013;8:e59011. doi: 10.1371/journal.pone.0059011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandemakers W, Abuhatzira L, Xu H, Caromile LA, Hebert SS, Snellinx A, Morais VA, Matta S, Cai T, Notkins AL, De Strooper B. Co-regulation of intragenic microRNA miR-153 and its host gene Ia-2 beta: identification of miR-153 target genes with functions related to IA-2beta in pancreas and brain. Diabetologia. 2013;56:1547–1556. doi: 10.1007/s00125-013-2901-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JM, Ray B, Lahiri DK. MicroRNA-153 physiologically inhibits expression of amyloid-beta precursor protein in cultured human fetal brain cells and is dysregulated in a subset of Alzheimer disease patients. J Biol Chem. 2012;287:31298–31310. doi: 10.1074/jbc.M112.366336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Zhu H, Xu Y, Huang L, Ma C, Deng W, Liu Y, Qin C. MicroRNA-153 negatively regulates the expression of amyloid precursor protein and amyloid precursor-like protein 2. Brain Res. 2012;1455:103–113. doi: 10.1016/j.brainres.2011.10.051. [DOI] [PubMed] [Google Scholar]
- Doxakis E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J Biol Chem. 2010;285:12726–12734. doi: 10.1074/jbc.M109.086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Liao X, Wong C. Downregulations of B-cell lymphoma 2 and myeloid cell leukemia sequence 1 by microRNA 153 induce apoptosis in a glioblastoma cell line DBTRG-05MG. Int J Cancer. 2010;126:1029–1035. doi: 10.1002/ijc.24823. [DOI] [PubMed] [Google Scholar]
- Xu Q, Sun Q, Zhang J, Yu J, Chen W, Zhang Z. Downregulation of miR-153 contributes to epithelial-mesenchymal transition and tumor metastasis in human epithelial cancer. Carcinogenesis. 2013;34:539–549. doi: 10.1093/carcin/bgs374. [DOI] [PubMed] [Google Scholar]
- Rao SA, Arimappamagan A, Pandey P, Santosh V, Hegde AS, Chandramouli BA, Somasundaram K. miR-219-5p inhibits receptor tyrosine kinase pathway by targeting EGFR in glioblastoma. PLoS One. 2013;8:e63164. doi: 10.1371/journal.pone.0063164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarachana T, Zhou R, Chen G, Manji HK, Hu VW. Investigation of post-transcriptional gene regulatory networks associated with autism spectrum disorders by microRNA expression profiling of lymphoblastoid cell lines. Genome Med. 2010;2:23. doi: 10.1186/gm144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, Hoang T, Xu X, Mi QS, Xin M, Wang F, Appel B, Lu QR. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65:612–626. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Cuellar TL, Scholze A, Ason B, Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT, Barres BA. Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65:597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Shin JY, McManus MT, Ptacek LJ, Fu YH. Dicer ablation in oligodendrocytes provokes neuronal impairment in mice. Ann Neurol. 2009;66:843–857. doi: 10.1002/ana.21927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocerha J, Faghihi MA, Lopez-Toledano MA, Huang J, Ramsey AJ, Caron MG, Sales N, Willoughby D, Elmen J, Hansen HF, Orum H, Kauppinen S, Kenny PJ, Wahlestedt C. MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc Natl Acad Sci U S A. 2009;106:3507–3512. doi: 10.1073/pnas.0805854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- De Pietri TD, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Liu Y, Huang M, Zhao X, Cheng L. Wnt1-cre-mediated conditional loss of Dicer results in malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation in mice. J Mol Cell Biol. 2010;2:152–163. doi: 10.1093/jmcb/mjq008. [DOI] [PubMed] [Google Scholar]
- Schaefer A, O’Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, Greengard P. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204:1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase-Koga Y, Low R, Otaegi G, Pollock A, Deng H, Eisenhaber F, Maurer-Stroh S, Sun T. RNAase-III enzyme Dicer maintains signaling pathways for differentiation and survival in mouse cortical neural stem cells. J Cell Sci. 2010;123:586–594. doi: 10.1242/jcs.059659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicker S, Schratt G. microRNAs: tiny regulators of synapse function in development and disease. J Cell Mol Med. 2008;12:1466–1476. doi: 10.1111/j.1582-4934.2008.00400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore R, Siegel G, Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochim Biophys Acta. 2008;1779:471–478. doi: 10.1016/j.bbagrm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, Wan X, Pavlidis P, Mills AA, Karayiorgou M, Gogos JA. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- Godlewski J, Newton HB, Chiocca EA, Lawler SE. MicroRNAs and glioblastoma; the stem cell connection. Cell Death Differ. 2010;17:221–228. doi: 10.1038/cdd.2009.71. [DOI] [PubMed] [Google Scholar]
- Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK, Richardson JC, Saunders AM, Roses AD, Richards CA. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- Zeng Y. Regulation of the mammalian nervous system by microRNAs. Mol Pharmacol. 2009;75:259–264. doi: 10.1124/mol.108.052118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnerty JR, Wang WX, Hebert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402:491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Kole AJ, Swahari V, Hammond SM, Deshmukh M. miR-29b is activated during neuronal maturation and targets BH3-only genes to restrict apoptosis. Genes Dev. 2011;25:125–130. doi: 10.1101/gad.1975411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. 2005;64:479–489. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- Koshkin PA, Chistiakov DA, Nikitin AG, Konovalov AN, Potapov AA, Usachev DY, Pitskhelauri DI, Kobyakov GL, Shishkina LV, Chekhonin VP. Analysis of expression of microRNAs and genes involved in the control of key signaling mechanisms that support or inhibit development of brain tumors of different grades. Clin Chim Acta. 2014;9(430C):55–62. doi: 10.1016/j.cca.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Lai NS, Dong QS, Ding H, Miao ZL, Lin YC. MicroRNA-210 overexpression predicts poorer prognosis in glioma patients. J Clin Neurosci. 2013. Sep 11, doi:10.1016/j.jocn.2013.06.024. [DOI] [PubMed]
- Wang S, Lu S, Geng S, Ma S, Liang Z, Jiao B. Decreased expression of microRNA-206 correlates with poor clinical outcome in patients with malignant astrocytomas. Pathol Oncol Res. 2014. doi: 10.1007/s12253-013-9701-6. [DOI] [PubMed]
- Yang TQ, Luo XJ, Wu TF, Ding DD, Zhao ZH, Chen GL, Xie XS, Li B, Wei YX, Guo LC, Zhang Y, Huang YL, Zhou YX, Du ZW. miR-16 inhibits glioma cell growth and invasion through the suppression of BCL2 and NF-kappaB1/MMP-9 signaling pathway. Cancer Sci. 2014. doi:10.1007/s12253-013-9701-6. [DOI] [PMC free article] [PubMed] [Retracted]
- Zhang Y, Chen X, Lian H, Liu J, Zhou B, Han S, Peng B, Yin J, Liu W, He X. MicroRNA-503 acts as a tumor suppressor in glioblastoma for multiple antitumor effects by targeting IGF-1R. Oncol Rep. 2013. [DOI] [PMC free article] [PubMed]
- Chen Z, Li D, Cheng Q, Ma Z, Jiang B, Peng R, Chen R, Cao Y, Wan X. MicroRNA-203 inhibits the proliferation and invasion of U251 glioblastoma cells by directly targeting PLD2. Mol Med Rep. 2014;9:503–508. doi: 10.3892/mmr.2013.1814. [DOI] [PubMed] [Google Scholar]
- Wu Z, Wu Y, Tian Y, Sun X, Liu J, Ren H, Liang C, Song L, Hu H, Wang L, Jiao B. Differential effects of miR-34c-3p and miR-34c-5p on the proliferation, apoptosis and invasion of glioma cells. Oncol Lett. 2013;6:1447–1452. doi: 10.3892/ol.2013.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai DW, Lu Q, Wang LX, Zhao WY, Cao YQ, Li YN, Han GS, Liu JM, Yue ZJ. Decreased miR-106a inhibits glioma cell glucose uptake and proliferation by targeting SLC2A3 in GBM. BMC Cancer. 2013;13:478. doi: 10.1186/1471-2407-13-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XR, Luo H, Li HL, Cao L, Wang XF, Yan W, Wang YY, Zhang JX, Jiang T, Kang CS, Liu N, You YP. Chinese Glioma Cooperative Group (CGCG) Overexpressed let-7a inhibits glioma cell malignancy by directly targeting K-ras, independently of PTEN. Neuro Oncol. 2013;15:1491–1501. doi: 10.1093/neuonc/not107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Huang Q, Chen K, Liu L, Lin C, Dai T, Yu C, Wu Z, Li J. miR-218 inhibits the invasive ability of glioma cells by direct downregulation of IKK-beta. Biochem Biophys Res Commun. 2010;402:135–140. doi: 10.1016/j.bbrc.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Glasgow SM, Laug D, Brawley VS, Zhang Z, Corder A, Yin Z, Wong ST, Li XN, Foster AE, Ahmed N, Deneen B. The miR-223/nuclear factor I-A axis regulates glial precursor proliferation and tumorigenesis in the CNS. J Neurosci. 2013;33:13560–13568. doi: 10.1523/JNEUROSCI.0321-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SZ, Hu YY, Zhao J, Zhao YB, Sun JD, Yang YF, Ji CC, Liu ZB, Cao WD, Qu Y, Liu WP, Cheng G, Fei Z. MicroRNA-34a induces apoptosis in the human glioma cell line, A172, through enhanced ROS production and NOX2 expression. Biochem Biophys Res Commun. 2014. [DOI] [PubMed]
- Rani SB, Rathod SS, Karthik S, Kaur N, Muzumdar D, Shiras AS. MiR-145 functions as a tumor-suppressive RNA by targeting Sox9 and adducin 3 in human glioma cells. Neuro Oncol. 2013;15:1302–1316. doi: 10.1093/neuonc/not090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Lin X, Tan X, Yin B, Han W, Zhao J, Yuan J, Qiang B, Peng X. Circadian gene Clock contributes to cell proliferation and migration of glioma and is directly regulated by tumor-suppressive miR-124. FEBS Lett. 2013;587:2455–2460. doi: 10.1016/j.febslet.2013.06.018. [DOI] [PubMed] [Google Scholar]
- Bier A, Giladi N, Kronfeld N, Lee HK, Cazacu S, Finniss S, Xiang C, Poisson L, de Carvalho AC, Slavin S, Jacoby E, Yalon M, Toren A, Mikkelsen T, Brodie C. MicroRNA-137 is downregulated in glioblastoma and inhibits the stemness of glioma stem cells by targeting RTVP-1. Oncotarget. 2013;4:665–676. doi: 10.18632/oncotarget.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S, Huang D, Yin D, Li F, Li X, Kung HF, Peng Y. Suppression of tumorigenicity by microRNA-138 through inhibition of EZH2-CDK4/6-pRb-E2F1 signal loop in glioblastoma multiforme. Biochim Biophys Acta. 1832;2013:1697–1707. doi: 10.1016/j.bbadis.2013.05.015. [DOI] [PubMed] [Google Scholar]
- Chen SM, Chen HC, Chen SJ, Huang CY, Chen PY, Wu TW, Feng LY, Tsai HC, Lui TN, Hsueh C, Wei KC. MicroRNA-495 inhibits proliferation of glioblastoma multiforme cells by downregulating cyclin-dependent kinase 6. World J Surg Oncol. 2013;11:87. doi: 10.1186/1477-7819-11-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Cen D, Luo X, Li D, Li P, Liang L, Meng Z. Downregulation of miR-383 promotes glioma cell invasion by targeting insulin-like growth factor 1 receptor. Med Oncol. 2013;30:557. doi: 10.1007/s12032-013-0557-0. [DOI] [PubMed] [Google Scholar]
- Peng B, Hu S, Jun Q, Luo D, Zhang X, Zhao H, Li D. MicroRNA-200b targets CREB1 and suppresses cell growth in human malignant glioma. Mol Cell Biochem. 2013;379:51–58. doi: 10.1007/s11010-013-1626-6. [DOI] [PubMed] [Google Scholar]
- Hui W, Yuntao L, Lun L, WenSheng L, ChaoFeng L, HaiYong H, Yueyang B. MicroRNA-195 inhibits the proliferation of human glioma cells by directly targeting cyclin D1 and cyclin E1. PLoS One. 2013;8:e54932. doi: 10.1371/journal.pone.0054932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Liu H, Liu Y, Wu J, Wang C, Hou X, Chen X, Yang G, Zhao L, Che H, Bi Y, Wang H, Peng F, Ai J. miR-143 inhibits glycolysis and depletes stemness of glioblastoma stem-like cells. Cancer Lett. 2013;333:253–260. doi: 10.1016/j.canlet.2013.01.039. [DOI] [PubMed] [Google Scholar]
- Chen L, Chen XR, Zhang R, Li P, Liu Y, Yan K, Jiang XD. MicroRNA-107 inhibits glioma cell migration and invasion by modulating Notch2 expression. J Neurooncol. 2013;112:59–66. doi: 10.1007/s11060-012-1037-7. [DOI] [PubMed] [Google Scholar]
- Wang S, Lu S, Geng S, Ma S, Liang Z, Jiao B. Expression and clinical significance of microRNA-326 in human glioma miR-326 expression in glioma. Med Oncol. 2013;30:373. doi: 10.1007/s12032-012-0373-y. [DOI] [PubMed] [Google Scholar]
- Ying Z, Li Y, Wu J, Zhu X, Yang Y, Tian H, Li W, Hu B, Cheng SY, Li M. Loss of miR-204 expression enhances glioma migration and stem cell-like phenotype. Cancer Res. 2013;73:990–999. doi: 10.1158/0008-5472.CAN-12-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Tan X, Zhao A, Zhu L, Yin B, Yuan J, Qiang B, Peng X. microRNA-214-mediated UBC9 expression in glioma. BMB Rep. 2012;45:641–646. doi: 10.5483/BMBRep.2012.45.11.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Lan J, Ge J, Nie Q, Mao Q, Qiu Y. miR-708 acts as a tumor suppressor in human glioblastoma cells. Oncol Rep. 2013;30:870–876. doi: 10.3892/or.2013.2526. [DOI] [PubMed] [Google Scholar]
- Fox JL, Dews M, Minn AJ, Thomas-Tikhonenko A. Targeting of TGFbeta signature and its essential component CTGF by miR-18 correlates with improved survival in glioblastoma. RNA. 2013;19:177–190. doi: 10.1261/rna.036467.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel T, Karginov FV, Xie S, Parker JS, Wang YD, Gajjar A, He L, Ellison D, Gilbertson RJ, Hannon G, Roussel MF. The miR-17 92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci U S A. 2009;106:2812–2817. doi: 10.1073/pnas.0809579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL, Cullen BR. Reduced expression of brain-enriched microRNAs in glioblastomas permits targeted regulation of a cell death gene. PLoS One. 2011;6:e24248. doi: 10.1371/journal.pone.0024248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Li KK, Pang JC, Ching AK, Wong CK, Kong X, Wang Y, Zhou L, Chen Z, Ng HK. miR-124 is frequently down-regulated in medulloblastoma and is a negative regulator of SLC16A1. Hum Pathol. 2009;40:1234–1243. doi: 10.1016/j.humpath.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang Y, Yu L, Sun C, Cheng D, Yu S, Wang Q, Yan Y, Kang C, Jin S, An T, Shi C, Xu J, Wei C, Liu J, Sun J, Wen Y, Zhao S, Kong Y. miR-146b-5p inhibits glioma migration and invasion by targeting MMP16. Cancer Lett. 2013;339:260–269. doi: 10.1016/j.canlet.2013.06.018. [DOI] [PubMed] [Google Scholar]
- Jiang J, Yang J, Wang Z, Wu G, Liu F. TFAM is directly regulated by miR-23b in glioma. Oncol Rep. 2013;30:2105–2110. doi: 10.3892/or.2013.2712. [DOI] [PubMed] [Google Scholar]
- Hu X, Chen D, Cui Y, Li Z, Huang J. Targeting microRNA-23a to inhibit glioma cell invasion via HOXD10. Sci Rep. 2013;3:3423. doi: 10.1038/srep03423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delic S, Lottmann N, Stelzl A, Liesenberg F, Wolter M, Gotze S, Zapatka M, Shiio Y, Sabel MC, Felsberg J, Reifenberger G, Riemenschneider MJ. MiR-328 promotes glioma cell invasion via SFRP1-dependent Wnt-signaling activation. Neuro Oncol. 2014;16:179–190. doi: 10.1093/neuonc/not164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Gong J, Huang W, Wang Z, Wang M, Yang J, Wu C, Wu Z, Han B. MicroRNA-106b-5p boosts glioma tumorigensis by targeting multiple tumor suppressor genes. Oncogene. 2013. [DOI] [PubMed]
- Ling N, Gu J, Lei Z, Li M, Zhao J, Zhang HT, Li X. microRNA-155 regulates cell proliferation and invasion by targeting FOXO3a in glioma. Oncol Rep. 2013;30:2111–2118. doi: 10.3892/or.2013.2685. [DOI] [PubMed] [Google Scholar]
- Sun B, Pu B, Chu D, Chu X, Li W, Wei D. MicroRNA-650 expression in glioma is associated with prognosis of patients. J Neurooncol. 2013;115:375–380. doi: 10.1007/s11060-013-1243-y. [DOI] [PubMed] [Google Scholar]
- Wang K, Wang X, Zou J, Zhang A, Wan Y, Pu P, Song Z, Qian C, Chen Y, Yang S, Wang Y. miR-92b controls glioma proliferation and invasion through regulating Wnt/beta-catenin signaling via Nemo-like kinase. Neuro Oncol. 2013;15:578–588. doi: 10.1093/neuonc/not004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z, Wang K, Wang G, Zhang A, Pu P. MiR-30a-5p antisense oligonucleotide suppresses glioma cell growth by targeting SEPT7. PLoS One. 2013;8:e55008. doi: 10.1371/journal.pone.0055008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Guessous F, Alvarado-Velez M, Marcinkiewicz L, Zhang Y, Kim J, Heister S, Kefas B, Godlewski J, Schiff D, Purow B, Abounader R. Oncogenic effects of miR-10b in glioblastoma stem cells. J Neurooncol. 2013;112:153–163. doi: 10.1007/s11060-013-1047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Zhang Z, Tu Y, Jin T, Liang H, Cui G, He S, Gao G. Correlation of microRNA-372 upregulation with poor prognosis in human glioma. Diagn Pathol. 2013;8:1. doi: 10.1186/1746-1596-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Sasayama T, Tanaka K, Nakamizo S, Nishihara M, Mizukawa K, Kohta M, Koyama J, Miyake S, Taniguchi M, Hosoda K, Kohmura E. MicroRNA-183 upregulates HIF-1alpha by targeting isocitrate dehydrogenase 2 (IDH2) in glioma cells. J Neurooncol. 2013;111:273–283. doi: 10.1007/s11060-012-1027-9. [DOI] [PubMed] [Google Scholar]
- Song L, Lin C, Gong H, Wang C, Liu L, Wu J, Tao S, Hu B, Cheng SY, Li M, Li J. miR-486 sustains NF-kappaB activity by disrupting multiple NF-kappaB-negative feedback loops. Cell Res. 2013;23:274–289. doi: 10.1038/cr.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Wang S, Geng S, Ma S, Liang Z, Jiao B. Increased expression of microRNA-17 predicts poor prognosis in human glioma. J Biomed Biotechnol. 2012;2012:970761. doi: 10.1155/2012/970761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z, Wang K, Zhang A, Wang G, Kang C, Han L, Pu P. miR-19a and miR-19b overexpression in gliomas. Pathol Oncol Res. 2013;19:847–853. doi: 10.1007/s12253-013-9653-x. [DOI] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Gabriely G, Yi M, Narayan RS, Niers JM, Wurdinger T, Imitola J, Ligon KL, Kesari S, Esau C, Stephens RM, Tannous BA, Krichevsky AM. Human glioma growth is controlled by microRNA-10b. Cancer Res. 2011;71:3563–3572. doi: 10.1158/0008-5472.CAN-10-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Li G, Feng D, Qin H, Gong L, Zhang J, Zhang Z. MicroRNA-21 expression is associated with overall survival in patients with glioma. Diagn Pathol. 2013;8:200. doi: 10.1186/1746-1596-8-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaby O, Lakomy R, Fadrus P, Hrstka R, Kren L, Lzicarova E, Smrcka M, Svoboda M, Dolezalova H, Novakova J, Valik D, Vyzula R, Michalek J. MicroRNA-181 family predicts response to concomitant chemoradiotherapy with temozolomide in glioblastoma patients. Neoplasma. 2010;57:264–269. doi: 10.4149/neo_2010_03_264. [DOI] [PubMed] [Google Scholar]
- Zhou X, Ren Y, Moore L, Mei M, You Y, Xu P, Wang B, Wang G, Jia Z, Pu P, Zhang W, Kang C. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest. 2010;90:144–155. doi: 10.1038/labinvest.2009.126. [DOI] [PubMed] [Google Scholar]
- Venkataraman S, Alimova I, Fan R, Harris P, Foreman N, Vibhakar R. MicroRNA 128a increases intracellular ROS level by targeting Bmi-1 and inhibits medulloblastoma cancer cell growth by promoting senescence. PLoS One. 2010;5:e10748. doi: 10.1371/journal.pone.0010748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JG, Zhao Y, Sethi P, Li YY, Mahta A, Culicchia F, Lukiw WJ. Micro-RNA-128 (miRNA-128) down-regulation in glioblastoma targets ARP5 (ANGPTL6), Bmi-1 and E2F-3a, key regulators of brain cell proliferation. J Neurooncol. 2010;98:297–304. doi: 10.1007/s11060-009-0077-0. [DOI] [PubMed] [Google Scholar]
- Fasano CA, Dimos JT, Ivanova NB, Lowry N, Lemischka IR, Temple S. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Roth P, Wischhusen J, Happold C, Chandran PA, Hofer S, Eisele G, Weller M, Keller A. A specific miRNA signature in the peripheral blood of glioblastoma patients. J Neurochem. 2011;118:449–457. doi: 10.1111/j.1471-4159.2011.07307.x. [DOI] [PubMed] [Google Scholar]
- Abu-Elneel K, Liu T, Gazzaniga FS, Nishimura Y, Wall DP, Geschwind DH, Lao K, Kosik KS. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics. 2008;9:153–161. doi: 10.1007/s10048-008-0133-5. [DOI] [PubMed] [Google Scholar]
- Saba R, Goodman CD, Huzarewich RL, Robertson C, Booth SA. A miRNA signature of prion induced neurodegeneration. PLoS One. 2008;3:e3652. doi: 10.1371/journal.pone.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ST, Chu K, Im WS, Yoon HJ, Im JY, Park JE, Park KH, Jung KH, Lee SK, Kim M, Roh JK. Altered microRNA regulation in Huntington’s disease models. Exp Neurol. 2011;227:172–179. doi: 10.1016/j.expneurol.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly MB, Wang Y, Qian Z, Jin J, Zhang Y, Bohlander SK, Le Beau MM, Larson RA, Golub TR, Rowley JD, Chen J. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci U S A. 2007;104:19971–19976. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- Bottoni A, Zatelli MC, Ferracin M, Tagliati F, Piccin D, Vignali C, Calin GA, Negrini M, Croce CM, Degli Uberti EC. Identification of differentially expressed microRNAs by microarray: a possible role for microRNA genes in pituitary adenomas. J Cell Physiol. 2007;210:370–377. doi: 10.1002/jcp.20832. [DOI] [PubMed] [Google Scholar]
- Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AW, Klijn JG, Wiemer EA, Martens JW. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci U S A. 2008;105:13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss GJ, Bemis LT, Nakajima E, Sugita M, Birks DK, Robinson WA, Varella-Garcia M, Bunn PA Jr, Haney J, Helfrich BA, Kato H, Hirsch FR, Franklin WA. EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann Oncol. 2008;19:1053–1059. doi: 10.1093/annonc/mdn006. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Ménard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XH, Wang Q, Chen JS, Fu XH, Chen XL, Chen LZ, Li W, Bi J, Zhang LJ, Fu Q, Zeng WT, Cao LQ, Tan HX, Su Q. Bead-based microarray analysis of microRNA expression in hepatocellular carcinoma: miR-338 is downregulated. Hepatol Res. 2009;39:786–794. doi: 10.1111/j.1872-034X.2009.00502.x. [DOI] [PubMed] [Google Scholar]
- Kotani A, Ha D, Hsieh J, Rao PK, Schotte D, den Boer ML, Armstrong SA, Lodish HF. miR-128b is a potent glucocorticoid sensitizer in MLL-AF4 acute lymphocytic leukemia cells and exerts cooperative effects with miR-221. Blood. 2009;114:4169–4178. doi: 10.1182/blood-2008-12-191619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Sevignani C, Calin GA, Siracusa LD, Croce CM. Mammalian microRNAs: a small world for fine-tuning gene expression. Mamm Genome. 2006;17:189–202. doi: 10.1007/s00335-005-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- Khan AP, Poisson LM, Bhat VB, Fermin D, Zhao R, Kalyana-Sundaram S, Michailidis G, Nesvizhskii AI, Omenn GS, Chinnaiyan AM, Sreekumar A. Quantitative proteomic profiling of prostate cancer reveals a role for miR-128 in prostate cancer. Mol Cell Proteomics. 2010;9:298–312. doi: 10.1074/mcp.M900159-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Yu F, Jiao Y, Feng J, Tang W, Yao H, Gong C, Chen J, Su F, Zhang Y, Song E. Reduced miR-128 in breast tumor-initiating cells induces chemotherapeutic resistance via Bmi-1 and ABCC5. Clin Cancer Res. 2011;17:7105–7115. doi: 10.1158/1078-0432.CCR-11-0071. [DOI] [PubMed] [Google Scholar]
- Myatt SS, Wang J, Monteiro LJ, Christian M, Ho KK, Fusi L, Dina RE, Brosens JJ, Ghaem-Maghami S, Lam EW. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer Res. 2010;70:367–377. doi: 10.1158/0008-5472.CAN-09-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A, Harris AW, Bath ML, Cory S. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature. 1990;348:331–333. doi: 10.1038/348331a0. [DOI] [PubMed] [Google Scholar]
- Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- Wu N, Wu GC, Hu R, Li M, Feng H. Ginsenoside Rh2 inhibits glioma cell proliferation by targeting microRNA-128. Acta Pharmacol Sin. 2011;32:345–353. doi: 10.1038/aps.2010.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlakha YK, Saini N. miR-128 exerts pro-apoptotic effect in a p53 transcription-dependent and -independent manner via PUMA-Bak axis. Cell Death Dis. 2013;4:e542. doi: 10.1038/cddis.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S, Shao G, Lv X, Liu Y, Fan Y, Wu A, Hu H. Downregulation of miRNA-128 sensitises breast cancer cell to chemodrugs by targeting Bax. Cell Biol Int. 2013;37:653–658. doi: 10.1002/cbin.10100. [DOI] [PubMed] [Google Scholar]
- Donzelli S, Fontemaggi G, Fazi F, Di Agostino S, Padula F, Biagioni F, Muti P, Strano S, Blandino G. MicroRNA-128-2 targets the transcriptional repressor E2F5 enhancing mutant p53 gain of function. Cell Death Differ. 2012;19:1038–1048. doi: 10.1038/cdd.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Portes V, Pinard JM, Billuart P, Vinet MC, Koulakoff A, Carrie A, Gelot A, Dupuis E, Motte J, Berwald-Netter Y, Catala M, Kahn A, Beldjord C, Chelly J. A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell. 1998;92:51–61. doi: 10.1016/S0092-8674(00)80898-3. [DOI] [PubMed] [Google Scholar]
- Tissir F, Goffinet AM. Reelin and brain development. Nat Rev Neurosci. 2003;4:496–505. doi: 10.1038/nrn1113. [DOI] [PubMed] [Google Scholar]
- Messi E, Florian MC, Caccia C, Zanisi M, Maggi R. Retinoic acid reduces human neuroblastoma cell migration and invasiveness: effects on DCX, LIS1, neurofilaments-68 and vimentin expression. BMC Cancer. 2008;8:30. doi: 10.1186/1471-2407-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone G, Vincenzi B, Zagami M, Santini D, Panteri R, Flammia G, Verzi A, Lepanto D, Morini S, Russo A, Bazan V, Tomasino RM, Morello V, Tonini G, Rabitti C. Reelin expression in human prostate cancer: a marker of tumor aggressiveness based on correlation with grade. Mod Pathol. 2007;20:344–351. doi: 10.1038/modpathol.3800743. [DOI] [PubMed] [Google Scholar]
- Seigel GM, Hackam AS, Ganguly A, Mandell LM, Gonzalez-Fernandez F. Human embryonic and neuronal stem cell markers in retinoblastoma. Mol Vis. 2007;13:823–832. [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Lu J, Yang C, Wang X, Cheng L, Hu G, Sun Y, Zhang X, Wu M, Liu Z. CASK and its target gene Reelin were co-upregulated in human esophageal carcinoma. Cancer Lett. 2002;179:71–77. doi: 10.1016/S0304-3835(01)00846-1. [DOI] [PubMed] [Google Scholar]
- Shi ZM, Wang J, Yan Z, You YP, Li CY, Qian X, Yin Y, Zhao P, Wang YY, Wang XF, Li MN, Liu LZ, Liu N, Jiang BH. MiR-128 inhibits tumor growth and angiogenesis by targeting p70S6K1. PLoS One. 2012;7:e32709. doi: 10.1371/journal.pone.0032709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI0215593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaven SW, Tontonoz P. Nuclear receptors in lipid metabolism: targeting the heart of dyslipidemia. Annu Rev Med. 2006;57:313–329. doi: 10.1146/annurev.med.57.121304.131428. [DOI] [PubMed] [Google Scholar]
- Adlakha YK, Khanna S, Singh R, Singh VP, Agrawal A, Saini N. Pro-apoptotic miRNA-128-2 modulates ABCA1, ABCG1 and RXRalpha expression and cholesterol homeostasis. Cell Death Dis. 2013;4:e780. doi: 10.1038/cddis.2013.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, Carazo JM, Pascual-Montano A. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37:W317–322. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi N, Alexander MS, Shimizu-Motohashi Y, Myers JA, Kawahara G, Kunkel LM. Regulation of IRS1/Akt insulin signaling by microRNA-128a during myogenesis. J Cell Sci. 2013;126:2678–2691. doi: 10.1242/jcs.119966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Liu Z, Phung S, Wang E, Yuan YC, Chen S. The role of microRNA-128a in regulating TGFbeta signaling in letrozole-resistant breast cancer cells. Breast Cancer Res Treat. 2010;124:89–99. doi: 10.1007/s10549-009-0716-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, Davidson BL. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16:495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]