Abstract
Molecular printing techniques, which involve the direct transfer of molecules to a substrate with submicrometre resolution, have been extensively developed over the past decade and have enabled many applications. Arrays of features on this scale have been used to direct materials assembly, in nanoelectronics, and as tools for genetic analysis and disease detection. The past decade has witnessed the maturation of molecular printing led by two synergistic technologies: dip-pen nanolithography and soft lithography. Both are characterized by material and substrate flexibility, but dip-pen nanolithography has unlimited pattern design whereas soft lithography has limited pattern flexibility but is low in cost and has high throughput. Advances in DPN tip arrays and inking methods have increased the throughput and enabled applications such as multiplexed arrays. A new approach to molecular printing, polymer-pen lithography, achieves low-cost, high-throughput and pattern flexibility. This Perspective discusses the evolution and future directions of molecular printing.
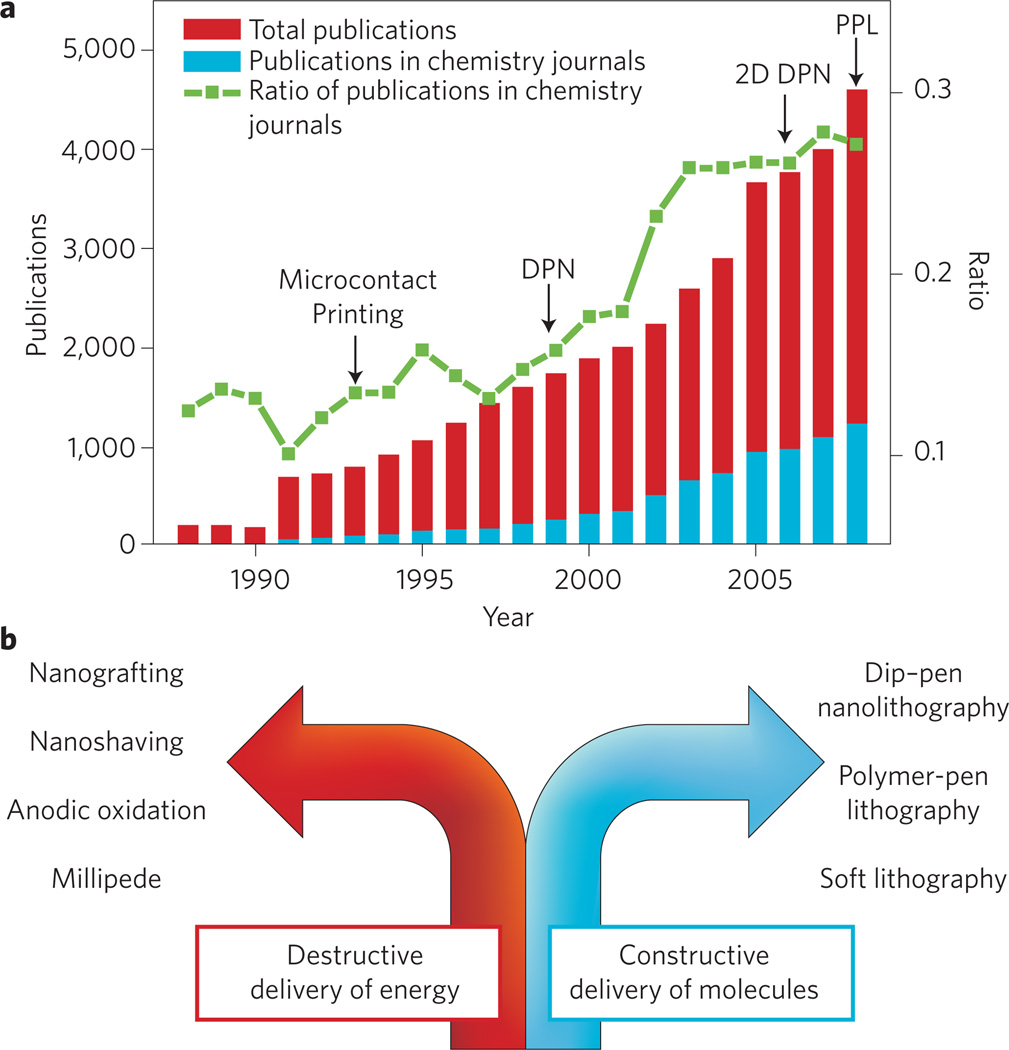
The past decade has witnessed the genesis and evolution of printing technologies capable of patterning surfaces with features smaller than 100 nm. These capabilities are a result of simultaneous advances in physics, chemistry, materials science and nanotechnology. The development of tools for reducing feature size is motivated primarily by (1) the semiconductor industry’s desire to continue increasing the number of transistors in a given area; (2) the central dogma of nanotechnology, which states that as feature sizes approach the nanoscale, new properties emerge that are not observed in bulk materials; and (3) biological studies and applications made possible by high-density bioarrays. As feature sizes approach the single-molecule limit, molecular transport, assembly and intermolecular interactions become dominant considerations and have shifted the science of patterning to the chemists’ domain (Fig. 1a).
Figure 1. Nanoscale printing.
a, The number of papers with the terms ‘nanolithography’, ‘printing’ and ‘patterning’ in the title published each year since 1988 (searched using Thomson Scientific Web of Knowledge). Red columns correspond to the total number of papers with any of the terms ‘nanolithography’, ‘printing’ and ‘patterning’ in their titles published in all science and engineering journals. Blue columns indicate the number of papers with the terms ‘nanolithograhpy’, ‘printing’ and ‘patterning’ in their titles published in chemistry journals. The green line shows that the ratio of those papers published in chemistry journals is increasing yearly. b, Diagram dividing molecular printing by constructive and destructive methods.
Solutions to the problem of nanoscale patterning being explored have traditionally included destructive, radiative techniques such as extreme UV lithography, soft X-ray lithography, electron-beam lithography and focused ion-beam writing1–2, as well as methods such as nanoimprint lithography3 and certain types of scanning-probe lithography (SPL)4. As nanopatterning techniques have become increasingly important to chemists, materials scientists and biologists, molecular printing capabilities, defined as processes where molecules or materials are directly transferred to a substrate of interest in the form of submicrometre features with at least one dimension on the molecular scale, have also become increasingly important. In this regard, two technologies — soft lithography5,6 and dip-pen nanolithography (DPN)7,8 — have emerged as the most widely used molecular printing methods for forming features on a wide variety of substrates. This perspective discusses the history and applications of molecular printing, focusing primarily on DPN and soft lithography as the first and most widely used techniques, and the recent development of polymer-pen lithography (PPL), a molecular printing method that combines the advantages of soft lithography and DPN into a single lithographic platform9.
Emergence of dip-pen nanolithography
In the early 1980s, new methods for shrinking feature sizes of patterns on silicon substrates were being actively pursued by the semiconductor industry as it became apparent that photolithography could not continue to indefinitely reduce the sizes of features as predicted by Moore’s Law and desired by electronics manufacturers. The brisk rate of feature-size reduction in CMOS devices demanded new ways to address and image these nanoscale structures as the existing technologies were no longer compatible with the smaller features. During this period, scanning probe microscopes (SPMs), specifically the scanning tunnelling microscope (STM)10 and atomic force microscope (AFM)11, emerged as imaging and spectroscopic tools that probe the topology of surfaces using nanosized tips that raster across them by piezoelectric actuation. SPMs have revolutionized surface science and even enabled the manipulation of surfaces at the single-atom level12,13. For the development of the SPM, Binnig and Rohrer were awarded the Nobel Prize in Physics in 1986.
Immediately following the invention of SPMs, researchers began using these machines for patterning surfaces, marking the birth of SPL14. A major milestone in SPL development occurred when Eigler and co-workers used an STM to pattern individual atoms on a surface, suggesting that SPMs could indeed be used for molecular printing or perhaps even manufacturing14. In this experiment14, the researchers repositioned individual Xe atoms on a single-crystal Ni surface to form the letters ‘IBM’ using an STM tip at 4 K. At this temperature, the van der Waals forces and electrostatic interactions between the atoms and the surface are greater than the interactions between the tip and the atom, thereby keeping the atoms anchored to the surface while they are dragged to their intended sites with the tip. Subsequently, the IBM group showed that this patterning technique could be used to study fundamental surface properties as a result of the atomic resolution afforded by the STM15. By arranging 48 Fe atoms in a circular structure on a Cu(111) surface, they were able to observe standing electron waves within the circular corral at 4 K. STM repositioning was extended to molecules as well, and a molecular counting device was fabricated by sequentially moving a row of 10 fullerenes on a Cu(111) surface16.
Although these impressive studies demonstrate the potential of SPMs as patterning tools, they also emphasize their limitations and impracticality. Specifically, this approach (1) requires controlled environments and low temperatures; (2) is painstakingly slow and indirect in nature, as it requires the picking up and subsequent movement of atoms; and (3) is not easily scaled. However, the work by Eigler, no matter how impractical, demonstrated the ultimate resolution of SPL techniques — atom-by-atom construction of nanopatterns — and established a challenge for the research community to develop rapid ways of printing atom- and molecule-based structures on a surface with nanoscale resolution.
For the next decade, scientists followed the model of the semiconductor industry and developed a series of indirect SPL methods, which focus on the delivery of energy rather than molecules to a surface to create functional patterns, typically with the aid of a resist material (Fig. 1b). Taking advantage of the registration enabled by piezo-actuation and the nanoscale radii of the tips, scientists were able to generate sub-50-nm features by scratching, etching and oxidizing surfaces5. For example, dynamic plough lithography can be used to scratch polymer-coated silicon surfaces, which can be subsequently processed using conventional silicon wet etching17. A process known as nanoshaving or nanografting uses the tip of an AFM and an applied force to remove a molecular monolayer on gold in a site-specific fashion18. Anodic oxidation of silicon was developed by Quate for patterning silicon substrates19,20. Sagiv et al. pioneered a related approach, which uses an applied bias between a conductive AFM tip and an n-octadecyltrichlorosilane monolayer, to electrochemically convert methyl groups to acids21. The reactive acid groups are used for subsequent chemistry and molecular immobilization. Such techniques have important applications but are limited by their indirect nature and challenges associated with parallelization.
A problem in developing any SPL or scanning probe imaging technique that functions in air, is dealing with the consequences of the capillary effect and meniscus formation. These factors convolute measurements taken with scanning probe instruments, and therefore many scientists rely on ultrahigh vacuum for their experiments. Surprisingly, the controlled direct transfer of material to a surface with nanoscale precision was not realized until 1999 when alkanethiols were deposited on gold substrates, with the use of humidity and the meniscus to facilitate transport and generate stable chemisorbed nanostructures, marking the invention of DPN22. Interestingly, a few years earlier, others had attempted what seemed to be a nearly identical experiment and concluded that transport did not occur23. The stronger interactions between the thiols and gold at room temperature, compared with the weak interactions between the tip and the ink, were key to forming stable patterns on the surface. The versatility of this method, using direct molecular transport from a tip to a surface to form patterns, was immediately apparent, and in the decade since its invention, DPN has been used to generate structures made of organometallic molecules24, polymers25, DNA26, proteins27,28, peptides29 and affinity templates, which allow for the subsequent immobilization of viruses30,31, colloidal nanoparticles32,33, metal ions34 and single-walled carbon nanotubes35 on many types of surfaces. Furthermore, DPN has enabled fundamental studies related to molecular transport36, and the fabrication of technological tools such as photomasks37, gas sensors38 and biological screening devices, including an assay for human immunodeficiency HIV-1 virus p24 antigen in serum samples39 and gene chips40.
DPN relies on the spontaneous formation of a meniscus between the tip and surface, which serves as a conduit for ink transport. The deposition rate is a function of tip–substrate contact time, the ink diffusion coefficient, and the ink coverage on the pen. Under appropriate conditions, feature sizes can be controlled on the sub-50-nm to many-micrometre length scale7,8. Patterns are formed by moving the tip across the surface at a controlled velocity, and piezo-actuation on three axes provides near-perfect registration. Because the piezoactuators are computer controlled, arbitrary patterns can be formed by the movement of the tip, which cannot be done by soft lithography, where the pattern is predetermined by a mould. By increasing the rastering speed of the tip across the surface, the transport of molecules is halted, and the same probe tip that is used for writing, acts instead as an imaging tool rather than a pen to provide feedback on the pattern quality. Finally, materials that are resistant to transport in a conventional DPN experiment can be dispersed in a hydrophilic or lipophilic carrier matrix that assists their movement through the meniscus41. Importantly, there is no need to expose the substrate to harsh ultraviolet, ion- or electron-beam radiation, characteristic of indirect patterning techniques, and therefore DPN can be used to print fragile or reactive organic and biological materials.
These advantages of DPN are responsible for its rapid development and dissemination over the past decade, however, in the early stages of its development, DPN faced many of the same drawbacks as surface-destructive indirect SPLs, primarily poor throughput, a concern that soon would become a major focus of DPN research.
High-throughput molecular printing with soft lithography
Soft lithography is a complementary molecular printing technique that overcomes the throughput problem associated with the early incarnations of DPN5,6. Specifically, microcontact printing (µCp), a form of soft lithography, uses an elastomer stamp, typically fabricated by conventional lithographic methods, to directly transfer materials to a surface of interest42. The µCp approach has flourished as a research-grade molecular printing technology because of its ease of use, low cost, and high throughput. It has become the poor man’s replacement for photolithography, allowing researchers in many fields to use micro- and nanofabrication. In µCp, elastomeric stamps are prepared by pouring an elastomer, including poly(dimethylsiloxane) (PDMS), polyurethanes, polyimides and natural polymers such as agarose, in a mould prepared by conventional photolithographic methods. The polymer is cured in the mould, thereby generating a relief structure in the elastomeric stamp. The stamp can then be used to pattern proteins, DNA, cells, alkanethiols, silanes, colloids and salts on a variety of flat as well as curved surfaces5,6. The type of elastomer used to form the stamps affects their surface properties, leading to many interesting systems. For example, biofilms (cells) were grown on an agarose stamp and could be repeatedly transferred to a surface as the films regenerated spontaneously on the stamp43.
The µCp technique, however, has several limitations. Although the stamps can print over large areas, they can only deposit a single pattern that is predetermined by the stamp. Therefore, a new mould must be fabricated each time a new pattern is desired. Feature size control is affected and limited by stamp swelling and shrinking, both during curing and stamp inking. The mechanical properties of the stamps can also limit design flexibility. For example, the softness of the elastomer limits the aspect ratio, height/length (h/l), of the features. If the feature aspect ratio on the stamp is not between 0.2 and 2, bending of the stamp can cause pattern defects, and, for features that are too widely separated (>0h), sagging of the stamp causes defects. Indeed, the fabrication of alkanethiol features smaller than 150 nm is a significant challenge using conventional methods44. By using DPN to introduce chemical modifications on a flat PDMS stamp and by passivating the surrounding areas with a perfluorinated monolayer, we have helped overcome this deficiency, and shown that features as small as 80 nm could be easily prepared with aspect ratios as low as 0.01. This study underscores the synergistic aspects of soft lithography and DPN44.
Today, µCp is primarily used for low-cost micro- and nanofabrication of physical templates (raised inorganic structures generated by using the patterned molecules as etch resists), the generation of arrays of soft, sometimes living architectures (for example, cells), or the generation of chemical affinity templates, which can be used subsequently to direct assembly events. It does have limitations, some of which can be solved with complementary printing methods such as DPN, but for preparing multicomponent materials and devices such as bioarrays, a convenient strategy for massive multiplexing with soft lithography still does not exist.
Parallel DPN
In the early stages of its development, the writing speed and cost of DPN were often cited as reasons limiting wider adoption by the scientific community and commercialization by industry. For example, it was claimed that45 “The serial nature of SPL results in a low sample throughput … [SPL] is probably restricted to research applications” and others noted that6 “… SPL patterning is expensive and difficult to adapt to large-scale patterns”. As a result, DPN research turned to increasing throughput, while maintaining its core attributes (Fig. 2). Solutions to this issue were suggested by two important studies that used parallel tip arrays. The Quate group had fabricated linear19 and 2D active20 arrays to perform anodic oxidation of silicon surfaces, and IBM fabricated 2D arrays for ultrahigh-density data-storage applications46. However, these designs were not suitable for molecular printing or massive parallelization, in part because of the demanding design requirements for electronically addressing each pen within the array. For DPN, both active and passive approaches to massive parallelization have been demonstrated. Initially, 1D arrays were prepared that increase throughput in DPN by a factor equal to the number of pens47. A 2D pen array required several innovations before affording flexibility and feature control. These include: (1) tips with low aspect ratio; (2) curved cantilevers; and (3) a straightforward alignment protocol used to bring all tips into contact with the surface simultaneously, thereby producing reproducible features over large areas48. As a result, massively parallel, 55,000-pen, 2D arrays were developed that are capable of molecular printing over square-centimetre areas with sub-100-nm resolution. This innovation increases the throughput of DPN (Fig. 3) by four orders of magnitude, and as many as 88,000,000 dots were fabricated in approximately 5 min (ref. 48). So far, DPN arrays with as many as 1.3 million cantilevers have been microfabricated.
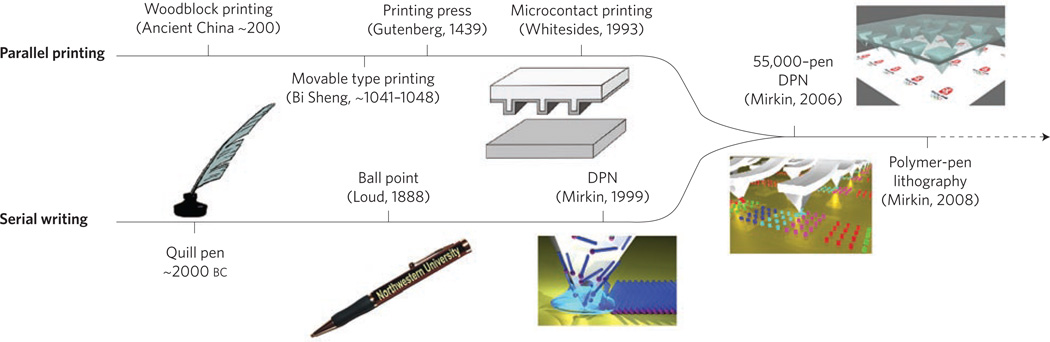
Figure 2. Timeline of the historical development of molecular printing or direct ink transfer to a surface.
Historically, methods of transferring ink directly to a substrate have evolved along two separate paths59: serial writing and parallel printing. Approximately 2000 bc, the quill pen — a serial writing tool that can create patterns with high registration and high resolution — was invented. However, the low throughput and poor reproducibility of serial writing tools could not keep abreast with the rapid development of human society and the demand for knowledge, resulting in the evolution of parallel printing. With the invention of woodblock printing in ancient China (~200 ad), and later, a more sophisticated printing press in Europe (1439) by Gutenberg, parallel printing emerged as an art that rapidly and reproducibly forms patterns. Serial writing and parallel printing complemented each other to help people communicate, record information and create art. A similar, dramatic transition is currently underway in the field of nanoscale printing. Microcontact printing — a parallel nanoprinting technique — provides high-throughput, large-area patterning; however, µCp can only duplicate patterns that are predefined by a template and thus cannot arbitrarily generate different patterns. In contrast, DPN — a serial writing technique — creates patterns with high resolution and registration but is limited in throughput. Recently, the two branches of nanoscale printing merged with the invention of massively parallel DPN and subsequently PPL. PPL combines many advantages of µCp with the attributes of DPN to provide high resolution, registration and throughput, soft-matter compatibility and affordability.
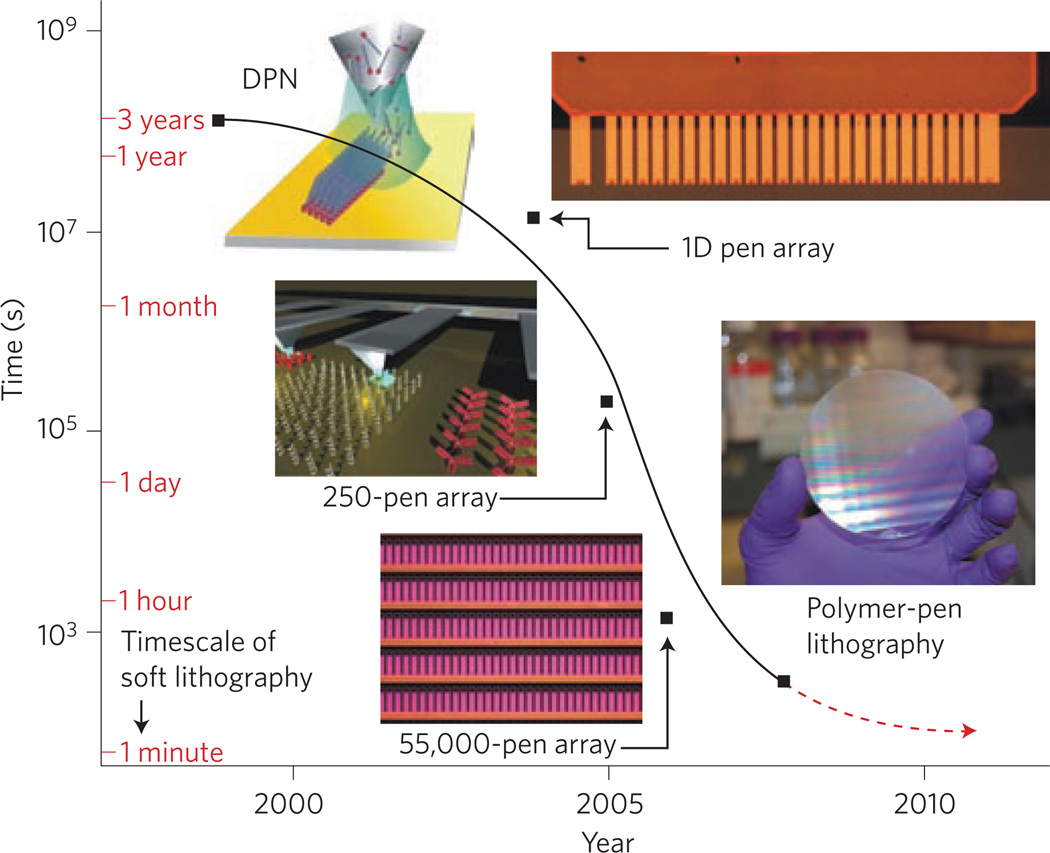
Figure 3. Advances in DPN throughput.
The time needed to pattern 1 cm2 with 1 billion 100-nm dot features. Using single-pen DPN, this process requires several years, which renders single-pen DPN unsuitable for commercial lithographic applications. The parallelization of DPN has increased throughput by several orders of magnitude over the past decade as a result of 1D and 2D pen arrays. PPL can be used to print a billion nanoscale features over 1 cm2 of substrate in 30 minutes. To complete the same task, µCp requires less than 1 minute. However, to fabricate a different pattern, soft lithography requires more than 1 day to produce a new mould and stamp, a process that is not required for parallel DPN and PPL. Figure reprinted with permission from ref. 9; © 2008 AAAS.
Multiplexed DPN
Massive parallelization of DPN opens the possibility of fabricating highly miniaturized, combinatorial libraries of nanostructures. Before molecular printing, these libraries have been the domain of microfabrication techniques such as photolithography, ink-jet printing, and robotic spotting. Microarrays of biological structures in particular have created many opportunities for studying cellular genetics, and have resulted in the development of rapid disease-screening assays49. By shrinking the size of these structures to the nanoscale, one could literally print on the scale of biology. This reduces the number of features per unit area and also allows single-particle structures such as proteins, viruses and cells to be manipulated. However, a challenge that must be overcome before multiplexed nanoarrays are commonplace is inking the tips within a pen array with different molecules.
Massively parallel DPN is opening the door to the routine creation of combinatorial libraries of molecule-based nanostructures. Commercial DNA arrays prepared by photolithography, ink-jet printing and spotting are currently limited in feature size to 1 µm, and more typically in the tens of micrometres range. The µCp techniques are not currently realistic for generating multicomponent structures on this scale because of a lack of registration during pattern alignment. Nanopippetting is a technique that, as a result of the nanopippette design, is capable of depositing two inks onto a substrate with nanometre resolution, but the number of inks has not been increased beyond two, which would create massively multiplexed patterns50.
The first DPN patterning of two different inks, 16-mercaptohexadecanoic acid (MHA) and 1-octadecanethiol (ODT), individually coated on two tips, relied on a serial ink-deposition process; this approach demonstrated the extraordinary registration capabilities of DPN but was not suitable for massive multiplexing51. Later, microscopic inkwells that could be filled through integrated microfluidic channels were developed, allowing up to eight different inks to be deposited simultaneously52. Inkwells, however, are not a route to massive multiplexing because with current designs a chip containing 55,000 inkwells would require a microfabricated area larger than 0.5 m2 just to accommodate the area occupied by the ink reservoirs. Our group has developed an inkjet methodology to individually coat pens within 1D and 2D pen arrays with chemically distinct materials41. The hydrophobicity of the tips, which was modified before inking, confines each droplet to a single tip, thereby avoiding cross contamination of inks on adjacent cantilevers separated by only 90 µm. Pen arrays inked in this fashion were used to form a multiplexed lipid array with sub-100-nm feature sizes. The approach is extendable to virtually any material that can be delivered via an ink-jet printer. As a result, DPN has taken a transformative step towards becoming a general nanofabrication tool that combines high throughput, high resolution and multiplexing capabilities. Although the multiplexing challenge has been initially addressed, commercialization and widespread adoption will lag until the cost of massively parallel DPN is reduced.
Polymer-pen lithography
Polymer-pen lithography is a new scanning-probe contact-printing method that promises to reduce the cost barriers associated with massively parallel DPN by combining the advantages of µCp and DPN to achieve high-throughput, flexible molecular printing that may open many new possibilities in the field of molecular printing and nanopatterning9. The PPL approach is cantilever-free, and uses elastomeric tips53 to print a digitized pattern, with spot sizes ranging from 90 nm to over 10 µm (Fig. 4), simply by changing the force and time over which ink is delivered. This feature-size dependence on force is a remarkably controllable parameter and distinguishes PPL from both DPN and conventional contact printing. A typical polymer-pen array contains many pyramidal elastomeric tips — from thousands to as many as 108 — which are made with a master prepared by conventional photolithography and subsequent chemical etching. Once the master has been prepared, the materials cost of each pen array is less than $1. When sharp tips are brought into contact with the substrate, ink is delivered at the points of contact. PPL can pattern features ranging in size from 100 µm to as small as 80 nm. Because the elastomeric tip array absorbs ink and acts as a reservoir, the same stamp can be used to print over large areas and for multiple printing tasks without re-inking.
Figure 4. Polymer-pen lithography.
a, Schematic of a PPL pen array. The array is glued to a piezoelectric scanner that is software-controlled. When the tips of the polymer pens are brought in contact with a substrate, ink is delivered at the points of contact. b, Three-dimensional representation demonstrates how the massively parallel nature of the pen array results in replication of a digitized pattern into nanoscale features over large lengthscales. c, Optical image of a representative region of ~15,000 miniaturized duplicates of the 2008 Beijing Olympic logo made by PPL. d, A magnified scanning electron microscope image of the logo made by PPL. The letters and numbers ‘Beijing 2008’ were generated from ~20,000 90-nm dots, whereas the picture and Olympic rings were made from ~4,000 600-nm dots. e, A representative optical microscope image of 100 gold circuits in 1 cm2 fabricated in a single writing stage by PPL, with features ranging from 100 µm to 500 nm. The inset shows a magnified SEM image of the circuit centre (red dashed square). f, A fluorescence microscopy image of an anti-mouse IgG array fabricated by PPL. Parts a, c, e and f reprinted with permission from ref. 9, © 2008 AAAS.
The PPL technique promises to make molecular printing broadly accessible to scientists who are not necessarily scanning-probe aficionados, such as chemists, biologists and materials scientists. This technique maintains many of the attributes of SPL — namely arbitrary pattern design, material flexibility and controllable feature size — while eliminating many of the drawbacks associated with other SPL methods. Already, PPL has proved effective for patterning small molecules and proteins and has been used to make patterns of electronic circuits9. Circuit patterns with feature sizes ranging from 100 µm to 500 nm were patterned in a PPL experiment by depositing MHA on a gold surface followed by etching away the gold where the MHA was absent. The resulting architectures, with features ranging over three orders of magnitude in size, were produced in a single writing step on centimetre-squared areas. This high throughput would be impossible to repeat by conventional DPN or electron-beam lithography, and this approach avoids the problems associated with mask production and processing common to photolithographic approaches.
Furthermore, protein nanoarrays have been generated by PPL. Tetramethylrhodamine 5-(and 6)-isothiocyanate (TRITC) conjugated anti-mouse IgG arrays were generated on a Codelink glass slide (GE Healthcare) by PPL. After washing, reproducible features were observed by fluorescence microscopy over centimetre-squared areas, demonstrating that PPL can also be used in biological investigations. In the future, we expect that PPL will be used to form combinatorial inorganic and biological nanoarrays for site-directed surface synthesis, and for many other applications in which electron- beam lithography or DPN are commonly used.
Summary and outlook
The past decade has witnessed the rapid development and dissemination of two complementary molecular printing tools: soft lithography and DPN. PPL is a recently developed technique that combines the pattern flexibility afforded by DPN and the low cost and high throughput inherent to soft lithography. DPN and PPL provide a flexible approach to patterning a variety of materials, but they are also platforms for studying the relationship between nanoscale molecular architectures and the chemical, material and biological properties of such architectures. These abilities arise directly from the feature shape and size control afforded by these methods as well as the ability to control the environment during deposition, which enables one to work with both soft and hard matter. As a result, DPN studies have provided fundamental insight into: (1) adsorbate self-assembly on surfaces36; (2) phase separation within multicomponent monolayers54; (3) nucleation and growth of biopolymers55; (4) virus cell infectivity at the single-cell and single-particle level56, cell–surface interactions28, nanoscale surface chemical transformations57, and the gas-diffusion characteristics of nanoscale sol–gel gas-sensor elements58. These patterning techniques have also led to important technological advances, including miniaturized oligonucleotiode and protein arrays, bio- and gas sensors, novel ways of encoding surfaces and storing information, ways of making master structures that can be used for replication, and molecule-based and nanoscale circuitry.
Although advances have been rapid and span many fields, several aspects of molecular printing continue to challenge researchers. In the case of DPN and PPL, individual tip actuation over large areas, feature size reduction to the single-molecule level (for example, individual proteins), and translation of the tips over large distances must be addressed before the full potential of this technology is realized. Additionally, the use of these techniques as tools for rapid and parallel site-specific chemical transformations is an emerging approach that could transform the manufacturing of gene chips by providing a route to site-specific combinatorial synthesis. Regardless of these challenges, molecular printing methods — µCp, DPN and PPL — have become affordable, reliable, technologies available to and extensively used by researchers throughout the world.
Acknowledgements
C.A.M. acknowledges AFOSR, NCI-CCNE, DARPA-SPAWAR, and the NSF-NSEC for support of this work. A.B.B. is grateful for an NIH Postdoctoral Fellowship.
References
- 1.Pease RFW. Nanolithography and its prospects as a manufacturing technology. J. Vac. Sci. Technol. B. 1992;10:278–285. [Google Scholar]
- 2.Rai-Choudhury P, editor. Handbook of Microlithography, Micromachining and Microfabrication. Vol. 1. SPIE Press; 1997. [Google Scholar]
- 3.Chou SY, Krauss PR, Renstrom PJ. Imprint lithography with 25-nanometer resolution. Science. 1996;272:85–87. [Google Scholar]
- 4.Wouters D, Hoeppener S, Schubert US. Local probe oxidation of self-assembled monolayers: Templates for the assembly of functional nanostructures. Angew. Chem. Int. Ed. 2009;48:1732–1739. doi: 10.1002/anie.200801013. [DOI] [PubMed] [Google Scholar]
- 5.Xia Y, Whitesides GM. Soft lithography. Angew. Chem. Int. Ed. 1998;37:550–575. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 6.Gates BD, et al. New approaches to nanofabrication: Molding, printing, and other techniques. Chem. Rev. 2005;105:1171–1196. doi: 10.1021/cr030076o. [DOI] [PubMed] [Google Scholar]
- 7.Ginger DS, Zhang H, Mirkin CA. The evolution of dip-pen nanolithography. Angew. Chem. Int. Ed. 2004;43:30–35. doi: 10.1002/anie.200300608. [DOI] [PubMed] [Google Scholar]
- 8.Salaita K, Wang Y, Mirkin CA. Applications of dip-pen nanolithography. Nature Nanotech. 2007;2:145–155. doi: 10.1038/nnano.2007.39. [DOI] [PubMed] [Google Scholar]
- 9.Huo F, et al. Polymer pen lithography. Science. 2008;321:1658–1660. doi: 10.1126/science.1162193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binnig G, Rohrer G, Gerber Ch, Weibel E. Surface studies by scanning tunneling microscopy. Phys. Rev. Lett. 1982;49:57–61. [Google Scholar]
- 11.Binnig G, Quate CF, Rohrer Ch. Atomic force microscope. Phys. Rev. Lett. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 12.Mccarty GS, Weiss PS. Scanning probe studies of single nanostructures. Chem. Rev. 1999;99:1983–1990. doi: 10.1021/cr970110x. [DOI] [PubMed] [Google Scholar]
- 13.Gimzewski JK, Joachim C. Nanoscale science of single molecules using local probes. Science. 1999;283:1683–1688. doi: 10.1126/science.283.5408.1683. [DOI] [PubMed] [Google Scholar]
- 14.Eigler DM, Schweizer EK. Positioning single atoms with a scanning tunneling microscope. Nature. 1990;344:524–526. [Google Scholar]
- 15.Crommie MF, Lutz CP, Eigler DM. Confinement of electrons to quantum corrals on a metal surface. Science. 1993;262:218–220. doi: 10.1126/science.262.5131.218. [DOI] [PubMed] [Google Scholar]
- 16.Cuberes MT, Schlittler RR, Gimzewski JK. Room-temperature repositioning of individual C60 molecules at Cu steps: Operation of a molecular counting device. Appl. Phys. Lett. 1996;69:3016–3018. [Google Scholar]
- 17.Heyde M, et al. Dynamic plowing nanolithography on polymethylmethacrylate using an atomic force microscope. Rev. Sci. Instrum. 1992;72:136–141. [Google Scholar]
- 18.Liu J-F, Cruchon-Dupeyrat S, Garno JC, Frommer J, Liu G-Y. Three-dimensional nanostructure construction via nanografting: Positive and negative pattern transfer. Nano Lett. 2002;2:937–940. [Google Scholar]
- 19.Minne SC, Manalis SR, Atalar A, Quate CF. Independent parallel lithography using the atomic force microscope. J. Vac. Sci. Tech. B. 1996;14:2456–2461. [Google Scholar]
- 20.Minne SC, et al. Centimeter scale atomic force microscope imaging and lithography. Appl. Phys. Lett. 1998;73:1742–1744. [Google Scholar]
- 21.Moaz R, Cohen SR, Sagiv J. Nanoelectrochemical patterning of monolayer surfaces: Toward spatially defined self-assembly of nanostructures. Adv. Mater. 1999;11:55–61. [Google Scholar]
- 22.Piner RD, et al. Dip-pen nanolithography. Science. 1999;283:661–663. doi: 10.1126/science.283.5402.661. [DOI] [PubMed] [Google Scholar]
- 23.Jaschke M, Butt H-J. Deposition of organic material by the tip of a scanning force microscope. Langmuir. 1995;11:1061–1064. [Google Scholar]
- 24.Ivanisevic A, et al. Redox-controlled orthogonal assembly of charged nanostructures. J. Am. Chem. Soc. 2001;123:12424–12425. doi: 10.1021/ja011933d. [DOI] [PubMed] [Google Scholar]
- 25.Nyamjav D, Ivanisevic A. Properties of polyelectrolyte templates generated by dip-pen nanolithography and microcontact printing. Chem. Mater. 2004;16:5216–5219. [Google Scholar]
- 26.Demers LM, Ginger DS, Park SJ, Li Z, Chung SW, Mirkin CA. Direct patterning of modified oligonucleotides on metals and insulators by dippen nanolithography. Science. 2002;296:1836–1838. doi: 10.1126/science.1071480. [DOI] [PubMed] [Google Scholar]
- 27.Lee K-B, et al. Protein nanoarrays generated by dip-pen nanolithography. Science. 2002;295:1702–1705. doi: 10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- 28.Lee KB, Lim JH, Mirkin CA. Protein nanostructures formed via direct-write dip-pen nanolithography. J. Am. Chem. Soc. 2003;125:5588–5589. doi: 10.1021/ja034236p. [DOI] [PubMed] [Google Scholar]
- 29.Cho Y, Ivanisevic A. TAT peptide immobilization on gold surfaces: A comparison study with a thiolated peptide and alkylthiols using AFM, XPS and FT-IRRAS. J. Phys. Chem. B. 2005;109:6225–6232. doi: 10.1021/jp045731q. [DOI] [PubMed] [Google Scholar]
- 30.Smith JC, et al. Nanopatterning the chemospecific immobilization of cowpea mosaic virus capsid. Nano Lett. 2003;3:883–886. [Google Scholar]
- 31.Vega RA, et al. Nanoarrays of single virus particles. Angew. Chem. Int. Ed. 2005;44:6013–6015. doi: 10.1002/anie.200501978. [DOI] [PubMed] [Google Scholar]
- 32.Liu X, et al. Arras of magnetic nanoparticles patterned via “dip-pen nanolithography”. Adv. Mater. 2002;14:231–234. [Google Scholar]
- 33.Gundiah G, et al. Dip-pen nanolithography with magnetic Fe2O3 nanocrystals. Appl. Phys. Lett. 2004;84:5341–5343. [Google Scholar]
- 34.Ding L, Li Y, Chu HB, Li XM, Liu J. Creation of cadmium sulfide nanostructures using AFM dip-pen nanolithography. J. Phys. Chem. B. 2005;109:22337–22340. doi: 10.1021/jp053389r. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, et al. Controlling the shape, orientation, and linkage of carbon nanotube features with nano affinity templates. Proc. Natl Acad. Sci. USA. 2006;103:2026–2031. doi: 10.1073/pnas.0511022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giam LR, Wang Y, Mirkin CA. Nanoscale molecular transport: The case of dip-pen nanolithography. J. Phys. Chem. A. 2009;113:3779–3782. doi: 10.1021/jp809061e. [DOI] [PubMed] [Google Scholar]
- 37.Jang JW, et al. Dip-pen nanolithography generated metal photomasks. Small. 2009 doi: 10.1002/smll.200801837. [DOI] [PubMed] [Google Scholar]
- 38.Tang Q, Shi SQ. Preparation of gas sensors via dip-pen nanolithography. Sens. Actuat. B. 2008;131:379–383. [Google Scholar]
- 39.Lee KB, et al. The use of nanoarrays for highly sensitive and selective detection of human immunodeficiency virus type 1 in plasma. Nano Lett. 2004;4:1869–1872. [Google Scholar]
- 40.Yapici MK, Zou J. Dip pen nanolithography functionalized electrical gaps for multiplexed DNA detection. Anal. Chem. 2008;80:5899–5904. doi: 10.1021/ac800445p. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, et al. A self-correcting inking strategy for cantilever arrays addressed by an inkjet printer and used for dip-pen nanolithography. Small. 2008;4:1666–1670. doi: 10.1002/smll.200800770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar A, Whitesides GM. Features of gold having micrometer to centimeter dimensions can be formed through a combination of stamping with an elastomeric stamp and an alkanethiol “ink” followed by chemical etching. Appl. Phys. Lett. 1993;63:2002–2004. [Google Scholar]
- 43.Weibel DB, et al. Bacterial printing press that regenerates its ink: Contact-printing bacteria using hydrogel stamps. Langmuir. 2005;21:6436–6342. doi: 10.1021/la047173c. [DOI] [PubMed] [Google Scholar]
- 44.Zheng Z, Jang J-W, Zheng G, Mirkin CA. Topographically flat, chemically patterned PDMS stamps made by dip-pen nanolithography. Angew. Chem. Int. Ed. 2008;47:9951–9954. doi: 10.1002/anie.200803834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suh KY, Khademhosseini A, Jon S, Langer R. Direct confinement of individual viruses within polyethylene glycol (PEG) nanowells. Nano Lett. 2006;6:1196–1201. doi: 10.1021/nl060571a. [DOI] [PubMed] [Google Scholar]
- 46.Bettiger P, et al. The “Millipede”- nanotechnology entering data storage. IEEE Trans. Nanotechnol. 2002;1:39–55. [Google Scholar]
- 47.Salaita K, et al. Sub-100 nm centimeter scale, parallel dip-pen nanolithography. Small. 2005;1:940–945. doi: 10.1002/smll.200500202. [DOI] [PubMed] [Google Scholar]
- 48.Salaita K, et al. Massively parallel dip-pen nanolithography with 55,000 pen two-dimensional arrays. Angew. Chem. Int. Ed. 2006;45:7220–7223. doi: 10.1002/anie.200603142. [DOI] [PubMed] [Google Scholar]
- 49.Schulze A, Downward J. Navigating gene expression with microarrays — a technology review. Nature Cell Biol. 2001;3:E190–E195. doi: 10.1038/35087138. [DOI] [PubMed] [Google Scholar]
- 50.Rodolfa KT, Bruckbauer A, Zhou D, Korchev YE, Klenerman D. Two-component graded deposition of biomolecules with a double-barreled nanopipette. Angew. Chem. Int. Ed. 2005;44:6854–6859. doi: 10.1002/anie.200502338. [DOI] [PubMed] [Google Scholar]
- 51.Hong SH, Zhu J, Mirkin CA. Multiple ink nanolithography: Toward a multiple-pen nano-plotter. Science. 1999;286:523–525. doi: 10.1126/science.286.5439.523. [DOI] [PubMed] [Google Scholar]
- 52.Hong S, Mirkin CA. A nanoplotter with both parallel and serial writing capabilities. Science. 2000;288:1808–1811. doi: 10.1126/science.288.5472.1808. [DOI] [PubMed] [Google Scholar]
- 53.Hong JM, Ozkeskin FM, Zou J. A micromachined elastomeric tip array for contact printing with variable dot size and density. J. Micromech. Microeng. 2008;18:015003. [Google Scholar]
- 54.Salaita K, et al. Spontaneous “phase separation” of patterned binary alkanethiol mixtures. J. Am. Chem. Soc. 2005;127:11283–11287. doi: 10.1021/ja042393y. [DOI] [PubMed] [Google Scholar]
- 55.Liu X, et al. The controlled evolution of a polymer single crystal. Science. 2005;307:1763–1766. doi: 10.1126/science.1109487. [DOI] [PubMed] [Google Scholar]
- 56.Vega RA, et al. Monitoring single-cell infectivity from virus-particle nanoarrays fabricated by parallel dip-pen nanolithography. Small. 2007;9:1482–1485. doi: 10.1002/smll.200700244. [DOI] [PubMed] [Google Scholar]
- 57.Braunschweig AB, Senesi AJ, Mirkin CA. Redox activating dip-pen nanolithography (RA-DPN) J. Am. Chem. Soc. 2009;131:922–923. doi: 10.1021/ja809107n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Su M, Li S, Dravid V. Miniaturized chemical multiplexed sensor array. J. Am. Chem. Soc. 2003;125:9930–9931. doi: 10.1021/ja035727c. [DOI] [PubMed] [Google Scholar]
- 59.DeVinne TL. The Invention of Printing. Francis Hart & Co; 1969. [Google Scholar]