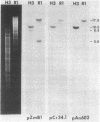
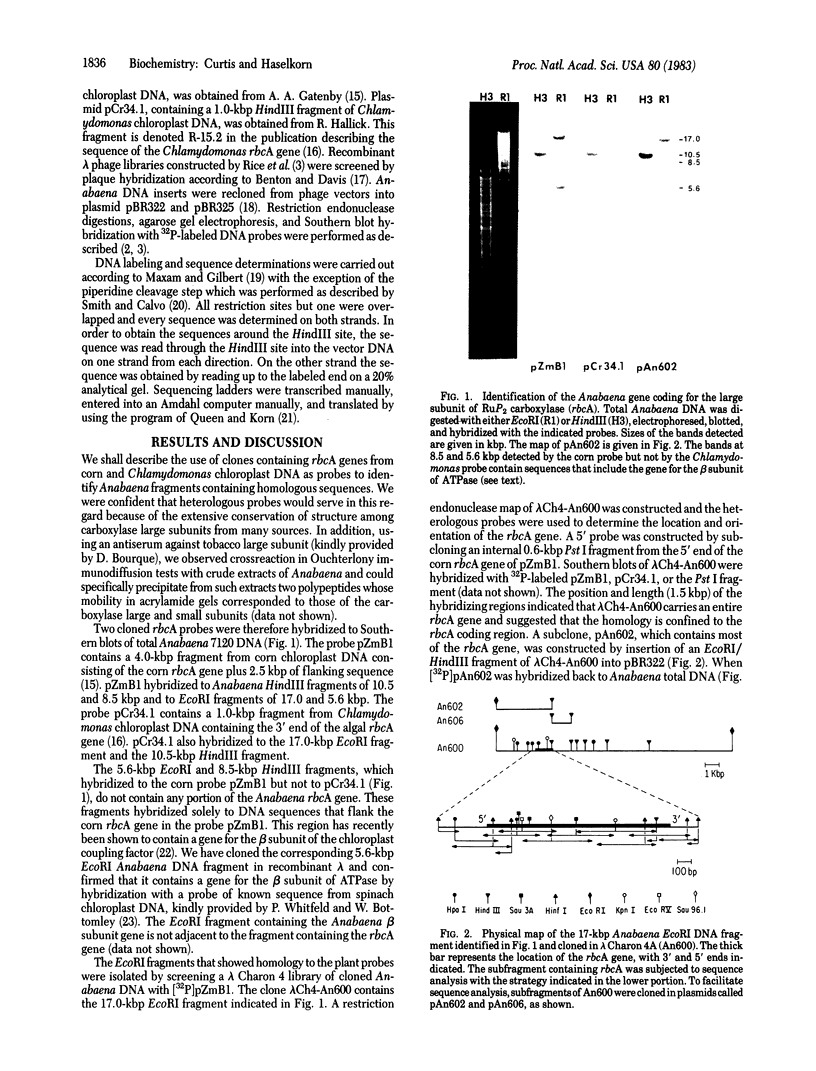
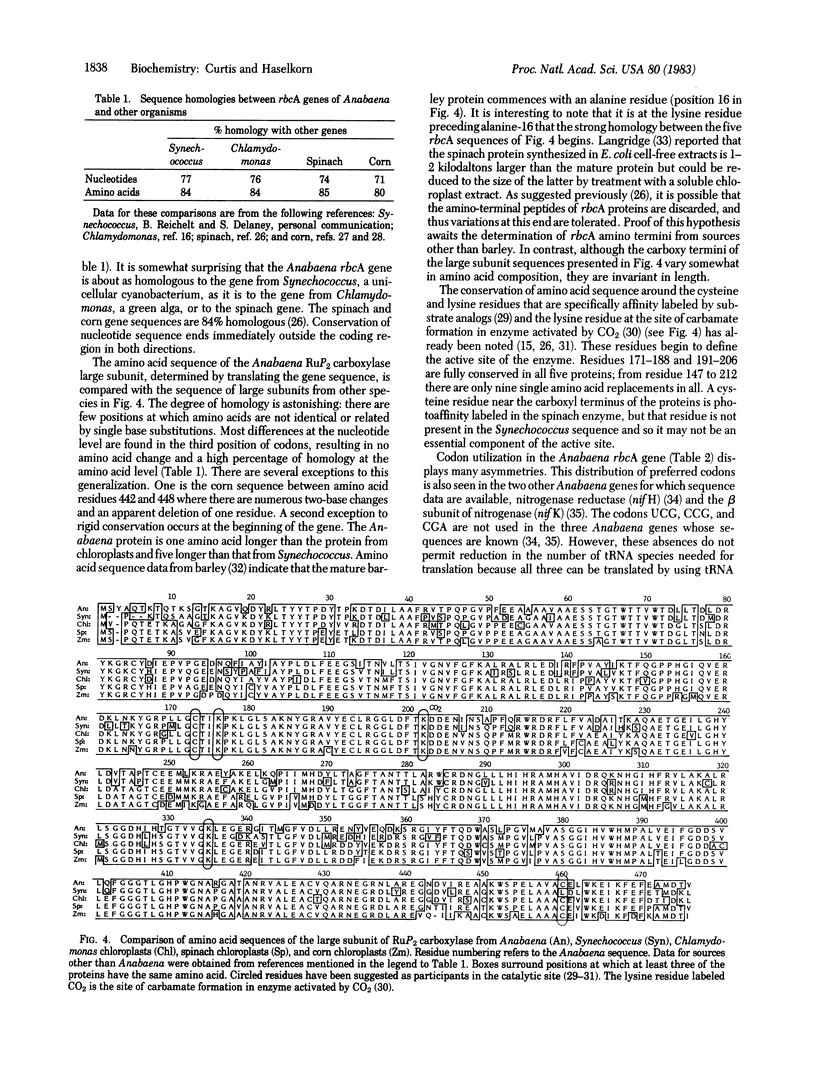
Abstract
Cloned DNA probes containing genes coding for the large subunit of ribulose-1,5-bisphosphate carboxylase (rbcA) of corn and of Chlamydomonas were used to identify, by heterologous hybridization, DNA fragments from Anabaena 7120 carrying the corresponding gene sequence. The same probes were used to isolate, from a recombinant λ library, a 17-kilobase-pair EcoRI Anabaena DNA fragment containing the coding sequence for the rbcA gene. The entire coding sequence, as well as 210 base pairs of 5′ flanking region and 210 base pairs of 3′ flanking region, was determined. Comparison of the nucleotide and amino acid sequences with those of corn, spinach, Chlamydomonas, and Synechococcus rbcA genes revealed homology of 71-77% at the nucleotide level and 80-85% at the amino acid level. Conservation of sequence is lost immediately outside the coding region on either side. Codon usage in the Anabaena rbcA gene is not significantly different from that in the Anabaena genes for nitrogenase reductase and nitrogenase β subunit.
Keywords: chloroplast DNA probes, sequence conservation, endosymbiont origin
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams D. G., Carr N. G. The developmental biology of heterocyst and akinete formation in cyanobacteria. Crit Rev Microbiol. 1981;9(1):45–100. doi: 10.3109/10408418109104486. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Backman K. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 1979;68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- Chan P. H., Wildman S. G. Chloroplast DNA codes for the primary structure of the large subunit of fraction I protein. Biochim Biophys Acta. 1972 Sep 14;277(3):677–680. doi: 10.1016/0005-2787(72)90115-3. [DOI] [PubMed] [Google Scholar]
- Chua N. H., Schmidt G. W. Post-translational transport into intact chloroplasts of a precursor to the small subunit of ribulose-1,5-bisphosphate carboxylase. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6110–6114. doi: 10.1073/pnas.75.12.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DORNER R. W., KAHN A., WILDMAN S. G. Proteins of green leaves. VIII. The distribution of fraction I Protein in the plant kingdom as detected by precipitin and ultracentrifugal analyses. Biochim Biophys Acta. 1958 Aug;29(2):240–245. doi: 10.1016/0006-3002(58)90180-x. [DOI] [PubMed] [Google Scholar]
- Dron M., Rahire M., Rochaix J. D. Sequence of the chloroplast DNA region of Chlamydomonas reinhardii containing the gene of the large subunit of ribulose bisphosphate carboxylase and parts of its flanking genes. J Mol Biol. 1982 Dec 25;162(4):775–793. doi: 10.1016/0022-2836(82)90547-2. [DOI] [PubMed] [Google Scholar]
- Fleming H., Haselkorn R. The program of protein synthesis during heterocyst differentiation in nitrogen-fixing blue-green algae. Cell. 1974 Oct;3(2):169–170. doi: 10.1016/0092-8674(74)90121-4. [DOI] [PubMed] [Google Scholar]
- Gatenby A. A., Castleton J. A., Saul M. W. Expression in E. coli of maize and wheat chloroplast genes for large subunit of ribulose bisphosphate carboxylase. Nature. 1981 May 14;291(5811):117–121. doi: 10.1038/291117a0. [DOI] [PubMed] [Google Scholar]
- Goebel W., Schrempf H. Replication of plasmid DNA in temperature-sensitivity DNA replication mutants of Escherichia coli. Biochim Biophys Acta. 1972 Feb 23;262(1):32–41. doi: 10.1016/0005-2787(72)90216-x. [DOI] [PubMed] [Google Scholar]
- Kieras F. J., Haselkorn R. Properties of ribulose-1,5-diphosphate carboxylase (carboxydismutase) from Chinese cabbage and photosynthetic microorganisms. Plant Physiol. 1968 Aug;43(8):1264–1270. doi: 10.1104/pp.43.8.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorimer G. H. Ribulosebisphosphate carboxylase: amino acid sequence of a peptide bearing the activator carbon dioxide. Biochemistry. 1981 Mar 3;20(5):1236–1240. doi: 10.1021/bi00508a028. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mazur B. J., Chui C. F. Sequence of the gene coding for the beta-subunit of dinitrogenase from the blue-green alga Anabaena. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6782–6786. doi: 10.1073/pnas.79.22.6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur B. J., Rice D., Haselkorn R. Identification of blue-green algal nitrogen fixation genes by using heterologous DNA hybridization probes. Proc Natl Acad Sci U S A. 1980 Jan;77(1):186–190. doi: 10.1073/pnas.77.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevarech M., Rice D., Haselkorn R. Nucleotide sequence of a cyanobacterial nifH gene coding for nitrogenase reductase. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6476–6480. doi: 10.1073/pnas.77.11.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen C. L., Korn L. J. Computer analysis of nucleic acids and proteins. Methods Enzymol. 1980;65(1):595–609. doi: 10.1016/s0076-6879(80)65062-9. [DOI] [PubMed] [Google Scholar]
- Rice D., Mazur B. J., Haselkorn R. Isolation and physical mapping of nitrogen fixation genes from the cyanobacterium Anabaena 7120. J Biol Chem. 1982 Nov 10;257(21):13157–13163. [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- SINGER S. J., EGGMAN L., CAMPBELL J. M., WILDMAN S. G. The proteins of green leaves. IV. A high molecular weight protein comprising a large part of the cytoplasmic proteins. J Biol Chem. 1952 May;197(1):233–239. [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Sugiura M. The nucleotide sequence of the tobacco chloroplast gene for the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Gene. 1982 Nov;20(1):91–102. doi: 10.1016/0378-1119(82)90090-7. [DOI] [PubMed] [Google Scholar]
- Smith D. R., Calvo J. M. Nucleotide sequence of the E coli gene coding for dihydrofolate reductase. Nucleic Acids Res. 1980 May 24;8(10):2255–2274. doi: 10.1093/nar/8.10.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G., Bottomley W., Whitfeld P. R. Structures of the genes for the beta and epsilon subunits of spinach chloroplast ATPase indicate a dicistronic mRNA and an overlapping translation stop/start signal. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6260–6264. doi: 10.1073/pnas.79.20.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G., Perrot B., Bottomley W., Whitfeld P. R. The structure of the gene for the large subunit of ribulose 1,5-bisphosphate carboxylase from spinach chloroplast DNA. Nucleic Acids Res. 1981 Jul 24;9(14):3251–3270. doi: 10.1093/nar/9.14.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]