Abstract
Egawa et al. recently showed the value of patient-specific induced pluripotent stem cells (iPSCs) for modeling amyotrophic lateral sclerosis in vitro. Their study and our work highlight the need for complementary assays to detect small, but potentially important, phenotypic differences between control iPSC lines and those carrying disease mutations.
In their recent study, Egawa et al. (1) analyse motor neurons differentiated from iPSC lines derived from patients with amyotrophic lateral sclerosis (ALS) carrying mutations in TDP-43 (1). Using multiple mutant TDP-43 iPSC lines, they confirm and extend in detail the in vitro recapitulation of ALS-associated phenotypes that we had shown previously (2). In contrast to our findings (2), Egawa et al. state that they do not observe a survival difference between cultured motor neurons derived from mutant TDP-43 iPSC lines versus control iPSC lines under basal conditions when using a lactate dehydrogenase (LDH) release assay to measure neuronal survival [Fig. S11C in (1)]. In our study (2), we did not make any claims about the survival of iPSC-derived mutant ALS and control motor neurons under basal conditions measured using the LDH assay. However, we did report a survival difference when using the LDH release assay in the presence of a stressor [Fig. 5C in (2)] and also when using real-time single-cell longitudinal survival analysis under basal conditions [Fig. 5B in (2)]. We would like to clarify this issue as well as discuss the value of using complementary survival analysis methods.
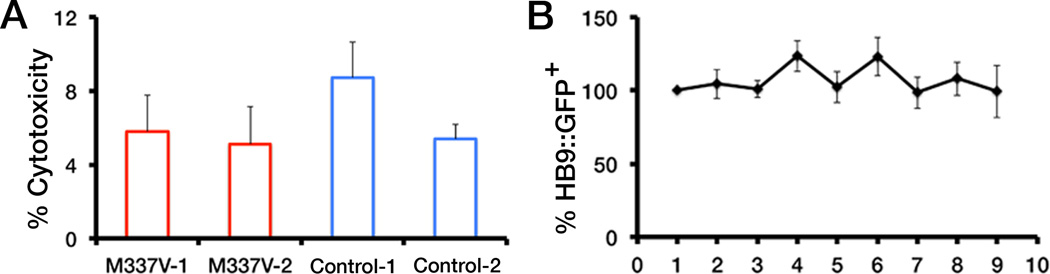
Specifically, in our study we aimed to address two issues regarding the effect of the M337V TDP-43 mutation on motor neuron survival (2). To address whether the M337V mutation caused an inherent neuronal vulnerability under basal conditions we used real-time single-cell longitudinal image analysis in which only the neurons that expressed a fluorescent reporter construct on the first day of the assay were followed serially and monitored for survival over 10 days [Fig. 5B in (2)]. Next, we investigated at the population level whether the TDP-43 mutation conferred a selective sensitivity to a stressor, which was assessed by a fluorometric cytotoxicity assay that measured LDH enzyme released into the culture medium by neurons that had lost membrane integrity [Fig. 5C in (2)]. Given the dynamic nature of iPSC-derived neuronal populations, a certain level of cell death and limited proliferation of residual neural progenitors are expected and this background activity can potentially mask subtle differences in survival. In fact, a breakdown of our control LDH release assay data (Fig. 1A, this letter), also does not reveal a difference in survival between M337V mutant or control iPSC-derived motor neurons under basal conditions, which is in agreement with Egawa et al.’s results [Fig. S11C in (1)].
Fig. 1. Survival of ALS mutant versus control iPSC-derived motor neurons.
(A) Comparison of cell death in motor neuronal cultures derived from two clones of mutant M337V TDP-43 iPSCs from one ALS patient (M337V-1 and M337V-2) and two control iPSC lines (from two different individuals; Control-1 and Control-2) under basal conditions using an LDH release assay (2). LDH release into the culture medium was normalized to total LDH after cell lysis for each well to determine percent cytotoxicity. Values are mean ± SEM. Data were analyzed by one-way ANOVA. There were no significant differences between the mean survival for motor neurons derived from mutant M337V TDP-43 iPSCs versus control iPSCs (n=4). (B) Cell survival of two independent control iPSC-derived motor neuronal cultures transfected with a HB9::GFP reporter construct. Cell survival was measured by counting the number of GFP-positive neurons over the course of nine days (2). The cell count on day 1 for each experiment was set to 100% and each time point after that was expressed as a percent of the value at day 1. Values are mean ± SEM, n=4. There were no significant differences in mean survival between day 1 and day 9.
The challenge of elucidating subtle but significant phenotypes in long-term neuronal cultures requires the application of multiple complementary readouts. Real-time single-cell longitudinal survival analysis using fluorescent reporter genes has enabled determination of differences in neuronal survival that may otherwise have been missed using conventional population-based assays such as the LDH release assay (3, 4). Another method to determine survival differences between different populations is to count cells that are positive for a particular reporter/marker at discrete time-points. Survival analysis performed by fluorescent reporter-based “snapshot” cell counts at fixed time points can also present challenges of interpretation because upon transient transfection fluorescent reporters can be expressed stochastically over the time course of the experiment. To test this possibility, we counted daily for nine days the number of green fluorescent protein (GFP)-positive cultured neurons derived from control iPSCs transfected with a motor neuron-specific HB9::GFP reporter construct. We used this approach instead of performing real-time survival analysis in which we only followed individual neurons that were GFP-positive on day 1 and recorded their time of death [Fig. 5B in (2)]. Our analysis revealed that the total number of GFP-positive motor neurons fluctuated over the course of nine days under basal conditions and did not differ significantly from day 1 (Fig. 1B; this letter). However, real-time single-cell longitudinal survival analysis using the same experimental setup did reveal an increased risk of neuronal death in control iPSC-derived motor neurons over time [Fig. 5B in (2)].
Rapid developments in iPSC technology now enable the comparison of cellular phenotypes between iPSC lines carrying a mutation versus control lines that do not. Real-time single-cell longitudinal survival analysis is a sensitive method for detecting phenotypes that may otherwise be masked due to variability arising from static measurements. The utility of this approach has recently been confirmed in Huntington’s disease patient-derived iPSC neuronal cultures (5). This study revealed a higher cumulative risk of death in neurons derived from iPSCs with CAG-repeat expansions compared to control iPSCs under basal conditions (5). We agree with Egawa et al. regarding the importance of independent multiple clonal line-based confirmation of phenotypic differences identified for any given disease mutation. Indeed, such studies would also benefit from the use of complementary sensitive assays to identify potentially important survival phenotypes.
This is a commentary on article Egawa N, Kitaoka S, Tsukita K, Naitoh M, Takahashi K, Yamamoto T, Adachi F, Kondo T, Okita K, Asaka I, Aoi T, Watanabe A, Yamada Y, Morizane A, Takahashi J, Ayaki T, Ito H, Yoshikawa K, Yamawaki S, Suzuki S, Watanabe D, Hioki H, Kaneko T, Makioka K, Okamoto K, Takuma H, Tamaoka A, Hasegawa K, Nonaka T, Hasegawa M, Kawata A, Yoshida M, Nakahata T, Takahashi R, Marchetto MC, Gage FH, Yamanaka S, Inoue H.Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci Transl Med. 2012;4(145):145ra104.
References
- 1.Egawa N, Kitaoka S, Tsukita K, Naitoh M, Takahashi K, Yamamoto T, Adachi F, Kondo T, Okita K, Asaka I, Aoi T, Watanabe A, Yamada Y, Morizane A, Takahashi J, Ayaki T, Ito H, Yoshikawa K, Yamawaki S, Suzuki S, Watanabe D, Hioki H, Kaneko T, Makioka K, Okamoto K, Takuma H, Tamaoka A, Hasegawa K, Nonaka T, Hasegawa M, Kawata A, Yoshida M, Nakahata T, Takahashi R, Marchetto MCN, Gage FH, Yamanaka S, Inoue H. Drug Screening for ALS Using Patient-Specific Induced Pluripotent Stem Cells. Sci Transl Med. 2012;4:145ra104–145ra104. doi: 10.1126/scitranslmed.3004052. [DOI] [PubMed] [Google Scholar]
- 2.Bilican B, Serio A, Barmada SJ, Nishimura AL, Sullivan GJ, Carrasco M, Phatnani HP, Puddifoot CA, Story D, Fletcher J, Park I-H, Friedman BA, Daley GQ, Wyllie DJA, Hardingham GE, Wilmut I, Finkbeiner S, Maniatis T, Shaw CE, Chandran S. Mutant induced pluripotent stem cell lines recapitulate aspects of TDP-43 proteinopathies and reveal cell-specific vulnerability. Proceedings of the National Academy of Sciences. 2012;109:5803–5808. doi: 10.1073/pnas.1202922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrasate M, Finkbeiner S. Automated microscope system for determining factors that predict neuronal fate. Proc Natl Acad Sci USA. 2005;102:3840–3845. doi: 10.1073/pnas.0409777102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 5.The HD iPSC Consortium. Induced Pluripotent Stem Cells from Patients with Huntington's Disease Show CAG-Repeat-Expansion-Associated Phenotypes. Cell Stem Cell. 2012;11:264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]