Abstract
Background
Human Rhinoviruses (HRV) are major causative agents of acute respiratory tract infections in all age group and important contributing factors of childhood morbidity and mortality. Clinical presentation is poorly specific and the great antigenic and genetic variability of HRVs renders the biological diagnosis complex. Here, we have evaluated several molecular diagnostic protocols, including Taqman probe-based and intercalating agent-based RT-PCR assays.
Methods
5,627 respiratory samples sent to the laboratory of Virology of the University Hospitals of Marseille, France, from March 2011 to February 2012, were tested using a real-time RT-PCR assay in the 5’NCR of the rhinoviral genome that associated a Taqman probe and the detection of DNA-BOXTO-dye complexes. A sample of 500 BOXTO-positive samples were further tested using the same probe assay (without BOXTO), and a SYBR Green assay (using the same amplification primers). The specific amplification of HRV sequences was assessed by NGS amplicon sequencing.
Results
The Taqman probe RT-PCR assay identified 696/5,627 samples (12,4%) as HRV-positive. BOXTO-positive samples included all probe-positive samples and 1,913 additional samples, of which only 24.3% were confirmed by sequencing. The SYBR Green assay was more specific (16/550 samples were probe-negative/SYBR Green-positive, all confirmed by 5′NCR sequencing), but 3/500 samples were probe-positive/SYBR Green-negative.
Conclusions
Our results highlight the difficulty in detecting HRVs in clinical samples using a single molecular detection system. Amongst the 3 systems tested, the best compromise was obtained with the SYBR Green assay, which, by comparison with our probe-based assay provided an improved sensitivity without altering the detection specificity. Interestingly, a majority of probe-negative/BOXTO- or SYBR Green-positive samples were not associated with mutations in the sequence targeted by the probe. Sequence-based modifications of the secondary structure of the HRV 5′NCR may be associated with a limited access to the probe hybridisation region. Further investigations may identify a test combining a probe based- and an intercalating agent-based detection, which will significantly improve the diagnosis of HRV infections.
Keywords: BOXTO, SYBR green, Taqman probe, Picornaviridae, Human rhinovirus, Respiratory infections, Molecular diagnosis
Background
Acute respiratory tract infections (ARIs) represent a major public health problem worldwide, a leading cause of human acute illnesses and an important contributing factor of childhood morbidity and mortality, especially in children under 5 years [1-3]. A majority of ARIs in children have a viral etiology, probably due to absent or incomplete immune protection, sustained viral shedding and high transmissibility amongst hosts [4]. Amongst a variety of RNA and DNA viruses that can infect the respiratory tract, Human Rhinoviruses (Picornaviridae family) [5], are recognized as the most prevalent in all age group worldwide [6]. Together with coronaviruses, they represent common causative agents of upper respiratory tract (URT) infections, traditionally defined as common cold [6,7], and thus a major cause of school and work absenteeism since children experience 8–12 and adults 2–3 URT episodes per year, on average [8]. Children are the major reservoir for HRV [9]. The mean age at the first symptomatic HRV infection is 4–6 months (vs >6 months for other viruses such as RSV [10]; more than 90% of children have experienced at least one HRV infection by the age of 2 years [11]. HRV infection is most often associated with a non-specific, self-limiting illness with clinical manifestations ranging from an asymptomatic presentation to fever, rhinorrhea, cough and wheezing. However, HRV infections are increasingly involved in otitis media, pneumonia, or chronic obstructive pulmonary disease, especially in infants, elderly and immunocompromised patients [12,13]. In addition, HRV represent a major viral aetiology of asthma exacerbations, with the highest incidence of all respiratory viruses in adults and children >2 years of age (60–65% of viral exacerbations) [14,15]. Although suspected in some studies, there is at the present day no proof of association between clinical severity and HRV species [16-20]. Altogether, these data highlight the predominant role of HRV as a respiratory pathogen especially in early life.
HRVs are small non-enveloped viruses with a single-stranded RNA genome of positive polarity; originally classified in the Rhinovirus genus, they have been integrated into the Enterovirus genus. Rhinoviruses share with Enteroviruses an identical genomic organization and have similar functional RNA secondary structures, but differ in their acid tolerance, receptor usage, and cell tropism [21]. The genome is approximately 7.2 kb long, and is composed of a 5′non-coding region (5′NCR), followed by a long open reading frame coding for four structural icosahedral capsid proteins (VP4, VP2, VP3 and VP1), and seven non-structural proteins 2A, 2B, 2C, 3A, 3B, 3C and 3D, and terminated by a short 3′UTR and poly A tract. HRVs are highly heterogeneous genetically and antigenically [22,23]. More than 140 serotypes have been described, and these fall into three species, HRV species A (HRV-A; 74 serotypes), HRV-B (25 serotypes) and a novel genetically distinct third genotype HRV-C, comprising 49 designated serotypes recognized in 2006 [24-27].
HRV infections occur throughout the year [28], usually with peaks in spring and autumn in temperate countries [29-31], the prevalence varying from 10% to 60% depending on the population or the period studied. Molecular studies suggest almost equal prevalence of HRVA and HRVC, with a under-representation of HRVB species [11,19,32-39]. A remarkably wide genetic diversity of HRV serotypes can be observed all year long [40].
The diagnosis of HRV infections is important for epidemiological purposes but also for optimising the medical management of patients (e.g., the opportunity for an antibiotic treatment). As clinical presentation is non-specific, it devolves to the diagnostic laboratory to confirm the presence of HRVs [41]. Detection of HRV by culture is slow and complex for HRVA and HRVB, whilst HRVC has been unculturable in vitro to date [23]. Serologic diagnosis is virtually impossible due to the number of serotypes, and rapid antigen test kits are not available [4]. Molecular methods such as real-time RT-PCR appear to be the most suitable method, combining short analysis time, high sensitivity, semi-quantification of viral load and the detection of the majority of respiratory viruses with multiplex methods [42-44]. Most of the published systems target the 5′NCR. However, the high genetic diversity of HRVs makes the detection of all variants difficult as evidenced by the alignment of available GenBank 5′NCR sequences. Indeed, a comparison of published HRV-specific PCR primers pairs showed that no single pair could detect all HRVs [45] and that more than one PCR is required for accurate description of HRV epidemiology.
The range of detection of HRV variants by a probe-based real-time PCR assay is predicted to be additionally restricted by mutations in the region of the probe. Accordingly, we have compared the performances of 3 tests for accurately detecting HRV in clinical samples: (i) a probe-based Taqman RT-PCR assay routinely used in our hospital laboratory; (ii) the same test used without the probe and in the presence of SYBR Green I, making theoretically possible to diagnose HRVs amplified by the primers but not detected by the probe; (iii) the same test used with both the probe and in the presence of the BOXTO intercalating agent, making theoretically possible to detect HRV detected and undetected by the probe in the same reaction.
Methods
Clinical Samples
5,627 samples (nasopharyngeal aspirates, swabs or saliva specimens) sent to the Laboratory of Virology, University Hospital La Timone (Marseille, France) from March 2011 to February 2012 and stored at -80°C upon reception were studied. They were predominantly from hospital inpatients or patients in emergency departments, who presented with respiratory symptoms.
All samples were analysed using real-time RT-PCR assay using an association of a specific-probe and BOXTO. 500 samples randomly chosen amongst those with a positive BOXTO result (see below) were secondarily tested using (i) the same specific-probe without BOXTO, and (ii) a SYBR Green assay.
RNA extraction
Each sample (200 μL) was spiked by a mix of T4 and MS2 phages used as internal controls [46]. Nucleic acids extraction was performed using the EZ1 Virus Mini Kit v2.0 and the EZ1 Advanced XL Biorobot (both from Qiagen). The final elution volume was 90 μL.
HRV molecular diagnosis
i) Taqman probe assay: a real-time one-step RT-PCR reaction was performed using the iScriptTM One-Step RT-PCR Kit (Biorad) and the Biorad CFX96 Real time System thermocycler. The 30 μL final reaction volume contained 10 μL of viral RNA, 0.3 μM of each primer (forward 1: WGCCYGCGTGGCKGCC, forward 2: AGCCYGCGTGGTGCCC; reverse: GAAACACGGACACCCAAAGTAGT) and 0.1 μM of specific probe (6FAM-CTCCGGCCCCTGAATGYGGCTAA-TAMRA); thermal cycling was: reverse transcription at 50°C for 10 min, initial denaturation at 95°C for 5 min, followed by 40 cycles of [95°C for 15 s, 60°C for 30 s]. This hospital diagnostic assay used a standardised hybridisation temperature and primers and probes concentrations were optimised using a classical matrix experimental procedure.
ii) BOXTO: the optimal concentration of BOXTO for was determined according to the manufacurer’s recommendation (TATAA Biocenter). In the case of the Taqman probe assay presented above, the best results were obtained with a 2.5 μM concentration. The protocol adapted from Lind et al. [47] was identical to that of the Taqman probe assay, with the addition of a final melting curve (65°C to 95°C, with an increase of 0.5°C every 5 s), visualised in the “HEX” reading canal.
iii) SYBR Green: the SYBR Green assay was conducted using the QuantiTect SYBR® Green RT-PCR Kit and comparable reaction parameters; thermal cycling was: reverse transcription at 50°C for 30 min, initial denaturation at 95°C for 15 min, followed by 40 cycles of [95°C for 15 s, 60°C for 30 s and 72°C for 30 s], and a final step with a melting visualised in the “HEX” reading canal.
5′NCR sequencing
5′NCR PCR products were purified (QIAquick Gel Extraction, QIAGEN). The 215 bp DNA products were tagged and analysed using the Ion Torrent NGS technology without any further fragmentation. Sequencing data were analysed using to the CLC Genomics Workbench bioinformatic software [48].
Structural analysis of Taqman probe
The on-line software M-fold v3.2 was used to predict the secondary structure of the Taqman probe sequence [49].
Statistical analysis
Statistical analysis was performed using the IBM SPSS Statistics software in this study.
Results
Between March 2011 and February 2012, 5,627 respiratory samples were received for detection of respiratory virus infections and analysed in this study.
i) Taqman probe/BOXTO assay:
Using the assay that combined a probe detection and a BOXTO detection:
– The Taqman probe identified 696 out of 5,627 samples (12,4%) as HRV positive ([P+] samples).
– The BOXTO intercalating agent detected all of the 696 [P+] samples with a specific melting curve peak between 84 and 86.5°C ([P + B+] samples). However, 1,913 out of the 4,931 [P-] samples also provided positive BOXTO results (total [B+]: 2,609 (46.3%)). A random sample of 258 PCR products from these [P-B+] samples was submitted to sequencing: only 24.3% (55/258) contained HRV sequences. Results are summarised in Figure 1.
Figure 1.
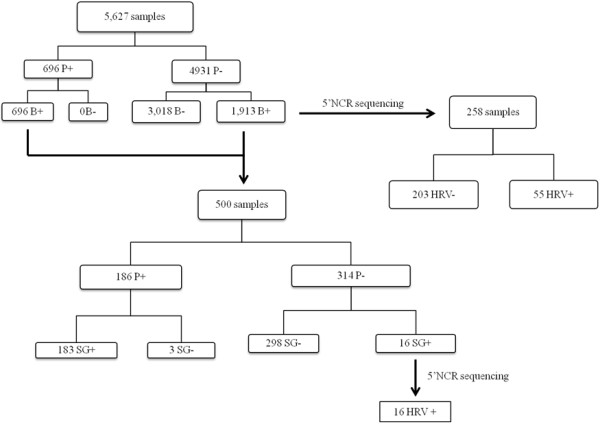
Evaluation of molecular diagnostic systems. P: Probe; B: BOXTO; SG: SYBR Green.
ii) SYBR Green assay:
In a random selection of 500 [B+] samples tested in parallel with the Taqman probe and the SYBR green assays, 183 (36.6%) were positive in both tests ([P + SG+] samples), 298 were [P-SG-], 3 were [P + SG-] and 16 [P-SG+]. SYBR green positive samples were associated with a melting curve peak between 81 and 84°C.
Ion Torrent sequencing of PCR products of the 16 [P-SG+] samples allowed identification of HRV sequences in all cases (Figure 1).
In the absence of reference test usable as a “gold standard” for the detection of HRV, it is difficult to provide a simple robust analysis of the performances of the different tests. In particular, there is most probably a proportion of our HRV-negative samples that would be identified as HRV-positive using other detection assays (e.g., other primers [46]) and therefore we are not able to provide a strict numbering of true and false negatives for our tests.
Here, we have used a few simple rules to compare the different assays and provide estimates of their performances:
(i) We have considered that [P+] samples were true positives, based on the considerable information available from the literature that suggests that probe-based real-time PCR systems are associated with a negligible rate of false positive results.
(ii) We have considered that the sequencing of amplicons was the reference test for determining if PCR products detected by the BOXTO and SYBR green intercalating agents resulted from the specific amplification of HRV genomes.
Accordingly, when the Taqman probe assay was compared with the SYBR Green assay, a total of 202 “true positive” samples was detected: 186 (92.1%) by the probe assay and 199 (94.9%) by the SYBR Green assay and confirmed by sequencing). Whatever the definition of “negatives”, this means that the sensitivity of the SYBR Green assay is better than that of the probe-based assay. As sequencing of SYBR Green amplicons did not identify any false positive, this also means that the specificity and positive predictive value of the SYBR Green assay are 100% in the sample investigated.
When the Taqman probe assay was compared with the BOXTO assay, 696 “true positive” samples were detected by the probe assay but a number of additional positives were detected by the BOXTO assay (of which an estimate of 24.3% were true positives, based on sequencing experiments). Whatever the definition of “negatives”, this means that the sensitivity of the BOXTO assay is better than that of the probe-based assay. However, sequencing of BOXTO amplicons from [P-B+] samples provided paradoxical results:
– it evidenced a large proportion (~75%) of false positives, which means that the specificity and positive predictive value of the BOXTO assay would be very low in the sample investigated (estimates are <70% and <50%, respectively, hypothesising the absence of false negative results).
– the proportion of confirmed positives (~25%) indicates that a significant proportion of HRV positive samples are not detected by the probe-based assay.
It was interesting to analyse the sequence corresponding to the probe in amplicons from [P-B+] and [P-SG+] samples. Interestingly, the alignment of 11 HRV sequences from [P-B+] samples showed only 3 single substitutions located in the region targeted by the probe, at position 8 (C → T), 15 (T → C), and 20 (C → T) respectively, whilst the remaining 8 sequences were identical to the expected sequence.
Similarly, alignment of 16 [P-SG+] sequences identified only one modified sequence in the region targeted by the probe (position 8, C → T), the remaining 11 sequences being identical to the expected sequence.
This suggested that different mechanisms could contribute to the inefficient probe-based detection of some HRV sequences. Besides the plausible role of specific mutations in the region targeted by the probe, other mechanisms, obviously independent from this sequence occurred. Using the on-line software M-fold v3.2, the secondary structure of the Taqman probe sequence was determined and showed a “loop-step” conformation (Figure 2), which may hamper the hybridisation process.
Figure 2.
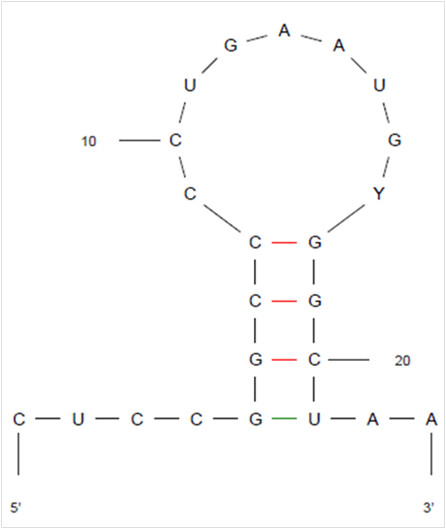
Predicted secondary structure of probe sequence, using the M-fold v3.2 on-line software.
Discussion
Despite the importance of ARIs in terms of morbidity as well as infant mortality, microbiological aetiologies of acute respiratory infections represent a complex, which has yet to be fully characterized in developed countries and remains largely unknown in developing countries. Dual viral infections are common, and a large proportion of children have evidence of viral-bacterial co-infection. In adults, viruses are the putative causative agents in a third of cases of community-acquired pneumonia, in particular influenza viruses, rhinoviruses, and coronaviruses [50]. Accordingly, much better characterization of the epidemiology of ARIs is still needed to improve understanding of the role of the different pathogens and of bacterial-viral interaction in the pathogenesis of ARIs. Since human rhinoviruses represent the most frequent cause of ARIs worldwide and in all age groups, improving the biological diagnosis of rhinoviral infections is a crucial issue.
In this study, we have tested and compared three different HRV molecular diagnostic assays. First, it is obvious that a proportion of samples truly infected by HRVs could not be detected using the Taqman probe test. This is evidenced by a number of probe-negative/BOXTO positive and probe-negative/SYBR green positive samples for which the presence of HRV amplicons was confirmed by direct sequencing of the PCR products. This phenomenon was shown to be highly reproducible and the same HRV positive samples were repeatedly detectable or undetectable using the probe-based assay (data not shown). Since all assays used the same amplification primers, the most likely explanation is a default in probe hybridisation. Analysis of the genomic sequence of HRVs that could not be detected by the probe-based assay showed that only a limited proportion of strains had mutations in the probe hybridisation region. Other mechanism(s) must therefore take place to explain the default of hybridisation of the probe. A possible explanation may lie in the highly structured form of the rhinoviral 5”NCR, which contains an Internal Ribosome Entry Site (IRES). On the one hand, the probe hybridisation region itself is located in a stem-loop region, and on the other hand mutations in different parts of the NCR may modify the secondary structure of the genomic RNA molecule and possibly limit the access to the probe hybridisation region, as previously reported [51]. This is, to our knowledge a rare example of inefficient probe-based detection that is not related to mutations in the probe hybridisation genomic region.
Second, as a number of HRV positive samples that could not be detected using a probe-based assay were identified by DNA-dye-complexes (using BOXTO or SYBR green molecules), it is interesting to consider the performances of these assays. Ideally, the probe-based detection and the intercalating agent-based reaction should be performed in the same reaction tube and detected in distinct reading canals. This is what was attempted by combining a FAM-TAMRA labelled probe and BOXTO. This system allowed the detection of a proportion of probe-negative samples, but the proportion of false positive results was so high that the use of this assay is not possible for clinical or epidemiological purposes (unacceptably low estimated specificity and positive predictive values). It remains from this experiment the important information that a significant proportion of Taqman-negative samples provided true positive results using the same primers and the BOXTO intercalating agent. Accordingly, we tested a number of BOXTO-positive samples using a standard Taqman assay and (separately) a standard SYBR green assay. The SYBR green assay was able to provide positive results in probe-negative samples, which, contrary to what was observed with BOXTO, were all confirmed by direct sequencing of amplicons, indicating that the SYBR green assay is associated with high specificity and positive predictive values. However, the assay failed to detect a few probe-positive samples. Finally, we investigated a new experimental procedure by combining the SYBR Green and a Texas-Red labelled probe. Preliminary experiments on 30 respiratory samples showed that the level of interference between Texas red-based and SYBR Green-based detections in a combined assay was lower than previously observed in the case of the couple (FAM-Boxto), but still an obstacle to routine use: (i) in 13samples testing negative using the probe or the SYBR Green alone, a combination of Probe + SG provided 11 P-SG- samples and 2 P + SG- samples (e.g., 2 false positives in 13 negative samples tested, which is too high for diagnostic use, but much lower than with the couple FAM-Boxto (circa 75%)); (ii) in 17 samples testing positive using the probe or the SG alone, the combination of Probe + SG provided 16 P + SG + samples and one P-SG + sample. The mean detection CT was slightly increased when detection was performed in the presence of SYBR Green and this was sufficient to make negative using the combined assay one sample that tested positive using the probe alone.
This final experiment reinforces the idea that it should be possible to decrease the level of interference between the probe-based and intercalating agent-based detection systems and the next steps should include a systematic investigation of the possible combinations that may allow to identify a couple usable for diagnostic purposes.
Conclusion
In conclusion, this study confirms the difficulty in detecting HRVs in clinical samples using a single molecular detection system. Besides the possible utilisation of distinct pairs of primers to adequately cover the natural genetic diversity of HRV isolates [46] our results suggest that the optimisation of the detection of amplicons produced by primers is an important issue. In our hands, a SYBR green-based assay was slightly more sensitive that a Taqman probe-based assay and constituted the best compromise amongst the techniques tested. Attempts to associate a probe-based detection with an intercalating agent-based detection should be renewed to identify a reliable couple (DNA dye/probe label) that could be used for routine diagnostic and epidemiological detection of human rhinoviruses.
Competing interests
The authors declare that they have no competing interest.
Authors’ contributions
JD performed the experiments, analysed the data and wrote the manuscript. LN conceived and designed the experiments, analysed the data and wrote the manuscript. VF performed the experiments, analysed the data and wrote the manuscript. OP performed the experiments and analysed the data. CG and LTP designed the experiments. XdL conceived and designed the experiments, analysed the data, wrote the manuscript and supervised the work. All authors read and approved the final manuscript.
Contributor Information
Julien Dupouey, Email: julien.dupouey@ap-hm.fr.
Laetitia Ninove, Email: laetitia.ninove@ap-hm.fr.
Vanessa Ferrier, Email: vanessa-ferrier@hotmail.fr.
Odile Py, Email: od314py@yahoo.fr.
Céline Gazin, Email: cgazin@yahoo.fr.
Laurence Thirion-Perrier, Email: laurencethirionperrier@gmail.com.
Xavier de Lamballerie, Email: xavier.de-lamballerie@univ-amu.fr.
Acknowledgments
The authors would like to thank Karine Barthelemy, Ludivine Molina and Raphael Subrini for their technical assistance.
References
- Williams BG, Gouws E, Boschi-Pinto C, Bryce J, Dye C. Estimates of world-wide distribution of child deaths from acute respiratory infections. Lancet Infect Dis. 2002 Jan;2(1):25–32. doi: 10.1016/S1473-3099(01)00170-0. [DOI] [PubMed] [Google Scholar]
- Mulholland K. Global burden of acute respiratory infections in children: implications for interventions. Pediatr Pulmonol. 2003 Dec;36(6):469–474. doi: 10.1002/ppul.10344. [DOI] [PubMed] [Google Scholar]
- Bryce J, Boschi-Pinto C, Shibuya K, Black RE. WHO estimates of the causes of death in children. Lancet. 2005;365:1147–1152. doi: 10.1016/S0140-6736(05)71877-8. [DOI] [PubMed] [Google Scholar]
- Kieninger E, Fuchs O, Latzin P, Frey U, Regamey N. Rhinovirus infections in infancy and early childhood. Eur Respir J. 2012;41(2):443–452. doi: 10.1183/09031936.00203511. [DOI] [PubMed] [Google Scholar]
- Whitton JL, Cornell CT, Feuer R. Host and virus determinants of picornavirus pathogenesis and tropism. Nat Rev Microbiol. 2005 Oct;3(10):765–776. doi: 10.1038/nrmicro1284. [DOI] [PubMed] [Google Scholar]
- Mackay IM. Human rhinoviruses: the cold wars resume. J Clin Virol. 2008 Aug;42(4):297–320. doi: 10.1016/j.jcv.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny FW Jr. The clinical impact of human respiratory virus infections. Am J Respir Crit Care Med. 1995 Oct;152(4 Pt 2):S4–S12. doi: 10.1164/ajrccm/152.4_Pt_2.S4. [DOI] [PubMed] [Google Scholar]
- Winther B, Gwaltney JM Jr, Mygind N, Hendley JO. Viral-induced rhinitis. Am J Rhinol. 1998;12(1):17–20. doi: 10.2500/105065898782102954. [DOI] [PubMed] [Google Scholar]
- Peltola V, Waris M, Osterback R, Susi P, Ruuskanen O, Hyypiä T. Rhinovirus transmission within families with children: incidence of symptomatic and asymptomatic infections. J Infect Dis. 2008 Feb 1;197(3):382–389. doi: 10.1086/525542. [DOI] [PubMed] [Google Scholar]
- Jartti T, Lee WM, Pappas T, Evans M, Lemanske RF Jr, Gern JE. Serial viral infections in infants with recurrent respiratory illnesses. Eur Respir J. 2008;32(2):314–320. doi: 10.1183/09031936.00161907. Epub 2008 Apr 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomqvist S, Roivainen M, Puhakka T, Kleemola M, Hovi T. Virological and serological analysis of rhinovirus infections during the first two years of life in a cohort of children. J Med Virol. 2002;66(2):263–268. doi: 10.1002/jmv.2140. [DOI] [PubMed] [Google Scholar]
- Peltola V, Waris M, Osterback R, Susi P, Hyypiä T, Ruuskanen O. Clinical effects of rhinovirus infections. J Clin Virol. 2008 Dec;43(4):411–414. doi: 10.1016/j.jcv.2008.08.014. [DOI] [PubMed] [Google Scholar]
- Turner RB. Rhinovirus: more than just a common cold virus. J InfectDis. 2007;195:765–766. doi: 10.1086/511829. [DOI] [PubMed] [Google Scholar]
- Corne JM, Marshall C, Smith S, Schreiber J, Sanderson G, Holgate ST, Johnston SL. Frequency, severity, and duration of rhinovirus infections in asthmatic and non-asthmatic individuals: a longitudinal cohort study. Lancet. 2002;359(9309):831–834. doi: 10.1016/S0140-6736(02)07953-9. [DOI] [PubMed] [Google Scholar]
- Rawlinson WD, Waliuzzaman Z, Carter IW, Belessis YC, Gilbert KM, Morton JR. Asthma exacerbations in children associated with rhinovirus but not human metapneumovirus infection. J Infect Dis. 2003;187(8):1314–1318. doi: 10.1086/368411. [DOI] [PubMed] [Google Scholar]
- Piralla A, Rovida F, Campanini G, Rognoni V, Marchi A, Locatelli F, Gerna G. Clinical severity and molecular typing of human rhinovirus C strains during a fall outbreak affecting hospitalized patients. J Clin Virol. 2009 Aug;45(4):311–317. doi: 10.1016/j.jcv.2009.04.016. [DOI] [PubMed] [Google Scholar]
- Bizzintino J, Lee WM, Laing IA, Vang F, Pappas T, Zhang G, Martin AC, Khoo SK, Cox DW, Geelhoed GC, McMinn PC, Goldblatt J, Gern JE, Le Souëf PN. Association between human rhinovirus C and severity of acute asthma in children. Eur Respir J. 2011 May;37(5):1037–1042. doi: 10.1183/09031936.00092410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Gonzalez R, Wang Z, Xiao Y, Chen L, Li T, Vernet G, Paranhos-Baccalà G, Jin Q, Wang J. Human rhinoviruses in Chinese adults with acute respiratory tract infection. J Infect. 2010 Oct;61(4):289–298. doi: 10.1016/j.jinf.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Gonzalez R, Xie Z, Xiao Y, Liu J, Chen L, Liu C, Zhang J, Ren L, Vernet G, Paranhos-Baccalà G, Shen K, Jin Q, Wang J. Human rhinovirus C infections mirror those of human rhinovirus A in children with community-acquired pneumonia. J Clin Virol. 2010 Oct;49(2):94–99. doi: 10.1016/j.jcv.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Carraro E, Kamikawa J, Leal E, Granato C, Bellei N. Rhinovirus species and their clinical presentation among different risk groups of non-hospitalized patients. J Med Virol. 2010 Dec;82(12):2110–2115. doi: 10.1002/jmv.21914. [DOI] [PubMed] [Google Scholar]
- Laine P, Savolainen C, Blomqvist S, Hovi T. Phylogenetic analysis of human rhinovirus capsid protein VP1 and 2A protease coding sequences confirms shared genus-like relationships with human enteroviruses. J Gen Virol. 2005;86(3):697–706. doi: 10.1099/vir.0.80445-0. [DOI] [PubMed] [Google Scholar]
- Huang T, Wang W, Bessaud M, Ren P, Sheng J, Yan H, Zhang J, Lin X, Wang Y, Delpeyroux F, Deubel V. Evidence of recombination and genetic diversity in human rhinoviruses in children with acute respiratory infection. PLoS One. 2009 Jul 27;4(7):e6355. doi: 10.1371/journal.pone.0006355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SK, Yip CC, Tsoi HW, Lee RA, So LY, Lau YL, Chan KH, Woo PC, Yuen KY. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J Clin Microbiol. 2007 Nov;45(11):3655–3664. doi: 10.1128/JCM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virus taxonomy list 2011, international committee on taxonomy of viruses. http://ictvonline.org/virusTaxonomy.asp?version=2011.
- Palmenberg AC, Spiro D, Kuzmickas R, Wang S, Djikeng A, Rathe JA, Fraser-Liggett CM, Liggett SB. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science. 2009 Apr 3;324(5923):55–59. doi: 10.1126/science.1165557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P, McIntyre C, Savolainen-Kopra C, Tapparel C, Mackay IM, Hovi T. Proposals for the classification of human rhinovirus species C into genetically assigned types. J Gen Virol. 2010;91(10):2409–2419. doi: 10.1099/vir.0.023994-0. [DOI] [PubMed] [Google Scholar]
- Wisdom A, Leitch EC, Gaunt E, Harvala H, Simmonds P. Screening respiratory samples for detection of human rhinoviruses (HRVs) and enteroviruses: comprehensive VP4-VP2 typing reveals high incidence and genetic diversity of HRV species C. J Clin Microbiol. 2009;47(12):3958–3967. doi: 10.1128/JCM.00993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi CD, Aebi C, Gorgievski-Hrisoho M, Mühlemann K, Barbani MT. Twelve years’ detection of respiratory viruses by immunofluorescence in hospitalised children: impact of the introduction of a new respiratory picornavirus assay. BMC Infect Dis. 2011 Feb 7;11:41. doi: 10.1186/1471-2334-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen T, Järvinen A. The common cold. Lancet. 2003 Jan 4;361(9351):51–59. doi: 10.1016/S0140-6736(03)12162-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winther B, Hayden FG, Hendley JO. Picornavirus infections in children diagnosed by RT-PCR during longitudinal surveillance with weekly sampling: association with symptomatic illness and effect of season. J Med Virol. 2006 May;78(5):644–650. doi: 10.1002/jmv.20588. [DOI] [PubMed] [Google Scholar]
- Jartti T, Lehtinen P, Vuorinen T, Osterback R, van den Hoogen B, Osterhaus AD, Ruuskanen O. Respiratory picornaviruses and respiratory syncytial virus as causative agents of acute expiratory wheezing in children. Emerg Infect Dis. 2004 Jun;10(6):1095–1101. doi: 10.3201/eid1006.030629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henquell C, Mirand A, Deusebis AL, Regagnon C, Archimbaud C, Chambon M, Bailly JL, Gourdon F, Hermet E, Dauphin JB, Labbé A, Peigue-Lafeuille H. Prospective genotyping of human rhinoviruses in children and adults during the winter of 2009–2010. J Clin Virol. 2012 Apr;53(4):280–284. doi: 10.1016/j.jcv.2011.10.009. [DOI] [PubMed] [Google Scholar]
- Moreira LP, Kamikawa J, Watanabe AS, Carraro E, Leal E, Arruda E, Granato CF, Bellei NC. Frequency of human rhinovirus species in outpatient children with acute respiratory infections at primary care level in Brazil. Pediatr Infect Dis J. 2011 Jul;30(7):612–614. doi: 10.1097/INF.0b013e31820d0de3. [DOI] [PubMed] [Google Scholar]
- Pogka V, Kossivakis A, Kalliaropoulos A, Moutousi A, Sgouras D, Panagiotopoulos T, Chrousos GP, Theodoridou M, Syriopoulou VP, Mentis AF. Respiratory viruses involved in influenza-like illness in a Greek pediatric population during the winter period of the years 2005–2008. J Med Virol. 2011 Oct;83(10):1841–1848. doi: 10.1002/jmv.22173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landa-Cardeña A, Morales-Romero J, García-Roman R, Cobián-Güemes AG, Méndez E, Ortiz-Leon C, Pitalúa-Cortés F, Mora SI, Montero H. Clinical characteristics and genetic variability of human rhinovirus in Mexico. Viruses. 2012 Feb;4(2):200–210. doi: 10.3390/v4020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyango CO, Welch SR, Munywoki PK, Agoti CN, Bett A, Ngama M, Myers R, Cane PA, Nokes DJ. Molecular epidemiology of human rhinovirus infections in Kilifi, coastal Kenya. J Med Virol. 2012 May;84(5):823–831. doi: 10.1002/jmv.23251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EK, Williams JV, Gebretsadik T, Carroll KN, Dupont WD, Mohamed YA, Morin LL, Heil L, Minton PA, Woodward K, Liu Z, Hartert TV. Host and viral factors associated with severity of human rhinovirus-associated infant respiratory tract illness. J Allergy Clin Immunol. 2011 Apr;127(4):883–891. doi: 10.1016/j.jaci.2010.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-García ML, Calvo C, Pozo F, Villadangos PA, Pérez-Breña P, Casas I. Spectrum of respiratory viruses in children with community-acquired pneumonia. Pediatr Infect Dis J. 2012 Aug;31(8):808–813. doi: 10.1097/INF.0b013e3182568c67. [DOI] [PubMed] [Google Scholar]
- Piralla A, Lilleri D, Sarasini A, Marchi A, Zecca M, Stronati M, Baldanti F, Gerna G. Human rhinovirus and human respiratory enterovirus (EV68 and EV104) infections in hospitalized patients in Italy, 2008–2009. Diagn Microbiol Infect Dis. 2012 Jun;73(2):162–167. doi: 10.1016/j.diagmicrobio.2012.02.019. [DOI] [PubMed] [Google Scholar]
- Savolainen C, Mulders MN, Hovi T. Phylogenetic analysis of rhinovirus isolates collected during successive epidemic seasons. Virus Res. 2002 Apr 23;85(1):41–46. doi: 10.1016/S0168-1702(02)00016-3. [DOI] [PubMed] [Google Scholar]
- Barrett B, Brown R, Voland R, Maberry R, Turner RB. Relations among questionnaire and laboratory measures of rhinovirus infection. Eur Respir J. 2006;28:358–363. doi: 10.1183/09031936.06.00002606. [DOI] [PubMed] [Google Scholar]
- Mahony JB. Detection of respiratory viruses by molecular methods. Clin Microbiol Rev. 2008 Oct;21(4):716–747. doi: 10.1128/CMR.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliendo AM. Multiplex PCR, and emerging technologies for the detection of respiratory pathogens. Clin Infect Dis. 2011 May;52(Suppl 4):S326–S330. doi: 10.1093/cid/cir047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Holloway B, Dare RK, Kuypers J, Yagi S, Williams JV, Hall CB, Erdman DD. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J Clin Microbiol. 2008 Feb;46(2):533–539. doi: 10.1128/JCM.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faux CE, Arden KE, Lambert SB, Nissen MD, Nolan TM, Chang AB, Sloots TP, Mackay IM. Usefulness of published PCR primers in detecting human rhinovirus infection. Emerg Infect Dis. 2011 Feb;17(2):296–298. doi: 10.3201/eid1702.101123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninove L, Nougairede A, Gazin C, Thirion L, Delogu I, Zandotti C, Charrel RN, De Lamballerie X. RNA and DNA bacteriophages as molecular diagnosis controls in clinical virology: a comprehensive study of more than 45,000 routine PCR tests. PLoS One. 2011 Feb 9;6(2):e16142. doi: 10.1371/journal.pone.0016142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind K, Ståhlberg A, Zoric N, Kubista M. Combining sequence-specific probes and DNA binding dyes in real-time PCR for specific nucleic acid quantification and melting curve analysis. Biotechniques. 2006 Mar;40(3):315–319. doi: 10.2144/000112101. [DOI] [PubMed] [Google Scholar]
- Merriman B, Rothberg JM. Ion Torrent R&D Team. Progress in ion torrent semiconductor chip based sequencing. Electrophoresis. 2012;33(23):3397–3417. doi: 10.1002/elps.201200424. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003 Jul 1;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuskanen O, Lahti E, Jennings LC, Murdoch DR. Viral pneumonia. Lancet. 2011 Apr 9;377(9773):1264–1275. doi: 10.1016/S0140-6736(10)61459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler JF, Kupfer B, Petersen N, Grotto RM, Rodrigues SM, Grywna K, Panning M, Annan A, Silva GF, Douglas J, Koay ES, Smuts H, Netto EM, Simmonds P, Pardini MI, Roth WK, Drosten C. A novel diagnostic target in the hepatitis C virus genome. PLoS Med. 2009 Feb 10;6(2):e31. doi: 10.1371/journal.pmed.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]


