Abstract
The promise of point-of-care medical diagnostics — tests that can be carried out at the site of patient care — is enormous, bringing the benefits of fast and reliable testing and allowing rapid decisions on the course of treatment to be made. To this end, much innovation is occurring in technologies for use in biodiagnostic tests. Assays based on nanomaterials, for example, are now beginning to make the transition from the laboratory to the clinic. But the potential for such assays to become part of routine medical testing depends on many scientific factors, including sensitivity, selectivity and versatility, as well as technological, financial and policy factors.
Recent technological advances have markedly improved the way in which we study disease and point towards new opportunities for diagnosing disease. Researchers now have tools to observe phenomena at the level of the atom, to sequence entire genomes and to understand the molecular basis of disease. In addition, new materials, especially nanostructures, are providing novel ways of detecting markers of disease at low concentrations, in complex sample media (such as serum) and with a wide variety of assay read-outs. But many of the latest innovations are not yet being used in routine diagnostic testing, especially when point-of-care issues are considerable, for example when the cost of deploying an assay and training staff at the point of care is high. As biodiagnostic applications based on these new materials continue to be developed, it will be important to be conscious of the key factors that drive this process so that new tests are more likely to reach the clinic. In this Perspective, we assess the factors of assay sensitivity, selectivity and versatility, and robustness, cost and portability. We also discuss some of the materials that are allowing new assays to be designed and the consequences of developing such technologies.
Sensitivity
The diagnosis of a disease on the basis of the presence or concentration of certain biomolecules requires assays that can detect molecules of interest (or targets) sensitively. In this post-genomic era, the targets are most commonly nucleic acids or proteins. Researchers have developed two general strategies to achieve high sensitivity: target-based amplification and signal-based amplification. In target-based amplification, a recognition event triggers a catalytic process that generates more of the target being recognized or surrogates for this target. The polymerase chain reaction (PCR) is a classic example of target amplification, and modern PCR techniques can reliably detect the presence of just a few copies of a nucleic acid sequence1. By contrast, in signal-based amplification, a catalytic entity is often used to increase the signal that results from a single binding event. A typical example is the enzyme-linked immunosorbent assay (ELISA)2, in which a target protein can be captured by an antibody and then sandwiched with a second antibody that incorporates (or is associated with) a catalytic, signal-generating entity. Certain techniques that do not involve amplification, for example single-molecule spectroscopy techniques, might seem to be sensitive; however, these types of spectroscopy typically require greater than nanomolar concentrations of the molecule to be present in order to find and probe it. Therefore, such approaches are not typically viewed as high-sensitivity methods in the context of medical diagnostics.
Target-based amplification is a more sensitive strategy than signalbased amplification and is generally considered to be a superior approach, because generating an exponential increase in target concentration leads to faster assay kinetics and pushes the thermodynamics of the probe–target capture reaction in favour of bound (detectable) target. In the short term, it seems that PCR will continue to be a benchmark for nucleic acid detection. But the instability and variability that are inherent in enzymatic processes limit its application outside an institutional setting, such as a research facility or a large central clinical lab. Another drawback to PCR is that lengthy optimization procedures are often required if several targets are to be amplified and detected at the same time, a process known as multiplexing, which is a desirable feature in the clinic, especially as panel assays (which test many disease markers simultaneously) grow in importance for diagnosing disease.
In the past decade, new materials and assays have been developed for signal-based amplification and detection, and assay sensitivities (Table 1) are now approaching those of target-based amplification. Many of these advances rely on nanoscale materials, which have attractive properties for such assays: they have unique and controllable size-dependent properties, have tunable chemical compositions, and in certain cases are chemically and physically robust structures3,4. The tailorable properties of nanomaterials, including their high surface-to-volume ratios, mean that target-binding events are often more easily transduced into detectable signals. An example is polyvalent nanoparticles that consist of gold particles modified with biomolecules; these can be used as diagnostic probes5. In one assay system, when the target binds to the biomolecules, the associated gold particles catalyse the reduction of silver, leading to a marked enhancement of signal, which can be read with a device that measures light scattered from the developed silver spots. This ‘scanometric’ strategy allows the detection of attomolar (10−18) concentrations of nucleic acids and proteins in complex biological samples6. Indeed, the properties of DNA-functionalized gold nanoparticles (including their optical, catalytic and binding properties) have been taken advantage of in a variety of detection methodologies, including colorimetric7, fluorescent8–10, chemiluminescent11, scanometric5, surface-plasmon-resonance-based12 and Raman-spectroscopy-based13 strategies. For protein detection, certain assays using protein-modified gold nanoparticles have detection limits that are many orders of magnitude lower than those possible with a conventional technique, such as an ELISA6. Detection systems based on other nanomaterials have also been evaluated. For example, biomolecule-modified carbon nanotubes14,15 and silicon nanowires16 are systems in which changes in electrical conductance on target binding can be translated into a spectroscopic or electrical signal. These strategies take advantage of properties of the nanomaterial, for example their conductance, to cause a measurable change in electrical signal and have resulted in detection methods of moderately high sensitivity17 (Table 1), which are likely to improve after further refinement.
Table 1.
Detection of protein biomolecules
| Tool or technique | Read-out | Illustration | Detection limit* | Molecules per drop (60 μl) | In clinical use |
|---|---|---|---|---|---|
| Colorimetry | Visual |
|
150 pM | 109 | Yes |
| Carbon nanotubes | Electrical |
|
100 pM | 109 | No |
| Chemiluminescence | Luminescence |

|
30 pM | 109 | Yes |
| ELISA | Luminescence |
|
1–10 pM | 107 | Yes |
| Quantum dots | Fluorescence |

|
500 fM | 107 | No |
| Silicon nanowires | Electrical |
|
1 fM | 104 | No |
| Metal nanoparticles (bio-barcode) | Scanometric and/or light scattering |

|
30 aM | 900 | Yes |
| Immuno-PCR | Fluorescence |
|
20 aM | 700 | Yes |
Detection limits are best-case examples from the literature and can vary substantially depending on the target and the assay conditions.
The ability to detect targets more sensitively brings its own set of challenges. Consider the example of Alzheimer’s disease. At present, markers for Alzheimer’s disease, such as amyloid-β-derived diffusible ligands18, are being identified through histological studies of brain tissue. However, a definitive diagnosis based on a brain biopsy is not a viable diagnostic option. Probes that allow imaging of the brain have been developed and might lead to powerful new biodiagnostic tools, but substantial additional work is needed to identify suitable markers and to correlate their presence and concentration with the state of the disease19. Importantly, some in vitro biodiagnostic tools based on new nanomaterials that have higher sensitivities than ELISAs might allow such markers to be identified in locations outside the brain, for example in the cerebrospinal fluid or blood, where the marker concentration is significantly lower20 (Fig. 1). But the detection of new biomarkers is just the first step towards a new treatment protocol. New biomarkers must be validated, and this process requires intensive prospective and retrospective correlative research. These studies are costly and laborious, and researchers who discover new biomarkers are often ill-equipped to test the utility of these biomarkers in a clinical setting. Indeed, even for biomarkers that have been validated in the laboratory, the process of approval by regulatory bodies such as the US Food and Drug Administration (FDA) can be long and complicated, and cannot keep pace with scientific discovery. Although these challenges present considerable barriers to the development and implementation of new biomarkers, the pay-off for doctors and patients will be enormous when it becomes possible to diagnose diseases that are not detectable with conventional biodiagnostic tools as a result of lack of sensitivity.
Figure 1. High-sensitivity detection can allow less invasive disease diagnosis: Alzheimer’s disease example.
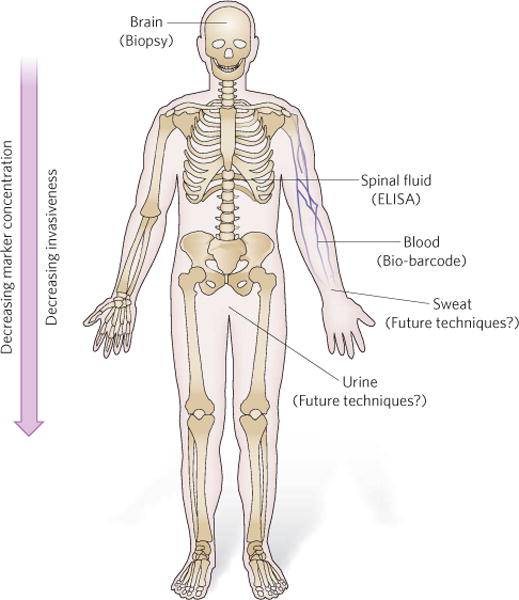
New technologies with higher sensitivities (lower limits of detection) allow markers to be detected at locations distant from the brain, for example in the cerebrospinal fluid, the blood, or even the urine or sweat. In this example, relevant technologies are indicated to illustrate the concept. Depending on the disease marker and the disease, the relevance of a given diagnostic tool will change.
When it becomes possible to detect a target at extremely low concentrations, barriers to sensitivity are no longer the motivating force behind the research and are superseded by more practical concerns about how the technology can be applied. Therefore, when researchers propose a new biomarker or reach a new detection limit, it is important for the medical community to determine the clinical utility of the biomarker. Important points to consider are the meaningful concentrations of a given biomarker, and how disease states and disease cure would be defined in the context of such previously undetectable biomarkers. In addition, it should be considered whether greater sensitivity allows biomarker detection in samples that are more distant from the source of biomarker production (such as blood, breath, urine and sweat) and what concentrations are clinically relevant (Fig. 1). Clearly, analytical benchmarks, which give clinical meaning to test results, will constantly need to be evaluated and redefined when this stage of the development process is reached.
An example from research in our laboratory highlights this challenge. We recently developed an ultrahigh-sensitivity biodiagnostic system called the bio-barcode assay. This assay involves the use of gold nanoparticle probes ‘decorated’ with DNA (the barcode DNA) that is end-functionalized with antibodies specific for a target of interest. The target in the assay is sandwiched between a gold particle probe and a magnetic particle probe. Isolation of the sandwich complex in a magnetic field, followed by chemical release of the barcode DNA, results in an amplification event (each protein target is ‘traded’ for hundreds of strands of barcode DNA). The barcode DNA is then identified using a scanometric assay and the Verigene System (Nanosphere, Northbrook, Illinois), a high-sensitivity, nanotechnology-enabled biodiagnostic platform for nucleic acid detection that is commercially available and has been cleared by the FDA. Using this assay, we have achieved unparalleled levels of sensitivity for detecting a variety of targets. Indeed, we have reached the point at which new definitions of disease states may be warranted as a result of the ability to detect previously unmeasurable concentrations of antigen.
At present, for example, the clinical limit of detection of prostate-specific antigen (PSA), which is often found in increased concentrations in the serum of men with prostate cancer, is 0.1 ng mL−1, using a conventional (ELISA) immunoassay. In men who have undergone radical prostatectomy (removal of the prostate gland), serum PSA concentrations are still monitored, because PSA is considered a valid marker for disease recurrence, but the main source of PSA has been removed. So it is often not possible to detect PSA in the serum of these men, because its concentration falls below 0.1 ng mL−1. These patients are therefore assigned the status ‘undetectable’, and disease recurrence is monitored in terms of PSA serum concentration increases above the 0.1 ng mL−1 cutoff. With the development of the bio-barcode assay, it was concluded that ‘undetectable’ PSA is a limitation of current technologies: using the bio-barcode assay, PSA is measurable in the serum of nearly all patients who have had a prostatectomy, a finding that might lead to a re-evaluation of the clinical definitions of prostate cancer cure and recurrence21. Accordingly, this redefinition could change the ways in which patients are monitored and treated after radical prostatectomy, and these findings present a challenge to the medical community to make appropriate use of the new technique for the benefit of their patients. In this case, many important questions remain. Can disease cure be defined as a patient having a consistent PSA concentration after prostatectomy? Can changes be made to the schedule of post-operative follow-up visits if a patient has a favourable and consistent PSA concentration? What increase in PSA concentration constitutes clinically significant disease recurrence, and how should such a patient be treated? Can new clinical trials be developed such that increasing PSA concentrations (below the conventional detection limit) are used to initiate and validate new therapy and, importantly, track an individual’s response to treatment?
This application of the bio-barcode assay highlights an interesting situation, in which it is possible to detect a disease marker at a much lower concentration than has traditionally been associated with a ‘disease-positive’ patient. The medical community now has a key role in further developing this assay and any related assays that might lead to changes in the definition of disease state, that might result in detection at an earlier stage, and that might improve the outcome for patients. Certainly, if doctors are to overcome the hurdles to the adoption of this technique and gain the advantage of more sensitive tests, the increase in sensitivity must provide meaningful information about their patients. Sceptics might ask, if the technologies for more sensitive assays are developed but no one knows what to do with the information, then why develop the tests? The answer is that enhancing sensitivity and changing analytical benchmarks are the first steps towards removing ‘blind spots’ in the ability to study, diagnose and treat disease, as well as in the ability to validate new therapeutics for diseases.
Selectivity and versatility
Not only do targets need to be detectable at low concentrations but, as a first step, they need to be identified specifically from among the numerous biomolecules present in each sample. Thus, one of the important drivers when developing new biodiagnostics is the need to recognize targets selectively, and for the next generation of biodiagnostics it will be crucial to develop methods that are versatile enough to be applicable to multiple diseases and disease states to allow their use in personalized diagnosis and medicine.
PCR is a prime example of a selective method that can be used to detect disease-specific targets present at a low copy number. PCR uses Watson–Crick base pairing to provide the selectivity needed for a nucleic acid probe to bind to a genomic DNA target and allow its subsequent amplification. PCR is also an excellent example of a technique that started as a research tool but was eventually translated into medical applications. Indeed, the versatility of PCR has allowed its use not only in clinical diagnostics but also in areas as diverse as forensics and archaeology.
Although PCR is highly selective, it lacks the versatility needed in medical diagnosis. It is clear that non-genetic targets, such as proteins, small molecules and ions, are also important biological indicators of disease. Recently, techniques such as immuno-PCR22 and the bio-barcode assay have been developed, and these allow proteins to be detected selectively with much greater sensitivity than in the past. Immuno-PCR, which involves the coupling of nucleic acids to antibodies and then PCR-based amplification of the nucleic acid label, provides a limit of detection comparable to the bio-barcode assay, although it is hampered by the limitations of PCR, which requires specialized assay conditions. In addition, both assays, when used for complex sample media, are limited by the selectivity of the antibodies used to capture the target. New materials can play a prominent part, at least in reducing assay complexity and in increasing the versatility of assay read-out. For example, nanoshells or noble-metal nanoparticles23 can be modified with agents that recognize biomolecules and used to detect proteins, elemental ions24,25 and small molecules26. Assays based on such materials rely on non-enzymatic amplification of the signal with colorimetric or spectroscopic outputs. These strategies are adaptable enough to be used for recognizing many types of analyte, and as mentioned earlier the properties of the nanostructures, including their size and shape, can be manipulated, providing the needed versatility.
In the case of proteins, the issue of selectivity is considerably more challenging than for nucleic acids. Indeed, despite advances, there are many challenges still associated with identifying and making antibodies with the desired properties for binding to a target of interest. In addition, these structures are more fragile than their DNA counterparts and require special handling during storage and use. These problems are exacerbated when using antibody-labelled probes in the context of multiplexing assays. When antibodies are adsorbed on surfaces and used to assay complex sample media, scientists must contend with poor target affinity and selectivity, which can result in significant crossreactivity with targets (that is, an antibody can bind nonspecifically to proteins in the sample). In developing next-generation biodiagnostic systems, an important step in increasing assay selectivity and versatility will be to replace antibody-based systems with chemical systems that do not have the current limitations of antibodies but have the same or greater selectivity. For example, short oligonucleotides with high selectivity and affinity, termed aptamers, provide an alternative route to highly selective recognition of targets, including small molecules and proteins27. Through multiple rounds of in vitro selection, aptamers can be chosen for their ability to bind to a particular target, resulting in high target selectivity28. With further development, aptamers may rival antibodies in their ability to recognize targets, including proteins and small molecules. Importantly, with nucleic acids (as opposed to antibodies) as a basis for recognition, it is conceptually simpler for researchers to modify sequences and structures chemically, making them potentially suitable for signal-based amplification technologies. Finding such new robust target-recognition agents with strong binding and amplification capacities (especially for protein targets) will continue to motivate the development of biodiagnostics.
Robustness and portability
At present, the medical diagnostics industry is highly centralized. Diagnostic testing is based mainly in laboratories that are remote from the site of patient care and are equipped with large and complex instruments that typically require highly skilled personnel to operate them. Therefore, a major challenge in the diagnostics field is the development of robust, portable and low-cost assays that will allow disease markers to be detected reliably in places as diverse as the battlefield, the developing world, community hospitals, the doctor’s office, the post office or perhaps even at home. For example, a home test may require the use of a dipstick assay to test urine, whereas in a hospital a more complex assay to test blood may be warranted. Although robustness, portability and affordability are necessary features of any technological development, they are not always considered at the initial stages of assay conception and proof-of-concept demonstration. Yet inability to meet these criteria can cause some technologies to hit a dead end in their application, while other technologies accelerate past.
High-density microarrays are a good example of a new technology that generates a large amount of information per unit of time, information that can aid in disease diagnosis29. Conceptually, a microarray provides a plethora of genetic information and meets the requirements for gathering data from complex sample media. A natural question to ask is what has impeded the translation of high-density microarrays to the clinic. And, although it may seem inane, why cannot such a microarray be picked up at a local pharmacy? Ultimately, the limited applicability of microarrays lies in their high cost and lack of robustness, as well as in the difficulty in interpreting and acting on the information generated from them. In this case, the ability to measure targets at the molecular level in the laboratory has outpaced the ability to translate the data into useful information for the patient. Consequently, the modern field of diagnostic medicine has moved towards using lower-density structures, which allow the detection of a more manageable number of targets and provide a predictive outcome for the user.
In developing technologies that are robust, portability must be considered. Technology cannot be physically bulky, expensive or cumbersome in other ways, or else diagnostic testing will remain mostly in centralized laboratories and in the hands of research scientists. As an example, issues of cost, safety and field use stymied the development of radioactivity-based assays, leading to a switch to fluorescence-based techniques when this technology became available. However, even fluorescence-based assays have drawbacks: photobleaching (fading of the fluorophore), ease of read-out and cost have presented challenges to achieving widespread point-of-care use. Recently, the use of nanomaterials such as quantum dots has overcome some of these problems: these materials have tunable emission profiles that yield bright signals in the visible to near-infrared spectrum and are less susceptible to photobleaching than their molecular counterparts30,31. Although quantum dots have already shown their utility in a variety of laboratory detection applications, other advantages (apart from a reduction in photobleaching) must be identified, and methods for making these nanomaterials in an environmentally acceptable manner will be required if these materials are to be translated into widespread, commercially viable medical diagnostic applications. Efforts in this area are under way with silicon quantum dots32. Furthermore, structures such as nanowires that can sense analyte binding (for example through changes in electrical conductivity) are being developed and will provide a major step towards new detection assays, as electrical detection systems are conceptually more portable and robust than many optical-based systems33. Although these technologies based on nanomaterials are still in the early stages, they are showing promise in the laboratory and, with further design and development, could have a major impact on medical diagnostics.
Looking forward
The established techniques of PCR and ELISA will eventually give way to methods based on new technologies that offer comparable amplification, greater multiplexing capabilities and the prospects of lower cost and portability. New materials — in particular noble-metal nanoparticles, quantum dots, carbon nanotubes and silicon nanowires — are now redefining analytical benchmarks for sensitivity, selectivity and versatility. When proteins and other non-nucleic-acid analytes can be detected as robustly as nucleic acids and with a technology as sensitive as PCR, the field of molecular diagnostics and medicine will be revolutionized.
When developing a new biodiagnostic test, however, there are many stakeholders, including researchers, industry, doctors, regulatory bodies and patients. Each group faces challenges with regard to both the invention and adoption of new technologies that are sensitive, selective, robust and portable. From the perspective of the researcher, the process for taking a scientific result or a breakthrough in materials development and moving it into the clinic is not straightforward. In addition, the time frame is long, and the process is expensive and not one that a conventional research laboratory is equipped to complete. This is the stage at which business enterprises have the financial and intellectual capital available to take promising technologies from the research laboratory into the clinic. However, not all promising technologies are marketable. From the perspective of the medical community, novel technologies can be a welcome addition to the doctor’s office. Nevertheless, change comes at a high cost. In addition to direct financial expenses, burdens include the time, infrastructure, regulatory issues, reimbursement considerations and experience needed to diagnose a disease on the basis of the latest technology. The disproportionate cost of adopting a technology relative to the immediate benefits to doctors and patients can prove high barriers to implementation. Regulatory agencies must find a way to keep pace with the rate of innovation and must provide incentives for generating new tests and validating disease markers. Finally, getting a technology into the hands of the patient (in a home test kit) comes with its own set of challenges, in which ease of use, cost and portability are key issues.
With such a diverse group of stakeholders with varied motivations, all of the drivers for the development of biodiagnostic assays need to be considered at each stage of development. Full understanding of these considerations is essential not only for gaining a fundamental scientific understanding of disease but also for designing and implementing innovative disease detection and treatment strategies. We predict that it will also yield the most successful biodiagnostic technologies in the future.
Acknowledgments
We thank P. Patel for assistance with preparing Table 1. We also thank the US National Science Foundation, the National Cancer Institute and the National Institutes of Health for research support.
Footnotes
Reprints and permission information is available at www.nature.com/reprints.
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature.
References
- 1.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 2.Engvall E, Perlmann P. Enzyme-linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry. 1971;8:871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- 3.Rosi NL, Mirkin CA. Nanostructures in biodiagnostics. Chem Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. This review highlights how nanomaterials contribute to diagnostics. [DOI] [PubMed] [Google Scholar]
- 4.Alivisatos P. The use of nanocrystals in biological detection. Nature Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 5.Taton TA, Mirkin CA, Letsinger RL. Scanometric DNA array detection with nanoparticle probes. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 6.Nam JM, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 7.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 8.You CC, et al. Detection and identification of proteins using nanoparticle–fluorescent polymer ‘chemical nose’ sensors. Nature Nanotechnol. 2007;2:318–323. doi: 10.1038/nnano.2007.99. [DOI] [PubMed] [Google Scholar]
- 9.Dubertret B, Calame M, Libchaber AJ. Single-mismatch detection using gold-quenched fluorescent oligonucleotides. Nature Biotechnol. 2001;19:365–370. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- 10.Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. Nano-flares: probes for transfection and mRNA detection in living cells. J Am Chem Soc. 2007;129:15477–15479. doi: 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Hu J, Jin Y, Yao X, Li J. In situ amplified chemiluminescent detection of DNA and immunoassay of IgG using special-shaped gold nanoparticles as label. Clin Chem. 2006;52:1958–1961. doi: 10.1373/clinchem.2006.071399. [DOI] [PubMed] [Google Scholar]
- 12.He L, et al. Colloidal Au-enhanced surface plasmon resonance for ultrasensitive detection of DNA hybridization. J Am Chem Soc. 2000;122:9071–9077. [Google Scholar]
- 13.Cao YC, Jin R, Mirkin CA. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Liu G, Jan MR. Ultrasensitive electrical biosensing of proteins and DNA: carbon-nanotube derived amplification of the recognition and transduction events. J Am Chem Soc. 2004;126:3010–3011. doi: 10.1021/ja031723w. [DOI] [PubMed] [Google Scholar]
- 15.Chen RJ, et al. Noncovalent functionalization of carbon nanotubes for highly specific electronic biosensors. Proc Natl Acad Sci USA. 2003;100:4984–4989. doi: 10.1073/pnas.0837064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui Y, Wei Q, Park H, Lieber CM. Nanowire nanosensors for highly sensitive and selective detection of biological and chemical species. Science. 2001;293:1289–1292. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]
- 17.Soman CP, Giorgio TD. Quantum dot self-assembly for protein detection with subpicomolar sensitivity. Langmuir. 2008;24:4399–4404. doi: 10.1021/la704078u. [DOI] [PubMed] [Google Scholar]
- 18.Catalano SM, et al. The role of amyloid-β derived diffusible ligands (ADDLs) in Alzheimer’s disease. Curr Top Med Chem. 2006;6:597–608. doi: 10.2174/156802606776743066. [DOI] [PubMed] [Google Scholar]
- 19.Perrin R, Fagan A, Holtzman D. Multimodal techniques for diagnosis and prognosis of Alzheimer’s disease. Nature. 2009;461:916–922. doi: 10.1038/nature08538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Georganopoulou DG, et al. Nanoparticle-based detection in cerebral spinal fluid of a soluble pathogenic biomarker for Alzheimer’s disease. Proc Natl Acad Sci USA. 2005;102:2273–2276. doi: 10.1073/pnas.0409336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thaxton CS, et al. Nanoparticle-based bio-barcode assay redefines ‘undetectable’ PSA and biochemical recurrence after radical prostatectomy. Proc Natl Acad Sci USA. 2009;106:18437–18442. doi: 10.1073/pnas.0904719106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sano T, Smith CL, Cantor CR. Immuno-PCR: very sensitive antigen detection by means of specific antibody–DNA conjugates. Science. 1992;258:120–122. doi: 10.1126/science.1439758. [DOI] [PubMed] [Google Scholar]
- 23.McFarland AD, Van Duyne RP. Single silver nanoparticles as real-time optical sensors with zeptomole sensitivity. Nano Lett. 2003;3:1057–1062. [Google Scholar]
- 24.Liu J, Lu Y. A colorimetric lead biosensor using DNAzyme-directed assembly of gold nanoparticles. J Am Chem Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- 25.Lee JS, Han MS, Mirkin CA. Colorimetric detection of mercuric ion in aqueous media using DNA-functionalized gold nanoparticles. Angew Chem Int Edn Engl. 2007;46:4093–4096. doi: 10.1002/anie.200700269. [DOI] [PubMed] [Google Scholar]
- 26.Hirsch LR, Jackson JB, Lee A, Halas NJ, West JL. A whole blood immunoassay using gold nanoshells. Anal Chem. 2003;75:2377–2381. doi: 10.1021/ac0262210. [DOI] [PubMed] [Google Scholar]
- 27.Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 28.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. This paper describes the identification of RNA aptamers. [DOI] [PubMed] [Google Scholar]
- 29.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. This study demonstrated the use of microarray analysis for profiling gene expression patterns. [DOI] [PubMed] [Google Scholar]
- 30.Chan WC, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 31.Bruchez M, Jr, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 32.Park JH, et al. Biodegradable luminescent porous silicon nanoparticles for in vivo applications. Nature Mater. 2009;8:331–336. doi: 10.1038/nmat2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nature Biotechnol. 2005;23:1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]


